Background: Maintenance of HSCs is challenged by DNA damage and oxidative stress.
Results: Fancd2 deficiency promoted cytoplasmic localization of Foxo3a in HSCs. Re-expression of Fancd2 restored nuclear Foxo3a localization and prevented HSC exhaustion.
Conclusion: Fancd2 is required for nuclear retention of Foxo3a and maintaining hematopoietic repopulation of HSCs.
Significance: Our results implicate an interaction between FA DNA repair and FOXO3a pathways in HSC maintenance.
Keywords: DNA Damage, DNA Repair, FOXO, Hematopoietic Stem Cells, Oxidative Stress, Fancd2
Abstract
Functional maintenance of hematopoietic stem cells (HSCs) is constantly challenged by stresses like DNA damage and oxidative stress. Here we show that the Fanconi anemia protein Fancd2 and stress transcriptional factor Foxo3a cooperate to prevent HSC exhaustion in mice. Deletion of both Fancd2 and Foxo3a led to an initial expansion followed by a progressive decline of bone marrow stem and progenitor cells. Limiting dilution transplantation and competitive repopulating experiments demonstrated a dramatic reduction of competitive repopulating units and progressive decline in hematopoietic repopulating ability of double-knockout (dKO) HSCs. Analysis of the transcriptome of dKO HSCs revealed perturbation of multiple pathways implicated in HSC exhaustion. Fancd2 deficiency strongly promoted cytoplasmic localization of Foxo3a in HSCs, and re-expression of Fancd2 completely restored nuclear Foxo3a localization. By co-expressing a constitutively active CA-FOXO3a and WT or a nonubiquitinated Fancd2 in dKO bone marrow stem/progenitor cells, we demonstrated that Fancd2 was required for nuclear retention of CA-FOXO3a and for maintaining hematopoietic repopulation of the HSCs. Collectively, these results implicate a functional interaction between the Fanconi anemia DNA repair and FOXO3a pathways in HSC maintenance.
Introduction
Hematopoietic stem cells (HSCs)2 are maintained in an undifferentiated quiescent state in a bone marrow (BM) niche (1). It appears that the HSC pool is maintained throughout the lifetime, and this lifelong persistence of HSCs is likely due to their balance between quiescence and cycling, as well as to their localization in specialized niches in BM (2, 3). Increased proliferation or differentiation ultimately may lead to premature exhaustion of the stem cell pool, which is defined by a decrease in the number of HSCs caused by their enhanced cell cycling (4). Whereas numerous molecular factors that contribute to quiescence exist in the HSCs and the BM niche, HSCs are inevitably exposed to stress such as accumulation of reactive oxygen species and DNA damage, which can drive HSCs into uncontrolled cell cycle entry and excessive proliferation. DNA damage or oxidative stress can actually result in HSC exhaustion (5, 6). Studies in ATM (ataxia telangiectasia mutated) deficient mice showed progressive BM failure resulting from a defect in HSC function that was associated with elevated reactive oxygen species that induce oxidative stress (7). Prolonged treatment of HSCs with an antioxidant can extend the life span of HSCs and protects against loss of self-renewal capacity (7).
Fanconi anemia (FA) is an inherited disorder characterized by progressive bone marrow failure, developmental defects, and predisposition to cancer (8, 9). Most FA patients demonstrate BM failure during childhood and are at risk of developing acute myelogenous leukemia (10, 11). FA is caused by a deficiency in any of the 16 FA genes (FANCA-Q) (12, 13), which cooperate in a DNA repair pathway for resolving DNA interstrand cross-link encountered during replication or generated by DNA-damaging agents (14, 15). Several FA proteins form a large nuclear complex that modifies the key downstream FA gene product FANCD2, by conjugation of a single ubiquitin molecule (monoubiquitination). This critical modification could be stimulated by DNA damage or genotoxic stress, resulting in the localization of the FANCD2 protein to sites of nuclear DNA lesions (16). Patients with FANCD2 mutations have earlier onset and more rapid progression of hematologic manifestations (17). It has been reported that Fancd2-deficient mice had multiple hematopoietic defects, including HSC and progenitor loss in early development, abnormal cell cycle status, and loss of quiescence in hematopoietic stem and progenitor cells, and compromised functional capacity of HSCs (5).
Mammalian FOXO (forkhead members of the class O) transcription factors, including FOXO1, FOXO3a, FOXO4, and FOXO6, are implicated in the regulation of diverse physiologic processes, including cell cycle arrest, apoptosis, DNA repair, stress resistance, and metabolism (18). Loss of FOXOs results in elevated reactive oxygen species, which in turn negatively regulates cellular responses (19). FOXO proteins are normally present in an active state in the nucleus of a cell. Activated AKT phosphorylates FOXO3a proteins at three consensus AKT phosphorylation sites (Thr-32, Ser-253, and Ser-315), triggering the inactivation of FOXO3a and its export from the nucleus into the cytoplasm (20). Mouse knockout studies have shown that Foxo factors, particularly Foxo3a, function to regulate the self-renewal of HSCs and contribute to the maintenance of the HSC pool during aging by providing resistance to oxidative stress (21).
We recently reported a functional interaction between FOXO3a and the FA protein FANCD2 in response to oxidative stress (22). However, the functional consequence of this interaction is not known. In this study, we have exploited the genetic relationship between the two proteins by generating Fancd2−/− Foxo3a−/− double-knockout (dKO) mice and demonstrating that Fancd2 cooperates with nuclear Foxo3a to prevent HSC exhaustion.
EXPERIMENTAL PROCEDURES
Animals
All protocols of animal experiments described in this study were approved by the Institutional Animal Care and Use Committee at Cincinnati Children's Hospital Medical Center. Fancd2+/− mice were provided by Dr. Markus Grompe (Oregon Health & Sciences University) (23). Double heterozygotes Fancd2+/− Foxo3a+/− mice were first generated from interbreeding Fancd2+/− heterozygotes with the Foxo3a+/− mice (18), and WT and dKO mice were generated from interbreeding of the double heterozygotes. All the animals including BoyJ (C57BL/6: B6, CD45.1+) mice were maintained in the animal barrier facility at Cincinnati Children's Hospital Medical Center.
Flow Analysis and Cell Sorting
Femurs and tibias were flushed to dissociate the BM fraction. Cells were resuspended in 5 ml of PBS, 0.5% BSA and filtered through a 70-μm filter (BD Biosciences). The mononuclear cells were isolated by Ficoll (GE Healthcare) gradient centrifugation. The following antibodies were used for flow cytometry analyses: APC-cy7-anti-c-Kit, PE-cy7-anti-Sca-1, Pacific Blue anti-CD150, FITC-anti-CD48, FITC-anti-CD34, PE-anti-CD45.1, APC-anti-CD45.2, APC-anti-Ki67, and APC-anti-BrdUrd antibodies. The lineage antibody mixture included the following biotin-conjugated anti-mouse antibodies: Mac1, Gr-1, Ter119, CD3e, and B220 (BD Biosciences). Secondary reagent used included streptavidin-PerCP-Cy5.5 (BD Biosciences). Initially, for LSK (Lineage− Sca-1+ c-Kit+) staining, cells were stained by using biotin-conjugated anti-lineage antibody mixture followed by staining with a secondary Percp-cy5.5-anti-streptavidin antibody (BD Biosciences), PE-cy7-anti-Sca-1 antibody (BD Biosciences), and APC-cy7-anti-c-Kit antibody (BD Biosciences). For the HSC (SLAM; Lin− c-Kit+ Sca-1+ CD150+ CD48−) subpopulation, cells were stained with antibodies for LSK cells in addition to Pacific Blue CD150 (Biolegend) and FITC-CD48 (Biolegend). Flow cytometry was performed on a FACS-LSR II (BD Biosciences), and analysis was done with FACSDiva version 6.1.2 software (BD Biosciences). For cell sorting, lineage negative cells were enriched using lineage depletion columns (StemCell Technologies) according to the manufacturer's instructions. The LSK population or HSCs (Lin− c-Kit+ Sca-1+ CD34−) were acquired by using the FACSAria II sorter (BD Biosciences).
Limiting Dilution Cobblestone Area-forming Cell (CAFC) Assay
Limiting dilution CAFC assay using five dilutions (0, 10, 30, 90, 270, and 810) of LSK cells was performed as described previously (24). Briefly, cells were plated on confluent OP9 stromal cells in 96-well plates with 10 wells/cell concentration, and numbers of CAFC were counted after 4 weeks.
BM Transplantation
For the limiting dilution assay, graded numbers of donor BM cells (CD45.2+) harvested from WT, Fancd2−/−, Foxo3a−/−, and dKO mice were mixed with 2 × 105 protector BM cells (CD45.1+) and transplanted into lethal irradiated (split dose of 700 and 475 Rad with 3 h apart) BoyJ mice (CD45.1+). Plotted are the percentages of recipient mice containing less than 1% CD45.2+ blood nucleated cells at 16 weeks after transplantation. The frequency of functional HSCs was calculated according to Poisson statistics.
For competitive repopulation assay, 50 LSK CD150+CD48+ (SLAM) cells or 1,000 LSK cells (CD45.2+) plus 4 × 105 recipient BM cells (CD45.1+) were transplanted into lethally irradiated BoyJ mice (CD45.1+). Five mice were transplanted for each genotyping group. Blood samples were collected from the recipients every 4 weeks after BM transplantation. The donor blood chimerism was determined by staining peripheral blood samples with APC-anti-CD45.2 (BD Biosciences) and analyzed on a FACS Canto instrument (BD Biosciences).
