Abstract
The present study aimed to investigate the association between epidermal growth factor receptor (EGFR) gene mutations and excision repair cross-complementing protein 1 (ERCC1) and ribonucleotide reductase subunit M1 (RRM1) mRNA expression in non-small cell lung cancer (NSCLC) tissue. The quantitative polymerase chain reaction was used to detect EGFR mutations, and ERCC1 and RRM1 mRNA expression in 257 cases of NSCLC. In the NSCLC samples the EGFR mutation rate was 49.03% (126/257). The rate was higher in females and non-smoking patients (P<0.05). High expression of ERCC1 mRNA was observed in 47.47% of the samples (122/257), while a high RRM1 mRNA expression was observed in 61.87% of the samples (159/257). In comparison with patients with NSCLC without EGFR mutations, patients with EGFR mutations had significantly lower levels of ERCC1 mRNA expression (P<0.05); however, EGFR mutations and expression levels of RRM1 mRNA were not correlated in NSCLC tissues (P>0.05). In addition, ERCC1 mRNA expression was not correlated with the expression levels of RRM1 mRNA (P>0.05). In conclusion, patients with NSCLC with EGFR mutations tend to have a low expression of ERCC1 mRNA and may potentially benefit from platinum-based chemotherapy.
Keywords: non-small cell lung cancer, epidermal growth factor receptor, excision repair cross-complementing protein 1, ribonucleotide reductase subunit M1, molecular detection, individualized treatment
Introduction
Globally, lung cancer has the highest rates of morbidity and mortality of all malignancies (1). Non-small cell lung cancer (NSCLC) accounts for 80–85% of lung cancer cases worldwide (2).
Human epidermal growth factor receptor (EGFR) belongs to the type I receptor family. This family has four cognate family members, including EGFR (HER1), HER2, HER3 and HER4, which mediate the following signal transduction pathways: Ras-Raf-mitogen-activated protein kinase kinase-extracellular signal-regulated kinase-mitogen-activated protein kinase, phospholipase C-γ, phosphatidylinositol-3-kinase/phosphoinositide-dependent kinase 1 and Janus kinase/signal transducers and activators of transcription (3). These receptors regulate cell proliferation, differentiation and apoptosis (4). EGFR mutations in patients with NSCLC occur in the intracellular tyrosine kinase (TK) region above the first four exons (18 to 21). A total of 30 different types of mutations have been identified in the TK region (5), the most common occurring in exons 19 and 21, which account for ~85% of all the mutations (6).
Excision repair cross-complementing gene 1 (ERCC1) is the key gene in two DNA repair pathways: nucleotide excision repair (NER) and chain crosslink repair (7). Overexpression of ERCC1 can rapidly repair damaged DNA arrest at the G2/M phase and cause cells to be resistant to platinum (8). Ribonucleotide reductase subunit M1 (RRM1) is involved in DNA synthesis and repair (9). Results from the Iressa® Pan-Asia Study (10) clinical trial indicated that, in Asian populations, patients with EGFR mutations were more responsive to chemotherapy than patients with wild-type EGFR. Patients with lung cancer with low expression of ERCC1 and RRM1 are more responsive to gemcitabine- and platinum-based chemotherapy, respectively (11,12). Patients with NSCLC are generally treated with chemotherapy drugs, including cisplatin and gemcitabine. In the present study, 257 patients with stages I-IV NSCLC from multiple hospitals were analyzed for the presence of EGFR mutations and expression levels of ERCC1 and RRM1 mRNA. The data were statistically analyzed to determine significant correlations between EGFR mutations and expression of these two chemotherapy resistance genes in patients with NSCLC. These data may prove useful in further identifying more effective individualized treatment plans for patients with EGFR mutations, particularly for patients with small-molecule EGFR-tyrosine kinase inhibitor (EGFR-TKI) primary or secondary resistance.
