Abstract
Ascorbic acid (AsA) is an important antioxidant in plants and an essential vitamin for humans. Extending the study of AsA-related genes from Arabidopsis thaliana to Brassica rapa could shed light on the evolution of AsA in plants and inform crop breeding. In this study, we conducted whole-genome annotation, molecular-evolution and gene-expression analyses of all known AsA-related genes in B. rapa. The nucleobase–ascorbate transporter (NAT) gene family and AsA l-galactose pathway genes were also compared among plant species. Four important insights gained are that: 1) 102 AsA-related gene were identified in B. rapa and they mainly diverged 12–18 Ma accompanied by the Brassica-specific genome triplication event; 2) during their evolution, these AsA-related genes were preferentially retained, consistent with the gene dosage hypothesis; 3) the putative proteins were highly conserved, but their expression patterns varied; and 4) although the number of AsA-related genes is higher in B. rapa than in A. thaliana, the AsA contents and the numbers of expressed genes in leaves of both species are similar, the genes that are not generally expressed may serve as substitutes during emergencies. In summary, this study provides genome-wide insights into evolutionary history and mechanisms of AsA-related genes following whole-genome triplication in B. rapa.
Keywords: AsA-related genes, Brassica rapa, evolutionary conservation, synteny analysis, gene dosage hypothesis, expression pattern
Introduction
Ascorbate or ascorbic acid (AsA), also known as vitamin C, is an important metabolite in many organisms. Since it was first isolated in the 1930s by Albert Szent-Györgyi, there have been numerous reports on the physiological and metabolic processes in which it is involved (Levine 1986). In plants, AsA is a multifunctional molecule with roles as antioxidant, redox signaling modulator, and enzyme cofactor, and it participates in processes such as pathogen defense, cell wall synthesis, growth regulation, and the modulation of plant morphology, flowering time, and the onset of senescence (Conklin and Barth 2004; Barth et al. 2006; Olmos et al. 2006; Cruz-Rus et al. 2012). Thus, AsA is indispensable in plants.
AsA-related genes involved in ascorbate biosynthesis, recycling, and transport weave into a complex network in plants. Several biosynthesis routes for AsA in plants have been proposed since 1998, involving l-galactose (D-Man/L-Gal) (Wheeler et al. 1998), l-gulose, galacturonate, and myo-inositol as initial precursors (fig. 1; Valpuesta and Botella 2004). The l-galactose pathway, commonly called the Smirnoff–Wheeler pathway, is considered the main route of AsA biosynthesis (Smirnoff et al. 2001). After ascorbate is metabolically oxidized in plants, some of the metabolic products can be recycled to the reduced state of ascorbate in what is called the recycling pathway (fig. 1; Arrigoni 1994; Chen et al. 2003). In addition, evidence for the transport of ascorbate has been also presented (Horemans et al. 2000; Franceschi and Tarlyn 2002; Maurino et al. 2006). All of the known AsA-related genes are fully characterized in Arabidopsis thaliana (see references in supplementary table S1, Supplementary Material online), a model plant that has provided valuable insights into angiosperm genome structure, function, and evolution. However, the evolution and duplication of AsA-related genes have not been discussed much.
Fig. 1.—
Proposed model for AsA biosynthesis and recycling pathways in plants. Four possible pathways produce AsA: the l-galactose (Smirnoff–Wheeler) (I), galacturonate (II), l-gulose (III), and myo-inositol (IV) pathways. Gray lines connect metabolites of substrates to products with the corresponding enzymes (named in boxes), and red lines indicate hypothetical reactions. Arrowheads denote directionality. Known enzymes are highlighted in blue boxes, and the color intensity reflects the corresponding enzyme gene numbers.
Angiosperm genome evolution is characterized by polyploidization through whole-genome duplication (WGD) followed by diploidization, which is typically accompanied by considerable homoeologous gene loss (Stebbins 1950). For example, the genome A. thaliana has experienced a paleohexaploidy (γ) duplication shared with most dicots and two subsequent genome duplications (α and β) since its divergence from Carica papaya, along with rapid DNA sequence divergence and extensive gene loss (fractionation; Bowers et al. 2003). Brassica rapa (A genome), a diploid species, shared this complex history and experienced an additional whole-genome triplication (WGT) event 13–17 Ma (Wang et al. 2011; Cheng et al. 2013). Thus, Brassica species afford an opportunity to study genome evolution.
Wang et al. (2011) used A. thaliana as an outgroup to investigate the structural and functional consequences of WGT. Specifically, B. rapa has undergone considerable fractionation since its divergence from A. thaliana; the approximately 42,000 genes in the B. rapa genome are considerably fewer than would be expected from a simple WGT of the approximately 27,000 genes in the A. thaliana genome (Wang et al. 2011). The extent of gene loss varies among the three genome segments. The least fractionated (LF) genome retains approximately 70% of the genes, whereas the medium fractionated (MF1) and most fractionated (MF2) genomes retain ∼46% and ∼36%, respectively (Wang et al. 2011; Tang et al. 2012). The expression of genes in these three subgenomes is also divergent. Cheng, Wu, Fang, Sun, et al. (2012) indicated that the dominantly expressed genes tended to be resistant to fractionation; genes in the LF were dominantly expressed over those in the MFs, whereas the genes in MF1 were slightly dominantly expressed over those in MF2. These traits make B. rapa a good species to use to study the evolutionary patterns of AsA-related genes during genome duplication events.
Genome duplication not only provided abundant genetic material for evolution, but also produced bulk genetic variation that allowed plants to adapt to diversified environments (Hittinger and Carroll 2007). AsA is an important antioxidant in plants and helps prevent oxidative stress in both plants and humans. Because humans have lost the ability to synthesize AsA, its content is an important breeding index for crop plants. Are AsA-related genes preferentially retained during fractionation after WGD? The gene dosage hypothesis predicts that genes in networks or that function in a dose-sensitive manner should be retained, because their products are required for stoichiometric balance (Freeling and Thomas 2006; Birchler and Veitia 2007; Lou et al. 2012). In this study, we tested this hypothesis using AsA-related genes from A. thaliana and B. rapa. The 102 AsA-related genes identified in B. rapa diverged mainly from 12 to 18 Ma, concurrent with the Brassica-specific WGT event. Importantly, fewer AsA-related genes have been completely lost in B. rapa than in three comparison gene sets (a set of neighboring genes, randomly chosen genes, and core eukaryotic genes). The gene structures of these AsA-related sequences are highly conserved in B. rapa, A. thaliana, and other species. However, plant AsA content and the numbers of expressed genes did not increase with the number of AsA-related genes during the WGT event.
