Abstract
The amphibian-killing chytrid fungus Batrachochytrium dendrobatidis (Bd) is one of the most generalist pathogens known, capable of infecting hundreds of species globally and causing widespread population declines and extinctions. However, some host species are seemingly unaffected by Bd, tolerating or clearing infections without clinical signs of disease. Variation in host immune responses is commonly evoked for these resistant or tolerant species, yet to date, we have no direct comparison of amphibian species responses to infection at the level of gene expression. In this study, we challenged four Central American frog species that vary in Bd susceptibility, with a sympatric virulent strain of the pathogen. We compared skin and spleen orthologous gene expression using differential expression tests and coexpression gene network analyses. We found that resistant species have reduced skin inflammatory responses and increased expression of genes involved in skin integrity. In contrast, only highly susceptible species exhibited suppression of splenic T-cell genes. We conclude that resistance to chytridiomycosis may be related to a species’ ability to escape the immunosuppressive activity of the fungus. Moreover, our results indicate that within-species differences in splenic proteolytic enzyme gene expression may contribute to intraspecific variation in survival. This first comparison of amphibian functional immunogenomic architecture in response to Bd provides insights into key genetic mechanisms underlying variation in disease outcomes among amphibian species.
Keywords: Batrachochytrium dendrobatidis, immunogenomics, comparative transcriptomics, immunosuppression, amphibian immunity
Introduction
A substantial proportion of pathogens are capable of infecting more than one host species, and of these some are considered “ecological generalists,” which can successfully infect and be transmitted by multiple hosts (Woolhouse et al. 2001). The ability of a pathogen to exploit multiple host species is a key factor in both their epidemiology (Paull et al. 2011) and evolution (Poulin et al. 2011; Walker et al. 2014). Generalism necessitates functional trade-offs that may limit the fitness of the pathogen in any one particular host (Leggett et al. 2013) and variation in host species abundance, exposure, and susceptibility makes it unlikely that each host species contributes equally to pathogen transmission (Brisson et al. 2008; Cronin et al. 2010; Streicker et al. 2013). Therefore, discovering the mechanisms by which variable host species respond, or not, to infection is a critical step in understanding how generalist emerging pathogens will spread and persist within multihost communities.
Sympatric host species, even within a single community, often exhibit remarkable differences in infection intensity and severity of disease-related clinical signs (Searle, Gervasi, et al. 2011; Johnson et al. 2012). Host life-history traits, such as habitat use and breeding behavior, influence the likelihood of pathogen exposure and undoubtedly play a role in observed species differences in infection (Bancroft et al. 2011; Hoverman et al. 2011). However, exposure alone is unlikely to account for host species variations in disease outcome, and host functional genomic variation can also be pivotal (Hahn et al. 2013; Sutherland et al. 2014). Likewise, pathogen fitness depends on its ability to evade, exploit, or manipulate host responses (Jiménez-López and Lorenz 2013; Boyett and Hsieh 2014; Cambier et al. 2014). Therefore, if a generalist pathogen differs in its ability to exploit host species, variation in disease outcomes will ultimately result from a complex combination of host traits that mediate exposure, and the functional genetic architecture underlying pathogen and host adaptations.
Batrachochytrium dendrobatidis (Bd), a fungus that causes chytridiomycosis, is a remarkable generalist, and has caused widespread amphibian declines and extinctions (Lips 1999; Lips et al. 2006). The fungal pathogen invades the skin of hundreds of amphibian species worldwide and causes death in many hosts (Berger et al. 1998; Longcore et al. 1999; Voyles et al. 2011). Initially, high Bd susceptibility was attributed to a lack of host immune responses to the fungus (Rosenblum et al. 2012), but recent transcriptomic studies revealed that highly susceptible species mount considerable immunogenetic responses, leading to the hypothesis that their immune responses are somehow ineffective against Bd (Ellison et al. 2014). Bd inhibits splenic lymphocyte proliferation and induces apoptosis of T cells in vitro (Fites et al. 2013) and in vivo gene expression supports this mechanism (Ellison et al. 2014). Nonetheless, some species appear to be less affected by Bd, tolerating infections without clinical signs of disease or clearing infections (Lips et al. 2003; Brem and Lips 2008). To date, studies of amphibian immune responses to Bd have focused primarily on highly susceptible species, because those species are of greatest concern in terms of current conservation efforts in the face of chytridiomycosis. However, characterization of immune pathway activation in resistant and tolerant species is essential. By identifying how resistant and susceptible species differ in their response to Bd, we can elucidate the mechanisms underpinning the variation in chytridiomycosis infections and, consequently, population decline among worldwide amphibian fauna.
In this study, we experimentally challenged four Central American frog species with varying Bd susceptibility (Agalychnis callidryas, Atelopus glyphus, Atelopus zeteki, and Craugastor fitzingeri) and compared transcriptome-wide gene expression in the skin and spleen of uninfected, infected, and self-cleared frogs. We used a Bd strain, JEL-423, that belongs to the Global Pandemic Lineage (GPL) that swept throughout Central America, devastating amphibian populations (Lips et al. 2008). We chose the four focal species because they are found in Central American tropical upland amphibian communities where Bd-induced declines are most prominent (Lips et al. 2008; Becker and Zamudio 2011) and because these species show marked variation in Bd susceptibility in the field. The Panamanian golden frog, At. zeteki, is highly susceptible to Bd, and the species is extinct in the wild (La Marca et al. 2005; Gewin 2008). Atelopus glyphus is one of the few species of Atelopus (out of 113) that had not experienced dramatic population declines in recent years (La Marca et al. 2005), but is currently listed as critically endangered and at high risk from chytridiomycosis (http://www.iucnredlist.org, last accessed January 2, 2015). Craugastor fitzingeri and A. callidryas are naturally infected with Bd, generally at low infection intensities (Puschendorf et al. 2006; García-Roa et al. 2014; Rebollar et al. 2014), and have not experienced Bd-induced population declines despite epizootic outbreaks of the pathogen in Central America (Puschendorf et al. 2006; Crawford et al. 2010). Here, for the first time, we use interspecific comparative transcriptomics to characterize and quantify differential gene expression responses to Bd infection in amphibian species with divergent Bd susceptibility. We test the hypothesis that species-specific immunogenomic architecture underlies the broad spectrum of chytridiomycosis susceptibility, tolerance, and resistance in global amphibian populations. To do so, we identify similarities and key differences in host functional genomic profiles and thus define crucial gene expression responses central to amphibians’ successful resistance to the deadly fungal pathogen.
Materials and Methods
Experimental Infections
The number, source of experimental animals, and Bd challenge assay protocols are summarized in supplementary table S1, Supplementary Material online. All animals were housed individually throughout the experiment in plastic aquaria, at 18–19 °C, 12:12 light cycle, and fed crickets weekly. Frogs in the infected treatment were challenged with Bd strain JEL-423 (GPL). All animals (controls and Bd challenged) were swabbed once a week to measure the growth rate of Bd (see below). Throughout the experiment, we used a fresh pair of gloves when handling each individual. Frogs were monitored daily for clinical signs of chytridiomycosis and we euthanized those that had lost righting abilities by applying 20% benzocaine to the venter. All other individuals (controls and those showing no clinical signs) were euthanized at the end of the experiments (table 1). Challenge experiments were performed with approval from and in accordance with the ethical standards of the US Institutional Animal Care and Use Committee under protocols 2013-0201-2016 (At. glyphus), R-12-98 (At. zeteki), and 2013-0401-2016-2 (A. callidryas, C. fitzingeri).
Table 1.
Summary of Treatment Group Sizes, De Novo Transcriptome Assembly Statistics, and Numbers of Differentially Expressed Genes
| Species | N controls | N infecteda | N clearedb | N chytridomycosisc | Average Bd load (SE) | Average reads per sampled | Assembly N50 | No. of annotated genes | No. of DEe orthologousf genes |
|
|---|---|---|---|---|---|---|---|---|---|---|
| Skin | Spleen | |||||||||
| Atelopus zeteki | 5 | 6 | 0 | 6 | 6,819,500 (2,431,929) | 25.9 × 106 | 1,596 | 12,056 | 3,849 | 2,291 |
| Atelopus glyphus | 3 | 9 (6) | 3 (2) | 2 | 3,353 (1,412) | 22.9 × 106 | 1,289 | 13,409 | 901 | 452 |
| Craugastor fitzingeri | 5 | 5 | 0 | 0 | 102,998 (57,652) | 20.3 × 106 | 1,704 | 12,409 | 1,413 | 2,660 |
| Agalychnis callidryas | 4 | 3 | 2 | 0 | 27 (23) | 27.4 × 106 | 1,230 | 12,929 | 6 | 182 |
aBd load of 1 or more zoospore genomical equivalents (GE) at death or end of experiment. Numbers in parentheses indicate N of spleen due to sample removal due to low quality RNA.
bBd load of 1 or more zoospore GE during experiment but was no longer infected at the end of the experiment. Numbers in parentheses indicate N of spleen due to sample removal due to low quality RNA.
cExhibited clinical signs of cytridiomycosis, lost righting reflex, and euthanized prior to the end of the experiment.
dAfter read quality control filtering and trimming.
eDifferential expression in control versus infected comparison, ≤0.05 FDR corrected P value.
fDefined by reciprocal best hits among all species pairs (total 8,732 genes).