Materials and methods
Specimens
For the detection of EGFR mutations, as well as ERCC1 and RRM1 mRNA expression levels, paraffin tissue specimens were collected from 257 patients from the General Military Hospital of Beijing PLA (Beijing, China; 103 cases), the Affiliated Zhongshan Hospital of Dalian University (Dalian, China; 58 cases) and the People’s Hospital of Weifang (Weifang, China; 96 cases). The patients had undergone surgery between 2004 and 2013; the pathological diagnosis was adenocarcinoma, and patients had not received preoperative chemotherapy, radiotherapy or biological immunotherapy. All protocols in the present study were approved by the Human Clinical and Research Ethics Committees of the General Military Hospital of Beijing PLA, the Affiliated Zhongshan Hospital of Dalian University (Dalian, China) and the People’s Hospital of Weifang (Weifang, China). Written informed consent was obtained from all of the patients.
Reagents and instruments
The DNA and RNA extraction kits were purchased from Qiagen (Hilden, Germany). The human EGFR mutation qualitative detection kit and the tumor-related gene expression relative quantification detection kit (ERCC1 and RRM1) were obtained from Amoy Diagnostics Co., Ltd. (Xiamen, China). The B-500 instrument to measure nucleic acid protein concentrations was purchased from Shanghai Chong Meng Biotechnology Co. Ltd. (Shanghai, China) and the ABI 7500 quantitative polymerase chain reaction (qPCR) instrument was purchased from Applied Biosystems® (Life Technologies, Foster City, CA, USA).
qPCR to detect EGFR mutations in NSCLC tissues
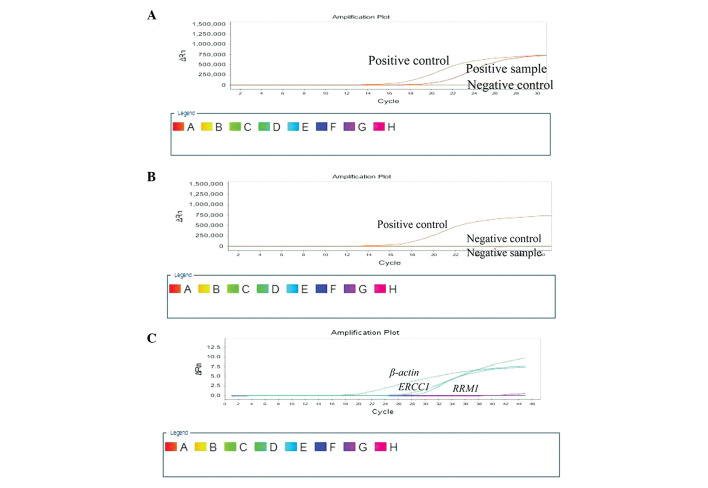
Between four and eight 4-μm-thick paraffin tissue sections were obtained and dewaxed. The genomic RNA extraction kit was used to extract RNA from the tissue samples according to the manufacturer’s instructions. A spectrophotometer was used to determine the purity and concentration of the extracted RNA, which was used as a template to synthesize the corresponding DNA. The human EGFR mutation qualitative detection kit, which contains 29 different fusion mutant primers and probes to amplify EGFR exons 18, 19, 20 and 21, was used (ADx-EG09; Amoy Diagnostics Co., Ltd.); the DNA was amplified in an ABI 7500 qPCR instrument (Fig. 1A and B). The qPCR cycling conditions were set as follows: 95°C for 5 min, follwed by 45 cycles of 95°C for 30 sec, 60°C for 30 sec and 72°C for 45 sec.
Figure 1.
Mutations of the EGFR gene and the expression of ERCC1 and RRM1 mRNA. (A) Positive control, positive sample and negative control of the EGFR gene. (B) Positive control, negative sample and negative control of the EGFR gene. (C) Expression of ERCC1 and RRM1 mRNA. EGFR, epidermal growth factor receptor; ERCC1, excision repair cross-complementing protein 1; RRM1, ribonucleotide reductase subunit M1.
qPCR to detect the expression of ERCC1 and RRM1 mRNA in NSCLC tissue
RNA was extracted from 4-μm-thick paraffin tissue sections in accordance with the aforementioned methods. The tumor-associated gene expression detection kit for ERCC1 (ADx-ER01) and RRM1 (ADx-RR01) (Amoy Diagnostics Co., Ltd.) was used to determine the mRNA expression level of these genes using the absolute quantitative method; β-actin was used as the reference gene. ERCC1 had a standard mean of 4.29×10−3, and RRM1 had a standard mean of 11.37×10−3 (Fig. 1C).