For serial BM transplantation, 2,000 sorted LSKs were transplanted into sublethally irradiated (6 grays) primary recipients. At 8 weeks, a portion of the primary recipient mice was sacrificed, and CD45.2+ LSKs were sorted again and transplanted into lethally irradiated secondary recipients at 1,500 cells/mouse.
Cell Cycle and Apoptosis Analysis
To analyze the cell cycle status of HSCs, BM cells were stained with antibodies against Lin+ cells, c-Kit, Sca-1, CD150, and CD48, followed by fixation and permeabilization with transcription factor buffer set (BD Biosciences) according to the manufacturer's instructions. After fixation, the cells were incubated with APC-anti-Ki67 (Biolegend), washed, and stained with PI (BD Bioscience). The cells were analyzed by flow cytometry.
For apoptosis detection, BM cells were stained with the SLAM antibodies described above and then stained with APC-annexin V (BD Biosciences) and PI. Annexin V-positive cells were determined as apoptotic cells using the FACS LSR II (BD Biosciences).
In Vivo BrdU Incorporation Assay
Mice were injected intraperitoneally with a single dose of BrdU (Sigma-Aldrich; 1 mg/6 g of mouse weight) 48 h prior to sacrifice. BM cells were harvested and stained with biotin-conjugated anti-lineage antibody mixture followed by staining with a secondary Percp-cy5.5-anti-streptavidin, PE-cy7-anti-Sca-1, APC-cy7-anti-c-Kit, and FITC-anti-CD34 (eBiosciences) antibodies and then fixed and stained with APC-anti-BrdU antibody (BD Biosciences) using the Cytofix/Cytoperm kit (BD Biosciences), according to the manufacturer's instructions. Analysis was performed on a FACS LSRII (BD Biosciences).
RNA Isolation, Quantitative PCR, and Microarray Analysis
Total RNA from SLAM cells isolated from mice with the indicated genotypes was prepared with RNeasy kit (Qiagen) following the manufacturer's procedure. Reverse transcription was performed with random hexamers and Superscript II RT (Invitrogen) and was carried out at 42 °C for 60 min and stopped at 95 °C for 5 min. First strand cDNA was used for real time PCR using primers listed in Table 1. Samples were normalized to the level of β-actin mRNA, and the relative expression levels were determined by the standard curve method.
TABLE 1.
Primer sequence
| Ccnd1 | Forward: 5′-TTTCTTGTAGCGGCCTGTTGT-3′ |
| Reverse: 5′-CGGAACACTAGAACCTAACAG-3′ | |
| Atr | Forward: 5′-TGCGCTCTGCTAGAGCACGGT-3′ |
| Reverse: 5′-AGTGCTGGCTGGCTGTGCTG-3′ | |
| Mafb | Forward: 5′-CAACAGCTACCCACTAGCCA-3′ |
| Reverse: 5′-GGCGAGTTTCTCGCACTTGA-3′ | |
| Msc | Forward: 5′-GGAGGACCGCTACGAGGACA-3′ |
| Reverse: 5′-ACCCACAGAAGGCTATGCT-3′ | |
| Apoe | Forward: 5′-GCCGACATGGAGGATCTACG-3′ |
| Reverse: 5′-CTTGTACACAGCTAGGCGCT-3′ | |
| Cat | Forward: 5′-CACTGACGAGATGGCACACT-3′ |
| Reverse: 5′-TGTGGAGAATCGAACGGCAA-3′ | |
| Rpa3 | Forward: 5′-CACAGTATATCGACCGGCCC-3′ |
| Reverse: 5′-TTTCCTCGTCAAGTGGCTCC-3′ | |
| Prdx3 | Forward: 5′-TGTCGTCAAGCACCTGAGTG-3′ |
| Reverse: 5′-TTGGCTTGATCGTAGGGGAC-3′ | |
| Ercc5 | Forward: 5′-TGTCTCTTGCTAGTGCCGTG-3′ |
| Reverse: 5′-TGTGTTCTTGTCGTCGGAGG-3′ | |
| Pink1 | Forward: 5′-GATGTCGTCCTGAAGGGAGC-3′ |
| Reverse: 5′-TGGAGGAACCTGCCGAGATA-3′ | |
| Id1 | Forward: 5′-CTGCAGGCCCTAGCTGTTC-3′ |
| Reverse: 5′-TTTGCTCCGACAGACCAAGT-3′ | |
| Cdkn1b | Forward: 5′-AGAAGCACTGCCGGGATATG-3′ |
| Reverse: 5′-ACCTCCTGCCACTCGTATCT-3′ | |
| β-actin | Forward: 5′-CGGTTCCGATGCCCTGAGGCTCTT-3′ |
| Reverse: 5′-CGTCACACTTCATGATGGAATTGA-3′ |
For microarray analysis, cDNA was synthesized from total RNA and hydridized to Affymetrix mouse gene 2.0 ST array. The RNA quality and quantity assessment, probe preparation, labeling, and hybridization were carried out in the Cincinnati Children's Hospital Medical Center Affymetrix Core using standard procedures. Hybridization data were sequentially subjected to normalization, transformation, filtration, and functional classification. Data analysis was performed with Genespring GX11 (Agilent Technologies). Gene set enrichment analysis was performed using GSEA v2.0 software as described (25). The microarray data can be found in the Gene Expression Omnibus of NCBI through accession number GSE64215.
Lentiviral Vector Construction, Virus Production, and Transduction
The SF-LV-cDNA-eGFP lentiviral vector (26) was generously provided by Dr. Lenhand Rudolph (Institute of Molecular Medicine and Max-Planck-Research Department of Stem Cell Aging). The CA-FOXO3a cDNA carrying alanine substitutions at T32A, S253A, and S315A (20) was amplified (forward primer, 5′-ATTACCGGTATGGCAGAGGCACCGGCTTC-3′, and reverse primer, 5′-AAAGTTAACTCAGCCTGGCACCCAGC-3′) from the Addgene plasmid 8361 (Addgene) and inserted into SF-LV-cDNA-eGFP. The SF-LV-cDNA-mCherry lentiviral vector was created by replacing the IRES-eGFP cassette with an IRES-mCherry cassette, which was amplified from the Addgene plasmid 45766 (Addgene) using the following primer sets: forward primer, 5′-ATAAGAATGCGGCCGCCCCCTCTCCCTCCCCCCCCCCTAAC-3′, and reverse primer, 5′-GCGACGCGTGTTCTGCATTTACTTGTACAGCTCGTCCATG-3′. The Flag-tagged mouse Fancd2 cDNA was amplified by two-step PCR using primers (forward primer, 5′-TTGCACCGGTATGATTTCCAAAAGACGTCGGCTAGATTC-3′; reverse primer 1, 5′-AAATATGCGGCCGCTCAAGCGTAGTCGGGCACGTCGTAAGGGTAGCTGGTGCCGCCCAGGCTCTTGTCATCGTCATCCTTGTAATCGA-3′; and reverse primer 2, 5′-TGTCATCGTCATCCTTGTAATCGATATCATGATCTTTATAATCACCGTCATGGTCTTTGTAGTCGGATCCACTGGAGCTGTCGTCACTTTCATCACTGTCC-3′) from a mouse Fancd2 cDNA clone (provided by Dr. Markus Grompe from Oregon Health & Sciences University) and inserted into SF-LV-cDNA-mCherry empty vector. The K559R mutant form of mouse Fancd2 plasmid was created with the QuikChange site-directed mutagenesis kit (Agilent Technologies).
Lentivirus was produced in 293 T cells after transfection of 20 μg of cDNA plasmid, 15 μg of pCMVΔR8.91 helper plasmid, and 6 μg of pMD.G, using standard calcium phosphate transfection procedures (27). Medium was replaced with fresh medium 12 h after transfection. To harvest viral particles, supernatants were collected 48 h after transfection, filtered through 0.45-μm-pore size filters and concentrated by the PEG-itTM virus precipitation solution (System Biosciences) according to the manufacturer's protocol. Virus pellet was resuspended in sterile PBS and stored at −80 °C.
For lentivirus transduction, the sorted CD34−LSK cells or LSK cells were maintained in StemSpanTM serum-free expansion medium (Stemcell Technologies) with 50 ng/ml SCF (Peprotech) and 50 ng/ml TPO (Peprotech) for 24 h before transduction. Transduction medium consisted of concentrated lentivirus suspension diluted 1:30 and 6 μg/ml polybrene (Millipore). Lentivirus transduction was performed in round-bottomed 96-well plates, using 50-μl reaction volumes for 24 h at 37 °C. The cells were then resuspended in 100 μl and incubated for 3 days.
Immunofluorescent Analyses
CD34−LSK cells were sorted from mice with the indicated genotype and then cytospun on slides, fixed by 4% paraformaldehyde, permeabilized with blocking solution (1× PBS, 0.25% Triton X-100, 5% BSA), and subsequently processed for anti-FOXO3a monoclonal antibody (2497; Cell Signaling) or anti-pS473AKT antibody (05-1003; Millipore). Nuclei were visualized using DAPI (Invitrogen). Images were collected on a Nikon C2 confocal microscope.
Fluorescence intensity in the nuclear and cytoplasmic regions was quantified using ImageJ. Background-corrected cytoplasmic to nuclear ratios were calculated from mean fluorescence intensity measured within a small square or circular region of interest placed within the nucleus, cytoplasm, and outside of each cell. 20 cells/sample and 3–4 samples per group were analyzed. Quantitative fluorescence data were exported from ImageJ generated histograms into GraphPad software for further analysis and presentation.