For all species, we swabbed the abdomen, drink patch, hands, and feet five times each with a sterile cotton tipped swab weekly, and stored samples in capped tubes (Hyatt et al. 2007). All animals were also swabbed prior to the start of the experiment. We tested swabs for Bd using PrepMan Ultra and DNeasy (Qiagen), running samples in singlicate Taqman quantitative PCR (qPCR) (Boyle et al. 2004). A subset of swabs were also run in triplicate to ensure consistency. We ran each plate with JEL-423 standards of 0.1, 1, 10, 100, 1,000, and 10,000 zoospore genomic equivalents (GEs) to determine Bd presence and infection intensity. We categorized individuals as Bd positive when qPCR results showed an infection load greater than or equal to 1 Bd zoospore GEs (Kriger et al. 2006).
Immediately after euthanasia, frogs were dissected using sterilized instruments, and skin (ventral thigh) and spleen tissue samples were harvested from each individual. These tissues were chosen because skin is the primary site of infection for Bd (Longcore et al. 1999) and the spleen is the major lymphoid organ in frogs (Tischendorf 1985). In addition, Bd suppresses splenic leukocytes in susceptible species (Fites et al. 2013; Ellison et al. 2014). Tissue samples were immediately placed in RNALater (Invitrogen) and stored at −80 °C prior to RNA extraction and library preparation.
Because inoculation doses and timing of tissue harvest differed for our four focal species, we cannot ascertain that all frogs were at the exact same stages of disease progression at the time of sampling. The number of Bd zoospores that infect individuals during inoculations is highly variable, causing natural differences in infection intensities and time to death among individuals and species (Carey et al. 2006). However, the infected frogs used in the transcriptomic analysis in this study had in common that all were at a “mature” infection stage, colonized with mature zoosporangia and shedding zoospores. At the point of tissue sampling, all frogs categorized as infected carried their highest individual Bd load and thus all could be considered still within the disease progression phase (supplementary fig. S1, Supplementary Material online).
Transcriptome Sequencing and Gene Orthology
Total RNA was extracted from each tissue sample separately using RNAdvance tissue kit (Beckman Coulter, Inc.). We quantified and assessed RNA integrity using a Bioanalyzer 2100 RNA nano assay (Agilent Technologies). Four spleen samples of At. glyphus did not pass RNA integrity quality controls and thus were not sequenced (table 1). All other samples had RNA integrity values above 8.0. Libraries were generated using the Illumina TruSeq RNA sample preparation kit v2 (low throughput protocol) according to the manufacturer’s instructions (Illumina, San Diego, CA). Briefly, 0.4–1.2 µg of RNA was subjected to messenger RNA (mRNA) selection using poly-T oligo-attached magnetic beads followed by chemical fragmentation (6 min, 94 °C). Cleaved RNA fragments were then copied into first strand cDNA using SuperScript II reverse transcriptase (Invitrogen) and Illumina proprietary random hexamer primers. After second strand synthesis using Illumina-supplied consumables, the cDNA was amplified with reagents of the same kit and ligated to barcoded adapters. The final libraries were amplified using 14 PCR cycles. We quantified and assessed library quality on a Bioanalyzer 2100 and, within each species, randomly pooled equimolar samples onto two or three lanes of the Illumina flowcell (8–10 samples per lane). All sequencing runs were 100-bp single-end reads.
After Illumina standard quality control filtering, read quality for each sample was visualized using FastQC version 0.10.0 (Andrews 2010). All samples had higher than average guanine and cytosine content in the first 15 bp due to use of random hexamer primers during library preparation. Therefore, we used Trimmomatic version 0.27 (Lohse et al. 2012) to trim or delete the following: 1) The first 15 bp of each read, 2) any Illumina adapter sequence, 3) the 5′ and/or 3′ end of reads where quality score dropped below Q20, 4) anywhere within each read where a 5-bp window drops below Q20, and 5) any trimmed reads less than 36-bp long. This ensured only the highest quality reads were used for subsequent de novo assembly.
For each species, reads from all individuals and tissues were pooled to assemble a consensus transcriptome. Assemblies were performed using Trinity (Grabherr et al. 2011) with default parameter settings on a high-performance cluster with 64 central processing units and 512 GB random access memory. We filtered out transcripts with expression support of less than two reads per million mappable reads in at least two samples, to eliminate low-level expression noise (Harrison et al. 2012; Moghadam et al. 2013).
The longest sequence of each Trinity component (roughly equivalent to a single gene) was extracted from the assembly using custom Perl scripts. Next, these sequences (transcripts) were aligned via a local BLASTX to the National Center for Biotechnology Information (NCBI) vertebrate nonredundant (nr) protein database, retaining up to 20 hits with a minimum E-value of 1 × 10−6 and minimum bit score of 55. Any transcript aligning to the Bd transcriptome (Bd Sequencing Project, Broad Institute of Harvard and MIT, www.broadinstitute.org, last accessed January 2, 2015) was removed from downstream analyses. BLAST2GO version 2.5.0 (www.blast2go.com, last accessed January 2, 2015) was used to functionally annotate the assembled transcriptomes. Gene ontology (GO; www.geneontology.org, last accessed January 2, 2015) mapping was performed, extracting the GO terms associated with homologies identified by BLASTX, and producing a list of GO annotations represented as hierarchical categories of increasing specificity. We retained annotations with a minimum E-value of 1 × 10−6, a minimum annotation cut-off of 55, and a GO weight of 5. GO annotations were enhanced using the annotation augmentation tool ANNEX (Myhre et al. 2006). Finally, we performed InterPro (Quevillon et al. 2005) searches remotely from BLAST2GO via the InterPro EBI web server and merged InterProScan GOs with the original GO annotations. To assess the completeness of the transcriptomes, we examined the proportion of assembled transcripts that were full length or near full length, based on the NCBI model amphibian (Xenopus tropicalis) reference protein data using Trinity Perl scripts.
We performed a reciprocal best-hit analysis to identify 1:1 orthologs between the four species. We used BLASTN with an E-value threshold of 1 × 10−6 to search all transcripts from one species against the other three. Only transcripts that were the reciprocal best hit between all species pairs were retained for further analyses.
Gene Expression Analyses
Gene expression levels were determined using the Trinity pipeline; utilizing BWA read mapping (Li and Durbin 2009) and RSEM read count normalization (Li and Dewey 2011). For each species, we analyzed differential gene expression (DGE) of control (uninfected) and infected individual orthologous gene sets using the edgeR (Robinson et al. 2010) R package (R version 2.15.2, R Development Core Team). This consisted of estimating tagwise dispersion and normalization factors and differentially expressed (DE) testing using an exact test. A false discovery rate (FDR)–corrected P value of less than 0.05 was considered to be evidence of DGE. Two species (A. callidryas and At. glyphus) had individuals that were infected during the course of the experiment but cleared infection (swabbed Bd negative) by the end of the trials (table 1, supplementary fig. S1, Supplementary Material online). In these species, DGE tests were performed to compare expression between infected, cleared, and control frogs.
To quantify the overlap of differentially expressed genes between species, we constructed four-way Venn diagrams for each tissue using VENNY (Oliveros 2007), separating significantly increased and decreased expressed genes. We tested for enrichment of biological process GO terms and InterPro protein domains in each group of DE genes (e.g., specific to one species or shared among two or more species) using DAVID (Huang et al. 2008) with all orthologous genes as the background reference.