Statistical analysis
The data were analyzed using the statistical software SPSS (version 19.0; IBM SPSS, Armonk, NY, USA) using the χ2 and Fisher’s exact test with a test level α=0.05. The P-value was set to bilateral distribution and P<0.05 was considered to indicate a statistically significant difference.
Results
Association between EGFR mutations, expression of ERCC1 and RRM1 mRNA, and patient clinical characteristics
Of the 257 cases of NSCLC, EGFR mutations were present in 126 cases (49.03%). The EGFR mutation rate was higher in non-smoking patients (92/158, 58.23%; P<0.001); however, the EGFR mutation rate was not associated with the age, tumor size, lymph-node metastasis or clinical stage of the patient. High expression of ERCC1 mRNA was observed in 122/257 cases (47.47%). ERCC1 mRNA expression levels were not associated with the gender, age, smoking status, tumor size, lymph-node metastasis or clinical stage of the patient. High expression of RRM1 mRNA was observed in 159/257 cases (61.87%). RRM1 mRNA expression levels were not associated with the gender, age, smoking status, tumor size, lymph-node metastasis or clinical stage of the patient (Table I).
Table I.
Association between EGFR mutations, ERCC1 and RRM1 mRNA expression, and patient clinical characteristics.
| EGFR | ERCC1 | RRM1 | |||||||
|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
|||||||
| Clinical features | Mutant (n) | Wild-type (n) | P-value | High (n) | Low (n) | P-value | High (n) | Low (n) | P-value |
| Gender | <0.001 | 0.898 | 0.088 | ||||||
| Male | 52 | 86 | 65 | 73 | 92 | 46 | |||
| Female | 74 | 45 | 57 | 62 | 67 | 52 | |||
| Age, years | 0.287 | 0.943 | 0.714 | ||||||
| ≥59 | 68 | 62 | 62 | 68 | 79 | 51 | |||
| <59 | 58 | 69 | 60 | 67 | 80 | 47 | |||
| Smoking history | <0.001 | 0.199 | 0.129 | ||||||
| Yes | 34 | 65 | 52 | 47 | 67 | 32 | |||
| No | 92 | 66 | 70 | 88 | 92 | 66 | |||
| Tumor diameter, cm | 0.282 | 0.687 | 0.292 | ||||||
| ≥5 | 56 | 67 | 60 | 63 | 72 | 51 | |||
| <5 | 70 | 64 | 62 | 72 | 87 | 47 | |||
| Lymph node metastasis | 0.903 | 0.470 | 0.188 | ||||||
| Yes | 51 | 54 | 47 | 58 | 70 | 35 | |||
| No | 75 | 77 | 75 | 77 | 89 | 63 | |||
| Clinical stage | 0.203 | 0.339 | 0.577 | ||||||
| I | 50 | 42 | 40 | 52 | 59 | 33 | |||
| II+III+IV | 76 | 89 | 82 | 83 | 100 | 65 | |||
EGFR, epidermal growth factor receptor; ERCC1, excision repair cross-complementing protein 1; RRM1, ribonucleotide reductase subunit M1.
Association between EGFR mutations and expression levels of ERCC1 mRNA
Of the 126 patients with NSCLC with an EGFR mutation, 79 (62.70%) showed low expression of ERCC1. Of the 131 patients with NSCLC with the wild-type EGFR gene, 56 (42.75%) had low expression of ERCC1 mRNA. These data indicate that patients with NSCLC with an EGFR mutation had significantly lower expression of ERCC1 mRNA (P<0.05) (Table II).
Table II.
Association between EGFR mutations and the expression level of ERCC1 mRNA.
| ERCC1 mRNA expression | |||
|---|---|---|---|
|
|
|||
| EGFR | High (n) | Low (n) | Total (n) |
| Mutant | 47 | 79 | 126 |
| Wild-type | 75 | 56 | 131 |
| Total (n) | 122 | 135 | 257 |
EGFR, epidermal growth factor receptor; ERCC1, excision repair cross-complementing protein 1.