Statistics
Statistical significance was assayed by Student's t test and one-way analysis of variance. The values are presented as means ± S.D. A p value of <0.05 was considered significant. Limiting dilution assay used a Poisson-based probability statistic to calculate frequencies through the use of serial dilutions.
RESULTS
Deletion of Fancd2 and Foxo3a Causes HSC Exhaustion
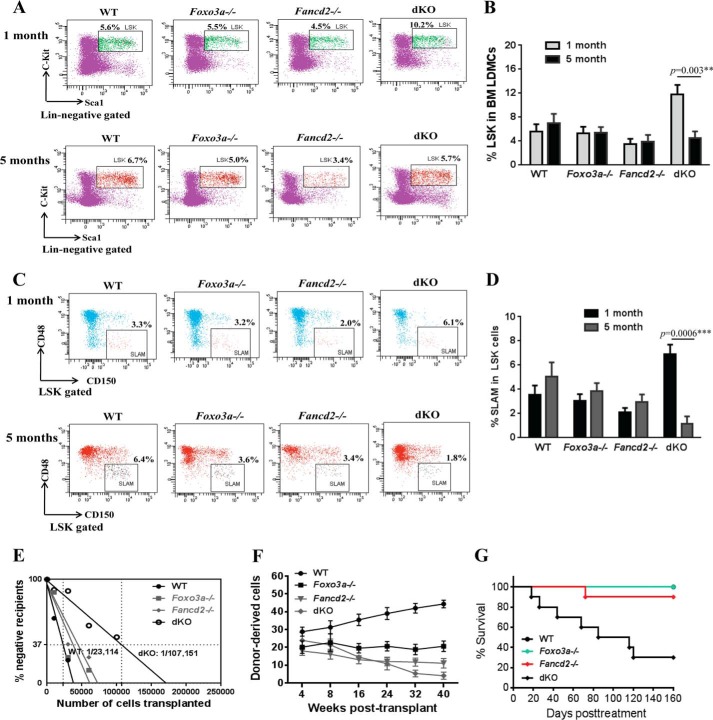
We previously reported an oxidative damage-specific interaction between FANCD2 and FOXO3a in human cells (22). To further investigate the genetic relationship between the two proteins, we generated Fancd2−/− Foxo3a−/− dKO mice. Because mice deficient for Fancd2 or Foxo3a show defects in HSC function (5, 21), we focused the effect of simultaneous loss of Foxo3a and Fancd2 on HSC maintenance. Surprisingly, deletion of both Fancd2 and Foxo3a in mice led to an initial expansion followed by a progressive decline of BM stem and progenitor cells. Specifically, at 1 month of age, dKO mice showed a significant increase in both progenitor (Lin− c-Kit+ Sca-1+ (LSK); Fig. 1, A and B) and HSCs (LSK CD150+ CD48−; SLAM; (28); Fig. 1, C and D). However, at 5 months of age, dKO BM stem and progenitor cells declined significantly (11.8 ± 1.6 at 1 month versus 4.53 ± 1.1 at 5 months for LSK; 6.9 ± 0.8 at 1 month versus 1.16 ± 0.60 at 5 months for SLAM; Fig. 1).
FIGURE 1.
Deletion of Fancd2 and Foxo3a causes HSC exhaustion. A, frequency of LSK (Lin− c-Kit+ Sca-1+) cells in the BM of WT, Foxo3a−/−, Fancd2−/−, and dKO mice at 1 and 5 months of age. Shown are representative flow plots. B, quantitation of BM LSK cells in WT, Foxo3a−/−, Fancd2−/−, and dKO mice at 1 and 5 months old. Each group comprises four to six mice. C, frequency of SLAM (LSK CD150+ CD48−) cells in BM LSK cells from WT, Foxo3a−/−, Fancd2−/−, and dKO mice at 1 and 5 months of age. Shown are representative flow plots. D, quantitation of BM SLAM cells in WT, Foxo3a−/−, Fancd2−/−, and dKO mice at 1 and 5 months old. Each group comprises four to six mice. E, competitive repopulating units determined by limiting dilution BM transplantation assay. Graded numbers of test BM cells (CD45.2+) were mixed with 2 × 105 protector BM cells (CD45.1+) and transplanted into irradiated congenic recipients (CD45.1+). Plotted are the percentages of recipient mice containing less than 1% CD45.2+ blood nucleated cells at 16 weeks after transplantation. Frequency of functional HSCs was calculated according to Poisson statistics. F, competitive repopulation assay. 50 SLAM test (CD45.2+) and 4 × 105 competitor (CD45.1+) whole BM cells were mixed and transplanted into irradiated CD45.1+ recipients. Donor-derived cells (CD45.2+) in the peripheral blood were determined at 4–40 weeks post-transplantation. The data are means ± S.E. (n = 8–10 from two independent experiments). G, Kaplan-Meier survival curves of secondary recipients (n = 8–10). 2,000 sorted LSKs from the indicated mice were transplanted into sublethally irradiated CD45.1+ mice, and 8 weeks later, the primary recipient mice were sacrificed, and CD45.2+ LSKs were sorted again and transplanted into lethally irradiated secondary recipients at 1,500 cells/mouse. The data shown are the survival rates expressed as a percentage. **, p < 0.01; ***, p < 0.001.
The results described above suggest that the dKO HSCs might have undergone replicative exhaustion. To test this notion, we determined the number of functional competitive repopulating units in aged (5 months old) dKO mice by performing limiting dilution BM transplantation assay. Graded numbers of test BM cells (CD45.2+) from WT, Fancd2−/−, Foxo3a−/−, or dKO mice were mixed with 2 × 105 protector BM cells (CD45.1+) and transplanted into lethally irradiated congenic recipients (CD45.1+). At 4 months after transplant, the frequency of functional competitive repopulating units dKO mice was 1 in 107,151 (p < 0.0001 versus WT), significantly lower than the frequencies in those of WT (1 in 23,114), Fancd2−/− (1 in 44,458; p = 0.0005 versus dKO), or Foxo3a−/− (1 in 37,428; p = 0.005 versus dKO) mice (Fig. 1E).
We next performed competitive repopulating experiments by transplanting 50 SLAM cells (CD45.2+) from WT, Fancd2−/−, Foxo3a−/−, or dKO mice, along with 4 × 105 recipient BM cells (CD45.1+) into lethally irradiated recipient mice (CD45.1+). We determined the repopulating ability of donor HSCs by periodically measuring the percentage of donor-derived cells in the peripheral blood of the transplant recipients. The percentage of donor-derived cells in the recipients of WT SLAM cells was steadily increased during the period of 4–40 weeks after transplantation (Fig. 1F). The donor chimerism in the peripheral blood of the recipients of Fancd2−/− or Foxo3a−/− cells was decreased 4 weeks after transplantation compared with that of recipients transplanted with the WT cells but remained relatively steady afterward. Significantly, donor chimerism in the recipients of dKO SLAM cells underwent progressive decline after 4 weeks post-transplantation (23.9 ± 2.58% at 4 weeks versus 14.5 ± 0.44% at 16 weeks and 4.07 ± 1.15% at 40 weeks; Fig. 1F). Furthermore, we performed serial transplantation to ascertain the phenomenon of dKO HSC exhaustion. As shown in Fig. 1G, 7 of 10 secondary recipients of dKO cells died within 4 months post-transplantation, whereas the majority of the recipients of WT, Fancd2−/−, or Foxo3a−/− cells survived beyond 160 days (Fig. 1G). Taken together, these results indicate that simultaneous loss of Foxo3a and Fancd2 results in HSC exhaustion.
Loss of Fancd2 and Foxo3a Increases Proliferation of HSCs
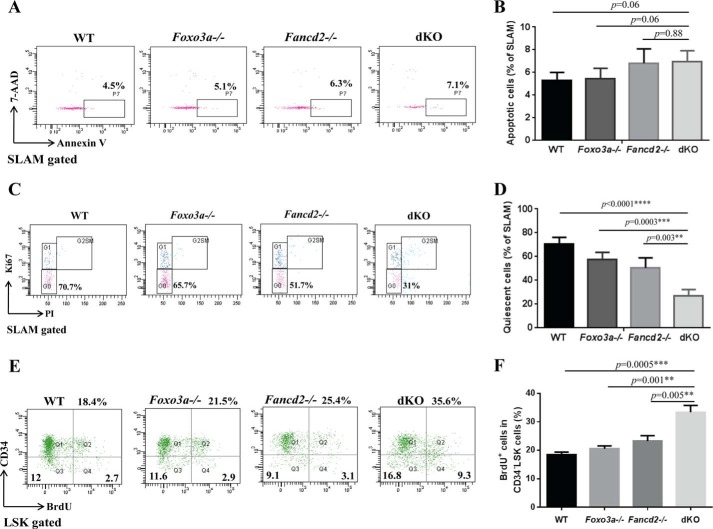
To determine the mechanism underlying dKO HSC exhaustion, we analyzed apoptosis and cell cycle of the phenotypic HSC (SLAM) cells from WT, Fancd2−/−, Foxo3a−/−, and dKO mice. We first examined whether increased apoptosis could be responsible for the observed HSC exhaustion in dKO mice. No increase in the frequency of annexin V-positive (apoptotic) cells was found in WT or Foxo3a−/− SLAM cells, although a moderate but statistically not significant increase in the apoptotic HSCs was detected in Fancd2−/− and dKO mice (Fig. 2, A and B). We next examined the cell cycle status of WT, Fancd2−/−, Foxo3a−/− and dKO SLAM cells using Ki67 and PI staining. Remarkably, we observed a significant decrease in the frequency of quiescent (G0) cells in dKO SLAM cells compared with that in WT, Fancd2−/−, or Foxo3a−/− SLAM cells (Fig. 2, C and D). Moreover, we observed a significant increase in the frequency of BrdU-positive cells in the CD34−LSK cells from dKO mice compared with those from WT, Fancd2−/−, or Foxo3a−/− mice, as detected by in vivo BrdU incorporation (Fig. 2, E and F). Together, these results indicate that Fancd2-Foxo3a deficiency leads to increased cycling in HSCs.