Weighted Gene Coexpression Network Analyses
Differential gene expression analyses consist of exact tests on each gene separately and thus necessitate stringent multiple test correction methods (e.g., FDR), and typically only genes with the largest differences in expression are identified. An alternative for quantifying systematic transcriptional responses of frogs to infection challenge by Bd is weighted gene coexpression network analysis (WGCNA), which identifies networks (modules) of coexpressed genes (i.e., genes that show consistent expression profiles across samples), and thus potentially identifies functionally important genes with only subtle changes in expression that may not be detected in typical DGE analyses. As A. callidryas is the most resistant species in this study, gene modules were defined in this species as the reference.
First, read counts were TMM normalized using a Trinity-provided Perl script to produce fragments per kilobase per million mapped expression values. Next, the R package WGNCA was used for network constructions (Langfelder and Horvath 2008). Briefly, WGCNA constructs networks using the absolute value of the Pearson’s correlation coefficient as the gene coexpression measure, which is raised to a power to create the adjacency matrix. The topological overlap distance calculated from the adjacency matrix is then clustered with the average linkage hierarchical clustering. Our modules were defined using the dynamicCutTree function and TOMType “signed” with a minimum module size of 100. A module eigengene distance threshold of 0.25 was also used to merge highly similar modules. One thousand permutations of randomly sampled modules were generated. Modules were considered robust if average module adjacencies were significantly higher than the randomly generated modules. Modules were then correlated with log-transformed Bd load to identify gene networks significantly involved in responses to Bd infection. GO term and protein domain enrichment tests of each gene module that significantly correlated with Bd load were performed using DAVID as described above.
Each gene within a module was ranked by its module membership (kME), calculated by WGCNA. Network hub genes were defined as those ranked in the top 100 module membership values and with the highest 150 network connection weights. Median log2 fold change of hub genes of each module was calculated for each species. Hub gene network connections were exported to Cytoscape (Shannon et al. 2003) for visualization.
To assess the degree to which A. callidryas modules were conserved across the other three species, module preservation statistics were computed using the modulePreservation function (500 permutations) (Langfelder et al. 2011). Network module preservation statistics quantify how density and connectivity patterns of modules defined in a reference data set are preserved in a test data set without the need to define modules in the test data set. A module can be significantly preserved in another species but not necessarily with the same direction of expression.
Results
Experimental Infections
Frogs of all four species were Bd negative at the start of the experiments. All control frogs remained Bd negative during the course of the experiment except one A. callidryas which we removed from subsequent analyses. Sample sizes, time course of experiments, and Bd loads are reported in supplementary information and supplementary table S1, Supplementary Material online.
For each species, we followed protocols used previously for At. zeteki infections (Ellison et al. 2014). Briefly, all Bd-challenged At. zeteki (N = 6) were euthanized on exhibiting signs of advanced chytridiomycosis (e.g., loss of righting reflex) between day 22 and 33 postinoculation. The average Bd load at death was 6,819,500 zoospore GEs (ZGEs, standard error [SE] = 2,431,929, table 1). Twelve At. glyphus were challenged with Bd. Two frogs were euthanized prior to the end of the experiment (day 46 and 55) due to severe signs of chytridiomycosis. All other individuals were euthanized on days 60–62 postinoculation. Average Bd load of infected At. glyphus was 3,353 ZGEs (SE = 1,412). The two individuals that were euthanized early did not possess the highest infection intensities (453 and 8,075, highest = 11,769 ZGEs). Three Bd-challenged frogs tested positive during the course of the experiment but had cleared infection by the end of the trial and so were grouped separately for subsequent analyses. Three of the six A. callidryas tested positive for Bd at the end of the experiment, but with low infection intensities (day 41 postinoculation, average ZGE = 27, SE = 23). The remaining three tested positive during the experiment but had cleared infection by day 41. Infected and cleared frogs were grouped separately for subsequent analyses. All five Bd-challenged C. fitzingeri tested positive for Bd by day 41 (average = 102,998, SE = 57,652).
Transcriptome Assemblies and Gene Orthologs
Average Illumina sequencing reads per sample and assembly statistics are summarized in table 1. All raw sequencing reads are deposited in NCBI Sequence Read Archive under submission accession numbers SRP029154 (At. zeteki), SRP046003 (At. glyphus), SRP045866 (C. fitzingeri), and SRP045871 (A. callidryas). The At. zeteki assembly (40,074 transcripts) had 15,252 sequences (38.1%) with at least one significant hit against the nonredundant NCBI protein database and of these, 12,056 (79.0%) were successfully annotated with GO terms. The At. glyphus assembly (34,947 transcripts) had 16,322 with hits (46.7%) and of those 13,409 (82.2%) were annotated. The C. fitzingeri assembly (29,257 transcripts) had 14,980 (51.2%) with hits and of those 12,409 (82.8%) were annotated. The A. callidryas assembly (36,645 transcripts) had 16,040 (43.8%) with hits and of those 12,929 (80.6%) were annotated. The top BLAST hit for the majority of all sequences (between 72.1% and 76.9%) were to Xenopus species. The majority of assembled transcripts (∼70% in all species) were considered full length or near full length (>90% coverage) based on matching to X. tropicalis reference protein data (supplementary fig. S2, Supplementary Material online). A total of 8,732 annotated sequences had reciprocal best hits in all four species and were deemed gene orthologs.
Host Species Overlap of Differentially Expressed Genes
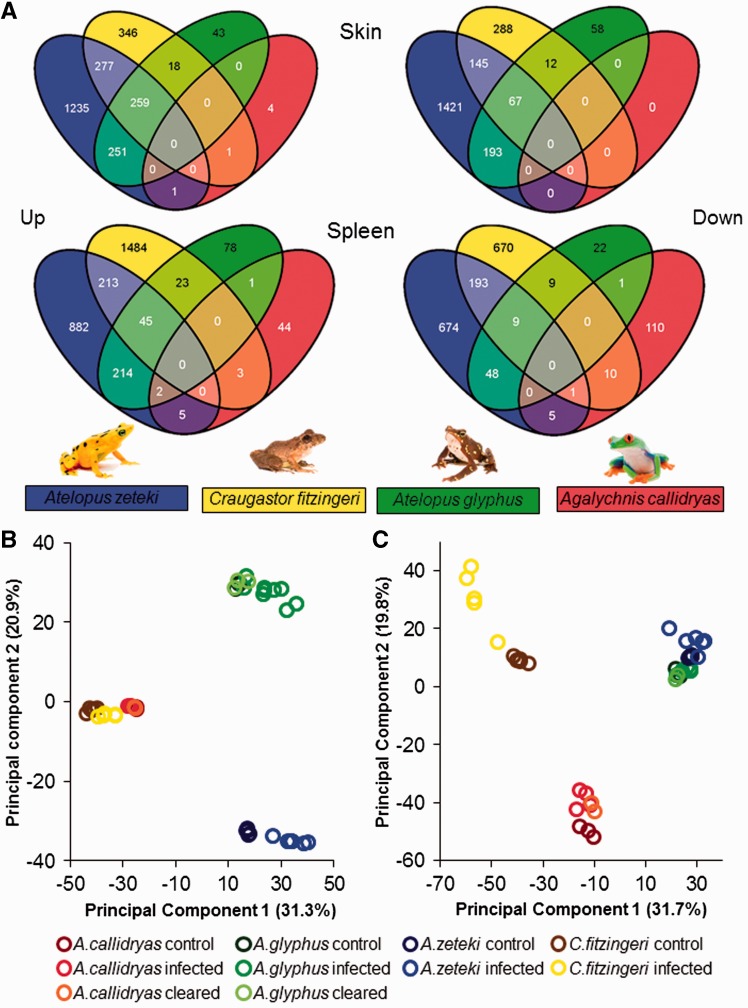
Figure 1A summarizes the number and overlap of genes found to be significantly differentially expressed (DE) between control (uninfected) and infected frogs in each of the four focal species. Figure 1B and C demonstrates overall separation of individual samples based on the entire orthologous gene set. Table 2 summarizes the groups of DE genes that were significantly enriched for GO (classifications of molecular function or biological process) terms. Complete lists of genes with significant differential expression in each species are provided in supplementary materials. For brevity, here we report groups of genes shared or unique to host species deemed as most relevant to Bd-infection responses.
Fig. 1.—
Venn diagrams of differentially expressed genes in spleen and skin tissues between control and infected frogs. GO term enrichment of groups is summarized in table 2. Photos courtesy of Brian Gratwicke. (A) Venn diagrams of differentially expressed genes in spleen and skin tissues between control and infected frogs. GO term enrichment of groups is summarized in table 2. Principal component analysis (PCA) of all orthologous gene expression data in (B) skin and (C) spleen.
Table 2.
