Abstract
Multiple transcontinental waves of drug resistance in Plasmodium falciparum have originated in Southeast Asia before spreading westward, first into the rest of Asia and then to sub-Saharan Africa. In vitro studies have suggested that hypermutator P. falciparum parasites may exist in Southeast Asia and that an increased rate of acquisition of new mutations in these parasites may explain the repeated emergence of drug resistance in Southeast Asia. This study is the first to test the hypermutator hypothesis using field isolates. Using genome-wide SNP data from human P. falciparum infections in Southeast Asia and West Africa and a test for relative rate differences we found no evidence of increased relative substitution rates in P. falciparum isolates from Southeast Asia. Instead, we found significantly increased substitution rates in Mali and Bangladesh populations relative to those in populations from Southeast Asia. Additionally we found no association between increased relative substitution rates and parasite clearance following treatment with artemisinin derivatives.
Keywords: Plasmodium falciparum, drug resistance, artemisinin, mutation rate, molecular evolution
1. INTRODUCTION
Resistance to chloroquine, an antimalarial drug used widely in population-level malaria control efforts, was first identified in western Cambodia in 1957, 20 years before its appearance in eastern Africa [1]. Following the introduction of other new antimalarial drugs, new resistance mutations have repeatedly originated in Southeast Asia, specifically western Cambodia [2], and subsequently spread westward toward Africa [3]. Most recently, clinical resistance to artemisinin, and mutations in the K13 propeller gene with which artemisinin resistance is strongly associated, were both first identified in western Cambodia [4,5], although it appears that K13 mutations have arisen in multiple independent foci in Southeast Asia [6].
Possible reasons for why resistance mutations in Plasmodium falciparum have repeatedly emerged in western Cambodia include antimalarial usage practices [7] and differences in transmission intensity of P. falciparum parasite populations, which in turn affect host immunity [8]. Another explanation for this phenomenon is suggested by in vitro evidence that P. falciparum isolates from Southeast Asia acquire new drug resistance mutations at higher rates than isolates from West Africa [9]. The identification of hypermutator P. falciparum lineages in the field, and evidence linking these lineages to emergent drug resistance mutations, would have important implications for malaria control and drug resistance containment strategies. Hypermutator phenotypes are common among some eubacterial pathogens under drug pressure [10]; however, to our knowledge, this phenomenon has never been observed in a eukaryotic parasite. Likewise, the evidence supporting the “hypermutator hypothesis” has come from in vitro studies on culture-adapted laboratory isolates, often using P. falciparum strains that have passed through thousands of generations of drug pressure. To date, there is no population-level evidence on mutation rate variation in P. falciparum isolates from human infections in the field.
In this study, we examined mutation rate variation in 177 P. falciparum isolates collected in clinical trials in Southeast Asia and in Mali, West Africa, using whole-genome sequencing data and a test of relative nucleotide substitution rates. In addition, we used clinical data on efficacy of artemisinin derivatives to examine the relationship between relative substitution rate and an emerging drug resistance phenotype.
2. METHODS
P. falciparum isolates were collected during artemisinin therapeutic efficacy trials in Bangladesh, Cambodia, Laos, Myanmar, Thailand, and Vietnam [6,11] and a Plasmodium population genetics study in Mali [12]. Isolates originated from a wide geographic range of the distribution of P. falciparum, and included those associated with known artemisinin-resistant phenotypes and lineages from areas where this phenotype is either uncommon or non-existent. We included all available samples from these sites for which complete sequencing data were available. ACTs are the recommended first-line treatment for confirmed uncomplicated P. falciparum infection in all six countries. During the study period, the reported maximum number of ACT treatment courses delivered per year was 2,842,500 (Mali, 2008) and the minimum was 51,425 (Laos, 2010) [13]. Compared to Bangladesh and Southeast Asia, P. falciparum transmission is significantly higher in Mali, where the parasite prevalence among children ages 2-10 years is >40% and the entomological inoculation rate is greater than 100 infective bites per individual [14].
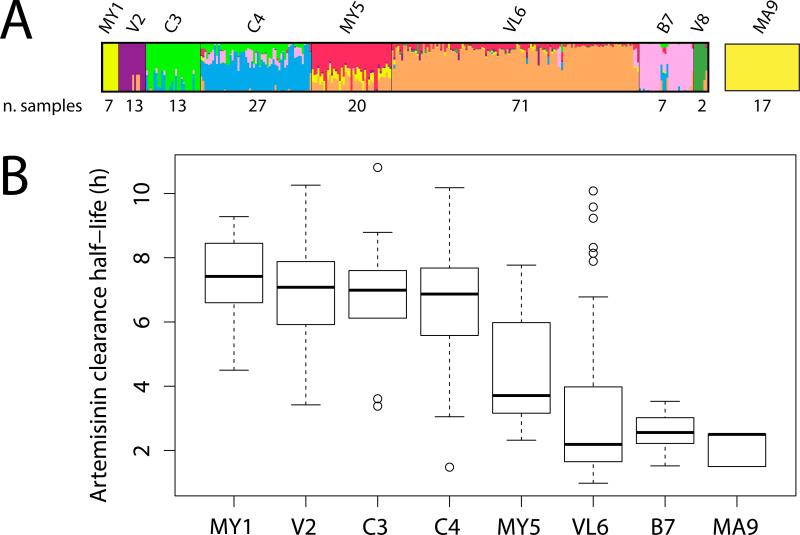
Sequencing, alignment, SNP calling, and quality filtering for these data have been described previously [15]. SNPs were detected using sequencing data for all isolates in the study that were amenable to sequencing, minimizing possible ascertainment bias in the identification of polymorphic sites. Population structure of isolates from Southeast Asia was estimated using the STRUCTURE software package [16], which employs Bayesian clustering methods to infer population number and membership from multilocus genotype data. The STRUCTURE run that achieved the highest estimated probability, without grouping samples by individual clone lineages, included eight populations (Figure 1). To exclude highly admixed samples, which could interfere with accurate relative rate comparisons between populations, isolates with membership coefficients in their assigned population of less than 0.50 were excluded from analysis. In addition, isolates with >5% missing SNP calls and those that may have represented polyclonal infections (defined as those samples with >0.005% heterozygous SNP calls) were excluded from subsequent analysis. Isolates from Mali were assumed to represent a separate population distinct from Southeast Asia isolates, without any significant substructure, consistent with previous analyses of P. falciparum population structure in Africa [17]. These steps yielded nine geographically distinct populations, of which eight included a sufficient number of samples for relative rate analysis.
Figure 1.
(A) Results of the STRUCTURE analysis with eight populations and number of samples after exclusion of polyclonal isolates and isolates with >5% missing SNP calls, for isolates from Bangladesh (B), Cambodia (C), Laos (L), Mali (MA), Myanmar (MY), Vietnam (V), and Vietnam-Laos (VL). Malian isolates were not included in the STRUCTURE analysis, but assumed to constitute a separate population, distinct from Southeast Asia, with no significant substructure. Following filtering to include only non-admixed, non-polyclonal samples with low amounts of missing data, the Vietnamese population V8 contained only two samples, and was excluded from subsequent analysis. (B) In vivo artemisinin parasite clearance half-lives by population. Parasite clearance half-lives for isolates in populations MY1-B7 were measured directly for each isolate. For isolates in population MA9, half-lives were extrapolated from previously published values for isolates collected in Mali between 2010-2011[32].
Nucleotide substitution rates were compared using the relative rate test of Tajima [18]. In brief, the test compares the number of unique nucleotide substitutions, m1 and m2, in two sequences, S1 and S2, (sampled from populations 1 and 2, respectively) versus an outgroup sequence. The sum of nucleotide sites where S1 ≠ S2 = outgroup are counted as unique substitutions for S1 and equals m1, and the sum of sites where S2 ≠ S1 = outgroup is m2. Sites in which S1 ≠ S2 ≠ outgroup or S1 = S2 ≠ outgroup are ignored. If S1 and S2 have the same rate of nucleotide substitution, the number of unique substitutions is expected to be equal between populations 1 and 2, such that E(m1) = E(m2). The mj value for each population was obtained by averaging over samples within that population. The values m1 and m2 were compared using a χ2 statistic. The probability of observed χ2 values was estimated by randomly permuting population assignments of individual samples. Whole-genome sequence data from Plasmodium reichenowi (Berriman et al., in press) was used as the outgroup sequence, and was aligned to known orthologous regions in the P. falciparum 3D7 reference genome using MAFFT version 7.154 [19]. Only SNPs in protein-coding regions were considered.
We applied this method to two sets of P. falciparum SNP data: (1) the complete set of all 418,463 synonymous and non-synonymous SNPs where the orthologous nucleotide in P. reichenowi could be determined; and (2) a subset of data set (1), which includes only synonymous SNPs, and excluding sites that fall in genomic regions with evidence of recent positive selection, as previously determined using analyses of extended haplotype homozygosity [11]. The first data set provided the maximum available power for detecting relative rate differences and the second yielded more conservative relative rate estimates based only on substitutions at 125,063 ostensibly neutral sites. With the large number of sites analyzed, the tests had high power to detect relative rate differences [20]. In addition, comparing mean rates among groups of isolates reduced variance through averaging, a well known statistical effect, and reduced error due to the stochastic distribution of substitutions among lineages [21].
3. RESULTS
We detected significant increases in both the mean rate of synonymous substitution and the mean rate of synonymous plus non-synonymous substitution in samples from Mali and Bangladesh versus those from Southeast Asia (Fig 2A). Large effect sizes are observed for all statistically significant comparisons in both data sets (Supplementary Figure 1). Furthermore, we observed little substitution rate variation among populations in Southeast Asia, including those comprised mostly of artemisinin-resistant and of artemisinin-sensitive parasites (Fig 2A, populations MY1-VL6). For none of the possible pairwise relative rate comparisons between Malian and Southeast Asian isolates did we observe a Southeast Asian isolate with a significantly higher relative rate than a Malian isolate (Figure 1, B), indicating that, among all samples analyzed, no individual parasite exists within the Southeast Asian populations for which the substitution rate is higher than that of a Malian parasite. There was no significant correlation between parasite clearance half-life after artemisinin treatment and the relative rate of synonymous substitution (Mantel test: r=0.003, p value=0.432, 6000 permutations).
Figure 2.
(A) Difference in mean number of unique nucleotide substitutions (mCOLUMN-mROW) and χ2 values for 418,463 synonymous and non-synonymous sites (below the diagonal of identity values) and 125,063 synonymous sites excluding SNPs in regions inferred to be under recent positive selection (above the diagonal of identity values). χ2 values for each population-population comparison were determined using methods described in the text, with P. reichenowi as the outgroup. 10,000 random permutations of population assignments were used to generate the null distribution of χ2 values and p-value for each population-population comparison. Relative rate comparisons with p-values less than the Bonferonni-corrected value of 0.0018 are marked with an asterisk. (B) Distribution of χ2 values for all individual relative rate comparisons between Malian isolates (population MA9) and isolates from Southeast Asia (Cambodia, Myanmar, Laos, or Vietnam) multiplied by d, a constant equal to 1 or -1 which reflects the directionality of the relative rate difference (where for each isolate i from Southeast Asia and isolate j from Mali). Values of > 3.85 (green plot region) indicate individual relative rate comparisons where the Malian isolate has a significantly greater relative rate (p < 0.05 for χ2 with 1 d.f.); values of < −3.85 (blue plot region) indicate relative rate comparisons where the isolate from Southeast Asia (SEA) has a significantly greater relative rate. Left panel: Relative rate comparisons based on 125,063 synonymous SNPs; Right panel: Relative rate comparisons based on 418,463 synonymous and non-synonymous SNPs.