FIGURE 2.
Loss of Fancd2 and Foxo3a increases proliferation of HSCs. A, flow cytometric analysis of apoptotic cells within the phenotypic HSC (SLAM) population. BM cells of WT, Foxo3a−/−, Fancd2−/−, and Foxo3a−/− Fancd2−/− dKO mice were gated for the SLAM population and analyzed for annexin V-positive cells. 7-AAD, 7-aminoactinomycin D. B, quantification of annexin V-positive cells within the SLAM population. Each group comprises six mice. C, cell cycle analysis of SLAM cells. BM cells of WT, Foxo3a−/−, Fancd2−/−, and Foxo3a−/− Fancd2−/− dKO mice were gated for the SLAM population and analyzed for cell cycle phases by flow cytometry. Representative dot plots of DNA content (PI) were plotted versus Ki-67 staining. G0, Ki-67− and 2n DNA; G1, Ki-67+ and 2n DNA; G2SM, Ki67+ and DNA>2n. D, quantification of quiescent (G0) cells within the SLAM population. Each group comprised six mice. E, BrdUrd incorporation analysis of CD34−LSK cells. WT, Foxo3a−/−, Fancd2−/−, and Foxo3a−/− Fancd2−/− dKO mice at the age of 4–6 weeks were injected with a single dose of BrdUrd. 48 h later, the mice were sacrificed, and BM cells were analyzed for BrdUrd positive cells by flow cytometry. F, quantification of BrdUrd positive cells in the CD34−LSK cells. Each group comprises four to six mice. **, p < 0.01; ***, p < 0.001; ****, p < 0.0001.
The Transcriptome of dKO HSCs Implicates Perturbation of Multiple Pathways in HSC Function
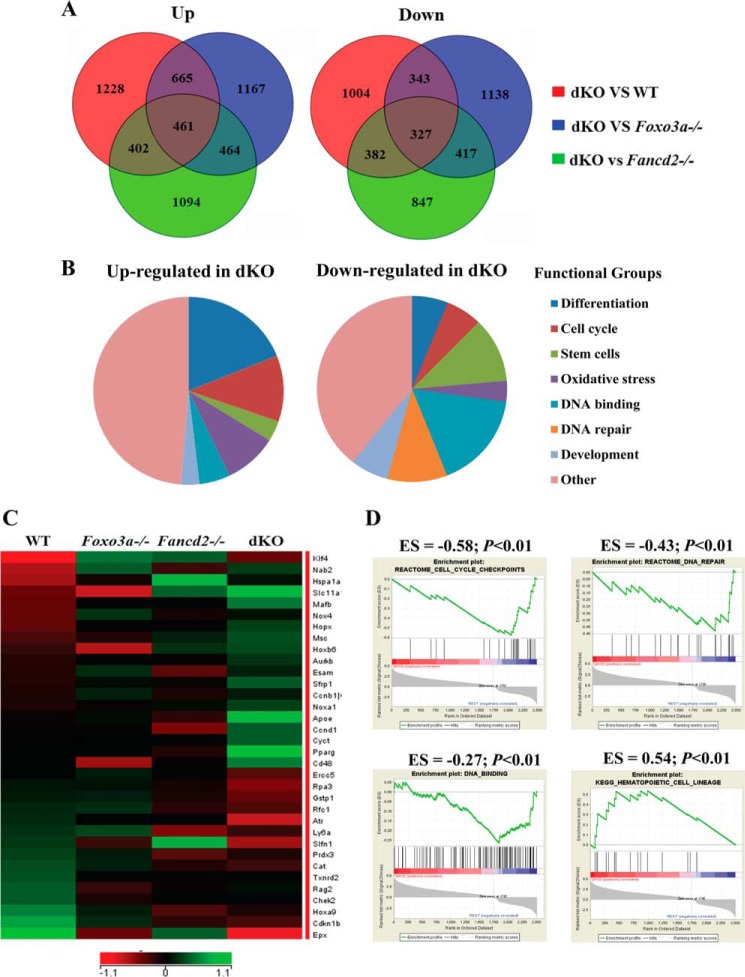
To identify the molecular mechanism underlying dKO HSC exhaustion, we performed global gene expression profiling analyses on phenotypic HSC (SLAM) cells freshly isolated from WT, Foxo3a−/−, Fancd2−/−, and dKO mice. Analysis of gene expression data revealed 788 unique differentially expressed genes, of which 461 were up-regulated and 327 down-regulated in dKO HSCs when compared with WT, Foxo3a−/−, and Fancd2−/− HSCs with fold changes larger than 1.3 (Fig. 3A). Microarray results were confirmed by quantitative RT-PCR for a subset of the genes (Table 2). Classification based on functional annotation revealed that a large proportion of differentially expressed genes in dKO HSCs belonged to cell cycle control (Ccnb1, Ccnd1, Aurkb, Atr, and Chek2) (Fig. 3, B and C), consistent with the observed increase in cycling HSCs. Another group of up-regulated genes was those involved in the differentiation of HSCs (Mafb, Msc, Apoe, and Hoxa9) (Fig. 3, B and C). Other differentially regulated transcripts included genes known to be involved in oxidative stress response (Cat, Txnrd2, and Prdx3) or to have roles in DNA repair (Ercc5, Rpa3, and Rfc1) (Fig. 3, B and C). In addition, Foxo3a target genes Cdkn1b, which is a cell cycle inhibitor protein, and Pink1, which encodes the PTEN-induced kinase 1, were down-regulated in the Fancd2 KO HSCs. In contrast, other Foxo3a target genes, such as Id1, that encodes the DNA-binding protein inhibitor ID-1, were up-regulated in Fancd2 KO HSCs (Table 2). These transcriptional changes suggest that multiple pathways might have been perturbed in HSC exhaustion caused by simultaneous loss of Foxo3a and Fancd2. To test this notion, we performed gene set enrichment analysis, which revealed a strong correlation of genes down-regulated in dKO SLAM cells with gene sets that identify cell cycle checkpoints, DNA repair, and DNA binding (Fig. 3D and Table 3). Conversely, the genes up-regulated in dKO HSCs strongly correlated with gene sets typical of hematopoietic lineage differentiation (Fig. 3D and Table 3).
FIGURE 3.
Global gene expression analysis of phenotypic HSCs. Whole genome microarray data were obtained from freshly isolated SLAM (LSK CD150+CD48−) cells from WT, Foxo3a−/−, Fancd2−/−, and dKO mice. A, Venn diagrams illustrating the overlap between genes up-regulated and down-regulated in WT, Foxo3a−/−, Fancd2−/−, and dKO SLAM cells. B, pie charts show the distribution of the 461 up-regulated and the 327 down-regulated genes in dKO SLAM cells into functional groups. C, heat map displays the expression of genes with cell cycle checkpoint, DNA repair, oxidative stress, and HSC differentiation-related functional annotations that are significantly down-regulated and up-regulated in dKO SLAM cells. The rows correspond to genes, and the columns correspond to samples. Gene expression values are indicated on a log2 scale according to the color scheme shown. Unregulated and down-regulated genes are presented in green and red, respectively. D, GSEA analyses are shown for gene sets identified for cell cycle checkpoints, DNA repair, DNA binding, and HSC differentiation pathways. For each GSEA, the p value and enrichment score (ES) are shown above each pathway graph.
TABLE 2.
Quantitative RT-PCR validation
| Gene name | Fold change |
||
|---|---|---|---|
| Foxo3a vs. WT | Fancd2 vs. WT | dKO vs. WT | |
| Ccnd1 | 0.89 ± 0.25 | 1.33 ± 0.1 | 1.84 ± 0.2 |
| Atr | 0.83 ± 0.08 | 0.97 ± 0.13 | 0.37 ± 0.14 |
| Mafb | 1.26 ± 0.11 | 1.25 ± 0.12 | 2.34 ± 0.58 |
| Msc | 1.29 ± 0.08 | 1.06 ± 0.08 | 1.82 ± 0.33 |
| Apoe | 1.07 ± 0.16 | 1.12 ± 0.16 | 2.58 ± 0.2 |
| Cat | 0.74 ± 0.08 | 0.95 ± 0.07 | 0.52 ± 0.13 |
| Rpa3 | 0.86 ± 0.04 | 1.08 ± 0.07 | 0.52 ± 0.13 |
| Prdx3 | 0.72 ± 0.07 | 0.92 ± 0.03 | 0.63 ± 0.14 |
| Ercc5 | 0.99 ± 0.09 | 1.01 ± 0.12 | 0.66 ± 0.1 |
| Pink1 | 0.64 ± 0.14 | 0.66 ± 0.13 | 0.58 ± 0.21 |
| Cdkn1b | 0.6 ± 0.07 | 0.62 ± 0.11 | 0.46 ± 0.01 |
| Id1 | 1.47 ± 0.04 | 1.40 ± 0.11 | 1.52 ± 0.19 |
TABLE 3.