Summary of Significant Biological Process GO Term Enrichment in Species-Specific and Shared Differentially Expressed Genes
| Group | No. of Genes | No. of Enriched GO Terms | Selected Biological Process GO Term Enrichment |
|---|---|---|---|
| Spleen up | |||
| Azet | 882 | 122 | Response to wounding, inflammatory response, apoptosis |
| Agly | 78 | 3 | Activation of phospholipase C activity |
| Cfit | 1,484 | 64 | Mitosis, transcription, regulation of apoptosis |
| Acal | 44 | 1 | Immune response |
| Azet-Agly | 214 | 5 | Cell adhesion, immune response, regulation of cell proliferation |
| Agly-Cfit | 23 | 10 | Cell cycle process, mitosis |
| Spleen down | |||
| Azet | 674 | 5 | Transcription, protein complex assembly |
| Cfit | 670 | 91 | Apoptosis, leukocyte activation, inflammatory response, cell adhesion |
| Azet-Cfit | 193 | 29 | Leukocyte differentiation, T-cell activation, metal ion homeostasis |
| Skin up | |||
| Azet | 1,235 | 57 | Apoptosis, endocytosis, positive regulation of I-κB/NF-κB cascade |
| Cfit | 346 | 3 | Mitotic cell cycle |
| Azet-Agly | 251 | 15 | Response to wounding, inflammatory response, regulation of apoptosis |
| Azet-Cfit | 277 | 18 | Inflammatory response, T-cell activation, apoptosis |
| Azet-Agly-Cfit | 259 | 51 | Inflammatory response, immunoglobulin immunity, complement activation |
| Skin down | |||
| Azet | 1,421 | 58 | Mitosis, cell adhesion, positive regulation of apoptosis |
| Cfit | 288 | 4 | Transcription |
| Azet-Agly | 193 | 5 | Skeletal system development, regionalization |
| Azet-Cfit | 145 | 1 | Epithelium development |
Note.—Azet = Atelopus zeteki; Agly = Atelopus glyphus; Cfit = Craugastor fitzingeri; Acal = Agalychnis callidryas. Full lists of GO terms are provided in supplementary materials.
We found 251 genes with increased expression in the skin of infected frogs (compared with controls) in both Atelopus species. These genes were enriched in 15 GO terms including “response to wounding,” “inflammatory response,” and a number of cell death–related terms. The 277 genes sharing increased expression in the skin of infected At. zeteki and C. fitzingeri were also enriched for similar inflammatory, wounding response and apoptosis-related terms. These shared genes also included leukocyte-related terms such as “T-cell activation” and “lymphocyte activation.” This gene group was also enriched in immunoglobulin protein domains (supplementary material). A substantial number of genes (259) exhibited significantly increased expression in infected individuals of both Atelopus species and C. fitzingeri. These were enriched for a wide variety of immune-related terms including inflammatory response, “B cell-mediated immunity,” “complement activation,” and “positive regulation of T-cell activation.” The majority of genes exhibiting decreased expression in the skin of infected At. glyphus were shared by At. zeteki (193) and enriched for terms related to bone development and regionalization.
The small number of genes showing increased expression in infected A. callidryas spleen were predominantly unique to the species (44; 80%) and enriched for the “immune response” GO term. A large proportion of genes with elevated expression in infected At. glyphus were shared with At. zeteki (214) and were enriched for five GO terms including “cell adhesion,” immune response, and “regulation of cell proliferation.” Twenty-three genes shared increased expression between infected At. glyphus and C. fitzingeri and were enriched for cell cycle process GO terms such as “mitosis” and “cell division.” Genes sharing decreased expression in infected At. zeteki and C. fitzingeri (193), enriched for 29 GO terms, include many related to lymphocyte terms such as T-cell activation and “lymphocyte differentiation.”
Cleared Versus Infected Frogs
During the course of Bd-challenge experiments, three individuals of A. callidryas and At. glyphus tested Bd positive but cleared infection by the end of the experiment. Comparisons of Bd-infected and Bd-cleared A. callidryas skin samples found 49 genes significantly differentially expressed (30 decreased and 19 increased in cleared frogs, relative to infected frogs). Genes with increased expression in the cleared group were enriched for “response to virus” and “defense response.” Cleared versus infected skin comparisons of At. glyphus found 69 genes differentially expressed (58 decreased and 11 increased in cleared frogs) but showed no significant GO term enrichment. In the spleen, 133 genes were differentially expressed between Bd-cleared and Bd-infected A. callidryas (30 decreased in cleared frogs, 103 increased). Genes with lower expression in cleared frogs were enriched for 11 GO terms primarily related to T-cell activation and differentiation. Nine genes were differentially expressed between At. glyphus cleared and infected frogs (4 increased, 5 decreased) but showed no significant GO term enrichment. No genes with differential expression between cleared and infected groups were shared between the two species. In comparisons of cleared and control frogs, A. callidryas showed 44 differentially expressed genes in the skin (cleared; 32 increased, 12 decreased) and six in the spleen (4 increased, 2 decreased). Atelopus glyphus cleared-control comparisons found 46 differentially expressed genes in the skin (35 increased, 11 decreased) and 14 in the spleen (10 increased, 4 decreased). No significant enrichment was identified in any cleared-control gene group. Full lists of differentially expressed genes and GO terms are provided in supplementary material.
Gene Coexpression Module Identification and Preservation
Gene coexpression modules—networks of genes with consistent expression patterns across samples—provide a complementary approach to individual gene DE tests for summarizing large gene expression data sets. In addition, module preservation statistics allow a more quantitative view of how well preserved or divergent changes in gene expression are among species in broad-scale comparative transcriptomic studies such as ours. Agalychnis callidryas—the most resistant focal species—was used as a reference. A total of 19 and 7 modules were defined in the skin and spleen gene coexpression networks of A. callidryas, respectively. All modules were considered “robust” as permutation testing confirmed that average module adjacency was always greater than the mean of 1,000 randomly sampled “modules” of equal size (all modules P < 0.001). In addition, one of the skin modules and three of the spleen modules were significantly associated with Bd load (table 3).
Table 3.
Summary of Gene Coexpression modules defined by WGCNA
| Module | No. of Genes | Bd Load Correlation | P Value | Hub Gene FCa | Top Enrichmentb | Top 5 Hub Genesc |
Craugastor fitzingeri |
Atelopus glyphus |
Atelopus zeteki |
|||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Preservationd | Hub Gene FCa | Preservationd | Hub gene FCa | Preservationd | Hub gene FCa | |||||||
| Skin | ||||||||||||
| SK1 | 947 | 0.88 | 9.00 × 10−4 | +0.08 | Cell adhesion | KTN1, CARM1, DMAP1, COL5A1, MTMR3 | 13 (2.17) | −0.26 | 6 (6.86) | −0.58 | 9 (3.86) | −0.48 |
| Spleen | ||||||||||||
| SP1 | 1,234 | −0.72 | 0.02 | −0.20 | Immune response | EXOC2, PPP2CA, CNDP2, SEC24D, GCSH | 2 (2.88) | −0.05 | 2 (9.13) | +0.31 | 3 (11.51) | +0.41 |
| SP2 | 154 | −0.77 | 0.009 | −0.22 | Immune response | KRT19, FAM82B, LDHA, C1S, HPSE | 7 (−2.38) | −0.34 | 6 (−1.20) | +0.38 | 4 (2.11) | +0.81 |
| SP3 | 145 | 0.84 | 0.002 | +1.00 | Regulation of T-cell activation | BACH2, LCK, CCR7, MBP, S1PR4 | 3 (0.00) | −1.01 | 1 (3.59) | −0.42 | 1 (5.19) | −0.50 |
Note.—Full lists of enriched GO terms are provided in supplementary materials. Significant values in italics.
aMedian log2 fold change of module hub genes (based on top 150 node connections).
bMost significantly enriched GO biological process term.
cFive most interconnected genes.
dMedian ranked preservation (Z summary preservation; ≥2 = weak to moderate preservation, ≥10 = high preservation).
The skin module SK1 (947 genes, supplementary fig. S4, Supplementary Material online) was positively correlated with Bd load (r = 0.83, P = 0.003) and predominantly enriched for cell adhesion (55% of genes). This module was also enriched for a number of GO terms related to skin structure such as “extracellular structure organization” and “epithelial cell differentiation.” This module had significant preservation in the other three species but with decreased expression in infected frogs.
