Background: Etiology of Huntington disease (HD) is related to the overproduction of mutant huntingtin (mHTT) protein.
Results: Amitriptyline improved motor symptoms via decreasing mHTT protein expression, improving neurotrophin signaling, and enhancing mitochondrial functions in HD mice.
Conclusion: Amitriptyline demonstrated beneficial effects in HD mice.
Significance: Amitriptyline has therapeutic potential in treating HD.
Keywords: Bioinformatics, Brain-derived Neurotrophic Factor (BDNF), Huntington Disease, Mitochondria, Neurodegenerative Disease, Amitriptyline, Motor Function, mHTT
Abstract
Huntington disease (HD) is a neurodegenerative disorder characterized by progressive motor impairment and cognitive alterations. Hereditary HD is primarily caused by the expansion of a CAG trinucleotide repeat in the huntingtin (Htt) gene, which results in the production of mutant huntingtin protein (mHTT) with an expanded amino-terminal polyglutamine (poly(Q)) stretch. Besides pathological mHTT aggregation, reduced brain-derived neurotrophic factor (BDNF) levels, impaired neurotrophin signaling, and compromised mitochondrial functions also contribute to the deleterious progressive etiology of HD. As a well tolerated Food and Drug Administration-approved antidepressant, amitriptyline (AMI) has shown efficacy in treating neurodegenerative murine models via potentiation of BDNF levels and amelioration of alterations in neurotrophin signaling pathways. In this study, we observed profound improvements in the motor coordination of AMI-treated N171-82Q HD model mice. The beneficial effects of AMI treatment were associated with its ability to reduce mHTT aggregation, potentiation of the BDNF-TrkB signaling system, and support of mitochondrial integrity and functionality. Our study not only provides preclinical evidence for the therapeutic potency of AMI in treating HD, but it also represents an important example of the usefulness of additional pharmacogenomic profiling of pre-existing drugs for novel therapeutic effects with often intractable pathological scenarios.
Introduction
Huntington disease (HD)4 is a devastating autosomal dominant neurodegenerative disease characterized by progressive motor dysfunction, emotional disturbances, dementia, and weight loss. Huntington disease occurs worldwide with a prevalence of 5–10 cases per 100,000 individuals (1). HD is primarily caused by the expansion of a CAG trinucleotide repeat in the Huntingtin (Htt) gene. The extended CAG repeat leads to the production of mutant huntingtin protein (mHTT) with an expanded polyglutamine (poly(Q)) stretch near the amino terminus of the protein. Multiple aspects of HD-related pathology are mediated by mHTT intracellular aggregation. mHTT protein aggregates are commonly present in both patients (2) and transgenic HD animal models (3). Mutant HTT protein exerts a toxic gain of function through aberrant protein folding and interaction, as well as a loss of normal HTT function (4). The diminution of mHTT aggregates in HD mouse models by selected therapies may be responsible for the significant improvement in the behavioral and neuropathological HD phenotype (3, 5). Soluble mHTT fragments may also mediate pathological activities in HD, and therefore reducing either mHTT aggregates and/or soluble fragments is vital for therapeutic remediation of HD (6, 7).
As a crucial pro-survival cellular factor, brain-derived neurotrophic factor (BDNF) protects striatal cells from excitotoxic insults in HD (8). Nonmutant endogenous HTT has been shown to induce BDNF gene expression in cortical neurons; although in contrast, mHTT can actively suppress BDNF expression (9). Potentially as a result of mHTT accumulation, reduced levels of BDNF have been found in both HD patient postmortem brains (10) and transgenic mouse models of HD (9); hence, it is likely that decreased expression of TrkB (the cognate receptor for BDNF) (11) and impaired BDNF-mediated neurotrophic signaling pathways may contribute to the pathogenesis of HD (12).
In addition to poor neurotrophic support, mitochondrial dysfunction is also regarded as a hallmark of HD pathophysiology (13). Studies of mitochondria isolated both from HD patients (14) and HD mouse models (15) suggest that mHTT itself may directly interact with mitochondria, leading to mitochondrial depolarization with decreased calcium ion levels (16). In addition, a growing body of evidence has demonstrated that the transcriptional repressive activity of mHTT upon one of the key regulators of mitochondrial biogenesis, peroxisome proliferator-activated receptor γ coactivator 1α (PGC-1α), leads to mitochondrial malfunction and advanced neurodegeneration in HD (17).
Currently, no effective treatment exists for HD, and there is a paucity of effective pharmacotherapeutic treatments for early and late term HD. Several rational strategies have been pursued in the development of HD disease-modifying drugs, including targeting mHTT aggregation inhibition, transcriptional regulation, and neuron preservation/neuroprotection and mitochondrial function modulation (18). Neuroprotective compounds that can slow down disease progression with few side effects have been studied for treating HD. For example, selective serotonin reuptake inhibitors (SSRIs), a class of drugs that is widely used for the treatment of patients with depression and severe anxiety disorders, have been shown to increase BDNF expression (19, 20) and stimulate neurogenesis responsive to BDNF. Duan et al. (21–23) have demonstrated that several SSRI compounds, including paroxetine and sertraline, elicited beneficial effects in HD mice. Grote et al. (24) also showed that fluoxetine, another SSRI drug, improved hippocampus-dependent cognitive and depressive-like behavioral symptoms that occur in HD mice by rescuing deficits of neurogenesis and volume loss in the dentate gyrus. Wang et al. (25) characterized the neuroprotective effects of nortriptyline, a tricyclic antidepressant, in mouse models of chronic neurodegeneration (amyotrophic lateral sclerosis and HD). They found that nortriptyline significantly delayed disease onset and extended the life span of HD mice by extending the presymptomatic portion of the disease without affecting mortality. In an established cellular model of HD, they also reported that nortriptyline inhibited mitochondrion-mediated cell death and decreased loss of mitochondrial membrane potential (25).
In contrast to the expensive, risk-overt, and time-consuming nature of de novo drug development, perhaps a more effective approach may be the “resourcing” of previously approved, and well tolerated, therapeutics in new pharmacogenomic settings. Seeking effective treatments in Food and Drug Administration-approved drugs has become a promising drug discovery route for HD, as well. Masuda et al. (26) showed Food and Drug Administration-approved drug tiagabine significantly extended survival, improved motor performance, and attenuated brain atrophy and neurodegeneration in N171-82Q HD mice. Based on above HD therapeutic research experience, in this study, we focused on the Food and Drug Administration-approved drug amitriptyline (Elavil).
Amitriptyline (AMI) is a member of the family of tricyclic antidepressants. In addition to its anti-depressant actions, AMI is also currently prescribed for the treatment of neuropathic pain, suggesting dose-dependent pluripotent actions of this drug (27). Interestingly, several studies have indicated that AMI elicits strong neurotrophic activity via a productive interaction with the BDNF neurotrophin tyrosine kinase receptor B (TrkB) system (28, 29). We have also demonstrated previously that AMI enhances cognitive function in aged Alzheimer disease mice, in a strongly BDNF/TrkB-dependent manner. This therapeutic activity was also closely associated with the activation of adult neurogenetic pathways (27). Based on the pathological features of HD, the close association between BDNF and HD, and the promising neuroprotective potency of AMI, we aimed to investigate the therapeutic potential of AMI in HD.
EXPERIMENTAL PROCEDURES
Animals, Drug Administration, and Tissue Collection
All animal procedures were approved by the Animal Care and Use Committee (ACUC) of the NIA. Male B6C3-Tg(HD82Gln)81Dbo/J (N171-82Q) mice and male wild-type (WT) littermates, 2 months of age, were maintained on a 12-h light/dark cycle in pathogen-free conditions. All animals received food and water ad libitum. The treatment group received 16 μg/g body weight (per day) of amitriptyline hydrochloride (AMI) per os in their drinking water (Sigma) for 6 weeks (n = 12/strain, AMI or WT-AMI), whereas the vehicle group (control) received plain animal facility drinking water (n = 12/strain, control and WT). Treatment was started when HD animals were pre-symptomatic (2 months of age) and continued for 6 weeks. Age-matched female B6C3-Tg(HD82Gln)81Dbo/J (N171-82Q) mice were also used for the body weight evaluations (n = 8/treatment). Body weight was measured weekly. At the end of the study, mice were euthanized via isoflurane inhalation and decapitation (Butler Animal Health Supply, Dublin, OH). Nine brains from each group were carefully dissected on ice, snap-frozen, and stored at −80 for further analysis. The remaining three brains from each group were fixed with 4% paraformaldehyde after perfusion, which is described in detail below. Blood samples for hormone measurements were collected as described previously (30). In brief, trunk blood was collected in an EDTA-coated plasma collection tube (VWR, Radnor, PA) and stored on wet ice for 1 h before being spun down at 3000 rpm for 30 min at 4 °C. The supernatant was removed immediately and stored at −80 °C until later analysis.
Motor Performance Assessments
Motor coordination was assessed using both an accelerating Rotarod and mouse beam walking assay as described previously (31). A Rotarod apparatus was used to test motor function on a weekly basis (Med Associates Inc., St. Albans, VT). Briefly, mice were trained to remain on the spinning Rotarod apparatus during a 2-min habituation trial (four revolutions per min (rpm)) on the day prior to the testing day. On test days, the mice were placed on the Rotarod, which gradually accelerated from 4 to 40 rpm over a 5-min time interval. The test was performed twice per day, and the latency to fall was measured and averaged. Rotarod performance was assessed weekly. The mouse beam walking assay is regarded as a more sensitive but less stressful motor coordination task. In this study, all animals were also evaluated with this test as described previously (31). Briefly, mice were placed on the beam at one end and allowed to walk to the goal box. Mice that fell were returned to the position from which they fell, with a maximum time of 60 s allowed on the beam. The measurements taken were time on beam and the number of foot slips (one or both hind limbs slipped from the beam). Prior to all experimental procedures, animals were habituated to the testing room environment for at least 30 min.
Metabolic Hormone Measurements
Plasma glucose levels were measured using the EasyGluco blood glucose system (US Diagnostics, Inc., New York). Plasma metabolic hormones, including insulin, leptin, peptide YY (PYY), gastric inhibitory peptide (GIP), and pancreatic peptide (PP), were measured using a rodent multiplex assay kit (Millipore, Billerica, MA) as described previously (32). Each sample was assayed in duplicate on a 96-well plate. Plasma lipid levels, including triglycerides and total ketone bodies, were assayed using enzymatic methods with commercial kits (Wako Chemicals USA, Inc., Richmond, VA) according to the manufacturer's instructions.
General Metabolic Status Evaluations
A comprehensive animal metabolic monitoring system (CLAMS; Columbus Instruments, Columbus, OH) was used to evaluate total energy expenditure and activity. On the test day, all animals were allowed to habituate to the testing environment for 30 min in their home cages. Additionally, once placed in the metabolic chambers, animals were given 2 h prior to formal data collection to acclimatize to the metabolic chamber environment. The respiratory exchange ratio (RER) is the ratio of VCO2 to VO2. VO2 and VCO2 represent the volume of O2 consumption and the volume of CO2 production, respectively (33). Activity was measured on the x and z axes by using infrared beams to count the beam breaks at consecutive intervals during a specified measurement period (24 h).