Gene sets with strong correlation to genes down-regulated and up-regulated in dKO HSCs
AML, acute myeloid leukemia.
| Gene set name | Gene set description | P value |
|---|---|---|
| Upregulated genes | ||
| Cell differentiation | ||
| KEGG_HEMATOPOIETIC_CELL_LINEAGE | Hematopoietic cell lineage | 2.28 e−4 |
| MATURE_B_LYMPHOCYTE_UP | Up-regulated genes in the B lymphocyte developmental signature | 5.44 e−9 |
| HEMATOPOIESIS_MATURE_CELL | Up-regulated in mature blood cell populations from adult bone marrow and fetal liver | 4.69 e−8 |
| DENDRITIC_CELL_MATURATION_UP | Genes up-regulated during in vitro maturation of CD14 + monocytes (day 0) into immature and mature dendritic cells | 6.18 e−7 |
| HEMATOPOIESIS_LATE_PROGENITOR | Up-regulated in hematopoietic late progenitor cells from adult bone marrow and fetal liver | 3.8 e−7 |
| REGULATION_OF_CELL_DIFFERENTIATION | Any process that modulates the frequency, rate, or extent of cell differentiation | 5.37 e−4 |
| Cell cycle | ||
| CELL_CYCLE | Occur in a cell during successive cell replication or nuclear replication events | 3.72 e−3 |
| POSITIVE_REGULATION_OF_CELL_CYCLE | Any process that activates or increases the rate or extent of progression through the cell cycle | 4.34 e−3 |
| Downregulated genes | ||
| Stem cell sets | ||
| LIM_MAMMARY_STEM_CELL_UP | Genes consistently up-regulated in mammary stem cells both in mouse and human species | 6.98 e−6 |
| JAATINEN_HEMATOPOIETIC_STEM_CELL_DN | Genes down-regulated in CD133+ cells (HSCs) compared to the CD133− cells | 4.87 e−7 |
| IVANOVA_HEMATOPOIESIS_LATE_PROGENITOR | Genes in the expression cluster up-regulated in hematopoietic late progenitor cells from adult bone marrow and fetal liver | 3.24 e−6 |
| GAL_LEUKEMIC_STEM_CELL | Genes down-regulated in leukemic stem cells from AML | 8.53 e−8 |
| WONG_ADULT_TISSUE_STEM_MODULE | Genes coordinately up-regulated in a compendium of adult tissue stem cells | 1.55 e−9 |
| BYSTRYKH_HEMATOPOIESIS_STEM_CELL_QTL | Transcripts in HSCs that are transregulated | 3.41 e−8 |
| DNA damage control | ||
| REACTOME_DNA_REPAIR | Genes involved in DNA repair | 1.07 e−4 |
| REACTOME_REPAIR_SYNTHESIS_FOR_GAP_FILL | Genes involved in repair synthesis for gap filling by DNA polymerase | 2.45 e−5 |
| REACTOME_NUCLEOTIDE_EXCISION_REPAIR | Genes involved in nucleotide excision repair | 6.08 e−5 |
| DNA binding | ||
| DNA_BINDING | Interacting selectively with DNA | 1.58 e−6 |
| TRANSCRIPTION_FACTOR_ACTIVITY | The function of binding to a specific DNA sequence to modulate transcription | 1.23 e−4 |
| TRANSCRIPTION_COFACTOR_ACTIVITY | The function that links a sequence-specific transcription factor | 3.95 e−4 |
| TRANSCRIPTION_FACTOR_BINDING | Interacting selectively with a transcription factor | 1.83 e−3 |
| Protein complex | ||
| REGULATION_OF_GENE_EXPRESSION | Any process that modulates the frequency, rate, or extent of gene expression | 2.07 e−3 |
| PROTEIN_COMPLEX | Any protein group composed of two or more subunits, which may or may not be identical | 1.66 e−4 |
| HISTONE_DEACETYLASE_COMPLEX | Complex that possesses histone deacetylase activity | 3.08 e−3 |
| TRANSCRIPTION_FACTOR_COMPLEX | Any complex, distinct from RNA polymerase, including one or more polypeptides capable of binding DNA at promoters regulatory sequences, and regulating transcription | 6.09 e−3 |
| PROTEIN_DNA_COMPLEX_ASSEMBLY | The aggregation, arrangement, and bonding together of proteins and DNA molecules to form a protein-DNA complex | 1.11 e−3 |
Fancd2 Is Required for Nuclear Localization of Foxo3a in HSCs
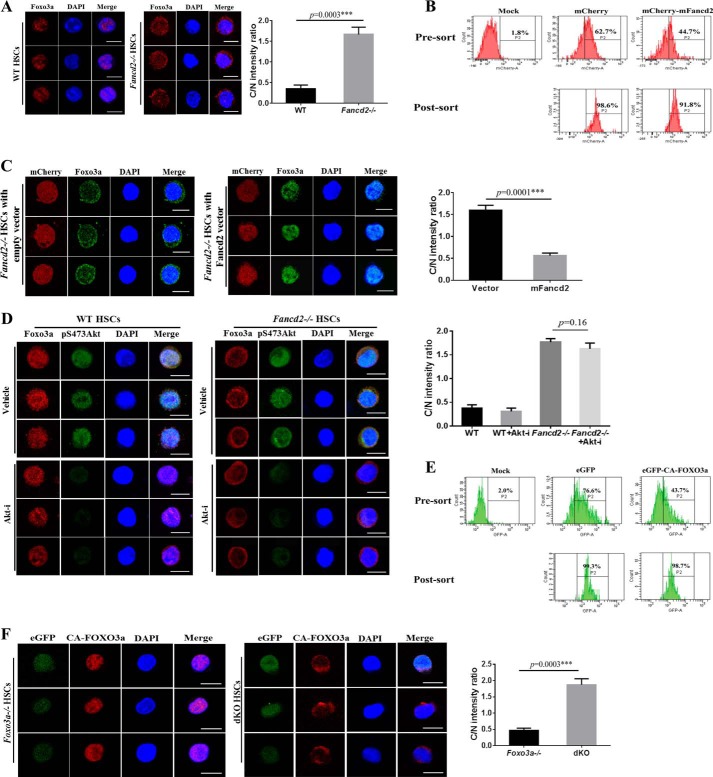
To understand how simultaneous loss of Fancd2 and Foxo3a affects transcriptional program in HSCs, we asked whether Fancd2 deficiency affected nuclear localization of Foxo3a in HSCs. We isolated BM CD34−LSK cells from the WT and Fancd2−/− mice and examined the distribution of Foxo3a by immunofluorescence. The majority of Foxo3a was present in the nucleus of WT CD34−LSK cells (Fig. 4A), consistent with a previous report (21). However, we observed increased cytoplasmic Foxo3a staining in Fancd2−/− CD34−LSK cells (Fig. 4A). To address whether this abnormal cytoplasmic localization of Foxo3a was due to the loss of Fancd2, we performed a rescue experiment with genetically corrected Fancd2−/− CD34−LSK cells by lentivirus expressing the WT mouse Fancd2 gene (Fig. 4B). It appeared that re-expression of the mouse Fancd2 could completely restore nuclear Foxo3a localization (Fig. 4C). These data suggest that a functional Fancd2 may be required for nuclear localization of Foxo3a in HSCs.
FIGURE 4.
Fancd2 is required for nuclear localization of Foxo3a in HSCs. A, increased cytoplasmic Foxo3a staining in Fancd2−/− HSCs. Left panel, freshly isolated CD34−LSK cells from WT and Fancd2−/− BM were immunostained to detect Foxo3a (red). Nuclei were visualized using DAPI (blue). Scale bar, 10 μm. Right panel, ratio of fluorescence intensity of anti-Foxo3a staining in cytoplasm (C) and the nucleus (N). B, flow cytometry of mCherry-positive cells before and after sorting. Mock, samples without virus. C, re-expression of Fancd2 restores Foxo3a nuclear localization. Left panel, mFancd2-mCherry or empty-mCherry lentivirus transduced Fancd2−/− CD34−LSK cells were stained with anti-Foxo3a antibody (green) and DAPI (blue). Scale bar, 10 μm. Right panel, ratio of fluorescence intensity of anti-Foxo3a staining in cytoplasm (C) and the nucleus (N). D, enhanced cytoplasmic localization of Foxo3a in Fancd2−/− HSCs is independent of Akt activation. Left panel, CD34−LSK cells from the WT and Fancd2−/− BM were treated with AKT inhibitor (5 μm) for 2 h. Foxo3a, pS473AKT, and nuclear DNA were visualized by red, green, and blue, respectively. Scale bar, 10 μm. Right panel, quantification of the fluorescence intensity of anti-Foxo3a staining in cytoplasm (C) and the nucleus (N). E, flow cytometry of GFP-positive cells before and after sorting. Mock, samples without virus. F, Fancd2 is required for nuclear retention of the constitutively active CA-FOXO3a in HSCs. Left panel, Foxo3a−/− and Fancd2−/− Foxo3a−/− dKO CD34−LSK cells were transduced with lentivirus expressing the active CA-FOXO3a and eGFP (green). Transduced CD34−LSK cells were staining by anti-FOXO3a antibody (red). Nuclei were visualized by DAPI (blue). Scale bar, 10 μm. Right panel, quantification of the fluorescence intensity of anti-FOXO3a staining in the nucleus (N) and cytoplasm (C). Each group comprises three to four mice and 20 cells per sample. Akt-i, Akt inhibitor; ***, p < 0.001.