Spleen module SP1 gene expression (1,234 genes, supplementary fig. S5, Supplementary Material online) was negatively correlated with Bd load (r = −0.72, P = 0.020). This module was enriched for immune response and other immune-related GO terms such as inflammatory response, “humoral immune response,” and response to wounding. This module was significantly preserved in all other species; however, hub gene expression patterns were reversed in the Atelopus species (i.e., increased expression in infected frogs). The spleen module SP2 (154 genes, supplementary fig. S6, Supplementary Material online) was negatively correlated with Bd load (r = −0.77, P = 0.009) and enriched for immune response. This module was only weakly preserved in At. zeteki but with increased hub gene expression in infected frogs. The spleen module SP3 (145 genes, fig. 2) was positively correlated with Bd load (r = 0.84, P = 0.002) and enriched for 17 GO terms related to lymphocyte and, more specifically, T-cell activation and differentiation. This module was not significantly preserved in C. fitzingeri, yet was the most preserved module in both Atelopus species but with reversed expression patterns (table 3).
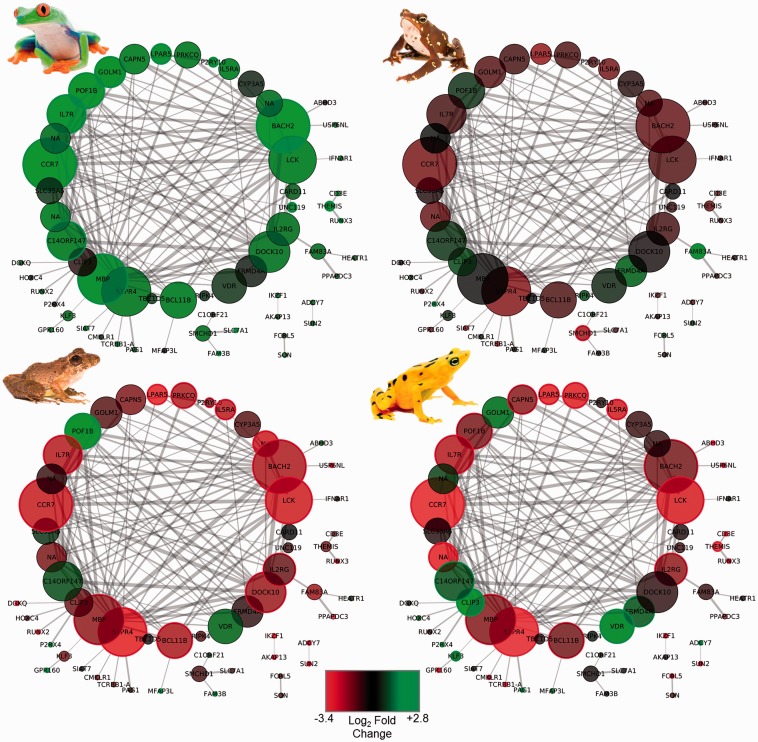
Fig. 2.—
Network view of Agalychnis callidryas SP3 spleen module (T-cell activation) hub genes. Nodes are labeled with official gene symbols when available. Edge line width represents connection strength (weight); thicker lines denote stronger connections. Node size is proportional to number connections. Node color represents the log2 fold change between control and infected frogs (clockwise from top left; A. callidryas, Atelopus glyphus, Atelopus zeteki, Craugastor fitingeri). Node borders represent differential gene expression results. Black = nonsignificant (after FDR correction); red = decreased expression in the infected; green = increased expression in the infected. Photos courtesy of Brian Gratwicke.
Discussion
Bd causes chytridiomycosis in hundreds of amphibians worldwide and is responsible for dramatic population declines and extinctions in many species (Lips et al. 2006; Skerratt et al. 2007). Yet some species carry infection with no apparent clinical signs and have not experienced declines due to the emergence of this fungal pathogen (Lips et al. 2003; Brem and Lips 2008). The underlying mechanisms for such wide species variation in disease outcome are not well understood, although differences in host immune function are commonly hypothesized (Richmond et al. 2009; Voyles et al. 2011). Here, using a comparative transcriptomic approach, we characterize shared patterns of response to Bd across divergent sympatric amphibian species and highlight key differences in immune pathway activation that likely underlie variation in susceptibility or resistance to chytridiomycosis.
Four species present in Central American tropical uplands (At. zeteki, At. glyphus, C. fitzingeri, and A. callidryas) were challenged with a virulent Bd strain (JEL-423) isolated from Panama (Lips et al. 2006). We observed considerable differences in infection intensities; At. zeteki became highly infected (on average 6,819,500 ZGEs), succumbing to chytridiomycosis within a month of inoculation. Craugastor fitzingeri harbored the next highest infection loads (table 1) yet did not exhibit clinical signs of chytridiomycosis after 41 days postinoculation. Atelopus glyphus, although cited at high risk of chytridiomycosis (http://www.iucnredlist.org, last accessed January 2, 2015), had relatively low to moderate infections (on the order of tens to thousands of zoospore GE), with only 2 of 12 inoculated exhibiting chytridiomycosis by the end of the experiment. Finally, A. callidryas developed the lowest level infections, with no more than 70 zoospore GE in any infected individual, and no Bd-induced mortality. These findings generally corroborate population changes observed in the field during the wave-like epizootic spread of virulent Bd through Central America (Lips et al. 2008). Atelopus zeteki is the only species in this study to have undergone severe declines (La Marca et al. 2005), while populations of A. callidryas and C. fitzingeri are apparently stable (Puschendorf et al. 2006; Crawford et al. 2010). However, although At. glyphus populations were considered stable (La Marca et al. 2005), Bd has recently been found in this species (unpublished data) and thus considered currently at high risk from chytridiomycosis.
Our aim in this study was to capture gene expression of infected frogs of all species at a broadly comparable point in chytridiomycosis disease progression. However, in addition to species differences in infection intensities (as seen above), the rate of Bd infection progression among and within species may also vary considerably (Carey et al. 2006). Thus, a degree of caution must be used when interpreting comparative gene expression data at a fixed time point such as this. That said, all infected frogs carried their highest individual load at the time of tissue harvest (supplementary fig. S1, Supplementary Material online) and therefore all species could be considered within the disease progression/growth phase (rather than disease retreat).
The overlap of genes with significantly higher expression in the skin of infected frogs (compared with controls) in both Atelopus species and C. fitzingeri was considerable (fig. 1). The shared increased skin expression gene groups were all enriched for inflammatory response. Inflammatory response genes increased in these species were particularly rich in components of the complement system (e.g., C1QA/B/C, C3, C5AR1, C7, and CFB). Decreased complement activation was observed earlier in infected Rana and Silurana species (Rosenblum et al. 2009, 2012), suggesting complement activity against Bd may be phylogenetically inconsistent. In addition, classic markers of inflammation such as TGF-β, interleukin (IL)-1B, and IL-8 were increased in the skin in C. fitzingeri and Atelopus species (supplementary fig. S7, Supplementary Material online). Curiously, IL-10 and its receptor exhibited increased expression only in the Atelopus species (supplementary fig. S7, Supplementary Material online). Although the anti-inflammatory role of IL-10 is well recognized (Mosser and Zhang 2008), prolonged high concentrations may actually have proinflammatory effects (Lauw et al. 2000). Moreover, expression of IL-10 in concert with IL-12 and IL-18 (both of which are also only increased in infected Atelopus, supplementary fig. S7, Supplementary Material online) enhances natural killer cell proliferation and cytotoxicity (Cai et al. 1999). Although the activity of interleukin-driven inflammatory responses are as yet poorly characterized in amphibians, their over expression or dysregulation can contribute significantly to disease pathology in mammals (Sears et al. 2011). Our results indicate that differences in the relative balance of these cytokines among species in response to Bd could be important in predicting the severity of infection in a particular species and clearly warrants further investigation.
Considering single gene differential expression (DE) tests alone, A. callidryas appears relatively “unresponsive” to Bd infection in the skin; only six genes were found to have significantly different expression patterns in infected frogs. However, DE testing necessitates conservative correction for multiple comparisons and so likely only detects the greatest changes in expression. We utilized a gene coexpression network approach to assess more subtle yet consistent changes in gene expression in infected frogs of this species. A single gene module (SK1) in the skin was positively associated with Bd load. The module has collagen (COL5A1) as one of the major hub genes of the network (table 3 and supplementary fig. S4, Supplementary Material online) and is enriched for several GO terms related to skin integrity including cell adhesion, extracellular structure organization, and epithelial cell differentiation. Preservation of the module in the other three species was significant but with reversed expression patterns; genes generally decreased expression with infection. In addition, genes sharing decreased expression in infected Atelopus and C. fitzingeri were enriched for terms such as “epithelium development” (At. zeteki and C. fitzingeri) and “skeletal system development” (At. zeteki and At. glyphus). One of the mechanisms by which Bd kills amphibians is through disruption of skin integrity (Voyles et al. 2009; Rosenblum et al. 2012). Our data indicate that the most resistant species, A. callidryas, actually increases expression of genes related to skin structure and thus potentially tempers the disruptive effects of chytrid infections.