Multiple highly drug-resistant P. falciparum isolates exhibit defective DNA mismatch repair phenotypes [22], suggesting a possible mechanistic link between increased substitution rate and the acquisition of drug resistance. Recently, Miotto et al. identified 32 SNPs in P. falciparum DNA repair genes that were highly differentiated in Cambodian isolates with delayed parasite clearance phenotypes, including polymorphisms in the DNA repair-related genes MLH1, pms1, and RAD50 [17]. We tested for relative rate differences between isolates carrying the reference versus non-reference allele for these SNPs in DNA repair genes (30 SNPs; for 2 SNPs there were no samples carrying the non-reference allele in our data). We found only a single instance where isolates bearing the non-reference SNP had a significantly higher number of unique mutations, a non-synonymous mutation in a hypothetical repair-related gene on chromosome 11 (MAL11:480718, p < 1.6 X 10−4 in the combined SNP data set); however, this rate difference was not observed for synonymous SNPs and the effect size for this difference was small (0.16, 95% CI:-0.14-0.46). Moreover, the effect sizes for all rate differences in this analysis were small, with 95% confidence intervals spanning both negative and positive values.
4. DISCUSSION
We found no evidence that P. falciparum parasites in Southeast Asia isolated from humans had elevated substitution rates when compared to parasites from West Africa, nor did we find elevated substitution rates among populations with slower parasite clearance half-lives. Indeed, we observed the opposite: isolates from Mali and Bangladesh, populations that both exhibit rapid parasite clearance half-lives after artemisinin treatment, had more substitutions than those from Southeast Asia when compared to the outgroup species, including those at synonymous sites located outside regions potentially affected by recent selective sweeps. Although these differences achieve statistical significance, their magnitude is small relative to the total number of polymorphic sites in our analysis. For example, the increase in proportion of substitutions per site observed in the Malian populations is 0.00017 - 0.00035 for synonymous and non-synonymous substitutions combined; and 0.00013 - 0.00039 for synonymous substitutions only (Figure 2). Consistent with this analysis, recent mutation accumulation experiments have shown that rates of mitotic evolution in P. falciparum do not vary significantly between 3D7-derived clones (a laboratory strain originally from west Africa) and clones derived from the highly drug-resistant Southeast Asian isolate Dd2 [23].
The small but statistically significant variation in substitution rates observed in our data may be attributable to underlying differences in mutation rates and/or differences in the fixation rate of new mutations. Linkage disequilibrium (LD) blocks are markedly shorter in African P. falciparum populations versus those in Southeast Asia [24], likely due to higher rates of effective sexual outcrossing and recombination in high transmission settings, increasing the ability of natural selection to act independently on linked mutations. In Southeast Asia, where haplotype blocks can extend over large chromosomal regions, new mutations are more likely to be affected by background selection [25]. The fact that the difference in substitution rate per site between countries is small and extends to synonymous sites suggest that the sites affected include slightly advantageous polymorphisms involved in processes such as mRNA stability or translation efficiency [26-28]. Underlying differences in mutation rate may also contribute to the observed variation in substitution rates, either due to inherent differences in DNA repair competency or the increased number of P. falciparum sexual generations per year that occur in areas of higher malaria transmission (for example, sub-Saharan Africa). P. falciparum has a high rate of recombination [29], which has been shown to be mutagenic on some eukaryotic taxa [30], and hence could differently impact populations that differ in rate of meiotic division.
Our results do not support the hypermutator hypothesis, and suggest that other explanations underlie the recurrent emergence in Southeast Asia of new drug resistance mutations. A distinct possibility is that the lower transmission intensity observed in Southeast Asia relative to Mali would be expected to result in a higher proportion of monoclonal infections and in lower infection rates in human populations in Southeast Asia. These conditions might increase the role of genetic drift relative to selection in the fate of new mutations, and therefore be more conducive to an increase in frequency, and within population fixation, of mutations that are deleterious [31] as many drug resistant mutations are known to be [3,8].
Supplementary Material
Supplementary Figure S1. Effect sizes (Hedge's g) for 28 population-population relative rate tests, calculated using the mean number of unique mutations in each population, for 418,463 synonymous and non-synonymous sites (lower triangular matrix) and 125,063 synonymous sites filtered to exclude SNPs in regions inferred to be under recent positive selection (upper triangular matrix).
HIGHLIGHTS.
-
(1)
We compared nucleotide substitution rates between P. falciparum subpopulations.
-
(2)
We compared 177 isolates from Mali, Bangladesh, and Southeast Asia.
-
(3)
Average rates of synonymous substitution were slightly higher in isolates from Mali
-
(4)
We found no association between faster substitution rates and artemisinin resistance.
Acknowledgements
We thank the Malaria Programme staff at the Wellcome Trust Sanger Institute, including Dominic Kwiatkowski, Olivo Miotto, Bronwyn MacInnis, Daniel Mead, Eleanor Drury, Susana Campino, Magnus Mankse, and James Stalker, who generated the whole genome sequencing data. In addition, we thank Dr. Timothy O'Connor for his constructive feedback on the manuscript. This work was supported by grants R03AI101680 (CVP) and R01AI101713 (ST-H) and U19AI10820 (JCS, CVP) from the National Institute of Allergy and Infectious Diseases and by the Howard Hughes Medical Institute. TSB was a Howard Hughes Medical Institute Medical Research Fellow.
ABBREVIATIONS
- ACT
artemisinin combination therapy
- SNP
single nucleotide polymorphism
- LD
linkage disequilibrium
Contributor Information
Tyler S. Brown, Johns Hopkins University School of Medicine, Baltimore, MD, USA and Howard Hughes Medical Institute / Center for Vaccine Development, University of Maryland School of Medicine, Baltimore, MD, USA
Christopher G. Jacob, Howard Hughes Medical Institute / Center for Vaccine Development, University of Maryland School of Medicine, Baltimore, MD, USA
Joana C. Silva, Institute for Genome Sciences and Department of Microbiology and Immunology, University of Maryland School of Medicine, Baltimore, MD, USA
Shannon Takala-Harrison, Howard Hughes Medical Institute / Center for Vaccine Development, University of Maryland School of Medicine, Baltimore, MD, USA.
Abdoulaye Djimdé, Malaria Research and Training Center, Department of Epidemiology of Parasitic Diseases, Faculty of Medicine and Pharmacy, University of Science, Techniques and Technology of Bamako, Bamako, Mali.
Arjen M. Dondorp, Mahidol-Oxford Tropical Medicine Research Unit, Faculty of Tropical Medicine, Mahidol University, Bangkok, Thailand and Center for Tropical Medicine, Nuffield Department of Medicine, Churchill Hospital, University of Oxford, Oxford, United Kingdom
Mark Fukuda, Armed Forces Research Institute of Medical Sciences, Bangkok, Thailand.
Harald Noedl, Institute of Specific Prophylaxis and Tropical Medicine, Medical University of Vienna, Austria and Malaria Research Initiative Bandarban, Bandarban, Bangladesh.
Myaing Myaing Nyunt, Howard Hughes Medical Institute / Center for Vaccine Development, University of Maryland School of Medicine, Baltimore, MD, USA.
Myat Phone Kyaw, Department of Medical Research (Lower Myanmar), Yangon, Myanmar.
Mayfong Mayxay, Lao-Oxford-Mahosot Hospital-Wellcome Trust Research Unit (LOMWRU), Microbiology Laboratory, Mahosot Hospital, Vientiane, Lao PDR and Faculty of Postgraduate Studies, University of Health Sciences, Vientiane, Lao PDR.
Tran Tinh Hien, Centre for Tropical Medicine Oxford University Clinical Research Unit Vietnam (OUCRU), Ho Chi Minh City, Vietnam.
Christopher V. Plowe, Howard Hughes Medical Institute / Center for Vaccine Development, University of Maryland School of Medicine, Baltimore, MD, USA
Michael P. Cummings, Laboratory of Molecular Evolution, Center for Bioinformatics and Computational Biology, University of Maryland, College Park, Maryland, USA
REFERENCES
- 1.Payne D. Spread of chloroquine resistance in Plasmodium falciparum. Parasitol Today. 1987;3:241–246. doi: 10.1016/0169-4758(87)90147-5. [DOI] [PubMed] [Google Scholar]
- 2.Anderson TJ, Roper C. The origins and spread of antimalarial drug resistance: lessons for policy makers. Acta Trop. 2005;94:269–280. doi: 10.1016/j.actatropica.2005.04.010. [DOI] [PubMed] [Google Scholar]
- 3.Plowe CV. The evolution of drug-resistant malaria. Trans R Soc Trop Med Hyg. 2009;103(Suppl 1):S11–14. doi: 10.1016/j.trstmh.2008.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dondorp AM, Nosten F, Yi P, Das D, Phyo AP, et al. Artemisinin resistance in Plasmodium falciparum malaria. N Engl J Med. 2009;361:455–467. doi: 10.1056/NEJMoa0808859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ariey F, Witkowski B, Amaratunga C, Beghain J, Langlois AC, et al. A molecular marker of artemisinin-resistant Plasmodium falciparum malaria. Nature. 2014;505:50–55. doi: 10.1038/nature12876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Takala-Harrison S, Jacob CG, Arze C, Cummings MP, Silva JC, et al. Independent Emergence of Artemisinin Resistance Mutations Among Plasmodium falciparum in Southeast Asia. J Infect Dis. 2014 doi: 10.1093/infdis/jiu491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Packard RM. The origins of antimalarial-drug resistance. N Engl J Med. 2014;371:397–399. doi: 10.1056/NEJMp1403340. [DOI] [PubMed] [Google Scholar]
- 8.Plowe CV, Kublin JG, Doumbo OK. P. falciparum dihydrofolate reductase and dihydropteroate synthase mutations: epidemiology and role in clinical resistance to antifolates. Drug Resist Updat. 1998;1:389–396. doi: 10.1016/s1368-7646(98)80014-9. [DOI] [PubMed] [Google Scholar]
- 9.Rathod PK, McErlean T, Lee PC. Variations in frequencies of drug resistance in Plasmodium falciparum. Proc Natl Acad Sci U S A. 1997;94:9389–9393. doi: 10.1073/pnas.94.17.9389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marvig RL, Johansen HK, Molin S, Jelsbak L. Genome analysis of a transmissible lineage of pseudomonas aeruginosa reveals pathoadaptive mutations and distinct evolutionary paths of hypermutators. PLoS Genet. 2013;9:e1003741. doi: 10.1371/journal.pgen.1003741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Takala-Harrison S, Clark TG, Jacob CG, Cummings MP, Miotto O, et al. Genetic loci associated with delayed clearance of Plasmodium falciparum following artemisinin treatment in Southeast Asia. Proc Natl Acad Sci U S A. 2013;110:240–245. doi: 10.1073/pnas.1211205110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Auburn S, Campino S, Miotto O, Djimde AA, Zongo I, et al. Characterization of within-host Plasmodium falciparum diversity using next-generation sequence data. PLoS One. 2012;7:e32891. doi: 10.1371/journal.pone.0032891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.World Health Organization World Malaria Report. 2010 [Google Scholar]
- 14.Gething PW, Patil AP, Smith DL, Guerra CA, Elyazar IR, et al. A new world malaria map: Plasmodium falciparum endemicity in 2010. Malar J. 2011;10:378. doi: 10.1186/1475-2875-10-378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Manske M, Miotto O, Campino S, Auburn S, Almagro-Garcia J, et al. Analysis of Plasmodium falciparum diversity in natural infections by deep sequencing. Nature. 2012;487:375–379. doi: 10.1038/nature11174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miotto O, Almagro-Garcia J, Manske M, Macinnis B, Campino S, et al. Multiple populations of artemisinin-resistant Plasmodium falciparum in Cambodia. Nat Genet. 2013;45:648–655. doi: 10.1038/ng.2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tajima F. Simple methods for testing the molecular evolutionary clock hypothesis. Genetics. 1993;135:599–607. doi: 10.1093/genetics/135.2.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bromham L, Penny D, Rambaut A, Hendy MD. The power of relative rates tests depends on the data. J Mol Evol. 2000;50:296–301. doi: 10.1007/s002399910034. [DOI] [PubMed] [Google Scholar]
- 21.Schwartz RS, Mueller RL. Variation in DNA substitution rates among lineages erroneously inferred from simulated clock-like data. PLoS One. 2010;5:e9649. doi: 10.1371/journal.pone.0009649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Castellini MA, Buguliskis JS, Casta LJ, Butz CE, Clark AB, et al. Malaria drug resistance is associated with defective DNA mismatch repair. Mol Biochem Parasitol. 2011;177:143–147. doi: 10.1016/j.molbiopara.2011.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bopp SE, Manary MJ, Bright AT, Johnston GL, Dharia NV, et al. Mitotic evolution of Plasmodium falciparum shows a stable core genome but recombination in antigen families. PLoS Genet. 2013;9:e1003293. doi: 10.1371/journal.pgen.1003293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Volkman SK, Sabeti PC, DeCaprio D, Neafsey DE, Schaffner SF, et al. A genome-wide map of diversity in Plasmodium falciparum. Nat Genet. 2007;39:113–119. doi: 10.1038/ng1930. [DOI] [PubMed] [Google Scholar]
- 25.Charlesworth B. The effects of deleterious mutations on evolution at linked sites. Genetics. 2012;190:5–22. doi: 10.1534/genetics.111.134288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chamary JV, Hurst LD. Evidence for selection on synonymous mutations affecting stability of mRNA secondary structure in mammals. Genome Biol. 2005;6:R75. doi: 10.1186/gb-2005-6-9-r75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Plotkin JB, Kudla G. Synonymous but not the same: the causes and consequences of codon bias. Nat Rev Genet. 2011;12:32–42. doi: 10.1038/nrg2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shabalina SA, Spiridonov NA, Kashina A. Sounds of silence: synonymous nucleotides as a key to biological regulation and complexity. Nucleic Acids Res. 2013;41:2073–2094. doi: 10.1093/nar/gks1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jiang H, Li N, Gopalan V, Zilversmit MM, Varma S, et al. High recombination rates and hotspots in a Plasmodium falciparum genetic cross. Genome Biol. 2011;12:R33. doi: 10.1186/gb-2011-12-4-r33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang SW, Friedman R, Yu N, Yu A, Li WH. How strong is the mutagenicity of recombination in mammals? Mol Biol Evol. 2005;22:426–431. doi: 10.1093/molbev/msi025. [DOI] [PubMed] [Google Scholar]
- 31.Hartl DL, Clark AG. Principles of Population Genetics. Sinauer Associates; Massachusetts: 2006. [Google Scholar]
- 32.Maiga AW, Fofana B, Sagara I, Dembele D, Dara A, et al. No evidence of delayed parasite clearance after oral artesunate treatment of uncomplicated falciparum malaria in Mali. Am J Trop Med Hyg. 2012;87:23–28. doi: 10.4269/ajtmh.2012.12-0058. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1. Effect sizes (Hedge's g) for 28 population-population relative rate tests, calculated using the mean number of unique mutations in each population, for 418,463 synonymous and non-synonymous sites (lower triangular matrix) and 125,063 synonymous sites filtered to exclude SNPs in regions inferred to be under recent positive selection (upper triangular matrix).