Western Blot Analysis in Mouse Striatum and Cortex
Striatum, cortex, and hippocampus tissues were processed using the QproteomeTM cell compartment kit according to the manufacturer's instructions (Qiagen, Valencia, CA). All protein extracts were quantified using BCA reagent (ThermoScientific, Rockford, IL) before resolution with SDS-PAGE and electrotransference to PVDF membranes (PerkinElmer Life Sciences). Membranes were blocked for Western blots as described previously (34), and primary antibody immune-reactive complexes were identified using alkaline phosphatase-conjugated secondary antisera (Sigma) with enzyme-linked chemifluorescence (GE Healthcare) and visualized with a Typhoon 9410 phosphorimager (GE Healthcare). Blots were probed with antibodies to mutant HTT (S830, gift from Dr. Gillian Bates), wild-type HTT (anti-mouse huntingtin protein antibody MAB2166, Millipore, Billerica, MA), DARPP32 (anti-DARPP32, Millipore), phospho-Thr34-DARPP32, phospho-Ser9-GSK3β, GSK3β, phospho-Ser473-AKT, AKT, phosphor-Ser133-CREB, CREB, VDAC, complex IV, UCP2, PGC1α, HSP70, HSP90, pro-BDNF, BDNF, phosphor-Tyr490-TrkB, TrkB, phospho-ERK1/2, ERK1/2, synaptophysin, and PSD95 (Cell Signaling Technology, Beverly, MA). β-Actin (A5316) antibody was purchased from Sigma. All reported values were normalized to actin expression.
Immunochemistry Evaluations of mHTT in Mouse Brain
Mice were deeply anesthetized with carbon dioxide and transcardially perfused with 4% paraformaldehyde in 0.1 m phosphate buffer (pH 7.4). Brains (n = 5/group) were postfixed, cryoprotected in 20% sucrose/phosphate buffer, and sectioned (40 μm, coronal) using a freezing microtome. The sections were processed for immunohistochemical localization of mutant huntingtin protein (1:200; mutant huntingtin protein antibody MAB5374, Millipore), using Vectastain Elite ABC kit (Vector Laboratories, Burlingame, CA) and developed using a Dako liquid diaminobenzidine substrate chromogen system (Dako Cytomation) as described previously (35, 36). A biotinylated secondary goat anti-mouse antibody was used at 1:500 (Vector Laboratories). To assess total huntingtin staining (diffuse and aggregate), a sample area of 0.85 × 0.70 mm2 was photographed using 10× objective with a video capture system (B/W CCD camera coupled to an Olympus IX51 upright microscope, Olympus, Center Valley, PA). For both diffuse and aggregate measures, with ImageJ (Version 1.61, National Institutes of Health), the digital grayscale images were converted into binary positive/negative data using constant threshold limit as described previously (27). The cell number of mHTT-positive cells (i.e. immunoreactive cells) was quantified for each image and averaged across images for each brain region in each animal to generate immunoreactive cell density (cell/mm2). Quantified fields were selected systematically using a predetermined pattern to maximize analysis of immunoreactivity in each brain region. The same image exposure time was applied to sections from all treatment groups within a given comparison. Following antigen retrieval with a 1× citrate buffer (Biogenex, San Ramon, CA) at 98 °C for 20 min, immunofluorescence analyses were performed as described previously (37). Brain sections were blocked in 5% bovine serum albumin (BSA; Sigma) and 0.1% Tween 20 in 1× Tris-buffered saline (TBS) (pH 7.4) for 1 h at room temperature, followed by incubation in a primary antibody (mHTT clone MAB5374, Millipore 1:200; NeuN MAB377, Millipore 1:200; MAP2, Abcam (Cambridge, MA), 1:200) in 1% BSA and 0.1% Tween 20 in TBS (pH 7.4) overnight at 4 °C. After washing, sections were incubated for 1 h in fluorescent secondary antibodies (1:1000 dilution; Invitrogen) along with DAPI (1:5000 dilution; Invitrogen) for nuclear staining. No fluorescent staining was observed in any sections when the primary antibodies were omitted.
Stereological Analysis
The optical fractionator probe of Stereoinvestigator software (MicroBrightField, Inc., Williston, VT) was used to obtain an unbiased estimate of NeuN-positive neurons in the striatum and associated cortex (located in the same section with striatum) as per the atlas of the mouse brain by Franklin and Paxinos (38). Stereological parameters were as follows: counting frame, 50 × 50 μm; optical dissector, 40 μm; grid size, 900 × 900 μm. For the population size estimate (number of sections per animal), a target coefficient of error (Gundersen's m = 1) of less than 0.10 was considered acceptable. Neuron counting was performed by the investigator blinded to treatment history.
Microarray and Bioinformatics Analyses
Microarrays were scanned using an Illumina BeadStation Genetic Analysis Systems scanner, and the image data were extracted using the Illumina BeadStudio Version 3.0. Quantitative Venn diagram distribution of significantly regulated transcripts was performed with VennPlex (39). Pathway annotation of these significantly regulated transcripts was then performed using specific parametric gene set enrichment analysis as described previously (40). In addition, we applied the use of a novel latent semantic analytical platform, Textrous! (41), to extract textual functional insights into the therapeutic activity of amitriptyline.
RNA Extraction and Real Time PCR Analysis
A two-step real time reverse transcription (RT) was performed to reverse transcribe total RNA into cDNA. Next, PCR was carried out using gene-specific primer pairs and SYBR Green PCR master mix (Applied Biosystems, Foster City, CA) in an ABI Prism 7000 sequence detection system (Applied Biosystems). The amplification conditions were 50 °C (2 min), 95 °C (10 min), and then 40 cycles at 95 °C (15 s) and 60 °C (1 min). The data were normalized to glyceraldehyde-3-phosphate dehydrogenase (Gapdh) mRNA. All real time PCR analyses are represented as the mean ± S.E. from at least three independent experiments, each performed in triplicate.
Statistical Analyses
Student's t tests were used for comparison between AMI-treated HD mice and control HD mice in all experiments. p < 0.05 was considered statistically significant throughout the study. Error bars on graphs represent the ±95% confidence interval. All data represent means ± S.E.
RESULTS
General Somatic Effects of Amitriptyline in N171-82Q Mice and WT Mice
Recent findings have elucidated the important role of whole-body metabolic disturbance in the pathogenesis of HD. To comprehensively capture the potential effects of AMI treatment in the physiological context of an HD condition, we evaluated the gross metabolic response of N171-82Q mice and their WT littermates to this pro-neurotrophic agent (27). We first analyzed the effects of AMI on body weight changes. Because the body weights of male HD mice and female HD mice are different and may respond differently to drug treatment, we evaluated the body weight changes for both genders. We found that a 6-week AMI treatment did not significantly alter body weight in both male and female HD mice, compared with their control HD mice (Fig. 1, A and B). However, in WT mice, AMI treatment slightly slowed the body weight gain (Fig. 1C); especially at week 6 (p < 0.05, WT versus WT-AMI). AMI treatment did not alter or extend the life span in HD mice (data not shown).
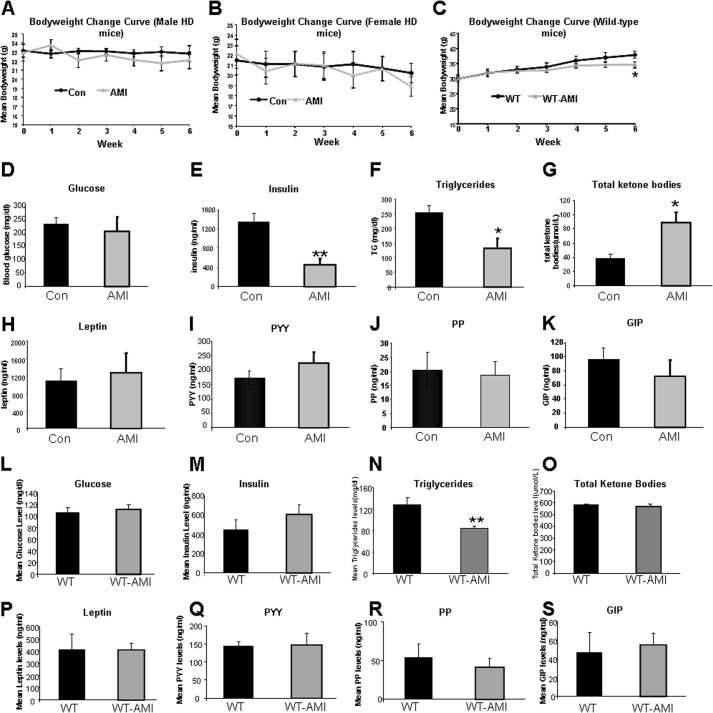
FIGURE 1.
AMI treatment alters circulating murine metabolic factors and body weight. A, alterations in body weight between AMI-treated and vehicle-treated male HD mice (vehicle-treated, black line, AMI-treated, gray line). B, alterations in body weight between AMI-treated and vehicle-treated female HD mice (vehicle-treated, black line; AMI-treated, gray line). C, alterations in body weight between AMI-treated and vehicle-treated male WT mice (vehicle-treated, black line; AMI-treated, gray line). D, AMI-mediated effects on fasting glucose in HD mice. E, AMI-mediated effects on fasting insulin in HD mice. F, AMI-mediated effects on the plasma triglyceride levels in HD mice. G, AMI-mediated effects on the plasma total ketone bodies levels in HD mice. Effects of AMI treatment on levels of leptin (H), peptide YY (PYY, I), pancreatic polypeptide (PP, J), and gastrointestinal inhibitory peptide (GIP, K) in HD mice. L, AMI-mediated effects on fasting glucose in WT mice. M, AMI-mediated effects on fasting insulin in WT mice. N, AMI-mediated effects on the plasma triglyceride levels in WT mice. O, AMI-mediated effects on the plasma total ketone bodies levels in WT mice. Effects of AMI treatment on levels of leptin (P), peptide YY (PYY, Q), pancreatic polypeptide (PP, R), and gastrointestinal inhibitory peptide (GIP, S) in WT mice. Data are means ± S.E. *, p ≤ 0.05, n = 12/group.
We further evaluated the effects of AMI on metabolic hormones in male HD and male WT mice. In HD mice, although AMI did not affect fasting glucose levels (Fig. 1D), circulating insulin levels were significantly reduced (Fig. 1E). AMI treatment also significantly attenuated circulating triglyceride levels (Fig. 1F), with a simultaneous potentiation of circulating ketone levels (Fig. 1G). No additional changes in other metabolic hormones (leptin, PYY, PP, and GIP) were found with AMI treatment (Fig. 1, H–K). In WT mice, expect a significant decrease of triglyceride levels (Fig. 1N), but all other above-mentioned metabolic parameters are comparable between AMI-treated and vehicle-treated animals (Fig. 1, L–S).