The skin is the point of Bd entry and proliferation in adult frogs, thus comparison of immune responses in the skin is clearly vital to our understanding of chytridiomycosis. However, recent work demonstrates that amphibian immunity to Bd—or the lack thereof—involves more than the defenses in the skin. The amphibian spleen is the primary lymphoid organ (Tischendorf 1985) and recent experiments indicate that Bd infection alters splenic lymphocyte functions in some species (Fites et al. 2013; Ellison et al. 2014). We found two gene coexpression modules in the spleen, enriched for immune responses, which were negatively correlated with Bd load in A. callidryas (SP1 and SP2; table 3). The SP1 module was highly preserved in all other species; however, both Atelopus species had reversed patterns of expression (table 3, supplementary fig. S5, Supplementary Material online). This module contains a number of important markers of inflammatory responses, such as complement (C1R, C3, C4A, CFB, CFH), tumor necrosis factor, and interleukins (IL-1RAP, IL-17, IL-10), indicating that, while A. callidryas and C. fitzingeri have a coordinated lowering of expression of certain proinflammatory genes (at least in late stage infections), Atelopus species, in contrast, have increased inflammatory responses in the spleen. The second spleen module (SP2) that negatively correlated with Bd in A. callidryas was not significantly preserved in C. fitzingeri and At. glyphus, and only weak preservation was observed in At. zeteki, suggesting that genes in this module exhibit decreased coexpression unique to A. callidryas (table 3). Of note, this module includes neutrophil markers (MMP8, CXCR1), proteinase inhibitors (SERPINB6, SERPINF2), genes linked to wound healing processes (HPSE, FAP), and inflammation (C1S, PTAFR) (supplementary fig. S6, Supplementary Material online).
We found remarkable overlap of genes with increased expression in the spleen of A. callidryas individuals that cleared infection in this study and infected At. zeteki from a prior study that had previously been Bd exposed (Ellison et al. 2014). In the earlier study, At. zeteki that survived infection with an attenuated strain of Bd (Langhammer et al. 2013) had highly increased expression of a small set of genes in the spleen when re-exposed to Bd compared with Bd-naïve individuals. A substantial number (38; approximately 37%) of these genes were also more highly expressed in A. callidryas individuals that cleared infection. These were primarily genes of proteolytic enzymes such as chymotrypsins (e.g., CTRL), elastases (e.g., CTRC), and carboxypeptidases (e.g., PCPA1). Chitinase (CHIT1), crucial for the degradation of the main constituent of fungal cell walls, also showed highly elevated expression in both species. These genes could provide important markers of either 1) innate individual differences in ability to cope with Bd infections and/or 2) prior exposure to Bd. Unfortunately, the A. callidryas in this study was captured from wild populations in an area that has been infected with Bd for several years (Woodhams et al. 2008), with unknown infection histories; thus, our data do not allow us to discriminate between the two hypotheses. However, this set of genes clearly warrant rigorous investigation in additional species as they are undoubtedly a key part of the amphibian response to repeated Bd exposure and likely integral to general amphibian defense mechanisms against fungal or other pathogens.
The most striking result of our study was that of the third spleen gene coexpression module that significantly correlated with Bd infections (SP3, table 3). Genes in this module had increased expression in infected A. callidryas, many of which were also detected in single gene DE tests (fig. 2). Coexpression was not well preserved in C. fitzingeri but was significant in both Atelopus species, albeit with substantial decreased gene expression patterns. Crucially, this module is rich in genes implicated in the regulation of T cells, including receptors (e.g., TCRB, CD3E), transcription factors (e.g., RUNX2, BACH2), inducers of activation and/or proliferation (e.g., CCR7, MBP, PRKCQ, IKZF1, CARD11), and signaling/migration pathways (e.g., LCK, S1PR4). Bd can inhibit amphibian splenic T-cell proliferation and induce apoptosis in culture (Fites et al. 2013) and in vivo (Ellison et al. 2014). However, this appears not to be the case for all species; our results indicate that Bd resistance of some hosts may be due in part to their ability to mount T-cell–mediated responses to infection, escaping the immunosuppressive action of the fungal pathogen. Moreover, in susceptible species, suppression appears not to be limited only to T cells, but include a deeper inhibition of T cell activation and signaling pathways.
Comparisons of skin defenses such as anti-microbial peptides (Rollins-Smith, Carey, et al. 2002; Rollins-Smith, Doersam, et al. 2002) and more recently skin microbiota (Harris et al. 2006; Lam et al. 2010; Woodhams et al. 2014) have shown promising correlations with resistance to chytridiomycosis. Undoubtedly, these defense mechanisms play an important role in the likelihood of Bd colonizing amphibian skin. However, on establishment of Bd within epidermal layers, our results indicate that chytridiomycosis resistance runs deeper than skin responses. We show that the spectrum of susceptibility to this fungal pathogen is mediated by a combination of both innate and acquired arms of the amphibian immune system, including the severity of inflammatory response to infection, the ability to maintain skin integrity during infection, and whether splenic lymphocyte activity is suppressed. More resistant species not only have less severe skin inflammation and can boost skin integrity maintenance during infection, but also apparently escape the immunosuppressive actions of Bd and increase splenic lymphocyte activity. Additionally, individual variation in expression of important proteolytic enzymes in the spleen may contribute to within-species differences in ability to reduce Bd infections.
This first comparison of immunogenetic responses of amphibian species with highly divergent susceptibility to chytridiomycosis provides a wealth of data to guide future studies and raises many important hypotheses about immunogenomic differences in amphibians responsible for variation in disease outcomes in nature. Our results also demonstrate the utility of broad-scale comparative functional genomics, providing promising functional gene categories, and even specific candidate genes that merit in-depth study at both species and population level. For example, further targeted studies of T-cell responses in more resistant species may help define a general mechanism of chytridiomycosis resistance. Furthermore, direct quantification of intraspecific splenic proteolytic enzyme activity against Bd is required to provide a causal link to the within-species patterns found in our data. Our results underscore that despite the importance of abiotic environmental factors (Becker and Zamudio 2011) and external skin defenses (Rollins-Smith, Doersam, et al. 2002; Woodhams et al. 2007, 2014) in shaping host response to the amphibian-killing fungus, the functional genetic architecture of a species provides the basis for chytridiomycosis resistance.
Our study has important implications for amphibian conservation efforts. Recent management efforts have focused on controlling exposure (Kriger and Hero 2007; Searle, Biga, et al. 2011), understanding abiotic control of fungal proliferation (Berger et al. 2004; Voyles et al. 2012), and manipulating commensal microbiota (Harris et al. 2008; Muletz et al. 2012; Bletz et al. 2013). We found that resistance to Bd infections varies naturally among species and within species, and that this variation has genomic underpinnings, not exclusively due to abiotic conditions and pathogen exposure levels. These results improve understanding of the functional genomics underlying individual and species differences in response to Bd infection and provides conservation biologists with potential mechanisms that explain variation in population or species declines. Our results offer new possibilities for conservation action, specifically we endorse practices that select for traits that provide resistance against Bd, which would improve the success rates of captive breeding programs and reintroduction programs in areas where Bd is endemic. Adaptive variation in hosts could arise at the molecular level, in genes and their expression, influenced by selection, and by variation in the way genes interact with the environment to produce phenotypes of varying plasticity. Therefore, we recommend that conservation efforts incorporate an evolutionary approach and be aimed at maintaining conditions necessary for natural selection to operate most efficiently on the components of resistance that are inherited.
Supplementary Material
Supplementary figures S1–S7, table S1, and materials are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
This study was supported by grants from the National Science Foundation (DEB-0815315, DEB-1136640, DEB-1136602, and DEB-1120249), the Cornell Center for Vertebrate Genomics, Cornell Center for Comparative and Population Genomics, and Cornell's Atkinson Center for Sustainable Future. The authors thank Daniel Medina, Molly Bletz, William Shoemaker, Marisa Riley, and Meredith Swartwout for assistance in the field and laboratory; Jorge Guerrel and Angie Estrada for rearing captive-bred frogs at Panama Amphibian Rescue and Conservation; Brian Gratwicke for photography; the Smithsonian Tropical Research Institute for logistical support; and Zamudio Laboratory members for their constructive comments.