Materials and Methods
Identification of AsA-Related Genes and Comparison Gene Sets
The coding sequences of 73 A. thaliana AsA-related genes were retrieved from previous reports for use as the set of reference genes in this study (supplementary table S1, Supplementary Material online). The gene and protein sequences were obtained from The Arabidopsis Information Resource (http://arabidopsis.org/index.jsp, last accessed January 8, 2015; Swarbreck et al. 2008). Sequences of B. rapa homologs to these AsA-related genes in A. thaliana were retrieved from the BRAD database (http://brassicadb.org/brad/, last accessed January 8, 2015) based on a BLASTp search (E-value ≤ 1e−20, identity ≥40%; Altschul et al. 1990; Xu et al. 2013). To rectify incorrect start codon predictions, splicing errors, and missed or extra exons, manual reannotation was performed using FGENESH (http://linux1.softberry.com/berry.phtml?topic=fgenesh&group=programs&subgroup=gfind, last accessed January 8, 2015) with parameters optimized for Arabidopsis. Sequences were then verified in the NCBI database (http://www.ncbi.nlm.nih.gov/, last accessed January 8, 2015). Core eukaryotic genes and random genes were downloaded from CEGMA (Parra et al. 2007) (http://korflab.ucdavis.edu/Datasets/cegma, last accessed January 8, 2015), and selected genes from microsyntenic regions corresponding to the AsA-related genes were used to BLAST search the Brassica database and for synteny analysis. These results were parsed with a Perl program.
Homologs of AsA-related genes of A. thaliana in Vitis vinifera, C. papaya, and Populus trichocarpa were retrieved from Phytozome v9.1 (http://www.phytozome.net/, last accessed January 8, 2015; Goodstein et al. 2012), and Amborella trichopoda genes were retrieved from the Amborella Genome Database (http://www.amborella.org/, last accessed January 8, 2015; Albert et al. 2013).
Synteny Analysis of AsA-Related Genes in A. thaliana and B. rapa
Synteny within and between A. thaliana and B. rapa was constructed by McScanX (http://chibba.pgml.uga.edu/mcscan2/, last accessed January 8, 2015) (MATCH_SCORE: 50, MATCH_SIZE: 5, GAP_SCORE: −3, E_VALUE: 1E−05) (Wang et al. 2012). An all-against-all BLASTP comparison provided the pairwise gene information and the P-value for a primary clustering. Then, paired segments were extended by identifying clustered genes using dynamic programming.
The position of each B. rapa AsA-related gene on the blocks was verified by searching for homologs between A. thaliana and the LF, MF1, and MF2 subgenomes of B. rapa at BRAD (http://brassicadb.org/brad/searchSynteny.php, last accessed January 8, 2015; Cheng, Wu, Fang, Wang, et al. 2012).
Ks Analysis
Coding sequences of A. thaliana AsA-related genes were aligned with those of B. rapa using ClustalW (Thompson et al. 2002). The coding-sequence alignments were regulated using an in-house Perl script. Ks values were calculated based on these alignments using the method of Nei and Gojobori as implemented in KaKs_calculator (Zhang et al. 2006). The Ks values of A. thaliana and C. papaya AsA-related genes were also analyzed.
Phylogenetic Analysis of AsA-Related Genes
For phylogenetic analysis, the protein sequences for AsA-related genes, including the nucleobase–ascorbate transporter (NAT) family and biosynthesis and recycling pathways were aligned using ClustalW2 with default parameters (Thompson et al. 2002). A phylogenetic tree was then constructed by the maximum likelihood method, and bootstrap values were calculated with 1,000 replications using MEGA5.2 (Tamura et al. 2011).
Identification of Conserved Motifs and Gene Ontology
To identify conserved motifs in the AsA-related genes of B. rapa, multiple expectation-maximization for motif elicitation (MEME) v. 4.9.0 (Bailey et al. 2009) was used with default parameters, except that optimum motif width was set to ≥10 and ≤100. The MEME motifs were annotated using SMART (Simple Motif Architecture Research Tool) v. 7.0 (http://smart.embl-heidelberg.de, last accessed January 8, 2015) and the Pfam database (Letunic et al. 2012; Punta et al. 2012). The gene ontology (GO) annotation information of all AsA-related genes in A. thaliana and B. rapa was analyzed by InterProScan program (Quevillon et al. 2005).
Expression Pattern Analysis
For expression profiling of the AsA-related genes in B. rapa, we used the Illumina RNA-seq data that were previously generated and analyzed by Tong et al. (2013). Six tissues of B. rapa accession Chiifu-401-42 (callus, root, stem, leaf, flower, and silique) were analyzed. Transcript abundance was expressed as fragments per kilobase of exon model per million mapped reads (FPKM). The A. thaliana development expression profiling was analyzed by AtGenExpress Visualization Tool with mean-normalized values (Schmid et al. 2005). The AsA-related gene-expression cluster from each tissue in A. thaliana and B. rapa was analyzed using Cluster 3.0 software (http://bonsai.hgc.jp/∼mdehoon/software/cluster/software.htm, last accessed January 8, 2015), and heat maps of the hierarchical clustering were visualized with TreeView (http://jtreeview.sourceforge.net/, last accessed January 8, 2015).
Plant Material and AsA Content
The Chinese cabbage and A. thaliana were used for the experiments. Plants were grown in pots containing a soil: vermiculite mixture (3:1) in the greenhouse of Nanjing Agricultural University, and the controlled-environment growth chamber was programmed for light 16 h/24 °C, dark 8 h/20 °C. AsA content was measured as described previously (Kampfenkel et al. 1995).
Results
AsA-Related Genes in A. thaliana and B. rapa
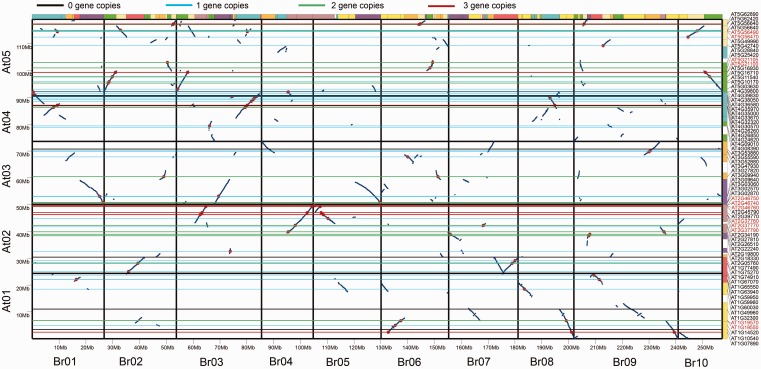
We obtained all 73 AsA-related genes in A. thaliana known from previous reports and the Kyoto Encyclopedia of Genes and Genomes (KEGG; supplementary table S1, Supplementary Material online) (Kanehisa et al. 2012). These sequences served as seeds to identify homoeologs in the B. rapa genome using a combination of BLAST searches and syntenic analysis with MCScanX (Altschul et al. 1990; Wang et al. 2012). The B. rapa genome has undergone WGT since it shared an ancestor with A. thaliana. The B. rapa genome had notably fewer than three times the number of genes in the A. thaliana genome, because some genes were lost after polyploidization (Wang et al. 2011). Here, we identified a total of 219 B. rapa regions syntenic to the A. thaliana AsA-related genes. Sixty-three (87%) A. thaliana AsA-related genes were in regions of three syntenic blocks in B. rapa, five were in two syntenic block regions, and the other five were in four syntenic block regions (fig. 2).