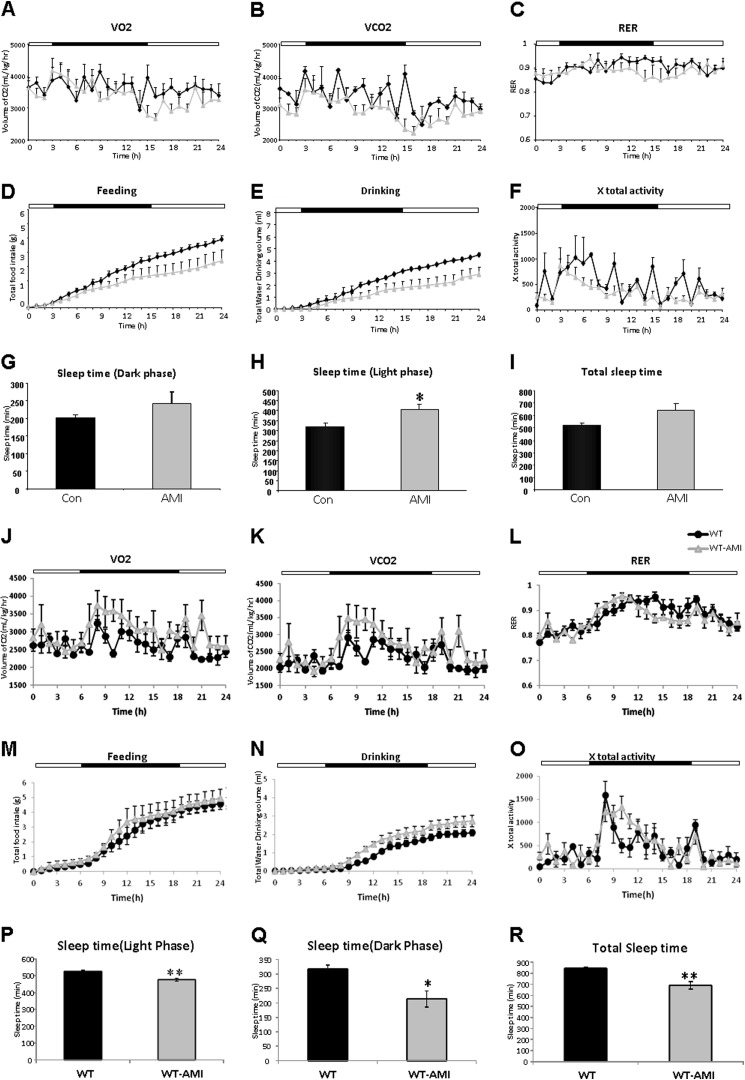
Complementing our analysis of circulating metabolic hormones, we also assessed the effects of AMI treatment upon whole-body metabolism (Fig. 2). No significant effects of AMI upon VO2, VCO2, or RER were found in the HD mice (Fig. 2, A–C). AMI did, however, cause nonsignificant reductions of food (Fig, 2D) and water (Fig. 2E) intake. There was also a trend for reduced ambulatory activity in the AMI-treated mice; however, this was not significant (Fig. 2F). Commensurate with this last finding, we found that there was a nonsignificant increase in dark-phase sleep time (Fig. 2G) and a significant increases in light-phase sleep time (Fig. 2H), resulting in an overall small nonsignificant increase in total sleep (Fig. 2I). In WT mice, AMI also did not affect VO2, VCO2, or RER (Fig. 2, J–L). The food (Fig. 2M) and water (Fig. 2N) intake of WT-AMI mice were also comparable with water-treated WT mice. However, there was a trend for increased ambulatory activity in the AMI-treated WT mice (Fig. 2O), which led to a significant decrease in light-phase sleep time (Fig. 2P), dark-phase sleep time (Fig. 2Q), and total sleep time (Fig. 2R).
FIGURE 2.
Comprehensive metabolic evaluations of AMI treatment. Whole-body metabolic status was assessed in AMI-treated HD mice evaluated using a CLAMS system across a variety of outputs (vehicle-treated, black line; AMI-treated, gray line), including oxygen consumption (VO2) (A), carbon dioxide production (VCO2) (B), RER (C), accumulated food intake (D), accumulated water intake (E), ambulatory total x axis activity (F), sleep time during the dark cycle (G), sleep time during the light cycles (H), and total sleep time during both light and dark cycles (I). Black line, vehicle-treated HD mice; gray line, AMI-treated HD mice. The same CLAMS system parameter assessment panel (J–R) was also performed in WT mice, both vehicle-treated (black line) and AMI-treated (gray line). Parameters in all panels were measured over a 24-h period. The dark cycle time is indicated by the black section of the pictogram bar in each CLAMS activity, A–F and J–O. Data are means ± S.E. *, p ≤ 0.05, n = 4/group.
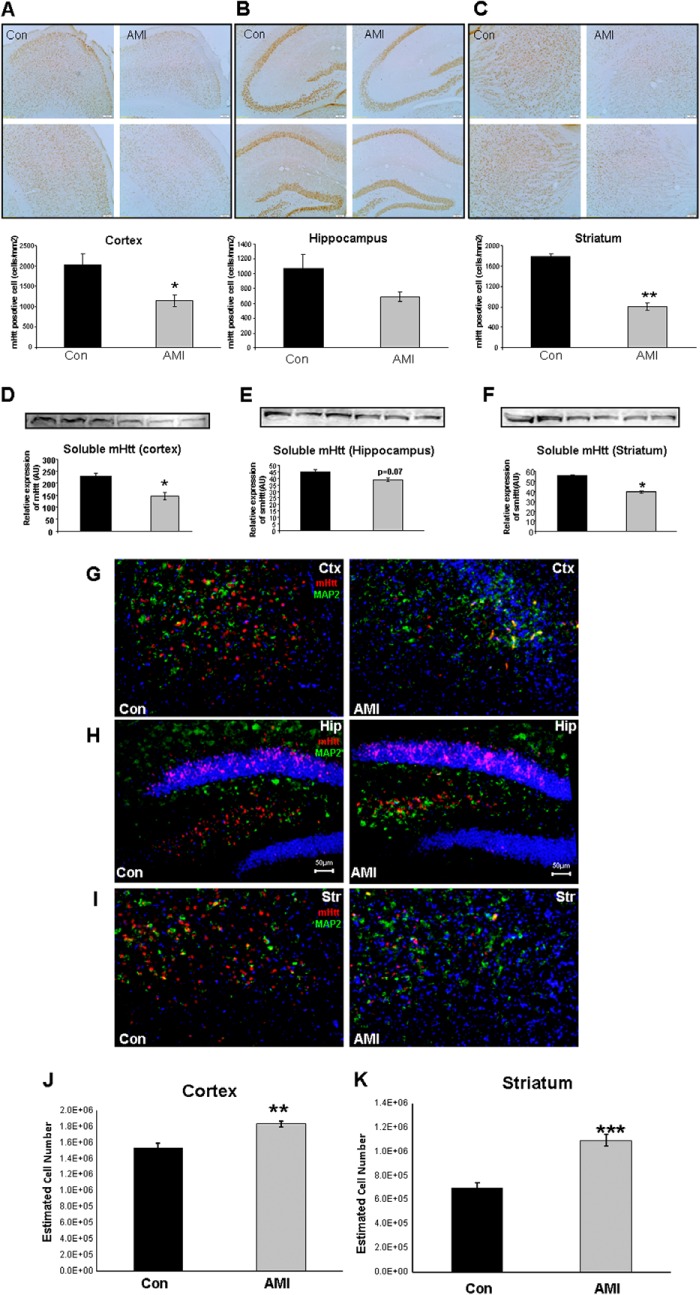
Amitriptyline Reduces Central Nervous Tissue Immunoreactive mHTT Levels and Increases NeuN-positive Cell Numbers in N171-82Q Mice
Reduction of mHTT protein expression in the brain is regarded as a potential therapeutic strategy in HD. We evaluated the effects of AMI treatment upon mHTT protein expression in tissue sections or tissues from multiple brain regions using diaminobenzidine staining, Western blot, and fluorescent immunostaining (Fig. 3). With diaminobenzidine staining, we found AMI treatment induced significant reductions of mHTT-positive cell density in both cortex (Fig. 3A) and striatum (Fig. 3C). A nonsignificant decrease of mHTT-positive cell density was also observed in the hippocampus (Fig. 3B). Using Western blot analysis, we verified changes of mHTT protein in three brain regions (Fig. 3, D–F). In addition, we also applied fluorescent immunostaining to evaluate this AMI-induced mHTT clearance. Once again, we found that AMI treatment effected a profound reduction in the number of mHTT-reactive cells in the cortex (Fig. 3G), hippocampus (Fig. 3H), and striatum (Fig. 3I) of the HD mice. To assess the potential neuroprotective effects of AMI, we also performed stereological analysis on striatal and cortical NeuN-positive cells. AMI-treated HD mice possessed significantly higher estimated NeuN-positive cell numbers than control HD mice, in both cortex (Fig. 3J) and striatum (Fig. 3K).
FIGURE 3.
AMI-induced alterations of mHTT protein expression, neuronal marker-MAP2, and NeuN-positive cell numbers in mouse brain. A–C, immunostaining (×10×) of mutant Huntingtin protein in both vehicle-treated (Control, Con) and AMI-treated HD mice (A, cortex; B, hippocampus; C, striatum). The associated histograms for A–C indicate the reductions in mHTT immunopositive areas mediated by AMI. Data from three animals of each group are expressed as means ± S.E. *, p ≤ 0.05; **, p ≤ 0.01. D–F, Western blot verifications of mHTT changes in cortex, hippocampus, and striatum. G–I, fluorescence immunostaining of mHTT (red), MAP2 (green), and nuclei (DAPI) in the cortex (Ctx, G), hippocampus (Hip, H), and striatum (Str, I) from control (Con, vehicle-treated) or AMI-treated mice. G–I are at ×10 magnification: scale bar, 50 μm. J and K, stereological analysis of cortical and striatal NeuN-positive cells. Data from five animals of each group are expressed as means ± S.E. *, p ≤ 0.05; **, p ≤ 0.01; ***, p ≤ 0.01.
Functional Bioinformatics Signatures of AMI Activity across Multiple Central Nervous Tissues
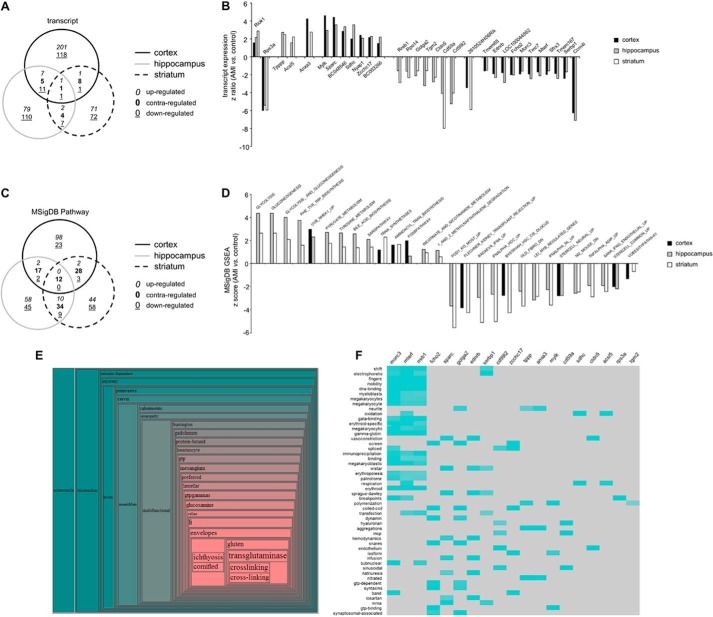
We next investigated the ability of AMI to effect potential therapeutic effects, i.e. mHTT clearance across the central nervous system (CNS) using an unbiased transcriptomic approach (42). Hence, we performed a quantitative assessment of the multidimensional transcriptomic effects of AMI in cortical (supplemental Table S1), hippocampal (supplemental Table S2), and striatal (supplemental Table S3) tissues of HD mice. To validate our transcriptomic data, we chose three random transcripts (Slc13A4, Slc6A13, and Cd59a) on which we performed real time PCR with gene-specific primers. All PCR results reliably recapitulated our transcriptomic data (data not shown). Using VennPlex analysis of the multitissue effects of AMI, we found that of the total of 752 significantly AMI-regulated transcripts in all three tissues (cortex, 355; hippocampus, 228; striatum, 169), 49 of these transcripts were common to at least two out of three tissues (Fig. 4A). Of these 49, we found that AMI coherently controlled the polarity of expression regulation of 31 of these transcripts in at least two tissues independently (Fig. 4B). Two of these transcripts, Riok1 and Rps3a, were significantly and coherently regulated in all three tissues involved. Using all the significantly regulated transcripts from each tissue, we performed parametric gene set enrichment analysis (PAGE: Broad Institute Molecular Signatures Database (MSigDB)) to identify potential complex signaling activities entrained by these transcriptomic effects in the three tissues (cortex, supplemental Table S4; hippocampus, supplemental Table S5; and striatum, supplemental Table S6). Using VennPlex, we were able to identify the presence of multiple signaling pathway collections that were coherently regulated by AMI across at least two different tissues. Coherent regulation polarity was only observed for signaling collections across two out of three tissues (Fig. 4C); however, consistent effects of AMI were seen for 28 PAGE MSigDB collections in at least two tissues (Fig. 4D). Considering the importance of metabolic regulation in HD, it was interesting to note that pathways influencing energy management were prominently featured and up-regulated by AMI, e.g. GLYCOLYSIS, PYRUVATE_METABOLISM and NICOTINATE_AND_NICOTINAMIDE_METABOLISM (Fig. 4D). In addition to PAGE MSigDB analysis, we applied our novel bioinformatics platform, Textrous!, to the set of transcripts reliably controlled by AMI across multiple brain regions (Fig. 4B). Using the collective processing function in Textrous! (creating a hierarchical word cloud), we found that this transcript subset was strongly associated with transglutaminase activity (Fig. 4E), a function that is currently appreciated as one of the most encouraging new avenues for neurodegenerative therapeutic research (43). It is interesting to note that we found that AMI significantly attenuated the expression of transglutaminase 2 in the hippocampus and the striatum. Using the Textrous! individual processing mode (Fig. 4F), we again found that multiple transcripts regulated by AMI were individually associated with functions related to stress resistance, energy balance, and neurodegeneration, e.g. DNA binding, oxidation, respiration, aggregations, and breakpoints.