Literature Cited
- Andrews S. FastQC. Version 0.10.0. [released 2012 May 3; next version 2012 June 2; cited 2015 January 2] 2010. Available from: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/: Babraham Bioinformatics.
- Bancroft B, et al. Species-level correlates of susceptibility to the pathogenic amphibian fungus Batrachochytrium dendrobatidis in the United States. Biodivers Conserv. 2011;20:1911–1920. [Google Scholar]
- Becker CG, Zamudio KR. Tropical amphibian populations experience higher disease risk in natural habitats. Proc Natl Acad Sci U S A. 2011;108:9893–9898. doi: 10.1073/pnas.1014497108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger L, et al. Chytridiomycosis causes amphibian mortality associated with population declines in the rain forests of Australia and Central America. Proc Natl Acad Sci U S A. 1998;95:9031–9036. doi: 10.1073/pnas.95.15.9031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger L, et al. Effect of season and temperature on mortality in amphibians due to chytridiomycosis. Aust Vet J. 2004;82:434–439. doi: 10.1111/j.1751-0813.2004.tb11137.x. [DOI] [PubMed] [Google Scholar]
- Bletz MC, et al. Mitigating amphibian chytridiomycosis with bioaugmentation: characteristics of effective probiotics and strategies for their selection and use. Ecol Lett. 2013;16:807–820. doi: 10.1111/ele.12099. [DOI] [PubMed] [Google Scholar]
- Boyett D, Hsieh MH. Wormholes in host defense: how helminths manipulate host tissues to survive and reproduce. PLoS Pathog. 2014;10:e1004014. doi: 10.1371/journal.ppat.1004014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle DG, Boyle DB, Olsen V, Morgan JAT, Hyatt AD. Rapid quantitative detection of chytridiomycosis (Batrachochytrium dendrobatidis) in amphibian samples using real-time Taqman PCR assay. Dis Aquat Organ. 2004;60:141–148. doi: 10.3354/dao060141. [DOI] [PubMed] [Google Scholar]
- Brem FMR, Lips KR. Batrachochytrium dendrobatidis infection patterns among Panamanian amphibian species, habitats and elevations during epizootic and enzootic stages. Dis Aquat Organ. 2008;81:189–202. doi: 10.3354/dao01960. [DOI] [PubMed] [Google Scholar]
- Brisson D, Dykhuizen DE, Ostfeld RS. Conspicuous impacts of inconspicuous hosts on the Lyme disease epidemic. Proc Biol Sci. 2008;275:227–235. doi: 10.1098/rspb.2007.1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai G, Kastelein RA, Hunter CA. IL-10 enhances NK cell proliferation, cytotoxicity and production of IFN-γ when combined with IL-18. Eur J Immunol. 1999;29:2658–2665. doi: 10.1002/(SICI)1521-4141(199909)29:09<2658::AID-IMMU2658>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- Cambier CJ, et al. Mycobacteria manipulate macrophage recruitment through coordinated use of membrane lipids. Nature. 2014;505:218–222. doi: 10.1038/nature12799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey C, et al. Experimental exposures of boreal toads (Bufo boreas) to a pathogenic chytrid fungus (Batrachochytrium dendrobatidis) Ecohealth. 2006;3:5–21. [Google Scholar]
- Crawford AJ, Lips KR, Bermingham E. Epidemic disease decimates amphibian abundance, species diversity, and evolutionary history in the highlands of central Panama. Proc Natl Acad Sci U S A. 2010;107:13777–13782. doi: 10.1073/pnas.0914115107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cronin JP, Welsh ME, Dekkers MG, Abercrombie ST, Mitchell CE. Host physiological phenotype explains pathogen reservoir potential. Ecol Lett. 2010;13:1221–1232. doi: 10.1111/j.1461-0248.2010.01513.x. [DOI] [PubMed] [Google Scholar]
- Ellison AR, et al. Fighting a losing battle: vigorous immune response countered by pathogen suppression of host defenses in the chytridiomycosis-susceptible frog Atelopus zeteki. G3 (Bethesda) 2014;4:1275–1289. doi: 10.1534/g3.114.010744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fites JS, et al. The invasive chytrid fungus of amphibians paralyzes lymphocyte responses. Science. 2013;342:366–369. doi: 10.1126/science.1243316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Roa R, Sunyer J, Fernández-Loras A, Bosch J. First record of Batrachochytrium dendrobatidis in Nicaragua. Herpetol J. 2014;24:65–68. [Google Scholar]
- Gewin V. Riders of a modern-day ark. PLoS Biol. 2008;6:e24. doi: 10.1371/journal.pbio.0060024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabherr MG, et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 2011;29:644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn DC, Summers S, Genovese K, He H, Kogut M. Obligate brood parasites show more functionally effective innate immune responses: an eco-immunological hypothesis. Evol Biol. 2013;40:554–561. [Google Scholar]
- Harris RN, James TY, Lauer A, Simon MA, Patel A. Amphibian pathogen Batrachochytrium dendrobatidis is inhibited by the cutaneous bacteria of amphibian species. Ecohealth. 2006;3:53–56. [Google Scholar]
- Harris RN, Lauer A, Simon MA, Banning JL, Alford RA. Addition of antifungal skin bacteria to salamanders ameliorates the effects of chytridiomycosis. Dis Aquat Organ. 2008;83:11–16. doi: 10.3354/dao02004. [DOI] [PubMed] [Google Scholar]
- Harrison PW, Mank JE, Wedell N. Incomplete sex chromosome dosage compensation in the Indian meal moth, Plodia interpunctella, based on de novo transcriptome assembly. Genome Biol Evol. 2012;4:1118–1126. doi: 10.1093/gbe/evs086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoverman J, Gray M, Haislip N, Miller D. Phylogeny, life history, and ecology contribute to differences in amphibian susceptibility to ranaviruses. Ecohealth. 2011;8:301–319. doi: 10.1007/s10393-011-0717-7. [DOI] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2008;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Hyatt AD, et al. Diagnostic assays and sampling protocols for the detection of Batrachochytrium dendrobatidis. Dis Aquat Organ. 2007;73:175–192. doi: 10.3354/dao073175. [DOI] [PubMed] [Google Scholar]
- Jiménez-López C, Lorenz MC. Fungal immune evasion in a model host–pathogen interaction: Candida albicans versus macrophages. PLoS Pathog. 2013;9:e1003741. doi: 10.1371/journal.ppat.1003741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson PTJ, et al. Living fast and dying of infection: host life history drives interspecific variation in infection and disease risk. Ecol Lett. 2012;15:235–242. doi: 10.1111/j.1461-0248.2011.01730.x. [DOI] [PubMed] [Google Scholar]
- Kriger KM, Hero JM. The chytrid fungus Batrachochytrium dendrobatidis is non-randomly distributed across amphibian breeding habitats. Divers Distrib. 2007;13:781–788. [Google Scholar]
- Kriger KM, Hines HB, Hyatt AD, Boyle DG, Hero JM. Techniques for detecting chytridiomycosis in wild frogs: comparing histology with real-time Taqman PCR. Dis Aquat Organ. 2006;71:141–148. doi: 10.3354/dao071141. [DOI] [PubMed] [Google Scholar]
- La Marca E, et al. Catastrophic population declines and extinctions in neotropical harlequin frogs (Bufonidae: Atelopus) Biotropica. 2005;37:190–201. [Google Scholar]
- Lam BA, Walke JB, Vredenburg VT, Harris RN. Proportion of individuals with anti-Batrachochytrium dendrobatidis skin bacteria is associated with population persistence in the frog Rana muscosa. Biol Conserv. 2010;143:529–531. [Google Scholar]
- Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfelder P, Luo R, Oldham MC, Horvath S. Is my network module preserved and reproducible? PLoS Comput Biol. 2011;7:e1001057. doi: 10.1371/journal.pcbi.1001057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langhammer P, et al. A fungal pathogen of amphibians, Batrachochytrium dendrobatidis, attenuates in pathogenicity with in vitro passages. PLoS One. 2013;8:e77630. doi: 10.1371/journal.pone.0077630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauw FN, et al. Proinflammatory effects of IL-10 during human endotoxemia. J Immunol. 2000;165:2783–2789. doi: 10.4049/jimmunol.165.5.2783. [DOI] [PubMed] [Google Scholar]
- Leggett HC, Buckling A, Long GH, Boots M. Generalism and the evolution of parasite virulence. Trends Ecol Evol. 2013;28:592–596. doi: 10.1016/j.tree.2013.07.002. [DOI] [PubMed] [Google Scholar]
- Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lips KR. Mass mortality and population declines of anurans at an upland site in western Panama. Conserv Biol. 1999;13:117–125. [Google Scholar]
- Lips KR, Diffendorfer J, Mendelson JR III, Sears MW. Riding the wave: reconciling the roles of disease and climate change in amphibian declines. PLoS Biol. 2008;6:e72. doi: 10.1371/journal.pbio.0060072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lips KR, Reeve JD, Witters LR. Ecological traits predicting amphibian population declines in Central America. Conserv Biol. 2003;17:1078–1088. [Google Scholar]
- Lips KR, et al. Emerging infectious disease and the loss of biodiversity in a neotropical amphibian community. Proc Natl Acad Sci U S A. 2006;103:3165–3170. doi: 10.1073/pnas.0506889103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohse M, et al. RobiNA: a user-friendly, integrated software solution for RNA-Seq-based transcriptomics. Nucleic Acids Res. 2012;40:W622–W627. doi: 10.1093/nar/gks540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longcore JE, Pessier AP, Nichols DK. Batrachochytrium dendrobatidis gen. et sp. nov. a chytrid pathogenic to amphibians. Mycologia. 1999;91:219–227. [Google Scholar]
- Moghadam HK, Harrison PW, Zachar G, Székely T, Mank JE. The plover neurotranscriptome assembly: transcriptomic analysis in an ecological model species without a reference genome. Mol Ecol Res. 2013;13:696–705. doi: 10.1111/1755-0998.12096. [DOI] [PubMed] [Google Scholar]
- Mosser DM, Zhang X. Interleukin-10: new perspectives on an old cytokine. Immunol Rev. 2008;226:205–218. doi: 10.1111/j.1600-065X.2008.00706.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muletz CR, Myers JM, Domangue RJ, Herrick JB, Harris RN. Soil bioaugmentation with amphibian cutaneous bacteria protects amphibian hosts from infection by Batrachochytrium dendrobatidis. Biol Conserv. 2012;152:119–126. [Google Scholar]
- Myhre S, Tveit H, Mollestad T, Lægreid A. Additional gene ontology structure for improved biological reasoning. Bioinformatics. 2006;22:2020–2027. doi: 10.1093/bioinformatics/btl334. [DOI] [PubMed] [Google Scholar]
- Oliveros JC. VENNY. An interactive tool for comparing lists with Venn diagrams. 2007. http://bioinfogp.cnb.csic.es/tools/venny/index.html.
- Paull SH, et al. From superspreaders to disease hotspots: linking transmission across hosts and space. Front Ecol Environ. 2011;10:75–82. doi: 10.1890/110111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulin R, Krasnov BR, Mouillot D. Host specificity in phylogenetic and geographic space. Trends Parasitol. 2011;27:355–361. doi: 10.1016/j.pt.2011.05.003. [DOI] [PubMed] [Google Scholar]
- Puschendorf R, Bolaños F, Chaves G. The amphibian chytrid fungus along an altitudinal transect before the first reported declines in Costa Rica. Biol Conserv. 2006;132:136–142. [Google Scholar]
- Quevillon E, et al. InterProScan: protein domains identifier. Nucleic Acids Res. 2005;33:W116–W120. doi: 10.1093/nar/gki442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebollar EA, et al. The lethal fungus Batrachochytrium dendrobatidis is present in lowland tropical forests of far Eastern Panamá. PLoS One. 2014;9:e95484. doi: 10.1371/journal.pone.0095484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richmond JQ, Savage AE, Zamudio KR, Rosenblum EB. Toward immunogenetic studies of amphibian chytridiomycosis: linking innate and acquired immunity. Bioscience. 2009;59:311–320. [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rollins-Smith LA, Carey C, et al. Activity of antimicrobial skin peptides from ranid frogs against Batrachochytrium dendrobatidis, the chytrid fungus associated with global amphibian declines. Dev Comp Immunol. 2002;26:471–479. doi: 10.1016/s0145-305x(01)00088-x. [DOI] [PubMed] [Google Scholar]
- Rollins-Smith LA, Doersam JK, et al. Antimicrobial peptide defenses against pathogens associated with global amphibian declines. Dev Comp Immunol. 2002;26:63–72. doi: 10.1016/s0145-305x(01)00041-6. [DOI] [PubMed] [Google Scholar]
- Rosenblum EB, Poorten TJ, Settles M, Murdoch GK. Only skin deep: shared genetic response to the deadly chytrid fungus in susceptible frog species. Mol Ecol. 2012;21:3110–3120. doi: 10.1111/j.1365-294X.2012.05481.x. [DOI] [PubMed] [Google Scholar]
- Rosenblum EB, et al. Genome-wide transcriptional response of Silurana (Xenopus) tropicalis to infection with the deadly chytrid fungus. PLoS One. 2009;4:e6494. doi: 10.1371/journal.pone.0006494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Searle CL, Biga LM, Spatafora JW, Blaustein AR. A dilution effect in the emerging amphibian pathogen Batrachochytrium dendrobatidis. Proc Natl Acad Sci U S A. 2011;108:16322–16326. doi: 10.1073/pnas.1108490108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Searle CL, Gervasi SS, et al. Differential host susceptibility to Batrachochytrium dendrobatidis, an emerging amphibian pathogen. Conserv Biol. 2011;25:965–974. doi: 10.1111/j.1523-1739.2011.01708.x. [DOI] [PubMed] [Google Scholar]
- Sears BF, Rohr JR, Allen JE, Martin LB. The economy of inflammation: when is less more? Trends Parasitol. 2011;27:382–387. doi: 10.1016/j.pt.2011.05.004. [DOI] [PubMed] [Google Scholar]
- Shannon P, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skerratt L, et al. Spread of chytridiomycosis has caused the rapid global decline and extinction of frogs. Ecohealth. 2007;4:125–134. [Google Scholar]
- Streicker DG, Fenton A, Pedersen AB. Differential sources of host species heterogeneity influence the transmission and control of multihost parasites. Ecol Lett. 2013;16:975–984. doi: 10.1111/ele.12122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutherland B, et al. Comparative transcriptomics of Atlantic Salmo salar, chum Oncorhynchus keta and pink salmon O. gorbuscha during infections with salmon lice Lepeophtheirus salmonis. BMC Genomics. 2014;15:200. doi: 10.1186/1471-2164-15-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tischendorf F. On the evolution of the spleen. Experientia. 1985;41:145–152. doi: 10.1007/BF02002606. [DOI] [PubMed] [Google Scholar]
- Voyles J, et al. Temperature alters reproductive life history patterns in Batrachochytrium dendrobatidis, a lethal pathogen associated with the global loss of amphibians. Ecol Evol. 2012;2:2241–2249. doi: 10.1002/ece3.334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voyles J, Rosenblum EB, Berger L. Interactions between Batrachochytrium dendrobatidis and its amphibian hosts: a review of pathogenesis and immunity. Microbes Infect. 2011;13:25–32. doi: 10.1016/j.micinf.2010.09.015. [DOI] [PubMed] [Google Scholar]
- Voyles J, et al. Pathogenesis of chytridiomycosis, a cause of catastrophic amphibian declines. Science. 2009;326:582–585. doi: 10.1126/science.1176765. [DOI] [PubMed] [Google Scholar]
- Walker DM, Castlebury LA, Rossman AY, Struwe L. Host conservatism or host specialization? Patterns of fungal diversification are influenced by host plant specificity in Ophiognomonia (Gnomoniaceae: Diaporthales) Biol J Linn Soc. 2014;111:1–16. [Google Scholar]
- Woodhams DC, et al. Resistance to chytridiomycosis varies among amphibian species and is correlated with skin peptide defenses. Anim Conserv. 2007;10:409–417. [Google Scholar]
- Woodhams DC, et al. Interacting symbionts and immunity in the amphibian skin mucosome predict disease risk and probiotic effectiveness. PLoS One. 2014;9:e96375. doi: 10.1371/journal.pone.0096375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodhams DC, et al. Chytridiomycosis and amphibian population declines continue to spread eastward in Panama. Ecohealth. 2008;5:268–274. doi: 10.1007/s10393-008-0190-0. [DOI] [PubMed] [Google Scholar]
- Woolhouse MEJ, Taylor LH, Haydon DT. Population biology of multihost pathogens. Science. 2001;292:1109–1112. doi: 10.1126/science.1059026. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.