Fig. 2.—
AsA-related homologous genes in segmental syntenic regions of the genomes of Brassica rapa and Arabidopsis thaliana. Conserved collinear blocks of genes (blue irregular lines) are shown between the ten B. rapa chromosomes (horizontal axis) and the five A. thaliana chromosomes (vertical axis). Red dots indicate AsA-related homologs in the two species. The colored horizontal lines denote copy number in B. rapa. The A. thaliana AsA genes are shown on their respective chromosomes.
Based on BLAST results and NCBI analysis, a total of 102 AsA-related gene homoeologs were identified in B. rapa (supplementary table S2, Supplementary Material online). Among them, 95 were located in the syntenic regions and seven homologs were identified at nonsyntenic sites (fig. 2). We identified four regions of homoeologs that had undergone tandem duplication and another that had undergone segmental duplication in B. rapa (supplementary fig. S1A, Supplementary Material online). Comparison of the AsA related gene homoeologs revealed that the position of each homolog on the conserved collinear block has been perfectly maintained throughout the divergent evolution of A. thaliana and B. rapa (fig. 1 and supplementary fig. S1, Supplementary Material online). Known enzymes involved in four possible biosynthesis pathways and the recycling pathway are indicated in figure 1.
Whole-genome analysis of the B. rapa genome has established that the three subgenomes can be distinguished by the degree of fractionation (Wang et al. 2011). To explore this variation, we assigned the AsA-related genes to the LF, MF1, and MF2 subgenomes (supplementary table S3, Supplementary Material online).
Differential Retention of AsA-Related Genes
The gene dosage hypothesis predicts that genes will be preferentially retained if their products are dose sensitive, interacting either with other proteins or in networks (Thomas et al. 2006; Birchler and Veitia 2007). Given the well-conserved synteny between A. thaliana and B. rapa (Cheng, Wu, Fang, Wang, et al. 2012), we compared the retention of the AsA-related genes relative to the set of 1,460 neighboring genes (ten on either side) flanking the 73 AsA-related genes (supplementary table S4, Supplementary Material online). Retention was also analyzed in two other gene sets, a set of 458 core eukaryotic genes and another set of 458 randomly selected genes from the microsyntenic regions corresponding to the AsA-related genes (fig. 3A). Similar numbers (45%) of AsA-related and core eukaryotic genes retained two or three copies, more than in the neighboring and randomly chosen gene sets (fig. 3B). Significantly, only 7 (9.6%) AsA-related genes in B. rapa were completely lost, which was less than that other three comparisons (fig. 3B). Among AsA-related genes, the AsA biosynthesis genes were loss less frequently than the ascorbate transporter and recycling pathway genes and retained mainly two or three copies (fig. 3C), indicating that they were preferentially retained. Among the biosynthetic pathway genes, those in the l-gulose pathway were more frequently lost than those in the l-galactose, galacturonate, and myo-inositol pathways (fig. 3D). However, l-gulose pathway genes were also more frequently retained in three copies. This might be due to the different types of duplication events, such as tandem duplication (supplementary table S3, Supplementary Material online). Interestingly, no gene of galacturonate pathway has been lost since the divergence from the last common ancestor with A. thaliana (fig. 3D).
Fig. 3.—
Retention of homologous copies in the syntenic region. (A) Collinear correlations of genes surrounding AsA genes in the Arabidopsis thaliana and Brassica rapa genomes. The B. rapa and A. thaliana chromosomes are colored according to the inferred ancestral chromosomes following an established convention. The lines representing AsA-related genes are red, those for 458 randomly selected genes are yellow, those for 458 core eukaryotic genes in the syntenic region are blue, and those for AsA-neighboring genes are gray. The figure was created using Circos software. (B) Retention of AsA-related genes and of neighboring, randomly selected, and core eukaryotic genes in the syntenic region after genome triplication and fractionation in Brassica rapa. (C) Retention rates of ascorbate transporter genes, AsA biosynthesis genes, AsA recycling pathway genes, and all AsA-related genes together. (D) Retention rates of genes in four possible AsA biosynthesis pathways, the l-galactose, galacturonate, l-gulose, and myo-inositol pathways.
The proportion of homoeologs retained varied among the LF, MF1, and MF2 subgenomes (supplementary fig. S2, Supplementary Material online). The LF subgenome retained more AsA-related genes (58.9%) found in A. thaliana than the MF1 (39.7%) and MF2 (31.5%) subgenomes (supplementary fig. S2A, Supplementary Material online). In these three subgenomes, AsA-related genes in LF subgenome showed higher retention rates (58.9% vs. 52.9%) than genes in the neighboring gene sets (supplementary fig. S2B, Supplementary Material online).
Analysis of Synonymous Substitution Rates
Using the syntenic orthologs identified for B. rapa and A. thaliana AsA-related genes, we calculated Ks (synonymous substitution rates) and Ka (nonsynonymous substitution rates) values for these homologous genes. In total, 91 syntenic gene pairs were analyzed. The results indicated the Ka/Ks ratios of all AsA-related syntenic orthologs were less than 1, representing purifying selection on the AsA-related genes (supplementary table S5, Supplementary Material online). The Ks values of the B. rapa AsA-related genes ranged from 0.3 to 0.6 and averaged 0.45 (∼15 Myr) (fig. 4A). Based on these calculations and a previous report of Ks values for B. rapa relative to A. thaliana (∼0.42–0.45, ∼14.5 Myr; Cheng et al. 2013), we can conclude that these AsA-related genes diverged following the WGT (13–17 Ma; Wang et al. 2011) in B. rapa and that the pairwise divergences among the three subgenomes are indistinguishable from one other (supplementary table S5, Supplementary Material online and fig. 4B). The different homologous members for one gene showed different evolutionary rates, but the divergence of the homologous members for one gene were mainly concurrent with the Brassica-specific WGT event (supplementary table S5, Supplementary Material online). Simultaneously, we also investigated the divergences of AsA-related genes; we found that the ascorbate transporter genes were less divergent than the biosynthetic and recycling pathway genes (fig. 4C).
Fig. 4.—
Pairwise comparison of Ks values for AsA-related genes. (A) The distribution of Ks values for AsA-related genes between Arabidopsis thaliana and Brassica rapa. The blue line indicates the divergence time (15 Ma). (B) The distribution of Ks values for AsA-related genes between each of the three B. rapa subgenomes and A. thaliana. (C) The distribution of Ks values for AsA transporter, biosynthetic pathway, and recycling pathway. The blue line indicates the main concentrated area of the Ks value (0.4–0.5).
