Abstract
Head and neck squamous cell carcinomas have distinct mutation and copy number profiles depending on human papillomavirus status. Although several challenges remain in biomarker implementation and clinical trial feasibility, incorporating available genomic data will expedite the development of novel therapeutics and predictive biomarker-driven clinical trials.
In this issue of Clinical Cancer Research, Seiwert and colleagues report mutation and copy number variation (CNV) analyses in 617 cancer-associated genes from 120 tumor-normal tissue pairs from head and neck squamous cell carcinoma (HNSCC) patients (1). They confirmed the presence of frequent mutations and CNVs in HNSCC and identified additional mutations differentially associated with human papillomavirus (HPV)-positive (+) tumors that were previously thought to be rare in HNSCCs. Further characterization of these mutational profiles may assist novel biomarker characterization and targeted therapeutic development benefiting HPV(+) and HPV− negative (−) patients.
The most common risk factors for HNSCC development are heavy tobacco use and alcohol consumption, as well as HPV infection (2, 3). HPV(+) and HPV(−) HNSCCs differ substantially in both epidemiology and clinical outcome. More specifically, HPV(+) patients have improved overall survival after initial diagnosis or following recurrent/metastatic disease (4, 5). Consistent with these notable clinical differences, recent comprehensive genetic analyses have revealed that the genetic landscapes of HPV(+) and (−) HNSCC differ greatly. There are three large genomic data sets available to date that evaluate these genetic variations using whole exome (WES) or whole genome sequencing (WGS) (6-9). However, earlier studies were dominated by HPV(−) samples, limiting the effective characterization HPV(+) tumors.
Building upon these previous studies, Seiwert and colleagues report three distinct findings; 1) HPV(+) and (−) tumors have a different profile of oncogenic mutations/CNVs, 2) HPV(+) tumors have frequent oncogene mutations previously described as rare in HNSCC, and 3) no significant difference is evident in the frequency of mutations between HPV(+) and HPV(−) tumors. These findings may have resulted from several key differences with previous three studies. First, HPV status was balanced between the two cohorts with HPV(+) tumors representing 42.5% of total tumor samples. Across the previous studies, HPV(+) tumors accounted for only ~13% of the samples. Secondly, tissue and clinical data collection occurred at a single institution for all samples included in the study. Additionally, concurrent chemoradiation was uniformly administered across all patients in the study (1, 6-9). Furthermore, utilizing a targeted sequencing approach increases the sensitivity for detecting somatic mutations in genes of interest rather than genome wide studies designed to maximize coverage (1). Exomic or genomic deep sequencing is time-consuming, labor intensive, informatically challenging, and relatively expensive. To avoid these issues, Seiwert, et al. opted for a targeted approach investigating 617 cancer specific genes to provide a sequencing depth nearly twice that of the Cancer Genome Atlas (TCGA) (6-9). Consequently, somatic mutations present at low allelic frequencies may have been missed in the previous studies due to insufficient coverage depth.
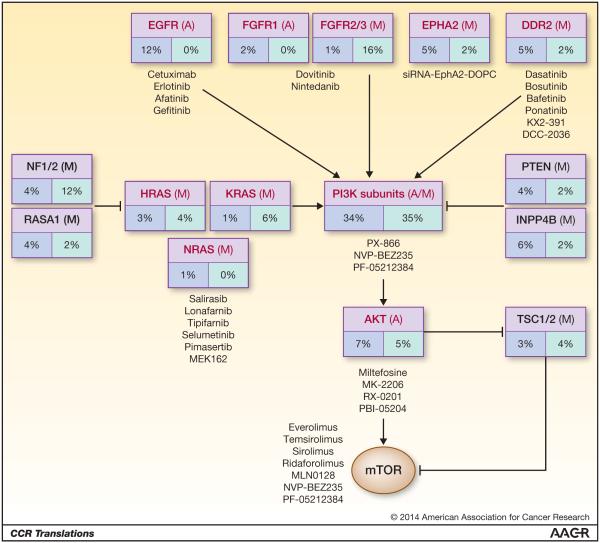
Identification of HPV-specific mutation profiles has far-reaching clinical implications as they represent potential therapeutic targets and predictive biomarkers. Currently, we rely on prognostic biomarkers (such as HPV-status) that are associated with outcome regardless of the treatment provided. If we intend to improve patient outcomes, a shift towards predictive biomarker-driven clinical trials must be undertaken to maximize clinical benefits. The most successful predictive biomarkers in recent years are activated oncogenes. These are commonly point mutations, such as c-Kit in gastrointestinal stromal tumors; however, copy number gain and gene upregulation, such as HER2 amplification in breast cancer, also represent powerful biomarkers. Based on current genome-wide sequencing and copy number data, several oncogenes in HNSCC are immediately targetable with agents in clinical development (Fig. 1). Current data suggest the frequency of oncogene mutations is relatively higher in HPV(+) compared to HPV(−) tumors, and the oncogene profiles differ based on the HPV status. For example, EGFR, CCND1 and FGFR1 amplifications are more common in HPV(−) tumors while FGFR2/3 mutations are more common in HPV(+) tumors.
Figure 1.
Therapeutic agents that are either FDA-approved or currently in clinical trials targeting the genetic aberrations found in HNSCC determined by Seiwert and colleagues (1). Red: oncogenes, Black: tumor suppressors, Blue box: HPV(−) tumors, Green box: HPV(+) tumors. The incidence of mutations (M) and amplifications (A) are separated by HPV status. Adapted from Seiwert et al. (1).
Currently, there are concerted efforts to conduct predictive biomarker driven clinical trials using targeted agents incorporating genomic, therapeutic, and clinical trial data in order to achieve the ultimate goal of personalized medicine. Because selecting patients with specific mutations results in small, fractionated cohorts, the use of site-specific phase II/III trials is no longer feasible. Consequently, novel designs are required such as multi-arm trials enrolling patients with a specific mutation in one of many simultaneous arms with a given targeted agent (also called umbrella, bucket, or basket trials). There are several multiple arm clinical trials currently underway, and HNSCC patients will be able to participate in the Molecular Analysis for Therapy Choice (MATCH) trial. In this study, patients with recurrent/metastatic cancers (regardless of organ site) will undergo pre- and post-treatment biopsies to sequence 200 pre-selected targetable genes in order to match with one of the ~25 targeted agents evaluated in the trial.
Although this technology is promising, significant challenges are inherent to mutational biomarker data. For example, previous studies suggested HPV(+) tumors have fewer mutations than HPV(−) tumors, while the current study failed to demonstrate this difference. This result suggests that mutations in HPV(+) tumors (including clinically relevant mutations) may be present at lower allelic frequencies than is observed in HPV(−) tumors, requiring increased sequencing depth to detect these key differences. If validated, this finding would help establish the ideal depth of coverage for routine mutation testing to be utilized for effective biomarker detection. To date, the threshold of cancer cells harboring a targetable mutation that is required for a clinically meaningful response is unknown. If we follow the current Response Evaluation Criteria In Solid Tumors (RECIST) criteria, an activating mutation may be required to occur at cancer cell fraction greater than 0.3 (corresponding to approximately >30% of tumor mass) to observe a positive therapeutic response. For the amplified genes, the threshold of copy number gain to consider significant is again unknown in HNSCC.
Accurate mutational and CNV detection aside, the therapeutic agents matched to these targets must be effective at inhibiting oncoprotein. There is significant variability even within the same class of targeted agents with respect to efficacy and toxicity. Additional issues include determining the most clinically significant oncogene in a landscape of aberrations, the emergence of resistant clones from heterogeneous tumors, obtaining tumors for testing from inaccessible anatomic locations, and access to drugs still in developmental pipeline in cases of trial ineligible patients. For HPV(−) patients, more creative approaches to target tumor suppressor loss or the relevant downstream consequences of negative regulator loss must be improved. Furthermore, while early therapeutic response can be assessed in multi-arm trials agnostic to disease site, confirmatory trials in patients with newly diagnosed or recurrent/metastatic HNSCC with survival benefit as the primary end point will be required. In the confirmatory trials, separate cohorts based on HPV status should be investigated, thus HNSCC provides significant, but not prohibitory challenges in future agent evaluation and trial feasibility. In conclusion, effectively incorporating sequencing data into the clinic will allow us to move beyond HPV status as our sole prognostic biomarker and necessitate novel clinical trial design and validation of biomarkers. However, the quest for personalized medicine will be easier when a comprehensive map of HNSCC is created which accurately encompasses the full breadth of heterogeneity provided by this challenging disease. We remain cautiously optimistic that we will be able to take advantage of this and future genomic studies to meaningfully benefit our patients.
Footnotes
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Seiwert TY, Zuo Z, Keck MK, Khattri A, Pedamallu CS, Stricker T, et al. Integrative and comparative genomic analysis of HPV-positive and HPV-negative head and neck squamous cell carcinomas. Clin Cancer Res. 2014;21 doi: 10.1158/1078-0432.CCR-13-3310. xxx-xxx. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Maier H, Dietz A, Gewelke U, Heller WD, Weidauer H. Tobacco and alcohol and the risk of head and neck cancer. Clin Investig. 1992;70:320–7. doi: 10.1007/BF00184668. [DOI] [PubMed] [Google Scholar]
- 3.D'Souza G, Kreimer AR, Viscidi R, Pawlita M, Fakhry C, Koch WM, et al. Case-control study of human papillomavirus and oropharyngeal cancer. N Engl J Med. 2007;356:1944–56. doi: 10.1056/NEJMoa065497. [DOI] [PubMed] [Google Scholar]
- 4.Chung CH, Schwartz DL. Impact of HPV-related head and neck cancer in clinical trials: opportunity to translate scientific insight into personalized care. Otolaryngol Clin North Am. 2012;45:795–806. doi: 10.1016/j.otc.2012.04.002. [DOI] [PubMed] [Google Scholar]
- 5.Fakhry C, Zhang Q, Nguyen-Tan PF, Rosenthal D, El-Naggar A, Garden AS, et al. Human papillomavirus and overall survival after progression of oropharyngeal squamous cell carcinoma. J Clin Oncol. 2014 Jun 23; doi: 10.1200/JCO.2014.55.1937. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Agrawal N, Frederick MJ, Pickering CR, Bettegowda C, Chang K, Li RJ, et al. Exome sequencing of head and neck squamous cell carcinoma reveals inactivating mutations in NOTCH1. Science. 2011;333:1154–7. doi: 10.1126/science.1206923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stransky N, Egloff AM, Tward AD, Kostic AD, Cibulskis K, Sivachenko A, et al. The mutational landscape of head and neck squamous cell carcinoma. Science. 2011;333:1157–60. doi: 10.1126/science.1208130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hayes DN, Grandis JR, El-Naggar AK. The Cancer Genome Atlas: integrated analysis of genome alterations in squamous cell carcinoma of the head and neck. J Clin Oncol. 2013;31 suppl; abstr 6009. [Google Scholar]
- 9.Kandoth C, McLellan MD, Vandin F, Ye K, Niu B, Lu C, et al. Mutational landscape and significance across 12 major cancer types. Nature. 2013;502:333–9. doi: 10.1038/nature12634. [DOI] [PMC free article] [PubMed] [Google Scholar]