Abstract
Identifying biomarkers in body fluids may improve the noninvasive detection of colorectal cancer. Previously, we identified N-Myc downstream-regulated gene 4 (NDRG4) and GATA binding protein 5 (GATA5) methylation as promising biomarkers for colorectal cancer in stool DNA. Here, we examined the utility of NDRG4, GATA5, and two additional markers [Forkhead box protein E1 (FOXE1) and spectrin repeat containing nuclear envelope 1 (SYNE1)] promoter methylation as biomarkers in plasma DNA. Quantitative methylation-specific PCR was performed on plasma DNA from 220 patients with colorectal cancer and 684 noncancer controls, divided in a training set and a test set. Receiver operating characteristic analysis was performed to measure the area under the curve of GATA5, NDRG4, SYNE1, and FOXE1 methylation. Functional assays were performed in SYNE1 and FOXE1 stably transfected cell lines. The sensitivity of NDRG4, GATA5, FOXE1, and SYNE1 methylation in all stages of colorectal cancer (154 cases, 444 controls) was 27% [95% confidence interval (CI), 20%–34%), 18% (95% CI, 12%–24%), 46% (95% CI, 38%– 54%), and 47% (95% CI, 39%–55%), with a specificity of 95% (95% CI, 93%–97%), 99% (95% CI, 98%–100%), 93% (95% CI, 91%–95%), and 96% (95% CI, 94%–98%), respectively. Combining SYNE1 and FOXE1, increased the sensitivity to 56% (95% CI, 48%–64%), while the specificity decreased to 90% (95% CI, 87%–93%) in the training set and to 58% sensitivity (95% CI, 46%–70%) and 91% specificity (95% CI, 80%–100%) in a test set (66 cases, 240 controls). SYNE1 overexpression showed no major differences in cell proliferation, migration, and invasion compared with controls. Overexpression of FOXE1 significantly decreased the number of colonies in SW480 and HCT116 cell lines. Overall, our data suggest that SYNE1 and FOXE1 are promising markers for colorectal cancer detection.
Introduction
Early detection of colorectal cancer offers opportunity to cure colorectal cancer (1). Colonoscopy is the gold standard for colorectal cancer screening, however this procedure is invasive, expensive, and not readily accessible or acceptable to the majority of age-eligible individuals (2). Therefore, the search for noninvasive screening methods has intensified. Currently, fecal immunochemical test (FIT) is used as a noninvasive and relatively inexpensive colorectal cancer screening modality. In addition, potential biomarkers in stool DNA have been described. Despite the high sensitivity and specificity of some of these markers in stool DNA, we hypothesized that a blood-based test, not depending on stool sampling, has the potential for better patient compliance and is better suited for systems without programmatic screening.
Epigenetic silencing of tumor-suppressor genes by aberrant promoter methylation frequently occurs in human cancers (3). Promoter methylation is suggested to be an early event in carcinogenesis and can be detected in biologic fluids in various cancers (4–7). Body fluids that have been used for cancer screening with methylation markers include urine (8), ejaculates of men (9), salivary rinses (10), sputum (11), peritoneal fluid (12), and ductal lavage and nipple fluid (13, 14) highlighting the potential for application in routine clinical practice.
For colorectal cancer, we and others have shown that detection of promoter methylation in fecal DNA holds promise as a colorectal cancer prescreening modality (7, 15–20). Genes known to be methylated, detected, and studied in tumor-derived DNA in blood of patients with colorectal cancer are ALX4 (21), TMEFF2 (22), CDKN2A (p16; ref. 23), CDH4 (24), NEUROG1 (25), NGFR (22), SEPT9 (22), MLH1 (26, 27), DAPK (28), THBD (29), SDC2 (30), and gene panels consisting of APC, MLH1, and HLTF (27), and APC, MGMT, RASSF2A, and Wif-1 (31). The sensitivity and specificity to detect colorectal cancer observed in these studies range from 21% to 86% and from 69% to 100%, respectively.
Our objective was to examine promoter methylation of two previously identified stool markers (NDRG4 and GATA5; refs. 15, 18) and two novel markers namely FOXE1 and SYNE1 (32), as potential biomarkers for the early detection of colorectal cancer in blood DNA. FOXE1 and SYNE1 were identified as frequently methylated genes using a transcriptome-wide approach to identify genes that are transcriptionally silenced by methylation in colorectal cancer (32). In addition, methylation of SYNE1 and FOXE1 has been described in a small cohort of patients with colitis-associated colorectal neoplasia (33).
Performance of the best combinatorial marker panel was evaluated by quantitative methylation analysis in two large sets of plasma samples from patients with colorectal cancer and controls. Furthermore, the currently unknown functional role of SYNE1 and FOXE1 in colorectal cancer was investigated.
Materials and Methods
Study population plasma samples
Two hundred and twenty plasma samples were prospectively collected from patients with colorectal cancer from multiple centers in Germany. Symptomatic patients were screened using colonoscopy and the clinical diagnosis of colorectal cancer was confirmed by histology. The trial started in 2007 and recruited patients with all disease stages. Included patients were diagnosed with colorectal cancer, had not been treated for colorectal cancer before blood collection, had not been treated for other malignancies during the previous 5 years and had surgery planned to assess the UICC stage and the involvement of lymph nodes.
Control blood samples (n = 664) were collected from 550 asymptomatic average risk and 134 symptomatic individuals, all without adenomas and/or colorectal cancer detected by colonoscopy screening. These individuals were enrolled in a multicenter colorectal cancer screening trial in Germany of average risk subjects. Participants underwent primary colonoscopy screening and blood samples were drawn before the procedure. Patient characteristics of patients with colorectal cancer and controls are shown in Supplementary Table S1. Plasma samples of patients with colorectal cancer and controls were randomized and divided in two different sets, one training and one test set, as depicted in Supplementary Fig. S1. Informed consent was obtained from all participants, adhering to ethics guidelines.
Collection and isolation of plasma samples
Nine milliliters of blood, using 10 mL EDTA Vacutainer tubes (BD Vacutainer; BD Hemogard, K2 EDTA spray-dried; cat no. 367525), was collected using standard venipuncture techniques. Plasma was separated by centrifugation at 1,500 × g for 15 minutes (double spin) within 2 hours of collection. DNA isolation from plasma was performed using the QIAamp Circulating Nucleic Acid Test Kit (Qiagen; cat no. 55114).
Sodium bisulfite treatment and quantitative methylation-specific PCR
Sodium bisulfite modification was performed using the EpiTect Bisulphite Kit (Qiagen; cat no. 59104) according to the manufacturer's instructions. Quantitative methylation-specific PCR on plasma samples was performed on a 7900HT real-time PCR system (Applied Biosystems). For the training set, two duplex PCRs NDRG4_FAM/FOXE1_TET and GATA5_FAM/SYNE1_TET were performed. Based on the performance of the first duplex reaction, only SYNE1 and FOXE1 were retained in the test set (one duplex PCR; SYNE1_TET/FOXE1_HEX). The PCR master mix was 30 µL QuantiTect Multiplex PCR with ROX dye (Qiagen), 10 µL template volume, forward primer (0.28 µmol/L) of both genes, reverse primer (0.28 µmol/L) of both genes and a single-stranded oligonucleotide hybridization probe (0.25 µmol/L) of both genes. The PCR program was: 15 minutes at 95°C; followed by 45 cycles of 30 seconds at 95°C, 30 seconds at 57°C, and 30 seconds at 72°C; followed by 5 minutes at 72°C. Serially diluted plasmids containing the target sequence were amplified to generate a standard curve against which the unknown samples are quantified by interpolation of their PCR cycle number (Ct value) to the corresponding plasmid copy. Because the yield showed very high intersubject variability, no direct control for DNA yield was incorporated. This standard curve ruled out any technical or reagent batch-related errors in methylation calls. Samples were handled and analyzed in a blinded fashion during storage, DNA isolation, and PCR analysis. Primer and probe sequences are provided in Supplementary Table S2.
Cell culture and transfections
Human colorectal cancer cell lines were cultured in Dulbecco's MEM (DMEM; Invitrogen) supplemented with 10% heat-inactivated fetal calf serum (FBS; HyClone). Cell lines (RKO, SW480, and HCT116) were purchased from ATCC (LGC standards). No authentication was done by the authors. Full-length FOXE1 cDNA was subcloned into the pIRES-neo3 expression vector (Clontech Laboratories Inc.). GFP-SYNE1 and GFP-SYNE1-KASH (Klarischt) were kindly provided by Dr. Zhang (Department of Medicine, Cambridge, United Kingdom).
HCT116 cells were transfected with the Nucleofector Kit V (Amaxa Biosystems) and RKO and SW480 cells were transfected using Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer's protocol. HCT116, RKO, and SW480 cells were transfected with a control construct (empty vector) or FOXE1-pIRESneo3, GFP-SYNE1, and GFP-SYNE1-KASH, selected for 10 days with G418 (HCT116 and SW480 400 µg/mL; RKO 1 mg/mL).
Colony formation assay
RKO, HCT116, and SW480 CRC cells were transfected in 6-well plates with control construct FOXE1-pIRESneo3, GFP-SYNE1, or GFP-SYNE1-KASH as described above. The next day, cells were diluted 1:20 and G418 (RKO 1 mg/mL; HCT116, SW480 400 µg/mL) was added. After 14 days of selection, colonies were stained and quantified.
In vitro cell proliferation, migration, and invasion assays
HCT116, RKO, and SW480 cells were seeded onto 96-well plates (5,000 cells per well) and after 96 hours, the cultures were pulsed for 6 hours with 0.3 µCi [methyl-3H] thymidine (Amersham Life Science) per well. Activity was measured using liquid scintillation. Cell migration and invasion assays were performed using 24-well Transwells (8-µmpore size) coated with (invasion) or without (migration) Matrigel (BD Biosciences). HCT116, RKO, and SW480 cells (20 × 104) in 1% FBS-DMEM were seeded into the upper chamber, and DMEM containing 20% FBS was placed in the lower chamber. After 48 hours, cells on the lower surface of the membrane were fixed with methanol and stained with 1% Toluine Blue in 1% borax.
Data analysis
For quantitative methylation-specific PCR analysis, receiver operating characteristics (ROC) curve analysis and the area under the curve (AUC) were determined to define the best markers with highest sensitivity and specificity. GATA5, NDRG4, SYNE1, and FOXE1 promoter methylation was considered positive if copies of any of these genes were detected. The measured methylated copy numbers were used as the basis for the age- and gender-association analyses, hence methylation was implemented as a continuous variable. Age was stratified in two groups, i.e., those patients younger than 65 and those age 65 and above. The Mann–Whitney rank-sum test was used to analyze the colony formation, migration, and invasion assays.
Results
NDRG4, GATA5, FOXE1, and SYNE1 methylation in blood DNA as a potential biomarker for colorectal cancer detection
The main objective of this study was to investigate the use of NDRG4, GATA5, SYNE1, and FOXE1 promoter methylation as biomarkers for detection of colorectal cancer–derived DNA in plasma by determining the optimal classifier in a training set and subsequently confirming this in a test set.
Training set
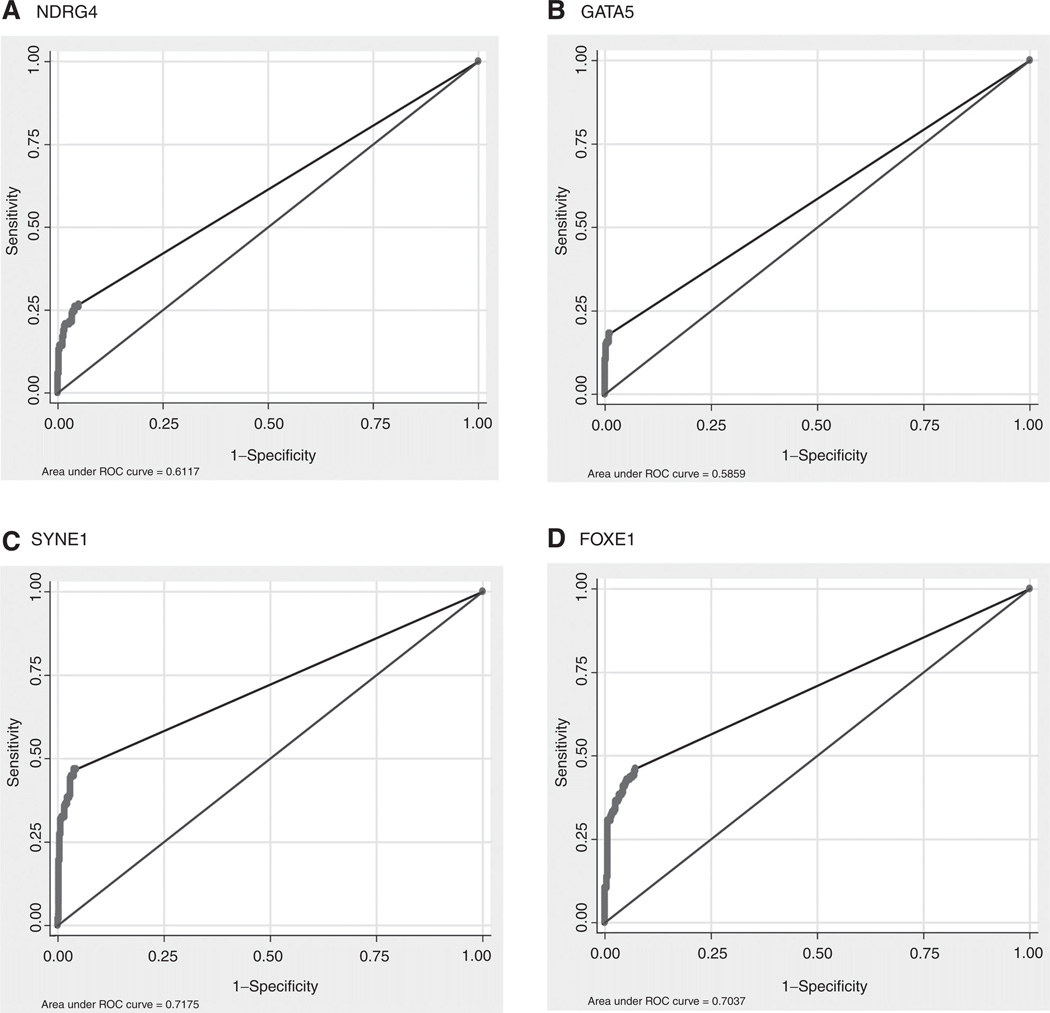
Using 444 normal control samples and 154 colorectal cancer samples of the training set, we evaluated NDRG4, GATA5, SYNE1, and FOXE1 as single marker candidates using ROC curve analysis. The AUC generated for plasma NDRG4, GATA5, SYNE1, and FOXE1 methylation was 0.61 (95% CI, 0.57–0.65, Fig. 1A), 0.59 (95% CI, 0.55–0.63; Fig. 1B), 0.72 (95% CI, 0.68– 0.75; Fig. 1C), and 0.70 (95% CI, 0.69–0.73; Fig. 1D), respectively, indicating that SYNE1 and FOXE1 have the highest performance as potential biomarkers.
Figure 1.
ROC curves. Sensitivity and specificity at various cutoff values for the training set to obtain a positive test for the single markers (A) NDRG4, (B) GATA5, (C) SYNE1, and (D) FOXE1 are shown in the ROC curve.
To study the sensitivity and specificity of these markers separately, we defined a cutoff of zero. The sensitivity of NDRG4 methylation in all stages of colorectal cancer was 27% (95% CI, 20%–34%) with a specificity of 95% (95% CI, 93%–97%). The overall sensitivity of GATA5 methylation was 18% (95% CI, 12%–24%) with a specificity of 99% (95% CI, 98%–100%). FOXE1 methylation has a sensitivity of 46% (95% CI, 38%–54%) in detecting patients with colorectal cancer and a specificity of 93% (95% CI, 91%–95%). Finally, SYNE1 methylation generated a sensitivity of 47% (95% CI, 39%–55%) for detection of colorectal cancer and a specificity of 96% (95% CI, 94%–98%). Early-stage colorectal cancers were detected at slightly lower rates than later stages, a phenomenon that occurred for three of the four markers analyzed (except SYNE1), as depicted in Table 1.
Table 1.
Training set plasma performance of single markers [NDRG4 and GATA5 (A); FOXE1 and SYNE1 (B)]
| A | ||||
|---|---|---|---|---|
| NDRG4 | GATA5 | |||
| Patient group | Positive | Sensitivity | Positive | Sensitivity |
| Stage I | 7/43 | 16% | 6/43 | 14% |
| Stage II | 5/44 | 11% | 4/44 | 9% |
| Stage III | 16/46 | 35% | 8/46 | 17% |
| Stage IV | 13/21 | 62% | 10/21 | 48% |
| Case total | 41/154 | 27% | 28/154 | 18% |
| Specificity | Specificity | |||
| Control | 22/442 | 95% | 5/444 | 99% |
| B | ||||
|---|---|---|---|---|
| FOXE1 | SYNE1 | |||
| Patient group | Positive | Sensitivity | Positive | Sensitivity |
| Stage I | 15/43 | 35% | 12/43 | 28% |
| Stage II | 19/44 | 43% | 23/44 | 52% |
| Stage III | 23/46 | 50% | 21/46 | 46% |
| Stage IV | 14/21 | 67% | 16/46 | 76% |
| Case total | 71/154 | 46% | 72/154 | 47% |
| Specificity | Specificity | |||
| Control | 32/444 | 93% | 19/444 | 96% |
The presence of methylated NDRG4 (P = 0.45, P = 0.12), GATA5 (P = 0.60; P = 0.13), FOXE1 (P = 0.88; P = 0.053), and SYNE1 (P = 0.39; P = 0.15) in plasma was not associated with age and gender, respectively. Given the low sensitivity of GATA5 and NDRG4 methylation, these markers did not add to the overall sensitivity (data not shown). In addition, combining SYNE1 and FOXE1 methylation increased the sensitivity to 56% (95% CI, 48%–64%) with a specificity of 90% (95% CI, 87%–93%; Table 2).
Table 2.
Training set results combined FOXE1 and SYNE1
| SYNE1 and FOXE1 | ||
|---|---|---|
| Patient group | Positive | Sensitivity |
| Stage I | 18/43 | 42% |
| Stage II | 25/44 | 57% |
| Stage III | 27/46 | 59% |
| Stage IV | 17/21 | 81% |
| Case total | 87/154 | 56% |
| Specificity | ||
| Control | 46/444 | 90% |
Test set
To confirm the clinical performance of SYNE1 and FOXE1 methylation as a multimarker panel, we tested plasma samples obtained from 66 patients with colorectal cancer and 240 healthy controls. Using the same threshold, we observed a sensitivity of 58% (95% CI, 46%–70%) and a specificity of 91% (95% CI, 80%–100%) for all stages of colorectal cancer for FOXE1 and SYNE1, respectively (Table 3). Stage I [37% (95% CI, 19%–55%)] and stage III [55% (95% CI, 33%–77%)] cancers were detected at lower rates than stage II [87% (95%CI,70%–100%)], and stage IV [100% (95%CI, NA)] cancers (Table 3). SYNE1 and FOXE1 promoter methylation was not associated with age (FOXE1, P = 0.36 and SYNE1, P = 0.38) and gender (FOXE1 P = 0.29 and SYNE1 P = 0.28).
Table 3.
Test set results combined FOXE1 and SYNE1
| SYNE1 and FOXE1 | ||
|---|---|---|
| Patient group | Positive | Sensitivity |
| Stage I | 10/27 | 37% |
| Stage II | 13/15 | 87% |
| Stage III | 11/20 | 55% |
| Stage IV | 4/4 | 100% |
| Case total | 38/66 | 58% |
| Specificity | ||
| Control | 21/240 | 91% |
FOXE1 and SYNE1 overexpression in colorectal cancer cell lines
The functional role of NDRG4 and GATA5 in vitro in colorectal cancer cell lines has been previously described (15, 18). Currently, nothing is known about the role of SYNE1 and FOXE1 in colorectal cancer.
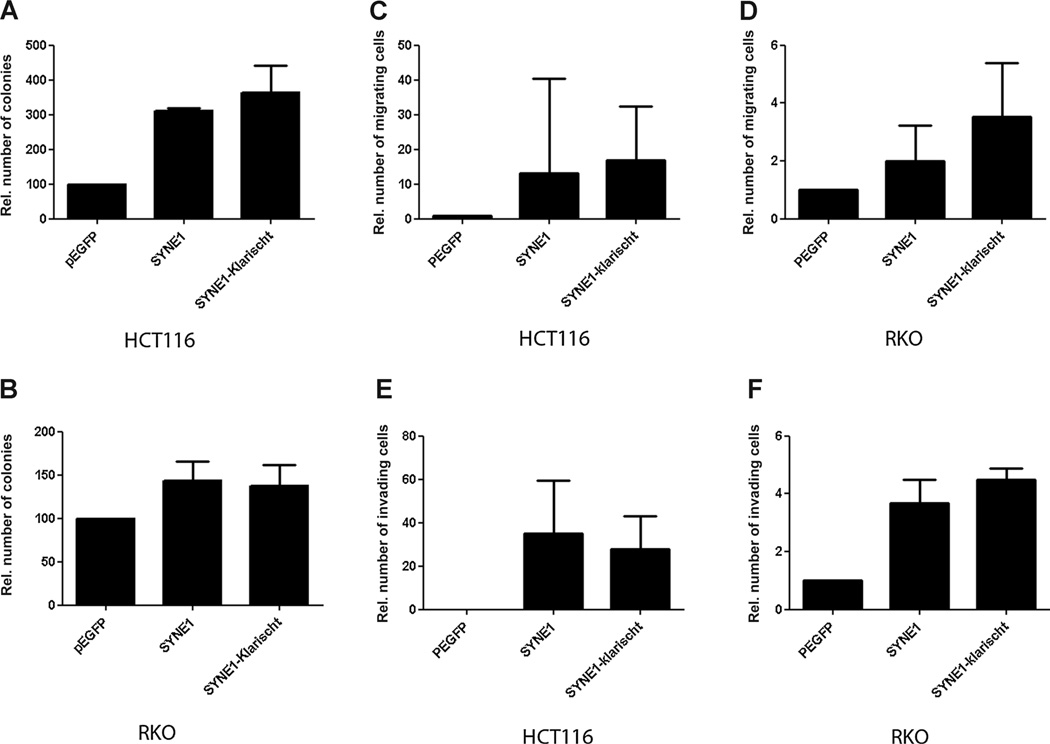
To examine whether SYNE1 has tumor-suppressor activities in colorectal cancer cells, HCT116 and RKO cells were transfected with SYNE1 expression constructs. Transfection with GFP-SYNE1 and GFP-SYNE1-KASH induced the number of G418-resistant colonies as compared with control transfectants in HCT116 (Fig. 2A, P = 0.001 and P = 0.07) and RKO (Fig. 2B, P = 0.13 and P = 0.17) cells. Migration (Fig. 2C; HCT116, P = 0.26 and P = 0.16 and 2D; RKO, P = 0.19 and P = 0.07) and invasion (Fig. 2E; HCT116, P = 0.29, and P = 0.20) and 2F; RKO, P = 0.27 and P = 0.03) of SYNE1-transfected cells was higher than that of control clones. However, although a trend was seen in all cell lines, most results were not statistically significant.
Figure 2.
Functional assays SYNE1. A and B, colony formation assay of cells transfected with SYNE1 expression vector or empty vector grown for 2weeks in medium containing antibiotics. Mean colony numbers relative to control transfectants are plotted (n = 3). Error bars, 95% CI. C and D, invasion assay through Matrigel-coated Transwells. Results represent mean number of SYNE1-transfected cells that passed through the Matrigel-coated membranes of the Transwell relative to control cells transfected with empty vector (n = 3). E and F, migration assay. Plotted are the mean numbers of SYNE1-transfected cells that migrated through Transwell membranes not coated with Matrigel relative to control cells transfected with empty vector (n = 3). Error bars, 95% CI.
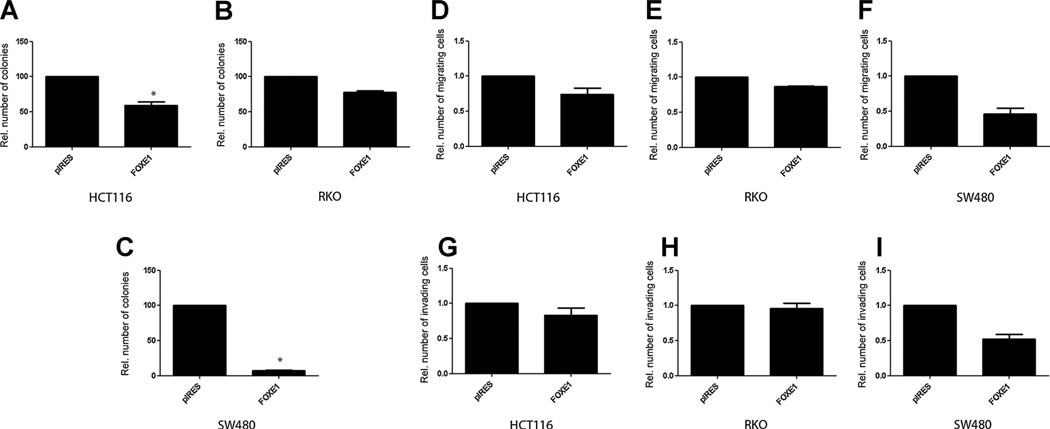
To investigate whether FOXE1 is a putative tumor suppressor in colorectal cancer, we first performed in vitro colony formation assays to determine the effects of full-length FOXE1 transfected into SW480, HCT116, and RKO cells. Overexpression with FOXE1 resulted in a significant reduction of G418-resistant colonies in HCT116 and SW480 cells [a nearly 2-fold reduction in HCT116 (Fig. 3A, P = 0.003) and more than 8-fold reduction in SW480 cells (Fig. 3C, P = 0.006)]. No significant effect was observed in RKO cells transfected with FOXE1 (Fig. 3B, P = 0.577). Overexpression of the FOXE1 gene had no significant effect on the migration [Fig. 3D (HCT116, P = 0.63), E (RKO, P = 0.82), F (SW480, P = 0.28)] and invasion [Fig. 3G HCT116, P = 0.79), H (RKO, P = 0.92), I (SW480, P = 0.38)] of these cells. Taken together, these data suggest that FOXE1 may have tumor-suppressive effects in human colorectal cancer cells.
Figure 3.
Functional assays FOXE1. A to C, colony formation assay of cells transfected with FOXE1 expression vector or empty vector grown for 2 weeks in medium containing antibiotics. Mean colony numbers relative to control transfectants are plotted (n = 3). Error bars, 95% CI. D to F, invasion assay through Matrigel-coated Transwells. Results represent mean number of FOX1-transfected cells that passed through the Matrigel-coated membranes of the Transwell relative to control cells transfected with empty vector (n = 3). G to I, migration assay. Plotted are the mean numbers of FOX1-transfected cells that migrated through Transwell membranes not coated with Matrigel relative to control cells transfected with empty vector (n = 3). Error bars, 95% CI. Melotte et al.
Discussion
We previously publishedNDRG4(18) and GATA4/GATA5 (15) promoter hypermethylation as potential sensitive and specific stool DNA markers and candidate tumor-suppressor genes in colorectal cancer. The potential of NDRG4 methylation for the detection of colorectal cancer was independently confirmed by Alhquist and colleagues and Imperiale and colleagues, who recently developed a multimarker diagnostic DNA test, including NDRG4 methylation, to screen for colorectal cancer (19, 34). This DNA test has shown to perform better compared with analyzing FIT alone. However, implementation of this test might be hampered in population-based screening programs due to the requirement of a complete stool sample affecting compliance and cost-effectiveness. Despite the high potential of these markers in stool DNA, we hypothesized that a blood-based test for colorectal cancer detection, not depending on stool sampling, has the potential for better patient compliance and is better suited for systems without programmatic screening. Therefore, we investigated the performance of our previously reported stool methylation markers (NDRG4 and GATA5) and two other promising methylation markers (SYNE1 and FOXE1) in blood. FOXE1 and SYNE1 were identified as frequently methylated genes in a transcriptome-wide approach to detect transcriptionally silenced genes by promoter CpG island methylation in colorectal cancer (32).
FOXE1 is a transcription factor that is characterized by a distinct forkhead domain, and plays a crucial role in thyroid morphogenesis. In thyroid cancer, FOXE1 is upregulated (35), although most cancers have a decreased expression of FOXE1 often due to promoter CpG island methylation as has been described in breast cancer (36), pancreatic cancer (37), and in cutaneous squamous cell carcinoma (38). Furthermore, FOXE1 has been reported as a sensitive (82%) and specific (98.5%) methylation marker for pancreatic cancer in pancreatic juice of patients (37).
We and others described DNA methylation of SYNE1 in colorectal cancer, suggesting a tumor-suppressor function in colorectal cancer (32, 39). SYNE1 promoter methylation and a possible tumor-suppressor function have been reported in lung cancer (40).However, the in vitro studies in this study show that SYNE1 overexpression in colorectal cancer cell lines induces cell proliferation, migration, and invasion indicating an oncogenic role for SYNE1 in colorectal cancer. Interestingly, in line with our functional data, gene expression data obtained from GEO from primary normal colon and colon cancer showed no decrease of SYNE1 expression in colorectal cancer tissues (data not shown), speculating that promoter methylation of SYNE1 in colorectal cancer does not result in downregulation of this gene.
We have studied the role of NDRG4 in colorectal cancer and found that NDRG4 significantly inhibited colony formation, cell proliferation, and invasion, and is frequently methylated (70% and 86% in two independent series) in colorectal cancer tissue samples (18). In parallel, we have published the potential tumor-suppressor role of GATA5 in colorectal cancer, which inhibited colony formation, cell growth, migration, invasion, and anchor-age-independent growth in vitro. Furthermore, promoter methylation of GATA5 was observed (79% and 74% in two independent series) in colorectal cancer tissues (15). GATA5 promoter methylation has been reported in other tumor types as well, such as small-cell lung cancer, pancreatic, esophageal, ovarian, and gastric cancer (41–45). In the present article, we investigated the performance of NDRG4, GATA5, SYNE1, and FOXE1 as methylation markers in blood DNA. Interestingly, although NDRG4 performed well as a methylation marker in stool (sensitivity of 61% and 53% for the training and test set, respectively), the detection rate in blood for colorectal cancer was only 27%. Molecular pathways and cellular mechanisms that underlie multistage processes of metastasis, including tumor invasion, tumor-cell dissemination through the bloodstream or the lymphatic system, colonization of distant organs and, outgrowth of metastases have been characterized (46). A difference in gene expression at the invasive front compared with the central area of the tumor and the luminal part of the tumor has been described for beta-catenin (47). This heterogeneity in gene expression probably indicates heterogeneity in the underlying mechanisms that are regulating gene expression. We therefore hypothesize that molecular or morphologic characteristics at the invasive front area of colorectal cancer, a part of the tumor that probably also easily invades blood vessels, may be different from the luminal part of the colorectal cancer that is shedding tumor cells in stool. An alternative explanation for differences in sensitivity in blood or fecal DNA could be the DNA isolation procedure. Diehl and colleagues demonstrated that the majority of mutated APC sequences in patients with colorectal cancer were detected in smaller size fragments of circulating DNA, whereas larger fragments tended to be wild-type (48). Therefore, improvement of isolation protocols of blood DNA, could, in theory, yield a blood DNA test for NDRG4 with higher sensitivity.
We here describe a biomarker panel of two genes able to discriminate colorectal cancer from non-colorectal cancer in plasma with a sensitivity of 56% and 58% and a specificity of 90% and 91%. Of all blood-based DNA markers, plasma septin-9 has been studied most extensively. Test sensitivity varies widely for both colorectal cancer (from 48% to 90%) and adenomas (from 11% to 29%), whereas specificity was more consistent ranging from 86% to 97% (49). Septin-9 was also evaluated in a screening setting, where sensitivity and specificity were 50% and 91%, respectively, for patients with colorectal cancer when two aliquots were tested and 64% and 88%, respectively, with three samples tested (49). Further research is necessary to investigate whether the sensitivity of SYNE1 and FOXE1 can be improved using different aliquots and whether it will be effective and cost-effective compared with no screening and compared with other screening test available. In addition, these data need to be validated in a large prospective colorectal cancer screening study and should be combined with other noninvasive screening test such as genetic or epigenetic DNA markers or the FIT to enhance sensitivity. Furthermore, sample collection, DNA isolation, bisulfite conversion, and assay sensitivity should be optimized. Detecting minute amounts of tumor DNA in blood, especially in the early-stage tumors, and defining the specificity of a DNA methylation marker in blood that contain DNA from many sources in the body are challenges for future research.
Supplementary Material
Acknowledgments
Grant Support
V. Melotte, G.A. Meijer, and M. van Engeland were supported by grant 03O-101 (DeCoDe, CTMM, the Center for Translational Molecular Medicine).
Footnotes
Note: Supplementary data for this article are available at Cancer Prevention Research Online (http://cancerprevres.aacrjournals.org/).
Disclosure of Potential Conflicts of Interest
L. Van Neste is the Director of Scientific Affairs at MDxHealth. J.G. Herman is a consultant/advisory board member for and reports receiving a commercial research grant from MDxHealth. No potential conflicts of interest were disclosed by the other authors.
Authors' Contributions
Conception and design: V. Melotte, J.M. Yi, N. Ahuja, M. van Engeland
Development of methodology: K.M. Smits, K.E. Schuebel, J.G. Herman, W. van Criekinge, N. Ahuja, M. van Engeland
Acquisition of data (provided animals, acquired and managed patients, provided facilities, etc.): H.E.C. Niessen, K.A.D. Wouters, J. Louwagie, N. Ahuja, M. van Engeland
Analysis and interpretation of data (e.g., statistical analysis, biostatistics, computational analysis): V. Melotte, M.H.F.M. Lentjes, K.M. Smits, L. Van Neste, W. van Criekinge, N. Ahuja, M. van Engeland
Writing, review, and/or revision of the manuscript: V. Melotte, M.H.F.M. Lentjes, K.M. Smits, L. Van Neste, J.G. Herman, W. van Criekinge, G.A. Meijer, N. Ahuja, M. van Engeland
Administrative, technical, or material support (i.e., reporting or organizing data, constructing databases): M.H.F.M. Lentjes, K.M. Smits, K.A.D. Wouters, S.B. Baylin, G.A. Meijer, N. Ahuja
Study supervision: N. Ahuja, M. van Engeland
References
- 1.Mandel JS, Bond JH, Church TR, Snover DC, Bradley GM, Schuman LM, et al. Reducing mortality from colorectal cancer by screening for fecal occult blood. Minnesota Colon Cancer Control Study. N Engl J Med. 1993;328:1365–1371. doi: 10.1056/NEJM199305133281901. [DOI] [PubMed] [Google Scholar]
- 2.Wee CC, McCarthy EP, Phillips RS. Factors associated with colon cancer screening: the role of patient factors and physician counseling. Prev Med. 2005;41:23–29. doi: 10.1016/j.ypmed.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 3.Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128:683–692. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Laird PW. The power and the promise of DNA methylation markers. Nat Rev Cancer. 2003;3:253–266. doi: 10.1038/nrc1045. [DOI] [PubMed] [Google Scholar]
- 5.Hoque MO, Begum S, Topaloglu O, Chatterjee A, Rosenbaum E, Van Criekinge W, et al. Quantitation of promoter methylation of multiple genes in urine DNA and bladder cancer detection. J Natl Cancer Inst. 2006;98:996–1004. doi: 10.1093/jnci/djj265. [DOI] [PubMed] [Google Scholar]
- 6.Jin Z, Olaru A, Yang J, Sato F, Cheng Y, Kan T, et al. Hypermethylation of tachykinin-1 is a potential biomarker in human esophageal cancer. Clin Cancer Res. 2007;13:6293–6300. doi: 10.1158/1078-0432.CCR-07-0818. [DOI] [PubMed] [Google Scholar]
- 7.Muller HM, Oberwalder M, Fiegl H, Morandell M, Goebel G, Zitt M, et al. Methylation changes in faecal DNA: a marker for colorectal cancer screening? Lancet. 2004;363:1283–1285. doi: 10.1016/S0140-6736(04)16002-9. [DOI] [PubMed] [Google Scholar]
- 8.Yu J, Zhu T, Wang Z, Zhang H, Qian Z, Xu H, et al. A novel set of DNA methylation markers in urine sediments for sensitive/specific detection of bladder cancer. Clin Cancer Res. 2007;13:7296–7304. doi: 10.1158/1078-0432.CCR-07-0861. [DOI] [PubMed] [Google Scholar]
- 9.Goessl C, Krause H, Muller M, Heicappell R, Schrader M, Sachsinger J, et al. Fluorescent methylation-specific polymerase chain reaction for DNA-based detection of prostate cancer in bodily fluids. Cancer Res. 2000;60:5941–5945. [PubMed] [Google Scholar]
- 10.Rosas SL, Koch W, da Costa Carvalho MG, Wu L, Califano J, Westra W, et al. Promoter hypermethylation patterns of p16, O6-methylguanine-DNA-methyltransferase, and death-associated protein kinase in tumors and saliva of head and neck cancer patients. Cancer Res. 2001;61:939–942. [PubMed] [Google Scholar]
- 11.Machida EO, Brock MV, Hooker CM, Nakayama J, Ishida A, Amano J, et al. Hypermethylation of ASC/TMS1 is a sputum marker for late-stage lung cancer. Cancer Res. 2006;66:6210–6218. doi: 10.1158/0008-5472.CAN-05-4447. [DOI] [PubMed] [Google Scholar]
- 12.Muller HM, Millinger S, Fiegl H, Goebel G, Ivarsson L, Widschwendter A, et al. Analysis of methylated genes in peritoneal fluids of ovarian cancer patients: a new prognostic tool. Clin Chem. 2004;50:2171–2173. doi: 10.1373/clinchem.2004.034090. [DOI] [PubMed] [Google Scholar]
- 13.Evron E, Dooley WC, Umbricht CB, Rosenthal D, Sacchi N, Gabrielson E, et al. Detection of breast cancer cells in ductal lavage fluid by methylation-specific PCR. Lancet. 2001;357:1335–1336. doi: 10.1016/s0140-6736(00)04501-3. [DOI] [PubMed] [Google Scholar]
- 14.Krassenstein R, Sauter E, Dulaimi E, Battagli C, Ehya H, Klein-Szanto A, et al. Detection of breast cancer in nipple aspirate fluid by CpG island hypermethylation. Clin Cancer Res. 2004;10:28–32. doi: 10.1158/1078-0432.ccr-0410-3. [DOI] [PubMed] [Google Scholar]
- 15.Hellebrekers DM, Lentjes MH, van den Bosch SM, Melotte V, Wouters KA, Daenen KL, et al. GATA4 and GATA5 are potential tumor suppressors and biomarkers in colorectal cancer. Clin Cancer Res. 2009;15:3990–3997. doi: 10.1158/1078-0432.CCR-09-0055. [DOI] [PubMed] [Google Scholar]
- 16.Chen WD, Han ZJ, Skoletsky J, Olson J, Sah J, Myeroff L, et al. Detection in fecal DNA of colon cancer-specific methylation of the nonexpressed vimentin gene. J Natl Cancer Inst. 2005;97:1124–1132. doi: 10.1093/jnci/dji204. [DOI] [PubMed] [Google Scholar]
- 17.Lenhard K, Bommer GT, Asutay S, Schauer R, Brabletz T, Goke B, et al. Analysis of promoter methylation in stool: a novel method for the detection of colorectal cancer. Clin Gastroenterol Hepatol. 2005;3:142–149. doi: 10.1016/s1542-3565(04)00624-x. [DOI] [PubMed] [Google Scholar]
- 18.Melotte V, Lentjes MH, van den Bosch SM, Hellebrekers DM, de Hoon JP, Wouters KA, et al. N-Myc downstream-regulated gene 4 (NDRG4): a candidate tumor suppressor gene and potential biomarker for colorectal cancer. J Natl Cancer Inst. 2009;101:916–927. doi: 10.1093/jnci/djp131. [DOI] [PubMed] [Google Scholar]
- 19.Ahlquist DA, Zou H, Domanico M, Mahoney DW, Yab TC, Taylor WR, et al. Next-generation stool DNA test accurately detects colorectal cancer and large adenomas. Gastroenterology. 2012;142:248–256. doi: 10.1053/j.gastro.2011.10.031. quiz e25–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Glockner SC, Dhir M, Yi JM, McGarvey KE, Van Neste L, Louwagie J, et al. Methylation of TFPI2 in stool DNA: a potential novel biomarker for the detection of colorectal cancer. Cancer Res. 2009;69:4691–4699. doi: 10.1158/0008-5472.CAN-08-0142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ebert MP, Model F, Mooney S, Hale K, Lograsso J, Tonnes-Priddy L, et al. Aristaless-like homeobox-4 gene methylation is a potential marker for colorectal adenocarcinomas. Gastroenterology. 2006;131:1418–1430. doi: 10.1053/j.gastro.2006.08.034. [DOI] [PubMed] [Google Scholar]
- 22.Lofton-Day C, Model F, Devos T, Tetzner R, Distler J, Schuster M, et al. DNA methylation biomarkers for blood-based colorectal cancer screening. Clin Chem. 2008;54:414–423. doi: 10.1373/clinchem.2007.095992. [DOI] [PubMed] [Google Scholar]
- 23.Zou H, Yu B, Zhao R, Wang Z, Cang H, Li D, et al. Detection of aberrant p16 methylation in the serum of colorectal cancer patients. Zhonghua Yu Fang Yi Xue Za Zhi. 2002;36:499–501. [PubMed] [Google Scholar]
- 24.Miotto E, Sabbioni S, Veronese A, Calin GA, Gullini S, Liboni A, et al. Frequent aberrant methylation of the CDH4 gene promoter in human colorectal and gastric cancer. Cancer Res. 2004;64:8156–8159. doi: 10.1158/0008-5472.CAN-04-3000. [DOI] [PubMed] [Google Scholar]
- 25.Herbst A, Rahmig K, Stieber P, Philipp A, Jung A, Ofner A, et al. Methylation of NEUROG1 in serum is a sensitive marker for the detection of early colorectal cancer. Am J Gastroenterol. 2011;106:1110–1118. doi: 10.1038/ajg.2011.6. [DOI] [PubMed] [Google Scholar]
- 26.Grady WM, Rajput A, Lutterbaugh JD, Markowitz SD. Detection of aberrantly methylated hMLH1 promoter DNA in the serum of patients with microsatellite unstable colon cancer. Cancer Res. 2001;61:900–902. [PubMed] [Google Scholar]
- 27.Leung WK, To KF, Man EP, Chan MW, Bai AH, Hui AJ, et al. Quantitative detection of promoter hypermethylation in multiple genes in the serum of patients with colorectal cancer. Am J Gastroenterol. 2005;100:2274–2279. doi: 10.1111/j.1572-0241.2005.50412.x. [DOI] [PubMed] [Google Scholar]
- 28.Yamaguchi S, Asao T, Nakamura J, Ide M, Kuwano H. High frequency of DAP-kinase gene promoter methylation in colorectal cancer specimens and its identification in serum. Cancer Lett. 2003;194:99–105. doi: 10.1016/s0304-3835(03)00055-7. [DOI] [PubMed] [Google Scholar]
- 29.Lange CP, Campan M, Hinoue T, Schmitz RF, van der Meulen-de Jong AE, Slingerland H, et al. Genome-scale discovery of DNA-methylation biomarkers for blood-based detection of colorectal cancer. PLoS One. 2012;7:e50266. doi: 10.1371/journal.pone.0050266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oh T, Kim N, Moon Y, Kim MS, Hoehn BD, Park CH, et al. Genome-wide identification and validation of a novel methylation biomarker, SDC2, for blood-based detection of colorectal cancer. J Mol Diagn. 2013;15:498–507. doi: 10.1016/j.jmoldx.2013.03.004. [DOI] [PubMed] [Google Scholar]
- 31.Lee BB, Lee EJ, Jung EH, Chun HK, Chang DK, Song SY, et al. Aberrant methylation of APC, MGMT, RASSF2A, and Wif-1 genes in plasma as a biomarker for early detection of colorectal cancer. Clin Cancer Res. 2009;15:6185–6191. doi: 10.1158/1078-0432.CCR-09-0111. [DOI] [PubMed] [Google Scholar]
- 32.Schuebel KE, Chen W, Cope L, Glockner SC, Suzuki H, Yi JM, et al. Comparing the DNA hypermethylome with gene mutations in human colorectal cancer. PLoS Genet. 2007;3:1709–1723. doi: 10.1371/journal.pgen.0030157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Papadia C, Louwagie J, Del Rio P, Grooteclaes M, Coruzzi A, Montana C, et al. FOXE1 and SYNE1 genes hypermethylation panel as promising biomarker in colitis-associated colorectal neoplasia. Inflamm Bowel Dis. 2014;20:271–277. doi: 10.1097/01.MIB.0000435443.07237.ed. [DOI] [PubMed] [Google Scholar]
- 34.Imperiale TF, Ransohoff DF, Itzkowitz SH, Levin TR, Lavin P, Lidgard GP, et al. Multitarget stool DNA testing for colorectal-cancer screening. New Engl J Med. 2014;370:1287–1297. doi: 10.1056/NEJMoa1311194. [DOI] [PubMed] [Google Scholar]
- 35.Nonaka D, Tang Y, Chiriboga L, Rivera M, Ghossein R. Diagnostic utility of thyroid transcription factors Pax8 and TTF-2 (FoxE1) in thyroid epithelial neoplasms. Mod Pathol. 2008;21:192–200. doi: 10.1038/modpathol.3801002. [DOI] [PubMed] [Google Scholar]
- 36.Weisenberger DJ, Trinh BN, Campan M, Sharma S, Long TI, Ananthnarayan S, et al. DNA methylation analysis by digital bisulfite genomic sequencing and digital MethyLight. Nucleic Acids Res. 2008;36:4689–4698. doi: 10.1093/nar/gkn455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Matsubayashi H, Canto M, Sato N, Klein A, Abe T, Yamashita K, et al. DNA methylation alterations in the pancreatic juice of patients with suspected pancreatic disease. Cancer Res. 2006;66:1208–1217. doi: 10.1158/0008-5472.CAN-05-2664. [DOI] [PubMed] [Google Scholar]
- 38.Venza I, Visalli M, Tripodo B, De Grazia G, Loddo S, Teti D, et al. FOXE1 is a target for aberrant methylation in cutaneous squamous cell carcinoma. Br J Dermatol. 2010;162:1093–1097. doi: 10.1111/j.1365-2133.2009.09560.x. [DOI] [PubMed] [Google Scholar]
- 39.Mokarram P, Kumar K, Brim H, Naghibalhossaini F, Saberi-firoozi M, Nouraie M, et al. Distinct high-profile methylated genes in colorectal cancer. PLoS ONE. 2009;4:e7012. doi: 10.1371/journal.pone.0007012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tessema M, Willink R, Do K, Yu YY, Yu W, Machida EO, et al. Promoter methylation of genes in and around the candidate lung cancer susceptibility locus 6q23–25. Cancer Res. 2008;68:1707–1714. doi: 10.1158/0008-5472.CAN-07-6325. [DOI] [PubMed] [Google Scholar]
- 41.De Jong WK, Verpooten GF, Kramer H, Louwagie J, Groen HJ. Promoter methylation primarily occurs in tumor cells of patients with non-small cell lung cancer. Anticancer Res. 2009;29:363–369. [PubMed] [Google Scholar]
- 42.Fu B, Guo M, Wang S, Campagna D, Luo M, Herman JG, et al. Evaluation of GATA-4 and GATA-5 methylation profiles in human pancreatic cancers indicate promoter methylation patterns distinct from other human tumor types. Cancer Biol Ther. 2007;6:1546–1552. doi: 10.4161/cbt.6.10.4708. [DOI] [PubMed] [Google Scholar]
- 43.Guo M, House MG, Akiyama Y, Qi Y, Capagna D, Harmon J, et al. Hypermethylation of the GATA gene family in esophageal cancer. Int J Cancer. 2006;119:2078–2083. doi: 10.1002/ijc.22092. [DOI] [PubMed] [Google Scholar]
- 44.Wakana K, Akiyama Y, Aso T, Yuasa Y. Involvement of GATA-4/-5 transcription factors in ovarian carcinogenesis. Cancer Lett. 2006;241:281–288. doi: 10.1016/j.canlet.2005.10.039. [DOI] [PubMed] [Google Scholar]
- 45.Akiyama Y, Watkins N, Suzuki H, Jair KW, van Engeland M, Esteller M, et al. GATA-4 and GATA-5 transcription factor genes and potential downstream antitumor target genes are epigenetically silenced in colorectal and gastric cancer. Mol Cell Biol. 2003;23:8429–8439. doi: 10.1128/MCB.23.23.8429-8439.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Christofori G. New signals from the invasive front. Nature. 2006;441:444–450. doi: 10.1038/nature04872. [DOI] [PubMed] [Google Scholar]
- 47.Brabletz T, Jung A, Hermann K, Gunther K, Hohenberger W, Kirchner T. Nuclear overexpression of the oncoprotein beta-catenin in colorectal cancer is localized predominantly at the invasion front. Pathol Res Pract. 1998;194:701–704. doi: 10.1016/s0344-0338(98)80129-5. [DOI] [PubMed] [Google Scholar]
- 48.Diehl F, Li M, Dressman D, He Y, Shen D, Szabo S, et al. Detection and quantification of mutations in the plasma of patients with colorectal tumors. Proc Natl Acad Sci U S A. 2005;102:16368–16373. doi: 10.1073/pnas.0507904102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Imperiale TF. Noninvasive screening tests for colorectal cancer. Dig Dis. 2012;30(Suppl 2):16–26. doi: 10.1159/000341884. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.