Footprint of the NAT Gene Family in Eudicots
The NAT gene family includes permeases for diverse substrates, such as xanthine, uracil, and vitamin C (de Koning and Diallinas 2000; Argyrou et al. 2001). In A. thaliana, 12 genes encode NAT proteins. They belong to clades I, II, III, and V, whereas the clade IV NAT genes are unique to monocots (Maurino et al. 2006). We characterized this gene family and identified 14 NAT homoeologs in B. rapa (supplementary table S2, Supplementary Material online). Similarly, by genome-wide analysis, we identified nine NATs each in V. vinifera and C. papaya (Jaillon et al. 2007; Ming et al. 2008), 15 in P. trichocarpa, and 7 in Am. trichopoda (Tuskan et al. 2006; Albert et al. 2013; supplementary table S6, Supplementary Material online). Vitis vinifera, P. trichocarpa, and C. papaya were included in our analysis because they did not undergo α and β duplications (Lee et al. 2013). In addition, Am. trichopoda, a basal angiosperm that did not undergo the γ duplication event (Jiao et al. 2011; Albert et al. 2013), was analyzed. To classify these NAT genes, phylogenetic trees were constructed for each species (B. rapa, C. papaya, P. trichocarpa, V. vinifera, and Am. trichopoda) by maximum likelihood using MEGA5 (supplementary fig. S3, Supplementary Material online). In each species, the NAT family was divided into four clades, which we will refer to as clades I–III and clade V, according to the classification of Maurino et al. (2006). Amborella trichopoda had these four NAT clades, indicating that these four clades originated from duplication events prior to the γ event (fig. 5 and supplementary fig. S3, Supplementary Material online).
Fig. 5.—
Copy number variation in the NAT family in eudicots. The phylogenetic tree of NAT genes is shown on the left, and the species tree is shown at the top. The α, β, γ, and salicoid duplications and the Brassica-specific triplication are indicated on the branches of the trees according to the Plant Genome Duplication Database. The NAT-family phylogenetic tree was constructed from protein sequences using maximum likelihood in MEGA5. Numbers are copy numbers of each gene in Brassica rapa (Bra), Arabidopsis thaliana (Ath), Carica papaya (Cpa), Populus trichocarpa (Ptr), Vitis vinifera (Vvi), and Amborella trichopoda (Atr).
However, the NAT genes that were duplicated in those events were mainly in clades II and III. For each of these two clades, only one gene was found in Am. trichopoda, suggesting that they had not duplicated prior to the γ event. The footprints of NAT4 and NAT6 appeared after the γ event, and those of NAT5, NAT8, and NAT9 were found after the α and β duplications in A. thaliana (fig. 5). Specifically, based on Ks values and the study by Bowers et al. (2003), AtNAT5 and AtNAT6 locus duplicated in the β duplication, AtNAT7 and AtNAT8 locus duplicated in the α duplication, whereas AtNAT9 and AtNAT10 locus may duplicated after the α duplication (supplementary table S7, Supplementary Material online). Interestingly, NAT8 and NAT9 were absent in Brassica. In clades I and V, the NAT genes had a high degree of retention, because all six species contained all members. Furthermore, P. trichocarpa and B. rapa contained more family members than did the other four species because of the salicoid duplication and Brassica WGT events, respectively (Tuskan et al. 2006; Wang et al. 2011).
Based on phylogenetic analysis, we inferred a possible evolutionary history of the NAT family (supplementary fig. S4, Supplementary Material online). Before the divergence of Brassicales, the family included all NAT genes except NAT5, NAT8, and NAT9. The gene family further expanded within Brassicaceae. Thus, the NAT family doubled in size in the B. rapa genome compared with that of Am. trichopoda through three duplications, one triplication, and fractionation.
Characteristics, Structure, and Expression Analysis of NAT Proteins
NATs are highly hydrophobic proteins and are predicted to possess membrane-spanning helices (Schwacke et al. 2003). To better understand the characteristics of A. thaliana and B. rapa NAT proteins, we identified all AtNAT and BraNAT proteins using the TMHMM server v. 2.0 (Krogh et al. 2001). The number of α-helical transmembrane helices ranged from 9 to 13 (supplementary table S8, Supplementary Material online), suggesting that their activities were related to substance transportation.
To discover motifs shared among the AtNAT and BraNAT proteins, we identified ten motifs using MEME (Bailey et al. 2009) and annotated them using SMART (Letunic et al. 2012). The annotations indicated that motifs 1–6 and 10 corresponded to the Xan_ur_permease domain, which has transporter activity. In the phylogenetic trees, the NAT proteins generally clustered in subgroups that shared similar motif compositions (supplementary fig. S5, Supplementary Material online), indicating functional similarities among members of the same subgroup. The A. thaliana and B. rapa NAT proteins had similar structures within each clade. Interestingly, the protein structure in clade V was significantly different from those in the other three clades (supplementary fig. S5C, Supplementary Material online). Protein sequence alignments revealed that NAT11 and NAT12 (present in clade V) possessed a highly hydrophilic N-terminal extension of about 120–130 amino acids (supplementary fig. S6, Supplementary Material online).
The tissue-specific (roots, stems, leaves, and flowers) expression patterns of AtNAT and BraNAT genes was studied by using AtGenExpress and B. rapa RNA-seq data (Schmid et al. 2005; Tong et al. 2013), respectively (supplementary fig. S5B, Supplementary Material online). The patterns were diverse, but homologous genes showed similar expression patterns. Notably, little or no signal for NAT10 was detected in either species (supplementary fig. S5B, Supplementary Material online).