It has been shown that FOXO3a nuclear localization and thus transcriptional activity is regulated by PI3K-mediated activation of AKT, which directly phosphorylates FOXO3a, leading to its nuclear export and inactivation (20). To exclude the possibility that increased cytoplasmic localization of Foxo3a in Fancd2−/− HSCs was a phenomenon secondary to AKT activation, we treated the WT and Fancd2−/− CD34−LSK cells with the AKT inhibitor (124005; Calbiochem; 5 μm) for 2 h and examined the phosphorylated AKT (pS473AKT) and Foxo3a subcellular localization. Treatment of WT CD34−LSK cells with the AKT inhibitor showed increased nuclear Foxo3a compared with vehicle controls (Fig. 4D). However, we did not observe a significant increase in nuclear localization of Foxo3a in Fancd2−/− CD34−LSK cells treated with the AKT inhibitor (Fig. 4D). Thus, increased cytoplasmic Foxo3a in Fancd2−/− HSCs is not driven by Akt-mediated phosphorylation.
A Role of Fancd2 in the Retention of Nuclear Foxo3a in HSCs
The observation that Fancd2 deficiency reduced Foxo3a in the nucleus of HSCs prompted us to investigate the status of an active form of FOXO3a (CA-FOXO3a), which is constitutively active because it cannot be inactivated by phosphorylation and thus is resistant to nuclear export (20), in Fancd2−/− HSCs. We transduced Foxo3a−/− and Fancd2−/−Foxo3a−/− dKO CD34−LSK cells with lentivirus expressing the active CA-FOXO3a and eGFP (Fig. 4E) and examined the subcellular localization of CA-FOXO3a in Foxo3a null cells with (Foxo3a−/−) or without (Fancd2−/− Foxo3a−/−) a Fancd2 gene. The transgene-encoded CA-FOXO3a was almost exclusively localized in the nucleus of the Fancd2-sufficient Foxo3a−/− CD34−LSK cells (Fig. 4F). However, Fancd2 deficiency strongly promoted cytoplasmic localization of CA-FOXO3a in the Foxo3a−/−Fancd2−/− CD34−LSK cells (Fig. 4F). Thus, it appears that Fancd2 plays a role in nuclear retention of the constitutively active CA-FOXO3a in HSCs.
Monoubiquitination Is Required for the Role of Fancd2 in FOXO3a Nuclear Retention
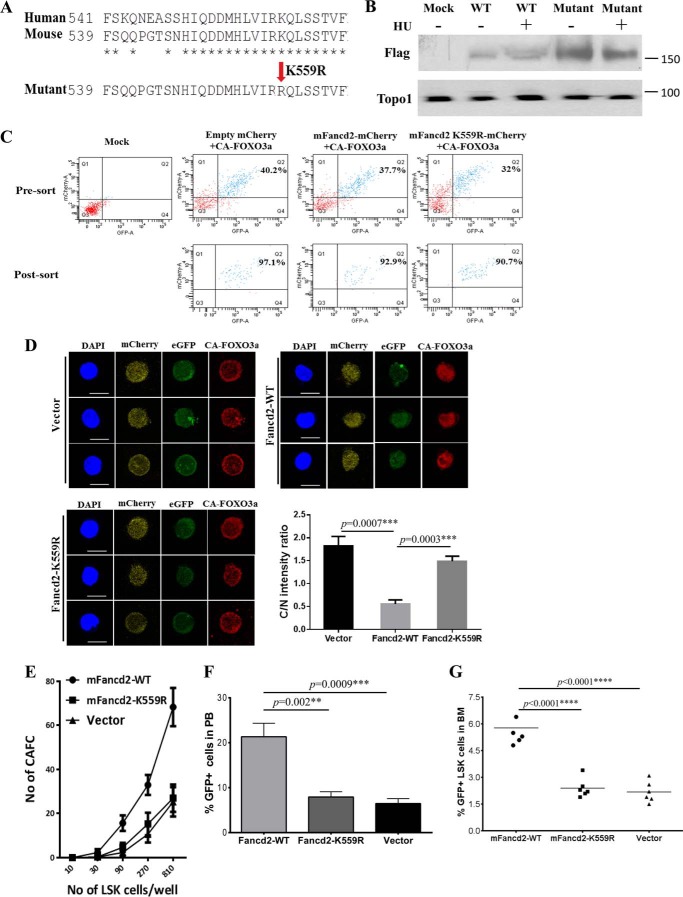
Monoubiquitination of human FANCD2 at Lys-561 is a critical step in the FA DNA repair pathway (29, 30). To determine whether FANCD2 monoubiquitination was required for FOXO3a nuclear retention, we identified the monoubiquitination site (Lys-559) in mouse Fancd2 and generated a mutant that had an arginine residue substituted for the lysine residue at position 559 (K559R; Fig. 5A). We confirmed the loss of monoubiquitination of the mutant Fancd2 in response to replicative stress induced by hydrourea (Fig. 5B). We co-transduced the dKO LSK cells with lentivirus expressing the constitutively active CA-FOXO3a with an eGFP marker (CA-FOXO3a-eGFP) and the WT mouse Fancd2 with a mCherry marker (mFancd2 WT-mCherry) or its nonmonoubiquitinated mutant (mFancd2 K559R-mCherry) and sorted for double-positive (eGFP+ mCherry+) CD34−LSK cells (Fig. 5C). Immunofluorescence analysis showed that expression of WT mFancd2 strongly promoted CA-FOXO3a localization in the nucleus (Fig. 5D). In contrast, co-transduction of CA-FOXO3a and the nonmonoubiquitinated mutant mFancd2 K559R or vector alone led to increased cytoplasmic localization of CA-FOXO3a in dKO CD34−LSK cells (Fig. 5D). Thus, monoubiquitination is essential for the effect of Fancd2 on FOXO3a nuclear retention.
FIGURE 5.
Fancd2 and Foxo3a cooperate to maintain HSC function. A, protein sequence alignment of human and mouse Fancd2 and generation of a mutant mFancd2 that had arginine residue substituted for the lysine residue at position 559. B, WT murine embryonic fibroblast cells were transduced with lentivirus carrying empty vector, mFancd2-WT, or mFancd2-K559R and then treated with or without hydrourea (HU) for 24 h. Cell lysis were separated by SDS-PAGE gel and immunoblotted with antibodies against Flag (mFancd2) and Topo1 (loading control). C, the dKO LSK cells were co-transduced with lentivirus expressing the CA-FOXO3a-eGFP and mFancd2 WT-mCherry or mFancd2 K559R-mCherry and then sorted for double-positive (eGFP+ mcherry+) CD34−LSK cells. Shown is flow cytometry analysis of the double-positive cells before and after sorting. Mock, samples without virus. D, monoubiquitination is essential for the effect of Fancd2 on FOXO3a nuclear retention. dKO LSK cells were co-transduced with lentivirus expressing the constitutively active CA-FOXO3a with an eGFP marker (CA-FOXO3a-eGFP) (green) and the WT mouse Fancd2 with a mCherry marker (mFancd2 WT-mCherry) or its nonmonoubiquitinated mutant (mFancd2 K559R-mCherry) (yellow). Transduced CD34−LSK cells were staining by anti-FOXO3a antibody (red). Nuclei were visualized by DAPI (blue). Scale bar, 10 μm. Right panel, quantification of the fluorescence intensity of anti-Foxo3a staining in the nucleus (N) and cytoplasm (C). Each group comprises three to four mice, and 20 cells per sample. E, limiting dilution CAFC assay. dKO LSK cells were co-transduced with lentivirus expressing the WT FOXO3a with an eGFP marker and the WT mouse Fancd2 with a mCherry marker (mFancd2-WT) or its nonmonoubiquitinated mutant (mFancd2-K559R) or an empty vector. Graded numbers of double-positive (eGFP+ mcherry+) LSK cells were plated on confluent OP9 stromal cells in 96-well plates, and numbers of CAFC were counted after 4 weeks. F and G, BM transplantation assay to determine the long term hematopoietic repopulating ability. 1,000 double-positive (eGFP+ mCherry+) LSK cells (CD45.2) co-transduced with FOXO3a-eGFP and mFancd2 WT-mCherry or mFancd2 K559R-mCherry or empty-mCherry lentivirus, along with 4 × 105 recipient BM cells (CD45.1) were transplanted into lethally irradiated recipient mice. The repopulating capacity of donor HSCs was monitored by measuring percentage of GFP positive cells in the peripheral blood of the transplant recipients at 4 months post-transplantation (F). The percentage of GFP-positive LSK cells in the BM of the transplant recipients was determined at 4 months post-transplantation (G). **, p < 0.01; ***, p < 0.001; ****, p < 0.0001.
Fancd2 and Foxo3a Cooperate to Maintain HSC Function
To evaluate the functional consequence of FANCD2 in FOXO3a nuclear retention, we carried out two sets of experiments to examine whether Fancd2 and Foxo3a cooperate to maintain HSC function. For in vitro experiments, we used limiting dilution CAFC assay (24) to evaluate the self-renewal capacity of the dKO LSK cells co-expressing the WT FOXO3a and the WT or mutant Fancd2. Graded numbers of double-positive (eGFP+ mcherry+) LSK cells were plated on confluent OP9 stromal cells in 96-well plates, and numbers of CAFC were counted after 4 weeks. There was a significant decrease in CAFC colonies formed by dKO LSK cells co-expressing the mutant Fancd2-K559R or vector alone and FOXO3a in wells plated with 90, 270, and 810 LSK cells compared with those co-expressing the WT Fancd2 and FOXO3a (Fig. 5E).