FIGURE 4.
Multitissue transcriptomic analysis of the AMI-induced functional signature. A, VennPlex analysis of AMI-regulated transcripts across the cortex, hippocampus, and striatum. B, core transcripts coherently regulated by AMI in at least two out of three experimental tissues. C, VennPlex analysis of AMI-regulated MSigDB signaling pathways across the cortex, hippocampus, and striatum. D, core signaling pathways coherently regulated by AMI in at least two out of three experimental tissues. E, collective Textrous! hierarchical word cloud created from the AMI signature transcript dataset (from B). F, individual Textrous! heatmap processing generated with the AMI signature transcript dataset (from B).
Although AMI appears to facilitate a therapeutic action, with respect to clearance of HTT immunoreactivity in multiple brain regions, especially in striatum and cortex, it appeared not to significantly affect the resting blood glucose in these mice, which is at a highly diabetic level. Therefore, we next investigated whether a phenotypic expression of a therapeutic action could be demonstrated in one of the most classical facets of HD, i.e. the disruption of motor coordination.
AMI-mediated Alteration of Striatal Function and Activity
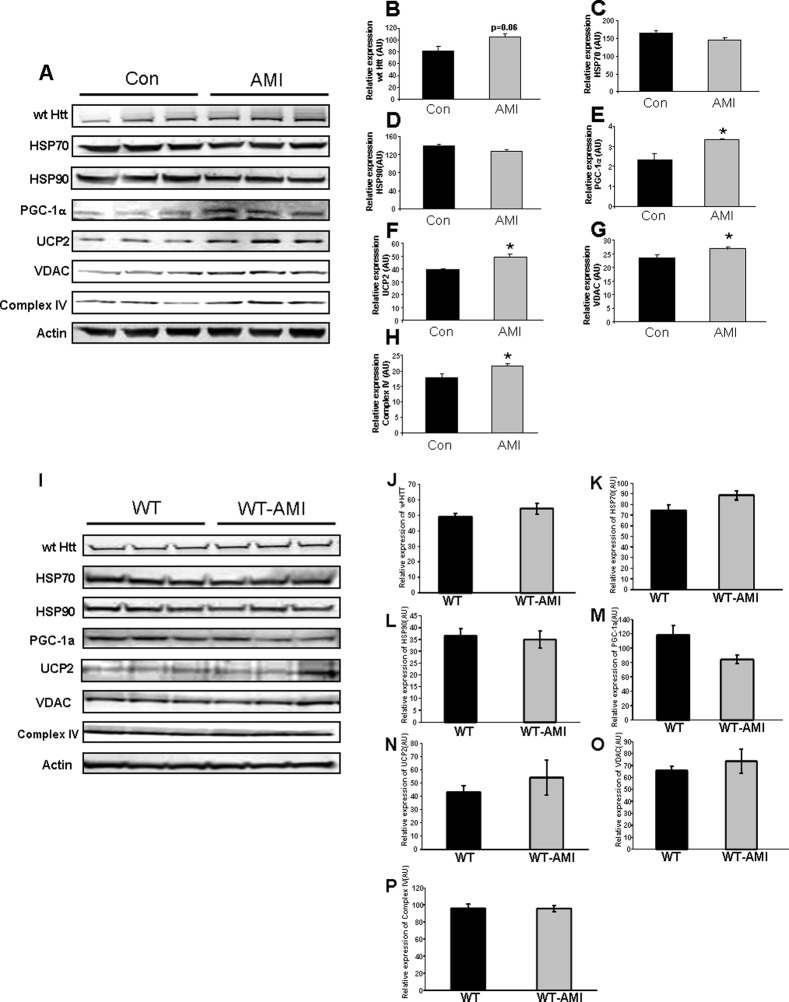
Fine motor coordination is mainly controlled through the striatum. In addition, it is thought that the striatum is also one of the centers of major metabolic disruption in HD (44). We therefore assessed, at the protein level, the effects of AMI treatment on this pivotal brain region. In HD mice, there was an AMI-induced nonsignificant increase in the levels of wild-type HTT (Fig. 5, A and B). AMI did not appear to exert any strong effect on HSP70/90 expression (Fig. 5, A, C, and D) but did significantly elevate striatal proteins linked to mitochondrial metabolic regulation, i.e. PGC-1α (Fig. 5, A and E), UCP2 (Fig. 5, A and F), VDAC (Fig. 5, A and G), and complex IV (Fig. 5, A and H). In WT mice, AMI treatment did not significantly alter the striatal expression levels of the above-mentioned proteins (Fig. 5, I–P).
FIGURE 5.
AMI-mediated alterations in striatal proteins involved in stress response and energy management. A, AMI-mediated alteration in the expression levels of striatal proteins associated with mutant huntingtin, mitochondrial functions, and stress responses measured using Western blot in HD mice as follows: B, wild-type HTT (wt HTT), C, HSP70; D, HSP90; E, PGC-1α; F, UCP2; G, VDAC; H, complex IV. I, AMI-mediated alteration in expression levels of striatal proteins associated with mutant huntingtin, mitochondrial functions, and stress responses measured using Western blot in WT mice as follows: J, wild-type HTT (wt HTT); K, HSP70; L, HSP90; M, PGC-1α; N, UCP2; O, VDAC; P, complex IV. Data from three animals of each group are expressed as means ± S.E. *, p ≤ 0.05; **, p ≤ 0.01; ***, p ≤ 0.001. (Control treatment, Con or WT; AMI treatment, AMI or WT-AMI.)
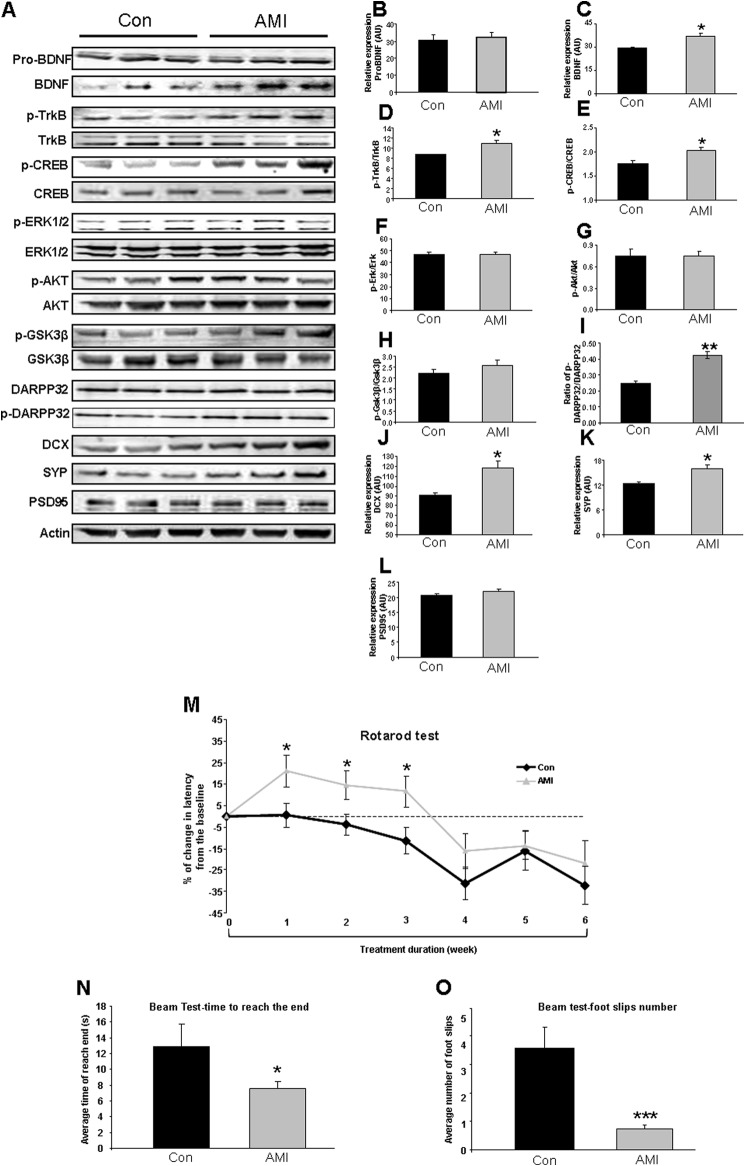
We also assessed the effect of AMI treatment on the neurosynaptic functionality of the striatum in both HD and WT mice (Figs. 6 and 7). In HD mice, consistent with our previous demonstration of the potent neurotrophic activity of AMI, we found that AMI treatment significantly increased striatal mature BDNF levels (Fig. 6, A and C), TrkB activity (Fig. 6, A and D), and phospho-CREB levels (Fig. 6, A and E). However, the pro-form of BDNF (pro-BDNF: Fig. 6, A and B) was not altered by AMI treatment. Striatal levels of phosphorylated ERK1/2 (Fig. 6, A and F), AKT (Fig. 6, A and G), and GSK3β (Fig. 6, A and H) were unaffected by AMI treatment. Although the levels of DARPP32 were unaffected by AMI treatment, we found AMI induced a significant increase in the levels of phosphorylated DARPP32, which resulted in a profound increase in the ratio of phospho-DARPP32/DARPP32 (Fig. 6, A and I). It suggests that without affecting the expression of DARPP32 (Fig. 6), AMI potentiated the dopamine signaling pathway by increasing the activation of DARPP32 (45, 46). We also found that the marker of de novo neuron synthesis, doublecortin (DCX), was significantly elevated by AMI (Fig. 6, A and J). It has recently been demonstrated that the striatum is an important region of potential neurogenesis (47). AMI therefore may facilitate the generation of new neurons to maintain striatal network connectivity. AMI was also found to selectively potentiate the expression of presynaptic and synaptophysin (Fig. 6, A and K) but not postsynaptic markers (PSD95, Fig. 6, A and L). Therefore, AMI appeared to generate a eumetabolic, neuroprotective, and pro-neurotrophic spectrum of actions in the murine HD striatum. To assess how these molecular actions converged on the physiological and behavioral activity of the striatum, we investigated the motor functions of the AMI-treated HD mice. Using the Rotarod performance test, we found that AMI treatment significantly improved the motor coordination of the mice over the course of the study (Fig. 6M). In addition to improved Rotarod performance, the AMI-treated mice demonstrated an improved completion time in a linear beam test (Fig. 6N) with significantly fewer foot slip events (Fig. 6O). Therefore, in two separate coordination tests, AMI significantly improved the fine motor control of the HD mice.