Triplication and Fractionation of AsA l-Galactose Pathway Genes
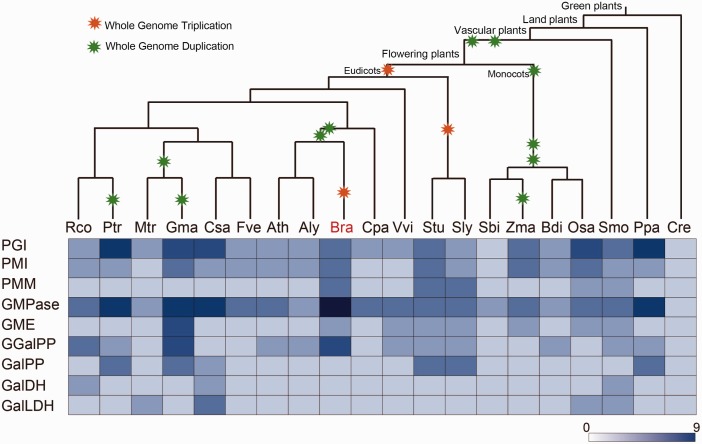
In 1998, the l-galactose (D-Man/L-Gal) pathway was proposed (Wheeler et al. 1998). Several other pathways have been subsequently reported, including the galacturonate, l-gulose, and myo-inositol pathways (fig. 1) (Valpuesta and Botella 2004). However, biochemical and molecular genetic evidence support that the l-galactose pathway is the main source of AsA in plants (Conklin et al. 1999; Tabata et al. 2001; Dowdle et al. 2007). To investigate the triplication and fractionation of the key biosynthesis pathway genes in different species, including Chlorophyta, bryophytes, and angiosperms, we collected relevant enzyme genes from KEGG and considered their evolutionary relationships according to the Plant Genome Duplication Database (supplementary table S9, Supplementary Material online; Kanehisa et al. 2012; Lee et al. 2013). Given that AsA is one of the most important antioxidants, it may be present in the common ancestor of all aerobic organisms. Using these data sets, all of the genes were identified in the common ancestor of embryophytes and some green algae species (Chlamydomonas reinhardtii, Volvox carteri, and Coccomyxa subellipsoidea). For green algae, 9 species were analyzed, including 2 chlorophyceae species, 5 prasinophytes species and 2 trebouxiophyceae species (supplementary table S9, Supplementary Material online). According to the KEGG annotation, except chlorophyceae species, all prasinophytes and one of trebouxiophyceae species lack some genes in this pathway (supplementary table S9, Supplementary Material online). Therefore, the AsA l-galactose biosynthesis pathway might have functioned in all higher plants and few green algae plants. There were no significant differences in the numbers of genes in these 20 plant species (fig. 6), although WGD events occurred, implying that these AsA-related synthase genes had high conservatism and retention.
Fig. 6.—
Deeply conserved AsA l-galactose pathway genes. l-Galactose pathway genes (rows) are conserved among plant families (columns), as indicated by species represented in Plant Genome Duplication Database. Boxes are highlighted if the enzyme-related genes were identified, and the color intensity reflects gene number according to KEGG.
Characteristics and Expression of AsA l-Galactose and Recycling Pathway Genes
In the l-galactose pathway, AsA is synthesized from the precursor d-glucose via nine enzymatic steps. The four upstream steps are responsible for glycolysis (ko00010) and for fructose and mannose metabolism (ko00051), which serves as the substrate for AsA biosynthesis (Kanehisa et al. 2012). The final five steps, starting with GDP-D-mannose, are unique to ascorbate biosynthesis (ko00053; supplementary fig. S7A and C, Supplementary Material online). The expression of these A. thaliana genes in root, stem, leaf, and flower were discussed in this study (supplementary fig. S7B, Supplementary Material online). It revealed that these genes were tissue-specific, but all genes were expressed in these four tissues, especially AtPMM and AtVTC4, implying that they play important roles in AsA biosynthesis. These genes were also analyzed in B. rapa (supplementary fig. S7B, Supplementary Material online). The B. rapa contained one to three homologs of the A. thaliana AsA genes, but the expression of homologous genes was different between both species, indicating the diversification of AsA-related gene regulating. The expression of three genes (BraPMI2, BraPMM.c, and BraVTC1.a) was lower in these tissues than other genes, implying these duplicated genes may be lost their function.
AsA metabolism-related enzymes, such as ascorbate oxidase (AO), ascorbate peroxidase (APX), dehydroascorbate reductase (DHAR), and monodehydroascorbate reductase (MDAR), are normally encoded by genes from multigene families (Chen et al. 2003). Their cycle can be depicted as a triangular loop (fig. 7). In this study, their phylogenetic relationships and expression patterns were analyzed (fig. 7). The proteins that shared a clade were closely related, indicating that they were functionally similar, but some of their expression patterns were different. Two homologous members for DHAR, MDAR, APX, and AO genes, respectively, in B. rapa were found with little expression (fig. 7). It indicated that their function may be lost during the Brassica-specific WGT event. The tissue-specific expression patterns of these genes were found in A. thaliana, whereas high expression in all tissues was found in 11 B. rapa genes (fig. 8). The different expression patterns in this multigene family may help plants adapt to different environments.
Fig. 7.—
Characteristics of the AsA recycling pathway genes and their expression patterns in Arabidopsis thaliana and Brassica rapa. The recycling pathway can be represented as a triangular loop. Gray arrows (reactions) connect metabolites of substrates to products via the corresponding enzymes. Maximum likelihood trees of each of four multigene families (APX, AO, DHAR, and MDAR) were built. Multiple sequence alignment of full-length proteins was performed using ClustalW2, and the phylogenetic trees were constructed using the MEGA5.2. Expression levels of these genes were determined in four tissues (root, stem, leaf, and flower).
Fig. 8.—
Expression patterns analysis of all AsA-related genes in Arabidopsis thaliana and Brassica rapa. Expression levels were analyzed in root, stem, leaf, and flower tissues. (A) The A. thaliana expression profiling was analyzed using the AtGenExpress Visualization Tool with mean-normalized values (supplementary table S10, Supplementary Material online). (B) Heat map of RNA-Seq data for Brassica rapa AsA-related genes. Gene expression FPKM values were analyzed. The bar at the bottom of each heat map represents relative expression values (supplementary table S11, Supplementary Material online). (C) Venn diagram showing the numbers of AsA-related genes with similar and different expression patterns in A. thaliana and B. rapa; those gene names are colored red in (A) and (B).
Function and Expression of AsA-Related Genes in A. thaliana and B. rapa
Based on sequence homology, all 73 AsA-related genes in A. thaliana and 101 such genes in B. rapa could be categorized into 13 functional groups (supplementary fig. S8A, Supplementary Material online); no GO annotation information was available for Bra019082 (BraVTC2.c). In each of the three main GO categories (biological process, cellular component, and molecular function), “binding,” “catalytic,” and “metabolic process” terms were dominant. Furthermore, the percentages of each classification for AsA-related genes were similar in A. thaliana and in B. rapa, indicating that AsA-related genes were highly conserved and may have similar functions in both species. One gene for “Organelle” was found in B. rapa (BraNAT10.a), but none occurred in A. thaliana; perhaps the protein structure of BraNAT10.a changed (supplementary fig. S8A and B, Supplementary Material online).
Through our analysis of gene function, we inferred that the AsA-related genes in A. thaliana and B. rapa may have similar functions. Our expression study revealed that only some of these homologs had similar expression patterns. We also analyzed the expression profile of all the AsA-related genes by cluster analysis in both species (fig. 8A and B). In particular, three genes have no or lower expression in A. thaliana (FPKM value <1.0), while there were 24 such genes in B. rapa. In four different tissues, the gene-expression numbers were similar in A. thaliana and B. rapa (70 and 79), whereas the AsA contents in leaves were also similar (supplementary fig. S9 and table S12, Supplementary Material online). We screened some genes with similar expression by comparing the homologs in these two species (fig. 8C). Approximately one-third of the AsA-related genes had similar expression in both species; 18 of 33 belonged to the LF subgenome of B. rapa. Cheng, Wu, Fang, Sun, et al. (2012) found that genes in the LF subgenome were dominantly expressed over those in the MF subgenomes, consistent with our results for AsA-related genes. We also compared the segmentally duplicated genes in the three subgenomes of B. rapa. Their expression patterns had significantly diverged, except in five gene pairs in the two subgenomes of B. rapa (supplementary fig. S10, Supplementary Material online).
Discussion
Most higher plant lineages have undergone polyploidization during their long evolutionary history. WGD events were important to the evolution of complexity in multicellular eukaryotes (Edger and Pires 2009). After duplication events, some gene copies are retained because they have important functions, while those that are functionally redundant may be lost (Lynch and Conery 2000; Qian et al. 2010). The gene balance hypothesis predicts that genes whose products participate in macromolecular complexes or in transcriptional or signaling networks are more likely to be retained, thus avoiding network instability caused by loss of one member (Birchler and Veitia 2007; Lou et al. 2012). Thus, genes that are highly connected within metabolic networks exhibit preferential retention in A. thaliana (Bekaert et al. 2011). The recycling and biosynthesis pathways involving AsA-related genes form a large network that affects plant growth, development, and stress responses (Arrigoni 1994; Wheeler et al. 1998). Brassica rapa has undergone WGT since it shared a common ancestor with A. thaliana and provides a resource for studying the evolution of polyploid genomes (Wang et al. 2011).
WGD results in gene duplication and is typically followed by substantial gene loss (Lee et al. 2013). To identify intra- or intergenome syntenic relationships among plant genes, we compared the whole-genome sequences of B. rapa with those of the model Brassicaceae species A. thaliana and identified the AsA-related genes. Most AsA-related genes in B. rapa were retained; only 9.6% of such genes in A. thaliana were not found in the B. rapa syntenic regions. Then, we compared the AsA-related genes with a randomly chosen gene set, a set of genes flanking the AsA-related genes, and core eukaryotic gene set; the AsA-related genes were retained at a higher frequency. This preferential retention was statistically significant for AsA-related genes as a whole, was especially evident for the AsA biosynthesis genes, and was weak for the ascorbate transporter genes. This finding may indicate the importance of the AsA biosynthesis genes, some of which are responsible for the biosynthesis of nucleotide sugar, which serves as the substrate not only for AsA biosynthesis but also for the biosyntheses of cell wall polysaccharides and glycoproteins.
The ascorbate transporters are nucleobase transporters. These NATs, also known as the nucleobase: cation symporter-2 family, have been identified in prokaryotes, fungi, plants, and mammals (Maurino et al. 2006). In this study, their numbers were steady in six angiosperm species and these orthologues of different species shared a higher similarity degree than the paralogues of one species (supplementary fig. S4, Supplementary Material online). A strong selection pressure against gene duplication and gene loss might exist in these NAT transporter genes. It was consistent with the ATP-binding cassette transporters (Gbelska et al 2006). Given that highly connected genes within metabolic networks are preferentially retained (Bekaert et al. 2011), the AsA biosynthetic and metabolic genes support the gene dosage hypothesis. Gene loss from the three subgenomes of B. rapa was biased; the LF subgenome preferentially retained AsA genes, similar to the report by Wang et al. (2011) that this subgenome retained more genes in B. rapa.
In addition to analyses of the evolutionary history of AsA-related genes, attempts have been made to predict their functions in diverse species based on sequence similarities and complete genome sequences (Huang et al. 2013; Xu et al. 2013). AsA-related proteins are highly conserved in eukaryotes, with nearly 60% identity among A. thaliana and B. rapa orthologs. These orthologs did not differ significantly among the three subgenomes, and the Ks values supported this conclusion. The orthologs had similar intron and exon numbers (supplementary tables S13 and S14, Supplementary Material online), indicating they may have similar gene structures. The proteins were analyzed by MEME, further proving that they were highly conserved.
During the evolution of higher plants, the basic enzymes of the AsA biosynthesis and recycling pathways have remained almost unchanged (fig. 1), but different numbers of the enzyme genes have yielded multiple interlocking feedback loops. One might logically infer that repeated WGD events facilitated that increase in complexity, highlighting the consequences for AsA content and function of more recent polyploidization events, such as those in B. rapa. In this study, 102 AsA-related genes were found in B. rapa, a species that is the evolutionary product of a Brassica-specific WGT. During evolution, the AsA-related gene network has become increasingly complex (fig. 9); repeated WGD events probably facilitated this progression from lower plants to higher plants. However, the expression patterns of these genes indicated that the numbers of genes expressed in roots, stems, leaves, and flowers were similar in A. thaliana and B. rapa. The AsA contents in leaves were also similar. We inferred that genes in the network that were not expressed were substitutes to prevent network outages caused by sudden failure of a gene or to adapt to the stress. It has been proposed that functionally redundant duplicate genes are used to backup important functions in the event of a severe mutation (Qian et al. 2010). Thus, plant AsA-related genes are highly conserved, and their architectural complexity may be a necessary byproduct of WGD and provided more flexibility to adapt to different environments.
Fig. 9.—
Interaction network of AsA-related genes in Arabidopsis thaliana and Brassica rapa. (A) Specific protein interactions of AsA-related genes in A. thaliana were constructed using STRING (Search Tool for the Retrieval of Interacting Genes/Proteins; http://string-db.org/, last accessed January 8, 2015). (B) The interaction network of AsA-related genes in B. rapa was based on the orthologs in A. thaliana. Ellipses represent AsA-related genes; green indicates genes with high expression levels in leaves, and white indicates those with no or low expression in leaves.
In summary, AsA, a significant antioxidant, protects plants against oxidative damage resulting from aerobic metabolism, photosynthesis, and a range of pollutants (Iqbal et al. 2009). AsA-related genes have been duplicated by WGT events. A total of 102 AsA-related genes were identified in B. rapa (supplementary table S2, Supplementary Material online), and 73 are known in A. thaliana (supplementary table S1, Supplementary Material online). In B. rapa, relatively few (9.6%) AsA-related genes were completely lost compared with three other gene sets (neighboring genes, randomly chosen genes, and core eukaryotic genes). The l-galactose pathway is the main route of AsA biosynthesis (Zhang 2013). Its genes may function in all higher plants, because they were found in all lineages higher than green algae. The expression patterns of homologs in B. rapa were not entirely consistent, indicating diversification of gene transcription regulation in AsA biosynthesis. Furthermore, AsA content and the number of expressed genes did not increase notably with the increase in AsA-related genes after the WGT event. AsA-related genes must be retained for plant growth and survival, especially to protect against oxidative stress, and the AsA content had not been as a breeding objective in B. rapa by humans. The AsA-related genes that are not expressed may act as substitutes during emergencies. Our analyses may provide new opportunities to discover AsA-related genes in A. thaliana and B. rapa, and the bioinformatics results also provided basic resources to examine the molecular regulation of the AsA-related genes in B. rapa. Our findings will help to select appropriate candidate genes for further functional characterization.
Supplementary Material
Supplementary figures S1–S11 and tables S1–S14 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
This work is supported by the National Program on Key Basic Research Projects (The 973 Programs, 2012CB113900, 2009CB119001), the National Natural Science Foundation of China (31272173, 31301782), and the Fundamental Research Funds for the Central Universities of China (KYZ201111), the Jiangsu Province Natural Science Foundation (BK20130673) and China Postdoctoral Science Foundation (2014M550294).
Literature Cited
- Albert VA, et al. The Amborella genome and the evolution of flowering plants. Science. 2013;342:1241089. doi: 10.1126/science.1241089. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Argyrou E, Sophianopoulou V, Schultes N, Diallinas G. Functional characterization of a maize purine transporter by expression in Aspergillus nidulans. Plant Cell. 2001;13:953–964. doi: 10.1105/tpc.13.4.953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrigoni O. Ascorbate system in plant development. J Bioenerg Biomembr. 1994;26:407–419. doi: 10.1007/BF00762782. [DOI] [PubMed] [Google Scholar]
- Bailey TL, et al. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009;37:W202–W208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barth C, De Tullio M, Conklin PL. The role of ascorbic acid in the control of flowering time and the onset of senescence. J Exp Bot. 2006;57:1657–1665. doi: 10.1093/jxb/erj198. [DOI] [PubMed] [Google Scholar]
- Bekaert M, Edger PP, Pires JC, Conant GC. Two-phase resolution of polyploidy in the Arabidopsis metabolic network gives rise to relative and absolute dosage constraints. Plant Cell. 2011;23:1719–1728. doi: 10.1105/tpc.110.081281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birchler JA, Veitia RA. The gene balance hypothesis: from classical genetics to modern genomics. Plant Cell. 2007;19:395–402. doi: 10.1105/tpc.106.049338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowers JE, Chapman BA, Rong J, Paterson AH. Unravelling angiosperm genome evolution by phylogenetic analysis of chromosomal duplication events. Nature. 2003;422:433–438. doi: 10.1038/nature01521. [DOI] [PubMed] [Google Scholar]
- Chen Z, Young TE, Ling J, Chang SC, Gallie DR. Increasing vitamin C content of plants through enhanced ascorbate recycling. Proc Natl Acad Sci U S A. 2003;100:3525–3530. doi: 10.1073/pnas.0635176100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng F, et al. Deciphering the diploid ancestral genome of the mesohexaploid Brassica rapa. Plant Cell. 2013;25:1541–1554. doi: 10.1105/tpc.113.110486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng F, Wu J, Fang L, Sun S, et al. Biased gene fractionation and dominant gene expression among the subgenomes of Brassica rapa. PLoS One. 2012;7:e36442. doi: 10.1371/journal.pone.0036442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng F, Wu J, Fang L, Wang X. Syntenic gene analysis between Brassica rapa and other Brassicaceae species. Front Plant Sci. 2012;3:198. doi: 10.3389/fpls.2012.00198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conklin P, Barth C. Ascorbic acid, a familiar small molecule intertwined in the response of plants to ozone, pathogens, and the onset of senescence. Plant Cell Environ. 2004;27:959–970. [Google Scholar]
- Conklin PL, et al. Genetic evidence for the role of GDP-mannose in plant ascorbic acid (vitamin C) biosynthesis. Proc Natl Acad Sci U S A. 1999;96:4198–4203. doi: 10.1073/pnas.96.7.4198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz-Rus E, Amaya I, Valpuesta V. The challenge of increasing vitamin C content in plant foods. Biotechnol J. 2012;7:1110–1121. doi: 10.1002/biot.201200041. [DOI] [PubMed] [Google Scholar]
- de Koning H, Diallinas G. Nucleobase transporters. Mol Membr Biol. 2000;17:75–94. doi: 10.1080/09687680050117101. [DOI] [PubMed] [Google Scholar]
- Dowdle J, Ishikawa T, Gatzek S, Rolinski S, Smirnoff N. Two genes in Arabidopsis thaliana encoding GDP-l-galactose phosphorylase are required for ascorbate biosynthesis and seedling viability. Plant J. 2007;52:673–689. doi: 10.1111/j.1365-313X.2007.03266.x. [DOI] [PubMed] [Google Scholar]
- Edger PP, Pires JC. Gene and genome duplications: the impact of dosage-sensitivity on the fate of nuclear genes. Chromosome Res. 2009;17:699–717. doi: 10.1007/s10577-009-9055-9. [DOI] [PubMed] [Google Scholar]
- Franceschi VR, Tarlyn NM. L-ascorbic acid is accumulated in source leaf phloem and transported to sink tissues in plants. Plant Physiol. 2002;130:649–656. doi: 10.1104/pp.007062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeling M, Thomas BC. Gene-balanced duplications, like tetraploidy, provide predictable drive to increase morphological complexity. Genome Res. 2006;16:805–814. doi: 10.1101/gr.3681406. [DOI] [PubMed] [Google Scholar]
- Gbelska Y, Krijger JJ, Breunig KD. Evolution of gene families: the multidrug resistance transporter genes in five related yeast species. FEMS Yeast Res. 2006;6:345–355. doi: 10.1111/j.1567-1364.2006.00058.x. [DOI] [PubMed] [Google Scholar]
- Goodstein DM, et al. Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 2012;40:D1178–D1186. doi: 10.1093/nar/gkr944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hittinger CT, Carroll SB. Gene duplication and the adaptive evolution of a classic genetic switch. Nature. 2007;449:677–681. doi: 10.1038/nature06151. [DOI] [PubMed] [Google Scholar]
- Horemans N, Foyer CH, Asard H. Transport and action of ascorbate at the plant plasma membrane. Trends Plant Sci. 2000;5:263–267. doi: 10.1016/s1360-1385(00)01649-6. [DOI] [PubMed] [Google Scholar]
- Huang S, et al. Draft genome of the kiwifruit Actinidia chinensis. Nat Commun. 2013;4:2640. doi: 10.1038/ncomms3640. doi:10.1038/ncomms3640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iqbal Y, Ihsanullah I, Shaheen N, Hussain I. Significance of vitamin C in plants. J Chem Soc Pak. 2009;31:169–170. [Google Scholar]
- Jaillon O, et al. The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature. 2007;449:463–467. doi: 10.1038/nature06148. [DOI] [PubMed] [Google Scholar]
- Jiao Y, et al. Ancestral polyploidy in seed plants and angiosperms. Nature. 2011;473:97–100. doi: 10.1038/nature09916. [DOI] [PubMed] [Google Scholar]
- Kampfenkel K, Vanmontag M, Inze D. Extraction and determination of ascorbate and dehydroascorbate from plant tissue. Anal Biochem. 1995;225:165–167. doi: 10.1006/abio.1995.1127. [DOI] [PubMed] [Google Scholar]
- Kanehisa M, Goto S, Sato Y, Furumichi M, Tanabe M. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res. 2012;40:D109–D114. doi: 10.1093/nar/gkr988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogh A, Larsson B, Von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- Lee TH, Tang H, Wang X, Paterson AH. PGDD: a database of gene and genome duplication in plants. Nucleic Acids Res. 2013;41:D1152–D1158. doi: 10.1093/nar/gks1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I, Doerks T, Bork P. SMART 7: recent updates to the protein domain annotation resource. Nucleic Acids Res. 2012;40:D302–D305. doi: 10.1093/nar/gkr931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine M. New concepts in the biology and biochemistry of ascorbic acid. New Engl J Med. 1986;314:892–902. doi: 10.1056/NEJM198604033141407. [DOI] [PubMed] [Google Scholar]
- Lou P, et al. Preferential retention of circadian clock genes during diploidization following whole genome triplication in Brassica rapa. Plant Cell. 2012;24:2415–2426. doi: 10.1105/tpc.112.099499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M, Conery JS. The evolutionary fate and consequences of duplicate genes. Science. 2000;290:1151–1155. doi: 10.1126/science.290.5494.1151. [DOI] [PubMed] [Google Scholar]
- Maurino VG, et al. Identification and expression analysis of twelve members of the nucleobase–ascorbate transporter (NAT) gene family in Arabidopsis thaliana. Plant Cell Physiol. 2006;47:1381–1393. doi: 10.1093/pcp/pcl011. [DOI] [PubMed] [Google Scholar]
- Ming R, et al. The draft genome of the transgenic tropical fruit tree papaya (Carica papaya Linnaeus) Nature. 2008;452:991–996. doi: 10.1038/nature06856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olmos E, Kiddle G, Pellny T, Kumar S, Foyer C. Modulation of plant morphology, root architecture, and cell structure by low vitamin C in Arabidopsis thaliana. J Exp Bot. 2006;57:1645–1655. doi: 10.1093/jxb/erl010. [DOI] [PubMed] [Google Scholar]
- Parra G, Bradnam K, Korf I. CEGMA: a pipeline to accurately annotate core genes in eukaryotic genomes. Bioinformatics. 2007;23:1061–1067. doi: 10.1093/bioinformatics/btm071. [DOI] [PubMed] [Google Scholar]
- Punta M, et al. The Pfam protein families database. Nucleic Acids Res. 2012;40:D290–D301. doi: 10.1093/nar/gkr1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian W, Liao BY, Chang AYF, Zhang J. Maintenance of duplicate genes and their functional redundancy by reduced expression. Trends Genet. 2010;26:425–430. doi: 10.1016/j.tig.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quevillon E, et al. InterProScan: protein domains identifier. Nucleic Acids Res. 2005;33:W116–W120. doi: 10.1093/nar/gki442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid M, et al. A gene expression map of Arabidopsis thaliana development. Nat Genet. 2005;37:501–506. doi: 10.1038/ng1543. [DOI] [PubMed] [Google Scholar]
- Schwacke R, et al. ARAMEMNON, a novel database for Arabidopsis integral membrane proteins. Plant Physiol. 2003;131:16–26. doi: 10.1104/pp.011577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smirnoff N, Conklin PL, Loewus FA. Biosynthesis of ascorbic acid in plants: a renaissance. Annu Rev Plant Biol. 2001;52:437–467. doi: 10.1146/annurev.arplant.52.1.437. [DOI] [PubMed] [Google Scholar]
- Stebbins CL., Jr . Variation and evolution in plants. London: Oxford University Press; 1950. [Google Scholar]
- Swarbreck D, et al. The Arabidopsis Information Resource (TAIR): gene structure and function annotation. Nucleic Acids Res. 2008;36:D1009–D1014. doi: 10.1093/nar/gkm965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabata K, Ôba K, Suzuki K, Esaka M. Generation and properties of ascorbic acid-deficient transgenic tobacco cells expressing antisense RNA for L-galactono-1, 4-lactone dehydrogenase. Plant J. 2001;27:139–148. doi: 10.1046/j.1365-313x.2001.01074.x. [DOI] [PubMed] [Google Scholar]
- Tamura K, et al. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H, et al. Altered patterns of fractionation and exon deletions in Brassica rapa support a two-step model of paleohexaploidy. Genetics. 2012;190:1563–1574. doi: 10.1534/genetics.111.137349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas BC, Pedersen B, Freeling M. Following tetraploidy in an Arabidopsis ancestor, genes were removed preferentially from one homeolog leaving clusters enriched in dose-sensitive genes. Genome Res. 2006;16:934–946. doi: 10.1101/gr.4708406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Gibson T, Higgins DG. Multiple sequence alignment using ClustalW and ClustalX. Curr Protoc Bioinform. 2002;00:2.3:2.3.1–2.3.22. doi: 10.1002/0471250953.bi0203s00. [DOI] [PubMed] [Google Scholar]
- Tong C, et al. Comprehensive analysis of RNA-seq data reveals the complexity of the transcriptome in Brassica rapa. BMC Genomics. 2013;14:689. doi: 10.1186/1471-2164-14-689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuskan GA, et al. The genome of black cottonwood, Populus trichocarpa (Torr. & Gray) Science. 2006;313:1596–1604. doi: 10.1126/science.1128691. [DOI] [PubMed] [Google Scholar]
- Valpuesta V, Botella MA. Biosynthesis of L-ascorbic acid in plants: new pathways for an old antioxidant. Trends Plant Sci. 2004;9:573–577. doi: 10.1016/j.tplants.2004.10.002. [DOI] [PubMed] [Google Scholar]
- Wang X, et al. The genome of the mesopolyploid crop species Brassica rapa. Nat Genet. 2011;43:1035–1039. doi: 10.1038/ng.919. [DOI] [PubMed] [Google Scholar]
- Wang Y, et al. MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012;40:e49. doi: 10.1093/nar/gkr1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler GL, Jones MA, Smirnoff N. The biosynthetic pathway of vitamin C in higher plants. Nature. 1998;393:365–369. doi: 10.1038/30728. [DOI] [PubMed] [Google Scholar]
- Xu Q, et al. The draft genome of sweet orange (Citrus sinensis) Nat Genet. 2013;45:59–66. doi: 10.1038/ng.2472. [DOI] [PubMed] [Google Scholar]
- Zhang Y. Ascorbic acid in plants. New York: Springer Press; 2013. Ascorbate biosynthesis in plants; pp. 35–43. [Google Scholar]
- Zhang Z, et al. KaKs_Calculator: calculating Ka and Ks through model selection and model averaging. Genomics Proteomics Bioinformatics. 2006;4:259–263. doi: 10.1016/S1672-0229(07)60007-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.