For in vivo experiments, we performed competitive BM transplantation assay to determine the long term hematopoietic repopulating ability of the test dKO LSK cells in lethally irradiated recipients. 1,000 double-positive (eGFP+ mcherry+)LSK cells (CD45.2+) co-transduced with FOXO3a-eGFP and mFancd2 WT-mCherry or mFancd2 K559R-mCherry lentivirus, along with 4 × 105 recipient BM cells (CD45.1+), were transplanted into lethally irradiated recipient mice. The repopulating capacity of donor HSCs was monitored by measuring donor-derived cells in the peripheral blood of the transplant recipients at 4 months post-transplantation. We observed significantly higher donor chimerism in the recipients of dKO LSK cells co-expressing Fancd2-WT and FOXO3a than those with cells co-expressing the mutant Fancd2-K559R or vector alone and FOXO3a (Fig. 5F). Furthermore, there was a concomitant decrease in donor stem and progenitor (LSK) cells in the BM of the recipients transplanted with cells co-expressing FOXO3a and the mutant Fancd2-K559R or empty vector at 4 months after transplantation (Fig. 5G). These results strongly argue a functional interaction between Fand2 and nuclear Foxo3a in HSC maintenance.
DISCUSSION
In this study, we demonstrated that deletion of both Fancd2 and Foxo3a in mice induced HSC exhaustion. Increased HSC cycling may be one of the underlying mechanisms. In supporting this notion, we provided several pieces of evidence: (i) cell cycle analysis showed a significant decrease in quiescent HSCs in dKO mice compared with not only WT but also Fancd2−/− or Foxo3a−/− mice (Fig. 2, C and D); (ii) in vivo BrdU incorporation assay revealed a marked increase in proliferation in dKO CD34−LSK cells compared with those from mice with other three genotypes (Fig. 2, E and F); (iii) global gene expression profiling on phenotypic HSC (SLAM) cells exhibited a deregulated cell cycle-associated transcriptional program that was unique to dKO HSCs (Fig. 3, B and C); and (iv) functionally, we demonstrated that a functional Fancd2 was required for nuclear retention of FOXO3a and for HSC maintenance (Figs. 4 and 5). These data suggest that a functional interaction between the FANCD2 and FOXO3a may be a novel mechanism by which HSCs maintain quiescence and avoid replicative exhaustion.
It has been reported that Foxo3a is not crucial for HSC differentiation but plays a role in HSC self-renewal (21). Deletion of Foxo3a impairs hematopoietic repopulating capacity of the HSCs, which is not caused by replicative exhaustion but at least in part by increased cellular accumulation of reactive oxygen species (21). FANCD2 plays critical roles in DNA damage repair of double-stranded breaks (31, 32). Long-lived quiescent HSCs may be particularly susceptible to the accumulation of double-stranded breaks over time (33). Mice deficient for key DNA repair pathway components exhibit diminished HSC self-renewal capacity with age and early functional exhaustion (34, 35). In this context, we showed that in Fancd2-deficient BM, the content of phenotypic HSCs was reduced, and their long term repopulating activity was impaired compared with WT HSCs (Fig. 1, C, D, and F), which is consistent with previously study (5). However, deletion of Fancd2 alone did not induce the phenotype typical of HSC exhaustion, as observed in dKO mice that showed an initial expansion followed by a progressive decline of both phenotypic HSCs and their long term repopulating activity (Fig. 1, C, D, and F). These results support the notion that Fancd2 and Foxo3a cooperate to prevent HSC exhaustion.
In quiescent WT murine HSCs, the majority of the Foxo3a protein is localized in the nucleus (36). It is also known that this nuclear localization is regulated by of Akt, which phosphorylates Foxo3a and induces the export of Foxo3a from the nucleus to the cytoplasm (20). We showed that there was a marked increase in cytoplasmic Foxo3a accompanied by reduced nuclear Foxo3a in phenotypic Fancd2−/− HSCs (Fig. 4A) and that this phenomenon was Akt-independent (Fig. 4D), suggesting a role for Fancd2 in nuclear Foxo3a retention. Indeed, genetic correction of the Fancd2−/− HSCs with a WT mouse Fancd2 completely rescued Foxo3a nuclear retention (Fig. 4C). Moreover, by co-expressing the constitutively active CA-FOXO3a and the nonmonoubiquitinated mutant mFancd2 K559R in dKO CD34−LSK cells, we showed that monoubiquitination is essential for the effect of Fancd2 on FOXO3a nuclear retention (Fig. 5D). This is consistent with previous observation that the nonmonoubiquitinated FANCD2 mutant failed to co-localize with FOXO3a on chromatin in response to oxidative DNA damage (22).
The mechanism responsible for FOXO3a sequestration in cytoplasm of the Fancd2−/− cells is not clear at this time. We speculate two possibilities. First, we previously showed that FOXO3a and FANCD2 associated as a nuclear complex and co-localized on chromatin DNA (22). In this context, FANCD2 might play a role in stabilizing FOXO3a in the nucleus. Consequently in Fancd2−/− HSCs, the loss of Fancd2 might lead to increased cytoplasmic CA-FOXO3a. Second, sequestration of CA-FOXO3a in cytoplasm might be caused by abnormal activation of ERK in Fancd2−/− HSCs. It is known that ERK regulates FOXO3a nuclear-cytoplasmic localization (37). ERK interacts with and phosphorylates FOXO3a predominantly at residues Ser-294, Ser-344, and Ser-425, which are different from the Akt phosphorylation sites at Thr-32, Ser-253, and Ser-315, leading to FOXO3a nuclear exportation. Several groups have reported constitutive activation of ERK in FA-deficient cells (38, 39). Thus, the constitutively activated Erk could act on the non-Akt phosphorylation sites leading to increased cytoplasmic localization of CA-FOXO3a in the Fancd2−/− HSCs.
In two well established assays designed to evaluate self-renewal (limiting dilution CAFC assay) and hematopoietic repopulating (competitive BM transplantation assay) ability of a HSC, we observed a significant decrease in both CAFC formation and long term repopulation by dKO LSK cells co-expressing the nonmonoubiquitinated Fancd2-K559R mutant and the WT FOXO3a compared with dKO LSK cells co-expressing the WT Fancd2 and FOXO3a (Fig. 5, E and F). Thus, it is tempting to speculate that a functional interaction between Fancd2 and FOXO3a is required for HSC maintenance. Moreover, that these assays (flow cytometry, limiting dilution CAFC assay, and competitive BM transplantation assay) observed an initial expansion followed by a progressive decline of both phenotypic HSCs and that their long term repopulating activity requires simultaneous inactivation of Foxo3a and Fancd2 indicate that the HSC exhaustion phenotype results from a synergistic effect. In this context, we speculate a coordinate regulation of HSC functions by the FOXO3 and FA pathways. Indeed, Foxo3a or Fancd2 single-knockout mice did not experience HSC exhaustion, as judged by progressive decline in HSC pool and HSC self-renewal capacity, suggesting that the defect in dKO HSCs was not simply additive but synergistic.
In summary, the present studies demonstrate that simultaneous inactivation of Fancd2 and Foxo3a in mice induces HSC exhaustion and identify increased HSC cycling as one of the underlying mechanisms for the defect. These findings reveal functional interaction between the FA DNA repair pathway and the FOXO3a stress response pathway in HSC maintenance and may suggest new targets for therapeutically exploring the pathogenic role of stress-induced HSC exhaustion in blood diseases.
Acknowledgments
We thank Dr. Markus Grompe (Oregon Health & Sciences University) for Fancd2+/− mice and the mouse Fancd2 cDNA and the Comprehensive Mouse and Cancer Core of the Cincinnati Children's Research Foundation (Cincinnati Children's Hospital Medical Center) for bone marrow transplantation service.
This work was supported, in whole or in part, by National Institutes of Health Grants R01 HL076712, R01 CA157537, and T32 HL091805.
- HSC
- hematopoietic stem cell
- BM
- bone marrow
- CAFC
- cobblestone area-forming cell
- dKO
- double-knockout
- FA
- Fanconi anemia.
REFERENCES
- 1. Wilson A., Trumpp A. (2006) Bone-marrow hematopoietic-stem-cell niches. Nat. Rev. Immunol. 6, 93–106 [DOI] [PubMed] [Google Scholar]
- 2. Orford K. W., Scadden D. T. (2008) Deconstructing stem cell self-renewal: genetic insights into cell-cycle regulation. Nat. Rev. Genet. 9, 115–128 [DOI] [PubMed] [Google Scholar]
- 3. Orkin S. H., Zon L. I. (2008) Hematopoiesis: an evolving paradigm for stem cell biology. Cell 132, 631–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wilson A., Laurenti E., Oser G., van der Wath R. C., Blanco-Bose W., Jaworski M., Offner S., Dunant C. F., Eshkind L., Bockamp E., Lió P., Macdonald H. R., Trumpp A. (2008) Hematopoietic stem cells reversibly switch from dormancy to self-renewal during homeostasis and repair. Cell 135, 1118–1129 [DOI] [PubMed] [Google Scholar]
- 5. Parmar K., Kim J., Sykes S. M., Shimamura A., Stuckert P., Zhu K., Hamilton A., Deloach M. K., Kutok J. L., Akashi K., Gilliland D. G., D'Andrea A. (2010) Hematopoietic stem cell defects in mice with deficiency of Fancd2 or Usp1. Stem Cells 28, 1186–1195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Yahata T., Takanashi T., Muguruma Y., Ibrahim A. A., Matsuzawa H., Uno T., Sheng Y., Onizuka M., Ito M., Kato S., Ando K. (2011) Accumulation of oxidative DNA damage restricts the self-renewal capacity of human hematopoietic stem cells. Blood 118, 2941–2950 [DOI] [PubMed] [Google Scholar]
- 7. Ito K., Hirao A., Arai F., Matsuoka S., Takubo K., Hamaguchi I., Nomiyama K., Hosokawa K., Sakurada K., Nakagata N., Ikeda Y., Mak T. W., Suda T. (2004)) Regulation of oxidative stress by ATM is required for self-renewal of haematopoietic stem cells. Nature 431, 997–1002 [DOI] [PubMed] [Google Scholar]
- 8. Bagby G. C., Jr. (2003) Genetic basis of Fanconi anemia. Curr. Opin. Hematol. 10, 68–76 [DOI] [PubMed] [Google Scholar]
- 9. D'Andrea A. D. (2010) Susceptibility pathways in Fanconi's anemia and breast cancer. N. Engl. J. Med. 362, 1909–1919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tischkowitz M., Dokal I. (2004) Fanconi anaemia and leukaemia: clinical and molecular aspects. Br. J. Haematol. 126, 176–191 [DOI] [PubMed] [Google Scholar]
- 11. Wilson D. B., Link D. C., Mason P. J., Bessler M. (2014) Inherited bone marrow failure syndromes in adolescents and young adults. Ann. Med. 46, 353–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kee Y., D'Andrea A. D. (2012) Molecular pathogenesis and clinical management of Fanconi anemia. J. Clin. Invest. 122, 3799–3806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Walden H., Deans A. J. (2014) The Fanconi anemia DNA repair pathway: structural and functional insights into a complex disorder. Annu. Rev. Biophys. 43, 257–278 [DOI] [PubMed] [Google Scholar]
- 14. Deans A. J., West S. C. (2011) DNA interstrand crosslink repair and cancer. Nat. Rev. Cancer 11, 467–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kottemann M. C., Smogorzewska A. (2013) Fanconi anaemia and the repair of Watson and Crick DNA crosslinks. Nature 493, 356–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kim H., D'Andrea A. D. (2012) Regulation of DNA cross-link repair by the Fanconi anemia/BRCA pathway. Genes Dev. 26, 1393–1408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kalb R., Neveling K., Hoehn H., Schneider H., Linka Y., Batish S. D., Hunt C., Berwick M., Callen E., Surralles J., Casado J. A., Bueren J., Dasi A., Soulier J., Gluckman E., Zwaan C. M., van Spaendonk R., Pals G., de Winter J. P., Joenje H., Grompe M., Auerbach A. D., Hanenberg H., Schindler D. (2007) Hypomorphic mutations in the gene encoding a key Fanconi anemia protein, FANCD2, sustain a significant group of FA-D2 patients with severe phenotype. Am. J. Hum. Genet. 80, 895–910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Castrillon D. H., Miao L., Kollipara R., Horner J. W., DePinho R. A. (2003) Suppression of ovarian follicle activation in mice by the transcription factor Foxo3a. Science 301, 215–218 [DOI] [PubMed] [Google Scholar]
- 19. Marinkovic D., Zhang X., Yalcin S., Luciano J. P., Brugnara C., Huber T., Ghaffari S. (2007) Foxo3 is required for the regulation of oxidative stress in erythropoiesis. J. Clin. Invest. 117, 2133–2144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Brunet A., Bonni A., Zigmond M. J., Lin M. Z., Juo P., Hu L. S., Anderson M. J., Arden K. C., Blenis J., Greenberg M. E. (1999) Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell 96, 857–868 [DOI] [PubMed] [Google Scholar]
- 21. Miyamoto K., Araki K. Y., Naka K., Arai F., Takubo K., Yamazaki S., Matsuoka S., Miyamoto T., Ito K., Ohmura M., Chen C., Hosokawa K., Nakauchi H., Nakayama K., Nakayama K. I., Harada M., Motoyama N., Suda T., Hirao A. (2007) Foxo3a is essential for maintenance of the hematopoietic stem cell pool. Cell Stem Cell 1, 101–112 [DOI] [PubMed] [Google Scholar]
- 22. Li J., Du W., Maynard S., Andreassen P. R., Pang Q. (2010) Oxidative stress-specific interaction between FANCD2 and FOXO3a. Blood 115, 1545–1548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Houghtaling S., Timmers C., Noll M., Finegold M. J., Jones S. N., Meyn M. S., Grompe M. (2003) Epithelial cancer in Fanconi anemia complementation group D2 (Fancd2) knockout mice. Genes Dev. 17, 2021–2035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. de Haan G., Ploemacher R. (2002) The cobblestone-area-forming cell assay. Methods Mol. Med. 63, 143–151 [DOI] [PubMed] [Google Scholar]
- 25. Subramanian A., Tamayo P., Mootha V. K., Mukherjee S., Ebert B. L., Gillette M. A., Paulovich A., Pomeroy S. L., Golub T. R., Lander E. S., Mesirov J. P. (2005) Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 102, 15545–15550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wang J., Sun Q., Morita Y., Jiang H., Gross A., Lechel A., Hildner K., Guachalla L. M., Gompf A., Hartmann D., Schambach A., Wuestefeld T., Dauch D., Schrezenmeier H., Hofmann W. K., Nakauchi H., Ju Z., Kestler H. A., Zender L., Rudolph K. L. (2012) A differentiation checkpoint limits hematopoietic stem cell self-renewal in response to DNA damage. Cell 148, 1001–1014 [DOI] [PubMed] [Google Scholar]
- 27. Schambach A., Galla M., Modlich U., Will E., Chandra S., Reeves L., Colbert M., Williams D. A., von Kalle C., Baum C. (2006) Lentiviral vectors pseudotyped with murine ecotropic envelope: increased biosafety and convenience in preclinical research. Exp. Hematol. 34, 588–592 [DOI] [PubMed] [Google Scholar]
- 28. Kiel M. J., Yilmaz O. H., Iwashita T., Yilmaz O. H., Terhorst C., Morrison S. J. (2005) SLAM family receptors distinguish hematopoietic stem and progenitor cells and reveal endothelial niches for stem cells. Cell 121, 1109–1121 [DOI] [PubMed] [Google Scholar]
- 29. Kennedy R. D., D'Andrea A. D. (2005) The Fanconi anemia/BRCA pathway: new faces in the crowd. Genes Dev. 19, 2925–2940 [DOI] [PubMed] [Google Scholar]
- 30. Smogorzewska A., Matsuoka S., Vinciguerra P., McDonald E. R., 3rd., Hurov K. E., Luo J., Ballif B. A., Gygi S. P., Hofmann K., D'Andrea A. D., Elledge S. J. (2007) Identification of the FANCI protein, a monoubiquitinated FANCD2 paralog required for DNA repair. Cell 129, 289–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kee Y., D'Andrea A. D. (2010) Expanded roles of the Fanconi anemia pathway in preserving genomic stability. Genes Dev. 24, 1680–1694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Unno J., Itaya A., Taoka M., Sato K., Tomida J., Sakai W., Sugasawa K., Ishiai M., Ikura T., Isobe T., Kurumizaka H., Takata M. (2014) FANCD2 binds CtIP and regulates DNA-end resection during DNA interstrand crosslink repair. Cell Rep. 7, 1039–1047 [DOI] [PubMed] [Google Scholar]
- 33. Mohrin M., Bourke E., Alexander D., Warr M. R., Barry-Holson K., Le Beau M. M., Morrison C. G., Passegué E. (2010) Hematopoietic stem cell quiescence promotes error-prone DNA repair and mutagenesis. Cell Stem Cell 7, 174–185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Nijnik A., Woodbine L., Marchetti C., Dawson S., Lambe T., Liu C., Rodrigues N. P., Crockford T. L., Cabuy E., Vindigni A., Enver T., Bell J. I., Slijepcevic P., Goodnow C. C., Jeggo P. A., Cornall R. J. (2007) DNA repair is limiting for haematopoietic stem cells during ageing. Nature 447, 686–690 [DOI] [PubMed] [Google Scholar]
- 35. Rossi D. J., Bryder D., Seita J., Nussenzweig A., Hoeijmakers J., Weissman I. L. (2007) Deficiencies in DNA damage repair limit the function of haematopoietic stem cells with age. Nature 447, 725–729 [DOI] [PubMed] [Google Scholar]
- 36. Yamazaki S., Iwama A., Takayanagi S., Morita Y., Eto K., Ema H., Nakauchi H. (2006) Cytokine signals modulated via lipid rafts mimic niche signals and induce hibernation in hematopoietic stem cells. EMBO J. 25, 3515–3523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Yang J. Y., Zong C. S., Xia W., Yamaguchi H., Ding Q., Xie X., Lang J. Y., Lai C. C., Chang C. J., Huang W. C., Huang H., Kuo H. P., Lee D. F., Li L. Y., Lien H. C., Cheng X., Chang K. J., Hsiao C. D., Tsai F. J., Tsai C. H., Sahin A. A., Muller W. J., Mills G. B., Yu D., Hortobagyi G. N., Hung M. C. (2008) ERK promotes tumorigenesis by inhibiting FOXO3a via MDM2-mediated degradation. Nat. Cell Biol. 10, 138–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Briot D., Macé-Aimé G., Subra F., Rosselli F. (2008) Aberrant activation of stress-response pathways leads to TNF-α oversecretion in Fanconi anemia. