Abstract
Introduction:
Enterococci are opportunistic pathogens which represent one of the leading agents of nosocomial infections, especially urinary tract infections (UTI) in hospitalized patients. The aim of the present study was to determine the resistance pattern and the type of resistance genes in vancomycin-resistant Enterococcus isolated from an educational hospital in Iran.
Materials and methods:
From February 2012 till February 2013, one hundred and eighty six clinical isolates from different department of educational hospitals were collected and identified as Enterococci and specified by biochemical tests. Identification was confirmed by specific PCR. Antibiotic resistance properties of strains were examined by Kerby-bauer method. PCR was performed for ddlE, ddlF, vanA and vanB genes.
Results:
One hundred and six (57%) isolates were identified as E. faecalis and 80 (43%) of the isolates were identified as E. faecium. 24 isolates had vanA gene and 19 isolates had vanB genes. In E. faecalis isolates, 15 isolates had vanB and 4 isolates had vanA gene. In E. faecium isolates, 20 isolates had vanA and 4 isolates had vanB gene. Prevalence of van genes between E. faecalis and E. faecium were significantly different for both vanA and vanB (p<0.01, p<0.041, respectively). VRE isolates were sensitive to Linezolid, Nitrofurantoin and Tigecyclin.
Discussion:
The overall prevalence of VRE was 23.65%, which shows an increase in VRE isolation in our region. Also, prevalence of E. faecium dramatically increased from 9% to 43% in the present study. Also increase in Gentamicin resistant isolates observed, but VRE isolates were sensitive to Linezolid, Tigecyclin and Nitrofurantoin. Stewardships for antibiotic usage in hospitals, especially for last option antibiotics, can prevent the spread of resistant isolates and losing all treatment options in the future.
Keywords: E. faecalis, E. faecium, vancomycin, VRE, resistance, Antibiotic, Iran
INTRODUCTION
Enterococci are Gram positive bacteria that are part of the normal intestinal flora of most humans (1). In the last 2 decades, several reports have documented that the two most important species, Enterococcus faecalis and Enterococcus faecium, are among the leading cause of several human infections, including bacteremia, septicemia, endocarditis, urinary tract infections, wound infections, neonatal sepsis and meningitis (2,3). In addition, the emergence of high-level aminoglycoside-resistant (HLAR) enterococci and vancomycin-resistant enterococci (VRE) causes great difficulties in clinical anti-infective therapy (4-6). The first VRE isolates were isolated in the UK and France in 1988 (7,8), because of rapid spread and limited options for VRE, these isolates has emerged as one of the most significant nosocomial pathogens worldwide, associated with high-level morbidity and mortality (9).
The mechanism of Vancomycin resistance in Enterococci is well understood. There are nine Vancomycin resistance containing van A, B, C, D, E,G, L, M, and vanN that vanA is the most predominant type worldwide (10-12). vanA confers a high degree of vancomycin and teicoplanin resistance and is mainly associated with vancomycin resistant Enterococcus faecium (13). vanB confers a high degree of vancomycin but susceptibility to other glycopeptides like teicoplanin since only the former antibiotic is capable of inducing the vanB resistance type (14). In this hospital-based study, a total number of 186 isolates collected from different departments of an educational hospital and identified to investigate the prevalence and antimicrobial resistance to antibiotics other than vancomycin to provide evidence for controlling inappropriate clinical use of antimicrobial agents and the further antimicrobial strategies for controlling enterococcal infections. ❑
MATERIAL AND METHOD
Sample collection and identification
One hundred and eighty six clinical isolates from different department of educational hospitals were collected and identified as Enterococci and specified by biochemical tests (12). Identification was confirmed by specific PCR for Enterococcus faecium and Enterococcus faecalis (15,16). Antibiotic resistance properties of strains were examined by Kerby-bauer method according to CLSI M100-S22 guideline 2012 (17), Antibiotic Discs were provided by Mast Group LTD (United Kingdom) and Staphylococcus aureus ATCC 25923 was used as Quality control for Disc diffusion.
DNA extraction
DNA extraction was done by CinnapureTM DNA extraction kit (Cinnagen, Iran). Bacterial pellet was resuspended in 100 µl G+ pre lysis buffer and was added 20 µl lysosyme and incubated at 37°C for at least 30 min. After adding lysis buffer and precipitation solution, the solution was transferred to a spin column and after washing the spin, DNA was eluted by elution buffer in 65°C (18,19).