FIGURE 6.
AMI regulates expression of neurotrophic proteins and improves motor function of HD mice. A, AMI-mediated alteration in expression levels of striatal proteins associated with BDNF-induced neurotrophic activity and synaptic structure measured using Western blot. B, pro-BDNF. C, mature BDNF. D, TrkB receptor activity, i.e. ratio of phosphorylated (p)-TrkB/TrkB; E, CREB activity, i.e. ratio of p-CREB/CREB in striatum; F, ERK activity, i.e. ratio of p-ERK/ERK; G, AKT activity, i.e. ratio of p-AKT/AKT; H, GSK3β activity, i.e. ratio of p-GSK3β/GSK3β; I, ratio of p-DARPP32/DARPP32; J, DCX; K, synaptophysin (SYP); L, PSD95. Data from three animals of each group are expressed as means ± S.E. *, p ≤ 0.05; **, p ≤ 0.01; ***, p ≤ 0.001. (Control treatment, Con; AMI treatment, AMI). M, rotarod performance change curves throughout 6-week AMI treatment (vehicle-treated, black line; AMI-treated, gray line). N, mean latency time to reach the goal box in the walking beam task. O, mean number of foot slips of HD mice (AMI-treated and vehicle-treated) in the walking beam task. Data from 12 animals of each group are expressed as means ± S.E. *, p ≤ 0.05; **, p ≤ 0.01; ***, p ≤ 0.001. (Control treatment, Con; AMI treatment, AMI).
FIGURE 7.
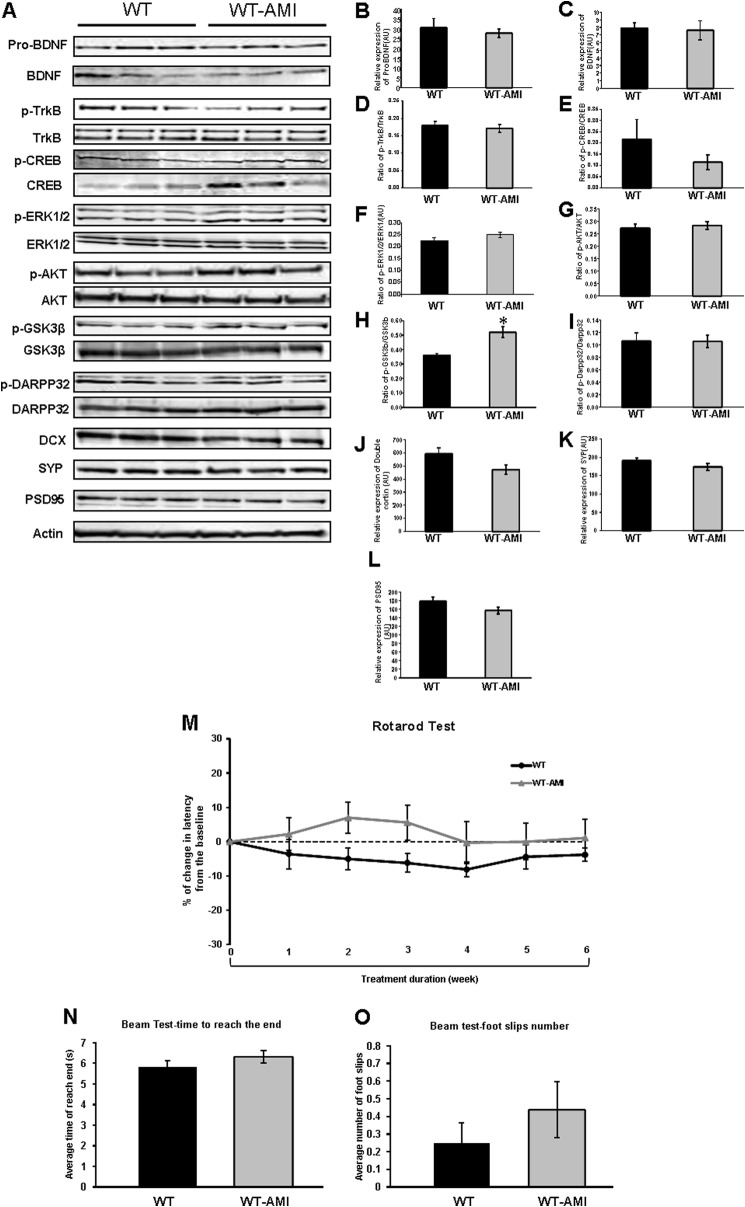
AMI regulates expression of neurotrophic proteins and improves motor function of WT mice. A, AMI-mediated alteration in expression levels of striatal proteins associated with BDNF-induced neurotrophic activity and synaptic structure measured using Western blot. B, ProBDNF. C, mature BDNF. D, TrkB receptor activity, i.e. ratio of phosphorylated (p)-TrkB/TrkB; E, CREB activity, i.e. ratio of p-CREB/CREB in striatum; F, ERK activity, i.e. ratio of p-ERK/ERK; G, AKT activity, i.e. ratio of p-AKT/AKT; H, GSK3β activity, i.e. ratio of p-GSK3β/GSK3β; I, ratio of p-DARPP32/DARPP32; J, DCX; K, synaptophysin (SYP); L, PSD95. Data from three animals of each group are expressed as means ± S.E. *, p ≤ 0.05; **, p ≤ 0.01; ***, p ≤ 0.001. (Control treatment, Con; AMI treatment, AMI). M, rotarod performance change curves throughout 6-week AMI treatment (vehicle-treated, black line; AMI-treated, gray line). N, mean latency time to reach the goal box in the walking beam task. O, mean number of foot slips of WT mice (AMI-treated and vehicle-treated) in the walking beam task. Data from 12 animals of each group are expressed as means ± S.E. *, p ≤ 0.05. (Control treatment, Con; AMI treatment, AMI).
To determine whether the AMI-induced changes in HD mice are a “true” rescue or a generalized effect, we performed similar analysis of striatal proteins and motor activity in WT animals (Fig. 7). Compared with vehicle-treated WT mice, WT-AMI mice displayed a significant induction only in the ratio of phospho-GSK3β/GSK3β (Fig. 7, A and H). With respect to other above-mentioned striatal proteins, AMI-treated WT mice shared similar expression levels with WT controls (Fig. 7, A–L). In the Rotarod behavioral test, AMI-treated WT mice showed comparable motor function with WT mice, except for a temporary nonsignificant trend of increase in latency at week 2 (Fig. 7M). However, in the mouse beam test, no significant difference was observed between two groups (Fig. 7, N and O).
DISCUSSION
Huntington disease is a progressive and ultimately fatal neurodegenerative disorder, which currently has no clinically proven therapy to either delay its onset or slow its progression (18). In this study, we evaluated the actions of AMI, a classic tricyclic antidepressant compound currently Food and Drug Administration-approved, on HD-related symptoms and pathologies in the N171-82Q mouse, a well characterized mouse model of HD.
In previous studies using a mouse model of Alzheimer disease, AMI has been shown to elevate BDNF levels (48, 49) and activate the TrkB-mediated neurotrophic signaling pathway (27), resulting in substantial improvements in neurological functions and behavioral performance. Our preliminary in vitro results also demonstrated that in SH-SY5Y cell model, the short term (within 3 h) AMI (500 nm) treatment could increase activations of TrkB and GSK3β in a time-dependent manner, which was reflected by the ratios of phospho-TrkB/TrkB and phospho-GSK3β/GSK3β (data not shown). In accordance with our results, similar small molecule pro-neurotrophics are also being developed for HD. For example, a recent study has reported that small molecule TrkB agonist 7,8-dihydroxyflavone and its synthetic derivative protect neurons, improve motor functions, and extend survival in the N171-82Q HD mouse model (50).
It is plausible that AMI may demonstrate similar beneficial effects upon other neurodegenerative disorders, as well as HD, given that most neurodegenerative disorders share similar common mechanisms such as impairments in the BDNF-TrkB signaling pathway. Indeed, in this study, we found that AMI treatment could attenuate cytotoxic mutant HTT accumulation, potentiate eumetabolic activity, and significantly improve the motor coordination of the HD mice, without any significant effects upon the life span of HD mice.
Currently, as multiple lines of evidence have demonstrated widespread pathophysiologies in peripheral systems of both HD patients and animal models (51), it is clear that HD is more than a purely neurological motor disorder (52, 53). Thus, in this study, we also evaluated potential effects of AMI on the general metabolic phenotype by measuring circulating metabolic hormones, lipid factors, and overall metabolic status. However, in terms of correcting metabolic dysfunctions, AMI displayed limited potency, which could be partly attributed to the lack of efficacy with regard to offsetting the weight loss (54) and hyperglycemia (55) commonly occurring in HD mice. However, some interesting findings were still obtained from AMI-treated HD mice in regard to metabolic functions, including reductions in plasma insulin and triglycerides levels (Fig. 1). Without affecting circulating glucose levels, a pronounced reduction of insulin might suggest that AMI could most likely affect the metabolism of insulin per se, instead of insulin sensitivity. Meanwhile, the mild elevations of total ketone bodies in the AMI-treated HD mice suggest a potential energy source shift from carbohydrate-based to lipid-based, which is consistent with the modest reductions in RER that we observed (Fig. 2). In general, compared with WT mice, HD mice also demonstrated a slightly disturbed circadian rhythm pattern (Fig. 2). Additionally, it seems that decreasing plasma triglycerides is the general action of AMI, because the reductions of triglycerides were observed in both HD-AMI and WT-AMI mice.
In HD, expanded poly(CAG) HTT leads to the production of huntingtin protein with an equally expanded poly(Q) stretch near the amino terminus. Despite a lack of consensus on the function of wild-type huntingtin (wtHTT), it is known that mutant poly(Q) huntingtin (mHTT) exerts a gain of toxic function through aberrant protein-protein interactions in human tissues, cell models, and animal models of HD (4). A number of studies have demonstrated that soluble mHTT fragments may also be engaged in the pathological processes of HD (56). We found AMI could effectively reduce mHTT expression in various brain regions, especially in the striatum and cortex. Intriguingly, instead of affecting aggregated mHTT, AMI treatment mainly caused a significant decrease of soluble mutant huntingtin fragments. This is consistent with the posit that specific neuronal vulnerability in HD may be dependent on the levels of diffuse, nonaggregated huntingtin protein found in affected neurons and provide an explanation for the pattern of striatal neuron loss (6, 7).
As a well known neurotrophic factor, BDNF plays a pivotal role in maintaining proper neuronal function (57). Reduced BDNF levels and impaired neurotrophic signaling pathway functioning is found in both HD patients and animal models of HD (10, 58). In the R6/1 HD mouse model, BDNF transgene expression relieved motor impairments and striatal neuropathology associated with HD (12); hence, enhancement of BDNF expression and its related neurotrophic signaling pathway is regarded as an important therapeutic strategy for HD. In this study, we found AMI treatment could locally elevate BDNF expression via activation of CREB in the striatum. In line with the profound up-regulation of BDNF, neurogenesis was also potentially enhanced by AMI, as indicated by a substantial increase of DCX expression in the striatum of AMI-treated HD mice. DCX is regarded as the cell marker for immature neuronal cells. Cho et al. (59) have demonstrated that overexpression of BDNF could induce neostriatal neurogenesis, which effectively slowed the disease progression in a transgenic HD murine model. Our previous study has demonstrated that AMI could profoundly induce neurogenesis in aged triple transgenic Alzheimer disease mice, which was associated with cognitive improvement (27). In this study, we mainly focused on the AMI-induced motor activity changes in HD mice, which is closely associated with pathological improvement in striatum. The cognitive effects of AMI in HD will be pursued in a following study.
Multiple lines of evidence have shown a strong relationship between BDNF-TrkB signaling and motor behaviors. Mutant mice with either forebrain-specific deletion of BDNF or TrkB deletion in the striatal progenitor cells display hind limb and fore limb clasping phenotypes, which have been observed in transgenic models with motor dysfunction or degeneration, including HD mouse models (60, 61). Most striatum functions are mediated by the medium sized spiny neurons. Ablation of the bdnf gene in the dopaminergic neurons also causes cell deaths of medium sized spiny neurons and leads to poor performance on the rotarod (62). Recently, Besusso et al. (63) also demonstrated that BDNF-TrkB signaling in striatopallidal neurons controls inhibition of locomotor behavior by modulating neuronal activity in response to excitatory input through the protein kinase C/MAPK pathway. Importantly, enhancing BDNF and TrkB in the striatum significantly improved motor coordination (12, 64) in HD animals. Our findings are highly consistent with the above-mentioned reports. Supporting the improvements of motor coordination in AMI-treated HD mice (Fig. 6), we showed the significant increases of striatal BDNF levels (Fig. 5) and activation of TrkB (Fig. 5), which is reflected by the ratio of p-TrkB/TrkB. The further mechanisms of the relationship have also been investigated in depth. The striatum is the largest component of the basal ganglia and is responsible for movement control. Li et al. (65) showed that ablation of either BDNF or TrkB in the striatum alone results in increased striatal neuron loss, reduced dendritic spines and enkephalin expression, diminished nigral dopaminergic projections, and severe deficits in striatal dopamine signaling through DARPP32. Plotkin et al. (66) reported that in the mouse model with early symptomatic HD the movement suppression in striatal neurons is caused by the failure of BDNF-TrkB engagements in postsynaptic signaling, which controls the induction of potentiation at corticostriatal synapses. In this study, in line with the improvement of BDNF-TrkB signaling, we also found the neuroprotective actions of AMI in both striatum and cortex (Fig. 3), by using the stereological analysis on NeuN-positive cells. Additionally, we demonstrated AMI treatment resulted in the enhancement of the DARPP32 signaling, reflected by an increase of p-DARPPR32/DARPP32 and potential synaptic functionality ameliorations, which is indicated by an increase of synaptic marker synaptophysin (Fig. 6).
Mitochondrial dysfunction is strongly implicated in the pathogenesis of multiple neurodegenerative disorders (67). In HD, the link to mitochondrial dysfunction has stemmed from post-mortem brain data (68) and animal models generated with mitochondrial toxins (69). Hence, preventing mitochondrial content loss and preserving intact mitochondrial function are also regarded as potentially effective strategies for HD treatment (70). With AMI treatment, we identified remedial effects for mitochondrial dysfunction in the striatum of HD mice, reflected by the increase in the expression of mitochondrial markers (VDAC and complex IV) and up-regulation of mitochondrial functionality indicators (PGC-1α and UCP2). VDAC mediates the high ionic permeability of the mitochondrial outer membrane (71). The electron transport complexes (I, II, III, and IV) and the Krebs cycle (tricarboxylic acid) are the mitochondrial metabolic pathways that are essential for generating the proton gradient across the inner membrane of the mitochondria that is used to produce ATP. Cytochrome c oxidase (complex IV) is an intracellular measure of oxidative energy metabolic capacity and respiration (72). Complex IV activity is decreased in the mitochondria from both HD patients (14) and HD mice (73). Both VDAC and complex IV are regarded as classic mitochondrial markers, suggesting that AMI treatment has provided considerable mitochondrial support in these mice. PGC-1α plays a critical role in regulating mitochondrial biogenesis and functionality (74), whereas UCP2 is known for its role in maintaining the mitochondrial membrane potential and preventing insults from free radicals (75). Further evidence has shown that UCP2 is capable of eliciting neuroprotective effects in neurodegenerative diseases (76) and traumatic central nervous system events such as stroke (77). The elevated striatal expression of PGC-1α and UCP2 could therefore be a significant component of multidimensional therapeutic efficacy of AMI.
Although we cannot exclude the involvements of other mechanisms, in our study it is highly plausible that reducing mHTT aggregates, enhancing BDNF-TrkB signaling, and ameliorating mitochondrial functions mainly contribute to the beneficial effects of AMI in the striatum, which led to improvement of motor behavior performance in HD mice.
To comprehensively appreciate the effects of AMI on HD pathophysiology, the transcriptomic signature of AMI was assessed in multiple regions of the mouse brain. Investigating the transcriptional response to AMI in multiple tissues allowed us to identify the core series of factors that could include the functional signature of AMI. We found a core series of transcripts that were coherently regulated across the cortex, hippocampus, and striatum (Fig. 4). Both Riok1 and Rps3a were both significantly altered by AMI in all three studied tissues. AMI treatment up-regulated Riok1 in all tissues studied, and pertinent to the remedial activities of AMI, it has been recently demonstrated that this kinase is a critical controller of AKT-mediated cell survival (78). The consistent AMI-mediated reduction of Rps3a may also indicate that this is linked to potential cell protection mechanisms (79) and anti-inflammatory behavior (80). In addition to these highly conserved AMI-response transcripts, many other factors consistently controlled across the brain are strongly implicated in potentially neuroprotective functions. For example, Tppp (tubulin polymerization-promoting protein) and Acsl5 (acyl-CoA synthetase long-chain family member 5) and Sdhc (succinate dehydrogenase complex, subunit C, integral membrane protein, 15 kDa) are strongly implicated in mitochondrial function and metabolic support (81–83). AMI also consistently up-regulated the expression of factors associated with neuronal development (Sparc, secreted protein, acidic, cysteine-rich (osteonectin)) (84), neuronal Akt-mediated protection (Nyap1, neuronal tyrosine-phosphorylated phosphoinositide-3-kinase adaptor 1), and attenuation of neurodegenerative cross-linking (Tgm2, transglutaminase 2) (43, 85). With our use of PAGE MSigDB bioinformatics analysis (Fig. 4, C and D), we also found that AMI strongly supports glycolytic pathways whose inhibition has been associated with HD disease progression (86). Interestingly, some of the most down-regulated PAGE MSigDB pathways were associated with the maintenance of stem cell properties, i.e. STEMCELL_NEURAL_UP (Fig. 4D). The population of this PAGE pathway with transcripts, reduced in their expression with AMI, corroborates our finding of increased DCX levels, suggesting a pro-neurogenetic activity of AMI in HD.
In summary, AMI appears to possess some promising therapeutic potential for the treatment of HD, both in terms of symptom management and pathology correction, in the murine N171-82Q Huntington disease model. Further mechanistic studies demonstrated that AMI is capable of eliciting beneficial effects in regard to multiple aspects of HD neuropathology, including relieving mutant HTT burdens in the cortex and striatum, protecting crucial proteins related to mitochondrial number and functionality in the striatum, and elevating BDNF levels and TrkB-mediated transcriptional regulation and pro-survival pathways. It is interesting, however, that AMI potently attenuated the motor deficits observed in HD but was relatively poor in lowering the pro-diabetic blood glucose levels observed in our mice. However, we did find that AMI was able to attenuate the excessive plasma levels of insulin, suggesting that it still does generate some anti-diabetic activity in this model. Therefore, a combination of an additional anti-diabetic agent such as exendin-4 (87) with AMI may prove to be even more efficacious in treating HD. Our study not only provides preclinical evidence for the therapeutic potential of AMI in treating HD, it also offers an example of a new drug development strategy focused on studying previously approved drugs in novel disease contexts.
Supplementary Material
Acknowledgments
We thank Drs. Kevin Becker, William H. Wood III, and Yongqing Zhang of the National Institutes of Health NIA Research Resources Branch for their assistance with transcriptomic analyses. We also thank Dr. Peter Rapp and Dr. Jeffrey Long of the National Institutes of Health NIA Laboratory of Behavioral Neuroscience for their assistance in stereological analysis. We also thank Dr. Gillian Bates from King's College London for generously donating the S830 antibody to us.
This work was supported, in whole or in part, by National Institutes of Health Intramural Research Program of the NIA.

This article contains supplemental Table S1–S6.
- HD
- Huntington disease
- BDNF
- brain-derived neurotrophic factor
- AMI
- amitriptyline
- RER
- respiratory exchange ratio
- SSRI
- selective serotonin reuptake inhibitor
- PYY
- peptide YY
- GIP
- gastric inhibitory peptide
- PP
- pancreatic peptide
- CREB
- cAMP-response element-binding protein
- VDAC
- voltage-dependent anion channel
- DCX
- doublecortin.
REFERENCES
- 1. Kremer B., Goldberg P., Andrew S. E., Theilmann J., Telenius H., Zeisler J., Squitieri F., Lin B., Bassett A., Almqvist E. (1994) A worldwide study of the Huntington's disease mutation: the sensitivity and specificity of measuring CAG repeats. N. Engl. J. Med. 330, 1401–1406 [DOI] [PubMed] [Google Scholar]
- 2. DiFiglia M., Sapp E., Chase K. O., Davies S. W., Bates G. P., Vonsattel J. P., Aronin N. (1997) Aggregation of Huntingtin in neuronal intranuclear inclusions and dystrophic neurites in brain. Science 277, 1990–1993 [DOI] [PubMed] [Google Scholar]
- 3. Dedeoglu A., Kubilus J. K., Jeitner T. M., Matson S. A., Bogdanov M., Kowall N. W., Matson W. R., Cooper A. J., Ratan R. R., Beal M. F., Hersch S. M., Ferrante R. J. (2002) Therapeutic effects of cystamine in a murine model of Huntington's disease. J. Neurosci. 22, 8942–8950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hoffner G., Djian P. (2002) Protein aggregation in Huntington's disease. Biochimie 84, 273–278 [DOI] [PubMed] [Google Scholar]
- 5. Ferrante R. J., Andreassen O. A., Dedeoglu A., Ferrante K. L., Jenkins B. G., Hersch S. M., Beal M. F. (2002) Therapeutic effects of coenzyme Q10 and remacemide in transgenic mouse models of Huntington's disease. J. Neurosci. 22, 1592–1599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sisodia S. S. (1998) Nuclear inclusions in glutamine repeat disorders: are they pernicious, coincidental, or beneficial? Cell 95, 1–4 [DOI] [PubMed] [Google Scholar]
- 7. Arrasate M., Mitra S., Schweitzer E. S., Segal M. R., Finkbeiner S. (2004) Inclusion body formation reduces levels of mutant huntingtin and the risk of neuronal death. Nature 431, 805–810 [DOI] [PubMed] [Google Scholar]
- 8. Pérez-Navarro E., Canudas A. M., Akerund P., Alberch J., Arenas E. (2000) Brain-derived neurotrophic factor, neurotrophin-3, and neurotrophin-4/5 prevent the death of striatal projection neurons in a rodent model of Huntington's disease. J. Neurochem. 75, 2190–2199 [DOI] [PubMed] [Google Scholar]
- 9. Zuccato C., Ciammola A., Rigamonti D., Leavitt B. R., Goffredo D., Conti L., MacDonald M. E., Friedlander R. M., Silani V., Hayden M. R., Timmusk T., Sipione S., Cattaneo E. (2001) Loss of Huntingtin-mediated BDNF gene transcription in Huntington's disease. Science 293, 493–498 [DOI] [PubMed] [Google Scholar]
- 10. Ferrer I., Goutan E., Marín C., Rey M. J., Ribalta T. (2000) Brain-derived neurotrophic factor in Huntington disease. Brain Res. 866, 257–261 [DOI] [PubMed] [Google Scholar]
- 11. Ginés S., Bosch M., Marco S., Gavaldà N., Díaz-Hernández M., Lucas J. J., Canals J. M., Alberch J. (2006) Reduced expression of the TrkB receptor in Huntington's disease mouse models and in human brain. Eur. J. Neurosci. 23, 649–658 [DOI] [PubMed] [Google Scholar]
- 12. Gharami K., Xie Y., An J. J., Tonegawa S., Xu B. (2008) Brain-derived neurotrophic factor over-expression in the forebrain ameliorates Huntington's disease phenotypes in mice. J. Neurochem. 105, 369–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bossy-Wetzel E., Petrilli A., Knott A. B. (2008) Mutant huntingtin and mitochondrial dysfunction. Trends Neurosci. 31, 609–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gu M., Gash M. T., Mann V. M., Javoy-Agid F., Cooper J. M., Schapira A. H. (1996) Mitochondrial defect in Huntington's disease caudate nucleus. Ann. Neurol. 39, 385–389 [DOI] [PubMed] [Google Scholar]
- 15. Tabrizi S. J., Workman J., Hart P. E., Mangiarini L., Mahal A., Bates G., Cooper J. M., Schapira A. H. (2000) Mitochondrial dysfunction and free radical damage in the Huntington R6/2 transgenic mouse. Ann. Neurol. 47, 80–86 [DOI] [PubMed] [Google Scholar]
- 16. Panov A. V., Gutekunst C. A., Leavitt B. R., Hayden M. R., Burke J. R., Strittmatter W. J., Greenamyre J. T. (2002) Early mitochondrial calcium defects in Huntington's disease are a direct effect of polyglutamines. Nat. Neurosci. 5, 731–736 [DOI] [PubMed] [Google Scholar]
- 17. Cui L., Jeong H., Borovecki F., Parkhurst C. N., Tanese N., Krainc D. (2006) Transcriptional repression of PGC-1a by mutant Huntingtin leads to mitochondrial dysfunction and neurodegeneration. Cell. 127, 59–69 [DOI] [PubMed] [Google Scholar]
- 18. Ryu H., Ferrante R. J. (2005) Emerging chemotherapeutic strategies for Huntington's disease. Expert Opin. Emerg. Drugs 10, 345–363 [DOI] [PubMed] [Google Scholar]
- 19. Nibuya M., Morinobu S., Duman R. S. (1995) Regulation of BDNF and trkB mRNA in rat brain by chronic electroconvulsive seizure and antidepressant drug treatments. J. Neurosci. 15, 7539–7547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Moltzen E. K., Bang-Andersen B. (2006) Serotonin reuptake inhibitors: the cornerstone in treatment of depression for half a century–a medicinal chemistry survey. Curr. Top. Med. Chem. 6, 1801–1823 [DOI] [PubMed] [Google Scholar]
- 21. Duan W., Guo Z., Jiang H., Ladenheim B., Xu X., Cadet J. L., Mattson M. P. (2004) Paroxetine retards disease onset and progression in Huntingtin mutant mice. Ann. Neurol. 55, 590–594 [DOI] [PubMed] [Google Scholar]
- 22. Duan W., Peng Q., Masuda N., Ford E., Tryggestad E., Ladenheim B., Zhao M., Cadet J. L., Wong J., Ross C. A. (2008) Sertraline slows disease progression and increases neurogenesis in N171-82Q mouse model of Huntington's disease. Neurobiol. Dis. 30, 312–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Peng Q., Masuda N., Jiang M., Li Q., Zhao M., Ross C. A., Duan W. (2008) The antidepressant sertraline improves the phenotype, promotes neurogenesis and increases BDNF levels in the R6/2 Huntington's disease mouse model. Exp. Neurol. 210, 154–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Grote H. E., Bull N. D., Howard M. L., van Dellen A., Blakemore C., Bartlett P. F., Hannan A. J. (2005) Cognitive disorders and neurogenesis deficits in Huntington's disease mice are rescued by fluoxetine. Eur. J. Neurosci. 22, 2081–2088 [DOI] [PubMed] [Google Scholar]
- 25. Wang H., Guan Y., Wang X., Smith K., Cormier K., Zhu S., Stavrovskaya I. G., Huo C., Ferrante R. J., Kristal B. S., Friedlander R. M. (2007) Nortriptyline delays disease onset in models of chronic neurodegeneration. Eur. J. Neurosci. 26, 633–641 [DOI] [PubMed] [Google Scholar]
- 26. Masuda N., Peng Q., Li Q., Jiang M., Liang Y., Wang X., Zhao M., Wang W., Ross C. A., Duan W. (2008) Tiagabine is neuroprotective in the N171-82Q and R6/2 mouse models of Huntington's disease. Neurobiol. Dis. 30, 293–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chadwick W., Mitchell N., Caroll J., Zhou Y., Park S.-S., Wang L., Becker K. G., Zhang Y., Lehrmann E., Wood W. H., 3rd, Martin B., Maudsley S. (2011) Amitriptyline-mediated cognitive enhancement in aged 3Tg Alzheimer's disease mice is associated with neurogenesis and neurotrophic activity. PLoS One 6, e21660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Xu H., Steven Richardson J., Li X.-M. (2003) Dose-related effects of chronic antidepressants on neuroprotective proteins BDNF, Bcl-2 and Cu/Zn-SOD in rat hippocampus. Neuropsychopharmacology 28, 53–62 [DOI] [PubMed] [Google Scholar]
- 29. Jang S.-W., Liu X., Chan C.-B., Weinshenker D., Hall R. A., Xiao G., Ye K. (2009) Amitriptyline is a TrkA and TrkB receptor agonist that promotes TrkA/TrkB heterodimerization and has potent neurotrophic activity. Chem. Biol. 16, 644–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Martin B., Golden E., Carlson O. D., Pistell P., Zhou J., Kim W., Frank B. P., Thomas S., Chadwick W. A., Greig N. H., Bates G. P., Sathasivam K., Bernier M., Maudsley S., Mattson M. P., Egan J. M. (2009) Exendin-4 improves glycemic control, ameliorates brain and pancreatic pathologies, and extends survival in a mouse model of Huntington's disease. Diabetes 58, 318–328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Stanley J. L., Lincoln R. J., Brown T. A., McDonald L. M., Dawson G. R., Reynolds D. S. (2005) The mouse beam walking assay offers improved sensitivity over the mouse Rotarod in determining motor coordination deficits induced by benzodiazepines. J. Psychopharmacol. 19, 221–227 [DOI] [PubMed] [Google Scholar]
- 32. Martin B., Pearson M., Kebejian L., Golden E., Keselman A., Bender M., Carlson O., Egan J., Ladenheim B., Cadet J. L., Becker K. G., Wood W., Duffy K., Vinayakumar P., Maudsley S., Mattson M. P. (2007) Sex-dependent metabolic, neuroendocrine, and cognitive responses to dietary energy restriction and excess. Endocrinology 148, 4318–4333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Scribner K. B., Pawlak D. B., Aubin C. M., Majzoub J. A., Ludwig D. S. (2008) Long-term effects of dietary glycemic index on adiposity, energy metabolism, and physical activity in mice. Am. J. Physiol. Endocrinol. Metab. 295, E1126–E1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chadwick W., Keselman A., Park S.-S., Zhou Y., Wang L., Brenneman R., Martin B., Maudsley S. (2011) Repetitive peroxide exposure reveals pleiotropic mitogen-activated protein kinase signaling mechanisms. J. Signal. Transduct. 2011, 636951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Simmons D. A., Casale M., Alcon B., Pham N., Narayan N., Lynch G. (2007) Ferritin accumulation in dystrophic microglia is an early event in the development of Huntington's disease. Glia 55, 1074–1084 [DOI] [PubMed] [Google Scholar]
- 36. Simmons D. A., Mehta R. A., Lauterborn J. C., Gall C. M., Lynch G. (2011) Brief ampakine treatments slow the progression of Huntington's disease phenotypes in R6/2 mice. Neurobiol. Dis. 41, 436–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Shin Y. K., Cong W. N., Cai H., Kim W., Maudsley S., Egan J. M., Martin B. (2012) Age-related changes in mouse taste bud morphology, hormone expression, and taste responsivity. J. Gerontol. A Biol. Sci. Med. Sci. 67, 336–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Franklin K. B., Paxinos G. (2008) The Mouse Brain In Stereotaxic Coordinates. 3rd Ed., pp. 34–75, Academic Press, New York [Google Scholar]
- 39. Cai H., Chen H., Yi T., Daimon C. M., Boyle J. P., Peers C., Maudsley S., Martin B. (2013) VennPlex–a novel Venn diagram program for comparing and visualizing datasets with differentially regulated datapoints. PLoS One 8, e53388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Martin B., Brenneman R., Golden E., Walent T., Becker K. G., Prabhu V. V., Wood W., 3rd, Ladenheim B., Cadet J. L., Maudsley S. (2009) Growth factor signals in neural cells: coherent patterns of interaction control multiple levels of molecular and phenotypic responses. J. Biol. Chem. 284, 2493–2511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Chen H., Martin B., Daimon C. M., Siddiqui S., Luttrell L. M., Maudsley S. (2013) Textrous!: extracting semantic textual meaning from gene sets. PLoS One 8, e62665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Gesty-Palmer D., Yuan L., Martin B., Wood W. H., 3rd, Lee M. H., Janech M. G., Tsoi L. C., Zheng W. J., Luttrell L. M., Maudsley S. (2013) β-Arrestin-selective G protein-coupled receptor agonists engender unique biological efficacy in vivo. Mol. Endocrinol. 27, 296–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Iannaccone M., Serretiello E., De Vivo G., Martin A., Stefanile A., Titta F., Gentile V. (2013) Transglutaminase inhibition as a possible therapeutical approach to protect cells from death in neurodegenerative diseases. Recent Pat. CNS Drug Discov. 8, 161–168 [DOI] [PubMed] [Google Scholar]
- 44. Oliveira J. M. (2010) Nature and cause of mitochondrial dysfunction in Huntington's disease: focusing on huntingtin and the striatum. J. Neurochem. 114, 1–12 [DOI] [PubMed] [Google Scholar]
- 45. Yuste J. E., Echeverry M. B., Ros-Bernal F., Gomez A., Ros C. M., Campuzano C. M., Fernandez-Villalba E., Herrero M. T. (2012) 7-Nitroindazole down-regulates dopamine/DARPP-32 signaling in neostriatal neurons in a rat model of Parkinson's disease. Neuropharmacology 63, 1258–1267 [DOI] [PubMed] [Google Scholar]
- 46. Fienberg A. A., Hiroi N., Mermelstein P. G., Song W., Snyder G. L., Nishi A., Cheramy A., O'Callaghan J. P., Miller D. B., Cole D. G., Corbett R., Haile C. N., Cooper D. C., Onn S. P., Grace A. A., Ouimet C. C., White F. J., Hyman S. E., Surmeier D. J., Girault J., Nestler E. J., Greengard P. (1998) DARPP-32: regulator of the efficacy of dopaminergic neurotransmission. Science 281, 838–842 [DOI] [PubMed] [Google Scholar]
- 47. Ernst A., Alkass K., Bernard S., Salehpour M., Perl S., Tisdale J., Possnert G., Druid H., Frisén J. (2014) Neurogenesis in the striatum of the adult human brain. Cell 156, 1072–1083 [DOI] [PubMed] [Google Scholar]
- 48. Hellweg R., Ziegenhorn A., Heuser I., Deuschle M. (2008) Serum concentrations of nerve growth factor and brain-derived neurotrophic factor in depressed patients before and after antidepressant treatment. Pharmacopsychiatry 41, 66–71 [DOI] [PubMed] [Google Scholar]
- 49. Lee B.-H., Myint A. M., Kim Y.-K. (2010) Psychotropic drugs on in vitro brain-derived neurotrophic factor production in whole blood cell cultures from healthy subjects. J. Clin. Psychopharmacol. 30, 623–627 [DOI] [PubMed] [Google Scholar]
- 50. Jiang M., Peng Q., Liu X., Jin J., Hou Z., Zhang J., Mori S., Ross C. A., Ye K., Duan W. (2013) Small-molecule TrkB receptor agonists improve motor function and extend survival in a mouse model of Huntington's disease. Hum. Mol. Genet. 22, 2462–2470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. van der Burg J. M., Björkqvist M., Brundin P. (2009) Beyond the brain: widespread pathology in Huntington's disease. Lancet Neurol. 8, 765–774 [DOI] [PubMed] [Google Scholar]
- 52. Cai H., Cong W. N., Ji S., Rothman S., Maudsley S., Martin B. (2012) Metabolic dysfunction in Alzheimer's disease and related neurodegenerative disorders. Curr. Alzheimer Res. 9, 5–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Cong W. N., Cai H., Wang R., Daimon C. M., Maudsley S., Raber K., Canneva F., von Hörsten S., Martin B. (2012) Altered hypothalamic protein expression in a rat model of Huntington's disease. PLoS One 7, e47240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ho D. J., Calingasan N. Y., Wille E., Dumont M., Beal M. F. (2010) Resveratrol protects against peripheral deficits in a mouse model of Huntington's disease. Exp. Neurol. 225, 74–84 [DOI] [PubMed] [Google Scholar]
- 55. Andreassen O. A., Dedeoglu A., Ferrante R. J., Jenkins B. G., Ferrante K. L., Thomas M., Friedlich A., Browne S. E., Schilling G., Borchelt D. R., Hersch S. M., Ross C. A., Beal M. F. (2001) Creatine increases survival and delays motor symptoms in a transgenic animal model of Huntington's disease. Neurobiol. Dis. 8, 479–491 [DOI] [PubMed] [Google Scholar]
- 56. Zala D., Benchoua A., Brouillet E., Perrin V., Gaillard M.-C., Zurn A. D., Aebischer P., Déglon N. (2005) Progressive and selective striatal degeneration in primary neuronal cultures using lentiviral vector coding for a mutant huntingtin fragment. Neurobiol. Dis. 20, 785–798 [DOI] [PubMed] [Google Scholar]
- 57. Bramham C. R., Messaoudi E. (2005) BDNF function in adult synaptic plasticity: The synaptic consolidation hypothesis. Prog. Neurobiol. 76, 99–125 [DOI] [PubMed] [Google Scholar]
- 58. Spires T. L., Grote H. E., Varshney N. K., Cordery P. M., van Dellen A., Blakemore C., Hannan A. J. (2004) Environmental enrichment rescues protein deficits in a mouse model of Huntington's disease, indicating a possible disease mechanism. J. Neurosci. 24, 2270–2276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Cho S. R., Benraiss A., Chmielnicki E., Samdani A., Economides A., Goldman S. A. (2007) Induction of neostriatal neurogenesis slows disease progression in a transgenic murine model of Huntington disease. J. Clin. Invest. 117, 2889–2902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Baquet Z. C., Gorski J. A., Jones K. R. (2004) Early striatal dendrite deficits followed by neuron loss with advanced age in the absence of anterograde cortical brain-derived neurotrophic factor. J. Neurosci. 24, 4250–4258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Baydyuk M., Russell T., Liao G. Y., Zang K., An J. J., Reichardt L. F., Xu B. (2011) TrkB receptor controls striatal formation by regulating the number of newborn striatal neurons. Proc. Natl. Acad. Sci. U.S.A. 108, 1669–1674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Baydyuk M., Xie Y., Tessarollo L., Xu B. (2013) Midbrain-derived neurotrophins support survival of immature striatal projection neurons. J. Neurosci. 33, 3363–3369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Besusso D., Geibel M., Kramer D., Schneider T., Pendolino V., Picconi B., Calabresi P., Bannerman D. M., Minichiello L. (2013) BDNF-TrkB signaling in striatopallidal neurons controls inhibition of locomotor behavior. Nat. Commun. 4, 2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Xie Y., Hayden M. R., Xu B. (2010) BDNF overexpression in the forebrain rescues Huntington's disease phenotypes in YAC128 mice. J. Neurosci. 30, 14708–14718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Li Y., Yui D., Luikart B. W., McKay R. M., Li Y., Rubenstein J. L., Parada L. F. (2012) Conditional ablation of brain-derived neurotrophic factor-TrkB signaling impairs striatal neuron development. Proc. Natl. Acad. Sci. U.S.A. 109, 15491–15496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Plotkin J. L., Day M., Peterson J. D., Xie Z., Kress G. J., Rafalovich I., Kondapalli J., Gertler T. S., Flajolet M., Greengard P., Stavarache M., Kaplitt M. G., Rosinski J., Chan C. S., Surmeier D. J. (2014) Impaired TrkB receptor signaling underlies corticostriatal dysfunction in Huntington's disease. Neuron 83, 178–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Beal M. F. (2005) Mitochondria take center stage in aging and neurodegeneration. Ann. Neurol. 58, 495–505 [DOI] [PubMed] [Google Scholar]
- 68. Rosenstock T. R., Duarte A. I., Rego A. C. (2010) Mitochondrial-associated metabolic changes and neurodegeneration in Huntington's disease–from clinical features to the bench. Curr. Drug Targets 11, 1218–1236 [DOI] [PubMed] [Google Scholar]
- 69. Brouillet E., Jacquard C., Bizat N., Blum D. (2005) 3-Nitropropionic acid: a mitochondrial toxin to uncover physiopathological mechanisms underlying striatal degeneration in Huntington's disease. J. Neurochem. 95, 1521–1540 [DOI] [PubMed] [Google Scholar]
- 70. Munoz-Sanjuan I., Bates G. P. (2011) The importance of integrating basic and clinical research toward the development of new therapies for Huntington disease. J. Clin. Invest. 121, 476–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. De Pinto V. D., Palmieri F. (1992) Transmembrane arrangement of mitochondrial porin or voltage-dependent anion channel (VDAC). J. Bioenerg. Biomembr. 24, 21–26 [DOI] [PubMed] [Google Scholar]
- 72. Villani G., Attardi G. (2000) In vivo control of respiration by cytochrome c oxidase in human cells. Free Radic. Biol. Med. 29, 202–210 [DOI] [PubMed] [Google Scholar]
- 73. Aidt F. H., Nielsen S. M., Kanters J., Pesta D., Nielsen T. T., Nørremølle A., Hasholt L., Christiansen M., Hagen C. M. (2013) Dysfunctional mitochondrial respiration in the striatum of the Huntington's disease transgenic R6/2 mouse model. PLoS Curr. PMC3614423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. McGill J. K., Beal M. F. (2006) PGC-1a, a new therapeutic target in Huntington's disease? Cell 127, 465–468 [DOI] [PubMed] [Google Scholar]
- 75. Mehta S. L., Li P. A. (2009) Neuroprotective role of mitochondrial uncoupling protein 2 in cerebral stroke. J. Cereb. Blood Flow Metab. 29, 1069–1078 [DOI] [PubMed] [Google Scholar]
- 76. Mattiasson G., Shamloo M., Gido G., Mathi K., Tomasevic G., Yi S., Warden C. H., Castilho R. F., Melcher T., Gonzalez-Zulueta M., Nikolich K., Wieloch T. (2003) Uncoupling protein-2 prevents neuronal death and diminishes brain dysfunction after stroke and brain trauma. Nat. Med. 9, 1062–1068 [DOI] [PubMed] [Google Scholar]
- 77. Deierborg T., Deierborg Olsson T., Wieloch T., Diano S., Warden C. H., Horvath T. L., Mattiasson G. (2008) Overexpression of UCP2 protects thalamic neurons following global ischemia in the mouse. J. Cereb. Blood Flow Metab. 28, 1186–1195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Read R. D., Fenton T. R., Gomez G. G., Wykosky J., Vandenberg S. R., Babic I., Iwanami A., Yang H., Cavenee W. K., Mischel P. S., Furnari F. B., Thomas J. B. (2013) A kinome-wide RNAi screen in Drosophila Glia reveals that the RIO kinases mediate cell proliferation and survival through TORC2-Akt signaling in glioblastoma. PLoS Genet. 9, e1003253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Russell L., Naora H., Naora H. (2000) Down-regulated RPS3a/nbl expression during retinoid-induced differentiation of HL-60 cells: a close association with diminished susceptibility to actinomycin D-stimulated apoptosis. Cell Struct. Funct. 25, 103–113 [DOI] [PubMed] [Google Scholar]
- 80. Lee H. Y., Park J. H., Lee C. H., Yan B., Ahn J. H., Lee Y. J., Park C. W., Cho J. H., Choi S. Y., Won M. H. (2012) Changes of ribosomal protein S3 immunoreactivity and its new expression in microglia in the mice hippocampus after lipopolysaccharide treatment. Cell. Mol. Neurobiol. 32, 577–586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Fjorback A. W., Sundbye S., Dächsel J. C., Sinning S., Wiborg O., Jensen P. H. (2011) P25α/TPPP expression increases plasma membrane presentation of the dopamine transporter and enhances cellular sensitivity to dopamine toxicity. FEBS J. 278, 493–505 [DOI] [PubMed] [Google Scholar]
- 82. Mashima T., Oh-hara T., Sato S., Mochizuki M., Sugimoto Y., Yamazaki K., Hamada J., Tada M., Moriuchi T., Ishikawa Y., Kato Y., Tomoda H., Yamori T., Tsuruo T. (2005) p53-defective tumors with a functional apoptosome-mediated pathway: a new therapeutic target. J. Natl. Cancer Inst. 97, 765–777 [DOI] [PubMed] [Google Scholar]
- 83. Ishii T., Miyazawa M., Onouchi H., Yasuda K., Hartman P. S., Ishii N. (2013) Model animals for the study of oxidative stress from complex II. Biochim. Biophys. Acta 1827, 588–597 [DOI] [PubMed] [Google Scholar]
- 84. Bhoopathi P., Chetty C., Dontula R., Gujrati M., Dinh D. H., Rao J. S., Lakka S. S. (2011) SPARC stimulates neuronal differentiation of medulloblastoma cells via the Notch1/STAT3 pathway. Cancer Res. 71, 4908–4919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Krzysztoń-Russjan J., Zielonka D., Jackiewicz J., Kusmirek S., Bubko I., Klimberg A., Marcinkowski J. T., Anuszewska E. L. (2013) A study of molecular changes relating to energy metabolism and cellular stress in people with Huntington's disease: looking for biomarkers. J. Bioenerg. Biomembr. 45, 71–85 [DOI] [PubMed] [Google Scholar]
- 86. Estrada-Sánchez A. M., Montiel T., Massieu L. (2010) Glycolysis inhibition decreases the levels of glutamate transporters and enhances glutamate neurotoxicity in the R6/2 Huntington's disease mice. Neurochem Res. 35, 1156–1163 [DOI] [PubMed] [Google Scholar]
- 87. Martin B., Chadwick W., Cong W. N., Pantaleo N., Daimon C. M., Golden E. J., Becker K. G., Wood W. H., 3rd, Carlson O. D., Egan J. M., Maudsley S. (2012) Euglycemic agent-mediated hypothalamic transcriptomic manipulation in the N171-82Q model of Huntington's disease is related to their physiological efficacy. J. Biol. Chem. 287, 31766–31782 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.