Association between EGFR mutations and the expression level of RRM1 mRNA
Of the 126 patients with NSCLC with an EGFR gene mutation, 51 (40.48%) had low expression of RRM1 mRNA, while 47 out of the 131 patients (35.88%) with the wild-type EGFR gene had low expression of RRM1 mRNA. These data indicate that there was no correlation between EGFR and RRM1 mutations in patients with NSCLC (P>0.05) (Table III).
Table III.
Association between EGFR mutations and the expression level of RRM1 mRNA.
| RRM1 mRNA expression | |||
|---|---|---|---|
|
|
|||
| EGFR | High (n) | Low (n) | Total (n) |
| Mutant | 75 | 51 | 126 |
| Wild-type | 84 | 47 | 131 |
| Total (n) | 159 | 98 | 257 |
EGFR, epidermal growth factor receptor; RRM1, ribonucleotide reductase subunit M1.
Association between the expression levels of ERCC1 and RRM1 mRNA
Of the 122 patients with NSCLC with high expression of ERCC1 mRNA, 72 cases (59.02%) had high expression of RRM1 mRNA. Of the 135 patients with NSCLC with low expression of ERCC1 mRNA, 48 cases (35.56%) had low expression of RRM1 mRNA. A total of 120 cases had low or high expression of both ERCC1 and RRM1 mRNA, and 137 cases had contrasting expression levels of ERCC1 and RRM1 mRNA. These data indicate that the expression of ERCC1 and RRM1 mRNA was not significantly correlated (χ2=0.800, P=0.371) in NSCLC tissue (Table IV).
Table IV.
Association between the expression levels of ERCC1 and RRM1 mRNA.
| RRM1 mRNA expression | |||
|---|---|---|---|
|
|
|||
| ERCC1 mRNA expression | High (n) | Low (n) | Total (n) |
| High | 72 | 50 | 122 |
| Low | 87 | 48 | 135 |
| Total (n) | 159 | 98 | 257 |
ERCC1, excision repair cross-complementing protein 1; RRM1, ribonucleotide reductase subunit M1.
Discussion
EGFR mutations and the responsiveness of NSCLC to the molecular targeted drugs gefitinib (trade name, Iressa) and erlotinib (trade name, Tarceva®) have a close association (13,14). Small-molecule TKIs have been shown to have a high efficiency in patients with an exon 19 deletion in the EGFR gene (15); however, patients with NSCLC with EGFR mutations in exon 20 are resistant to drug treatment with TKIs (16). Other studies have reported that the NER complexes prognosis is good but not suitable for receiving platinum-based chemotherapy (17,18).
The results of the present study showed that the EGFR mutation rate was 48.03% (126/257) in patients with stages I-IV NSCLC, which is consistent with previously reported data (14,19). A higher percentage of patients with NSCLC (47.47%; 122/257) showed high mRNA expression levels of ERCC1 compared with data from a previous study (20), while the percentage of patients with NSCLC with a high RRM1 mRNA expression level (61.87%; 159/257) was consistent with data presented in a previous study (21). The expression levels of ERCC1 and RRM1 mRNA were not associated with the gender, age, smoking status, tumor size, lymph node metastasis, pathological staging or other clinical characteristics of the patient.
The current study found that the mutational status of EGFR was associated with ERCC1 mRNA expression levels in patients with NSCLC; patients with EGFR mutations had a significantly lower expression of ERCC1 mRNA (P<0.05). EGFR mutations did not, however, significantly correlate with RRM1 expression levels (P>0.05) in patients with NSCLC. A previous study has shown that EGFR mutations in patients with NSCLC were correlated with expression levels of ERCC1 (P<0.001). Furthermore, EGFR mutations in the adenocarcinoma subgroup were correlated with ERCC1 expression levels (P=0.001) (22). It has also been shown that NER enzymes in cells can lead to cell damage, resulting in genomic instability and an increased mutation rate (23). Cancer cells with low expression of ERCC1 have a decreased ability to repair DNA damage, an increase in the number of EGFR mutations and increased sensitivity to platinum-based chemotherapy, which may be why patients with NSCLC with EGFR mutations have a higher response rate to chemotherapy.
The present study also found that, in NSCLC tissues, mRNA expression levels of ERCC1 and RRM1 were not correlated (P>0.05), which is inconsistent with data presented by Reynolds et al (24). The association between the expression of ERCC1 and RRM1 mRNA remains controversial and warrants further research. At present, numerous studies have confirmed that therapy can be selected based on patient expression levels of ERCC1 and RRM1, and this can be extended to patients with NSCLC (25,26).
In conclusion, the current study has demonstrated that patients with EGFR mutations tend to have lower expression levels of ERCC1 mRNA. We hypothesize that patients with EGFR mutations may be more responsive to cisplatin-based chemotherapy, although the molecular mechanisms require further study. No difference in the expression levels of RRM1 mRNA was observed between patients with NSCLC with EGFR mutations and those without mutations. In the present study, we have determined the first-line chemotherapy for tumors involving platinum and gemcitabine drug resistance genes and EGFR mutations but not the microtubule drug resistance gene TUBB3 or the thymidylate synthase resistance gene TYMS. Future studies, therefore, are likely to focus on EGFR, TUBB3 and TYMS mutations to better identify effective individualized treatment plans, particularly individualized treatment plans for EGFR-TKI (e.g. imatinib) primary or secondary resistance observed in certain patients.
Acknowledgements
This study was supported by the Wu Jieping Medical Foundation Clinical Research Special Project Fund (320.6750.1360).
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Sher T, Dy GK, Adjei AA. Small cell lung cancer. Mayo Clin Proc. 2008;83:355–367. doi: 10.4065/83.3.355. [DOI] [PubMed] [Google Scholar]
- 3.Cao C, Lu S, Sowa A, et al. Priming with EGFR tyrosine kinase inhibitor and EGF sensitizes ovarian cancer cells to respond to chemotherapeutical drugs. Cancer Lett. 2008;266:249–262. doi: 10.1016/j.canlet.2008.02.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Herbst RS. Review of epidermal growth factor receptor biology. Int J Radiat Oncol Biol Phys. 2004;59(2 Suppl):21–26. doi: 10.1016/j.ijrobp.2003.11.041. [DOI] [PubMed] [Google Scholar]
- 5.Sharma SV, Bell DW, Settleman J, Haber DA. Epidermal growth factor receptor mutations in lung cancer. Nat Rev Cancer. 2007;7:169–181. doi: 10.1038/nrc2088. [DOI] [PubMed] [Google Scholar]
- 6.Chan SK, Gullick WJ, Hill ME. Mutations of the epidermal growth factor receptor in non-small cell lung cancer - search and destroy. Eur J Cancer. 2006;42:17–23. doi: 10.1016/j.ejca.2005.07.031. [DOI] [PubMed] [Google Scholar]
- 7.Simon GR, Ismail-Khan R, Bepler G. Nuclear excision repair-based personalized therapy for non-small cell lung cancer: from hypothesis to reality. Int J Biochem Cell Biol. 2007;39:1318–1328. doi: 10.1016/j.biocel.2007.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rosell R, Lord RV, Taron M, Reguart N. DNA repair and cisplatin resistance in non-small-cell lung cancer. Lung Cancer. 2002;38:217–227. doi: 10.1016/S0169-5002(02)00224-6. [DOI] [PubMed] [Google Scholar]
- 9.Su C, Zhou S, Zhang L, et al. ERCC1, RRM1 and BRCA1 mRNA expression levels and clinical outcome of advanced non-small cell lung cancer. Med Oncol. 2011;28:1411–1417. doi: 10.1007/s12032-010-9553-9. [DOI] [PubMed] [Google Scholar]
- 10.Mok TS, Wu YL, Thongprasert S, et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med. 2009;361:947–957. doi: 10.1056/NEJMoa0810699. [DOI] [PubMed] [Google Scholar]
- 11.Olaussen KA, Dunant A, Fouret P, et al. DNA repair by ERCC1 in non-small-cell lung cancer and cisplatin-based adjuvant chemotherapy. N Engl J Med. 2006;355:983–991. doi: 10.1056/NEJMoa060570. [DOI] [PubMed] [Google Scholar]
- 12.Bepler G, Kusmartseva I, Sharma S, et al. RRM1 modulated in vitro and in vivo efficacy of gemcitabine and platinum in non-small-cell lung cancer. J Clin Oncol. 2006;24:4731–4737. doi: 10.1200/JCO.2006.06.1101. [DOI] [PubMed] [Google Scholar]
- 13.Lynch TJ, Bell DW, Sordella R, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 14.Paez JG, Jänne PA, Lee JC, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 15.Riely GJ, Pao W, Pham D, et al. Clinical course of patients with non-small cell lung cancer and epidermal growth factor receptor exon 19 and exon 21 mutations treated with gefitinib or erlotinib. Clin Cancer Res. 2006;12:839–844. doi: 10.1158/1078-0432.CCR-05-1846. [DOI] [PubMed] [Google Scholar]
- 16.Kobayashi S, Boggon TJ, Dayaram T, et al. EGFR mutation and resistance of non-small-cell lung cancer to gefitinib. N Engl J Med. 2005;352:786–792. doi: 10.1056/NEJMoa044238. [DOI] [PubMed] [Google Scholar]
- 17.Ceppi P, Volante M, Novello S, et al. ERCC1 and RRM1 gene expressions but not EGFR are predictive of shorter survival in advanced non-small-cell lung cancer treated with cisplatin and gemcitabine. Ann Oncol. 2006;17:1818–1825. doi: 10.1093/annonc/mdl300. [DOI] [PubMed] [Google Scholar]
- 18.Cobo M, Isla D, Massuti B, et al. Customizing cisplatin based on quantitative excision repair cross-complementing 1 mRNA expression: a phase III trial in non-small-cell lung cancer. J Clin Oncol. 2007;25:2747–2754. doi: 10.1200/JCO.2006.09.7915. [DOI] [PubMed] [Google Scholar]
- 19.Pao W, Miller V, Zakowski M, et al. EGF receptor gene mutations are common in lung cancers from ‘never smokers’ and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc Natl Acad Sci USA. 2004;101:13306–13311. doi: 10.1073/pnas.0405220101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Deng QH, Qiu Y, Mo MC, et al. EGFR gene copy number, ERCC1 and BRCA1 protein expression and their relationship in non-small cell lung cancer. Zhonghua Zhong Liu Za Zhi. 2011;33:508–512. (In Chinese) [PubMed] [Google Scholar]
- 21.Lin XY, Chen Y, Chen ZZ. Expression of ERCC1 and RRM1 in non-small cell lung cancer (NSCLC) and clinical prognosis. Fujian Yike Daxue Xuebao. 2011;45:10–14. (In Chinese) [Google Scholar]
- 22.Gandara DR, Grimminger P, Mack PC, et al. Association of epidermal growth factor receptor activating mutations with low ERCC1 gene expression in non-small cell lung cancer. J Thorac Oncol. 2010;5:1933–1938. doi: 10.1097/JTO.0b013e3181fd418d. [DOI] [PubMed] [Google Scholar]
- 23.Mitsudomi T, Morita S, Yatabe Y, et al. West Japan Oncology Group: Gefitinib versus cisplatin plus docetaxel in patients with non-small-cell lung cancer harbouring mutations of the epidermal growth factor receptor (WJTOG3405): an open label, randomised phase 3 trial. Lancet Oncol. 2010;11:121–128. doi: 10.1016/S1470-2045(09)70364-X. [DOI] [PubMed] [Google Scholar]
- 24.Reynolds C, Obasaju C, Schell MJ, et al. Randomized phase III trial of gemcitabine-based chemotherapy with in situ RRM1 and ERCC1 protein levels for response prediction in non-small-cell lung cancer. J Clin Oncol. 2009;27:5808–5815. doi: 10.1200/JCO.2009.21.9766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Simon GR, Schell MJ, Begum M, et al. Preliminary indication of survival benefit from ERCC1 and RRM1-tailored chemotherapy in patients with advanced nonsmall cell lung cancer: evidence from an individual patient analysis. Cancer. 2012;118:2525–2531. doi: 10.1002/cncr.26522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hu YY, Zhang DB, Dong YG, et al. The protein expression and the significance of RRM1 and ERCC1 in non-small cell lung cancer. Shandong Yi Yao. 2012;52:1–3. (In Chinese) [Google Scholar]