Genomic PCR
PCR was performed in 25 µl volumes that contained 20-200 ng DNA, 0.5 µM of specific primers for E. faecalis (ddlE1:ATCAAGTACAGTTAGTCTTTATTAG, ddlE2: ACGATTCAAAGCTAACTGAATCAGT) (20) E. faecium (ddlF1: TTGAGGCAGACCAGATTGACG, ddlF2: TATGACAGCGACTCCGATTCC) (21) and for vanA (vAF: AATACTGTTTGGGGGTTGCTC, vAR: TTTTTCCGGCTCGACTTCCT)(22), vanB (vBF: GCGGGGAGGATGGTGCGATACAG, vBR: GGAAGATACCGTGGCTCAAAC) (22) with 1.5 mM MgCl2, 200 µM of each dNTP, 1X PCR buffer and 2 U DNA polymerase (Cinnage, Iran). DNA was amplified by general PCR. An initial denaturation of 10 min at 94°C was followed by 35 cycles of denaturation at 94°C (1 min), annealing at 58°C for 1 min and extension at 72°C for 1 min, followed by a final extention at 72°C for 10 min. product length were 941bp for E. faecalis, 658 bp for E. faecium, 734bp for vanA and 420 bp for vanB. Positive controls for PCR were E. faecalis MMH594, E. faecium C38 and E. faecium ATCC 51559 (vanA) and E. faecalis ATCC 51299 (vanB). Negative controls consisted of the PCR components of the reaction mixtures lacking Enterococci DNA. PCR products were electrophoreses in 1.5% agarose gels and after staining with 0.5µg/ml ethidium bromide visualized under UV light. The size of the fragments was determined by comparing with 100 bp DNA ladder plus size marker (Fermentas, Germany).
Statistical analysis
Chi-square test (or Fisher exact test) was performed for data analysis. P values below 0.05 were considered to be significant. Statistical analysis was done by SPSS. 21 software. ❑
RESULTS
From February 2012 till February 2013, One hundred and eighty six isolates of Enterococci were collected from an educational hospital and were characterized for their type and species and antibiotic resistance properties. All isolates were examined for presence of van genes. One hundred and eleven isolates (59.67%) were from female patients and seventy five isolates (40.32%) were from male patients. The origins of the isolates were one hundred forty nine urine (80.1%), twenty wounds (10.75%), six bloods (3.2%), four phlegms (2.15%), three stools (1.61%), two asits (1.07%) and two tracheas (1.07%). By biochemical tests and PCR, 106 (57%) isolates identified as E. faecalis and 80 (43%) of isolates were E. faecium (Figure 1). Pattern of antibiotic resistance of isolates are presented in Figure 2 (for total isolates), Figure 3 (for E. faecalis) and Figure 4 (for E. faecium). PCR for detecting vanA and vanB genes was done for all isolates (Figure 5) which 24 isolates had vanA gene and 19 isolates had vanB genes. In E. faecalis isolates, 15 isolates (14.15%) had vanB and 4 isolates (3.7%) had vanA gene. In E. faecium isolates 20 isolates (25%) had vanA and 4 isolates (5%) had vanB gene. Prevalence of van genes between E. faecalis and E. faecium were significantly different for both vanA and vanB (p<0.01, p<0.041, respectively). Pattern of antibiotic resistance in isolates possess van genes are presented in Table 1, which shows the susceptibility of these isolates to Gentamicin, Linezolid, Nitrofurantoin and Tigecyclin. ❑
Figure 1. Gel electrophoresis for PCR product of isolates identification. Enterococcus faecalis (left gel) and Enterococcus faecium (right gel).
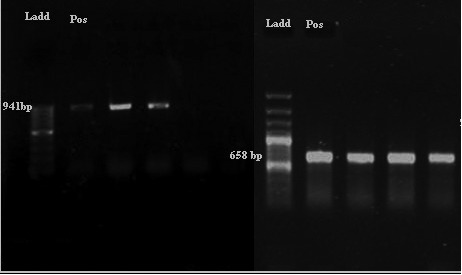
Figure 2. Antibiotic resistance of all Enterococci isolate collected in this study based on disc diffusion. Concentartion of disc presented after antibiotic names.
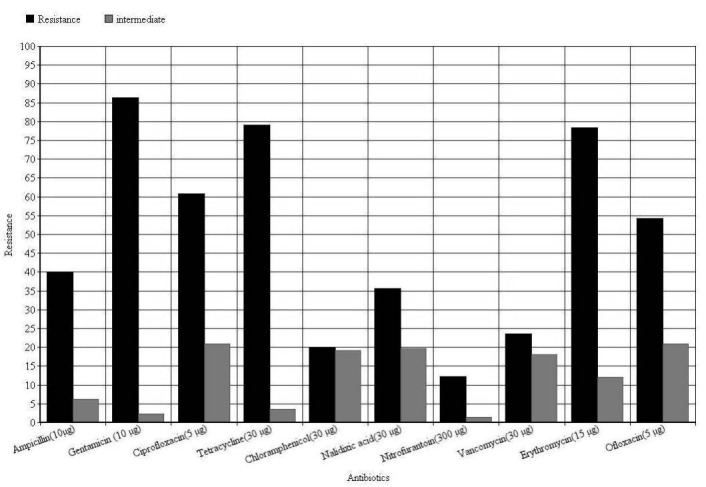
Figure 3. Antibiotic resistance of Enterococcus faecalis isolates based on disc diffusion. Concentration of disc presented after antibiotic names.
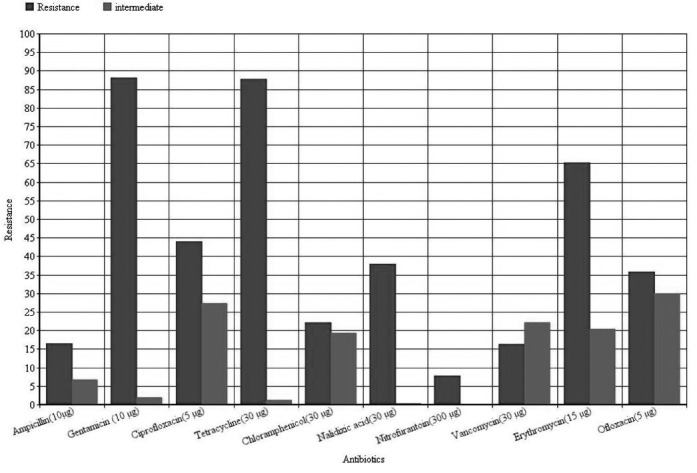
Figure 4. Antibiotic resistance of Enterococcus faecium isolates based on disc diffusion. Concentartion of disc presented after antibiotic names.
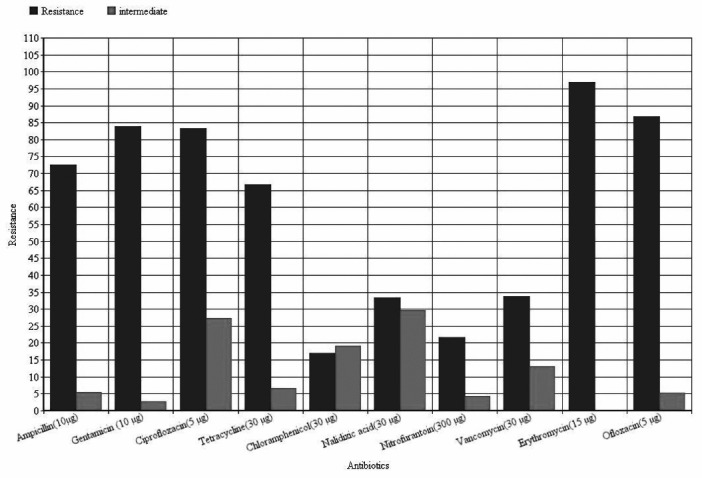
Figure 5. Gel electrophoresis of vanB(left) and vanA(right) PCR products in 0.8% Agarose gel.
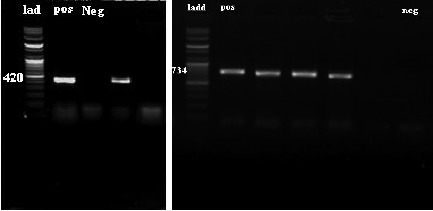
Table 1.
Pattern of Antibiotic resistance in isolates have van genes.
| Gentamicin | Linezolid | Nitrofurantoin | Tigecyclin | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Resistance | Sensitive | Resistance | Sensitive | Resistance | Sensitive | Resistance | Sensitive | ||
| E. faecalis | vanA | 1(25%) | 3(75%) | 4(100%) | 0 | 2(50%) | 2(50%) | 0 | 4(100%) |
| vanB | 8(53.33%) | 7(46.66%) | 0 | 15(100%) | 5(33.33%) | 10(66.66%) | 1(6.66%) | 14(93.33%) | |
| E.faecium | vanA | 16(80%) | 4(20%) | 0 | 20(100%) | 13(65%) | 7(35%) | 3(15%) | 17(85%) |
| vanB | 4(100%) | 0 | 0 | 4(100%) | 4(100%) | 0 | 0 | 4(100%) | |
DISCUSSION
Vancomycin-resistant enterococci have been increasingly reported worldwide since first described in 1987, although the epidemiology of these microorganisms varies widely in different geographical areas (23). In the present study the overall prevalence of VRE was 44/186 (23.65%) (Figure 2), which Vancomycin resistant E. faecalis were 17/106 (16.03%) (Figure 3) and in E. faecium were 27/80 (33.75%) (Figure 4). This was consistent with reports from Egypt with 25% VRE isolation (24) and lower than reports from Korea (4.5%) (25), Ethiopia (5.5%) (26) and Tehran-Iran (9.5%) (27). Comparing results of the present study with the earlier studies in Iran showed an increase in VRE isolation from 10-15% to 23.65% (27, 28), also, prevalence of E. faecium dramatically increased from 9% (27) or 19.8 % (28) to 43% in the present study. This high prevalence of E. faecium in our study can be the main reason of high VRE isolation at our investigated hospital. Of the 44 VRE isolates, in 43 isolates one of vanA or vanB genes were detected and in one isolate we couldn't find resistance related gene. Resistance in this isolate can be due to thicker cell wall production or other resistance mechanisms. Also vanA and vanB genes prevalence was significantly different between E. faecium and E. faecalis isolates, vanA was dominant resistance gene in E. faecium and vanB was dominant in E. faecalis. Although a high percentage of resistance against tetracycline (87.6%) and Ciprofloxacin (83.3%) (Figure 3) observed in E. faecalis isolates (p<0.05), no other significant difference observed between E. faecalis and E. faecium isolates. High percentage of resistance against Gentamicin (E. faecalis: 87.7%, E. faecium: 83.7%, Total: 86%) and Erythromycin (E. faecalis: 65%, E. faecium: 96.25%, Total: 78.5%) observed in isolates (Figure 1, 2 and 3). This high percentage of resistance was in agreement with recent studies in cross-sectional studies in Ethiopia and Egypt (24, 26) but it was higher than other recent studies in the same region of our study with Tehran (41.66%) (27), Tabriz (32.43%) (28), Tabriz (36.2%) (29). These results indicate a high increase in Gentamicin resistant isolates that reduces treatment options for enterococci. The same pattern of resistance was found in isolates possess van genes (Table 1), but fortunately the results for Linezolid and Tigecyclin showed no resistance to these antibiotics in VRE isolates (Table 1). Although 66.6% of E. faecalis vanA positive isolates were resistant to Nitrofurantoin, but other VRE isolates were sensitive to this antibiotic. This result introduces Nitrofurantoin for treating Urinary tract infections by VRE. ❑
CONCLUSION
Finding of this study shows increase prevalence of VRE isolates in Iran associated with increase in E. faecium isolation from hospitals. Also dramatically increase in Gentamicin resistance isolates observed, but VRE isolates were sensitive to Linezolid, Tigecyclin and Nitrofurantoin. Stewardships for antibiotic usage in hospitals, especially for last option antibiotics, can prevent the spread of resistant isolates and losing all treatment options in the future.
ACKNOWLEDGMENTS
The authors would like to thank Hossein Navidinia and Mohammad Momenian for their review of manuscript and all staff of Microbiology lab (Drug applied research Center) for their collaborations and helps. Also Authors thank Mr Nikmaram in Imam Reza Hospital- Tabriz for help on sample collection.
CONFLICT OF INTEREST
none declared.
FINANCIAL SUPPORT
This work was supported by Drug Applied Research Center, Tabriz University of Medical Sciences: [grant number 1393415].
References
- 1.Bourgogne A, Singh KV, Fox KA, et al. EbpR is important for biofilm formation by activating expression of the endocarditis and biofilm-associated pilus operon (ebpABC) of Enterococcus faecalis OG1RF. J Bacteriol. 2007;189:6490–6493. doi: 10.1128/JB.00594-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Giacometti A, Cirioni O, Schimizzi AM, et al. Epidemiology and microbiology of surgical wound infections. J Clin Microbiol. 2000;38:918–22. doi: 10.1128/jcm.38.2.918-922.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Higaki S, Morohashi M, Yamagishi T. Isolation of Enterococcus species from infectious skin lesions. Drugs Exp Clin Res. 2002;28:91–3. [PubMed] [Google Scholar]
- 4.Bonten MJ, Willems R, Weinstein RA. Vancomycin-resistant enterococci: Why are they here, and where do they come from? Lancet Infect Dis. 2001;1:314–325. doi: 10.1016/S1473-3099(01)00145-1. [DOI] [PubMed] [Google Scholar]
- 5.Adhikari L. High-level aminoglycoside resistance and reduced susceptibility to vancomycin in nosocomial enterococci. J Glob Infect Dis. 2010;2:231–235. doi: 10.4103/0974-777X.68534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cetinkaya Y, Falk P, Mayhall CG. Vancomycin-resistant enterococci. Clin Microbiol Rev. 2000;13:686–707. doi: 10.1128/cmr.13.4.686-707.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Leclercq R, Derlot E, Duval J, et al. Plasmid-mediated resistance to vancomycin and teicoplanin in Enterococcus faecium. N Engl J Med. 1988;319:157–161. doi: 10.1056/NEJM198807213190307. [DOI] [PubMed] [Google Scholar]
- 8.Uttley AH, Collins CH, Naidoo J, et al. Vancomycin-resistant enterococci. Lancet. 1988;1:57–58. doi: 10.1016/s0140-6736(88)91037-9. [DOI] [PubMed] [Google Scholar]
- 9.Murray BE. Vancomycin-resistant enterococcal infections. N Engl J Med. 2000;342:710–721. doi: 10.1056/NEJM200003093421007. [DOI] [PubMed] [Google Scholar]
- 10.Protonotariou E, Dimitroulia E, Pournaras S, et al. Trends in antimicrobial resistance of clinical isolates of Enterococcus faecalis and Enterococcus faecium in Greece between 2002 and 2007. J Hosp Infect. 2010;75:225–227. doi: 10.1016/j.jhin.2009.12.007. [DOI] [PubMed] [Google Scholar]
- 11.Sofianou D, Pournaras S, Giosi M, et al. Substantially increased E. faecalis carriage of vancomycin-resistant enterococci in a tertiary Greek hospital after a 4 year time interval. J Antimicrob Chemother. 2004;54:251–254. doi: 10.1093/jac/dkh293. [DOI] [PubMed] [Google Scholar]
- 12.Souli M, Sakka V, Galani I, et al. Colonisation with vancomycin- and linezolid-resistant Enterococcus faecium in a university hospital: molecular epidemiology and risk factor analysis. Int J Antimicrob Agents. 2009;33:137–142. doi: 10.1016/j.ijantimicag.2008.08.017. [DOI] [PubMed] [Google Scholar]
- 13.Kang M, Xie Y, He C, et al. Molecular characteristics of vancomycin-resistant Enterococcus faecium from a tertiary care hospital in Chengdu, China. Eur J Clin Microbiol Infect Dis. doi: 10.1007/s10096-013-2029-z. [DOI] [PubMed] [Google Scholar]
- 14.Werner G, Klare I, Fleige C, et al. Vancomycin-resistant vanB-type Enterococcus faecium isolates expressing varying levels of vancomycin resistance and being highly prevalent among neonatal patients in a single ICU. Antimicrob Resis Infect Control. 2012;1:21–21. doi: 10.1186/2047-2994-1-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Facklam RR. Recognition of group D streptococcal species of human origin by biochemical and physiological tests. Appl Microbiol. 1972;23:1131–1139. doi: 10.1128/am.23.6.1131-1139.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kafil HS, Mobarez AM, Moghadam MF. Adhesion and virulence factor properties of enterococci isolated from clinical samples in Iran. Indian J Pathol Microbiol. 2013;56:238–242. doi: 10.4103/0377-4929.120375. [DOI] [PubMed] [Google Scholar]
- 17.Clinical and Laboratory Standards Institute. M100-S22. Performance standards for antimicrobial susceptibility testing: 22nd informational supplement. 2012 Wayne, PA: CLSI
- 18.Toledo-Aran, Valle J, et al. The enterococcal surface protein, Esp, is involved in Enterococcus faecalis biofilm formation. Appl Environ Microbiol. 2001;67:4538–4545. doi: 10.1128/AEM.67.10.4538-4545.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Asgharzadeh M, Kafil HS, Roudsary AA, et al. Tuberculosis transmission in Northwest of Iran: Using MIRU-VNTR, ETR-VNTR and IS6110-RFLP methods. Infect Genet Evol. 2011;11:124–131. doi: 10.1016/j.meegid.2010.09.013. [DOI] [PubMed] [Google Scholar]
- 20.Sandoe JA, Witherden IR, Cove JH, et al. Correlation between enterococcal biofilm formation in vitro and medical-device-related infection potential in vivo. J Med Microbiol. 2003;52:547–550. doi: 10.1099/jmm.0.05201-0. [DOI] [PubMed] [Google Scholar]
- 21.Dupre I, Zanetti S, Schito AM, et al. Incidence of virulence determinants in clinical Enterococcus faecium and Enterococcus faecalis isolates collected in Sardinia (Italy). J Med Microbiol. 2003;52:491–498. doi: 10.1099/jmm.0.05038-0. [DOI] [PubMed] [Google Scholar]
- 22.Khana SA, Nawaza MS, Khana AA, et al. Molecular characterization of multidrug-resistant Enterococcus spp. from poultry and dairy farms: detection of virulence and vancomycin resistance gene markers by PCR. Mol Cell Probes. 2005;19:27–34. doi: 10.1016/j.mcp.2004.09.001. [DOI] [PubMed] [Google Scholar]
- 23.Gossens H, Habes D, Rossi R, et al. European survey of vancomycin resistant enterococci in at- risk hospital wards and in vitro susceptibility testing of ramoplanin against these isolates. J Antimicrob Chemother. 2003;51:iii5–iii12. doi: 10.1093/jac/dkg271. [DOI] [PubMed] [Google Scholar]
- 24.Al-Tonbary YA, Soliman OE, Sarhan MM, et al. Nosocomial infections and fever of unknown origin in pediatric hematology/oncology unit: a retrospective annual study. World J Pediatr. 2011;7:60–64. doi: 10.1007/s12519-010-0212-1. [DOI] [PubMed] [Google Scholar]
- 25.Kee SY, Park CW, Lee JE, et al. Western Dialysis Physical Association: Healthcare-associated risk factors of vancomycin-resistant Enterococci colonization among outpatients undergoing hemodialysis. Jpn J Infect Dis. 2012;65:57–60. [PubMed] [Google Scholar]
- 26.Abebe W, Endris M, Tiruneh M, et al. Prevalence of vancomycin resistant Enterococci and associated risk factors among clients with and without HIV in Northwest Ethiopia: a cross-sectional study. BMC Public Health. 2014;14:185–185. doi: 10.1186/1471-2458-14-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aleyasin A, Mobarez AM, Sadeghizadeh M, et al. Resistance to vancomycin in enterococcus faecium and faecalis clinical isolates. Pak J Med Sci. 2007;23:390–393. [Google Scholar]
- 28.Behnood A, Farajnia S, Moaddab SR, et al. Prevalence of aac(6')-Ie-aph(2")-Ia resistance gene and its linkage to Tn5281 in Enterococcus faecalis and Enterococcus faecium isolates from Tabriz hospitals. Iran J Microbiol. 2013;5:203–208. [PMC free article] [PubMed] [Google Scholar]
- 29.Balaei Gajan E, Shirmohammadi A, Aghazadeh M, et al. Antibiotic Resistance in Enterococcus faecalis Isolated from Hospitalized Patients. J Dent Res Dent Clin Dent Prospects. 2013;7:102–104. doi: 10.5681/joddd.2013.018. [DOI] [PMC free article] [PubMed] [Google Scholar]


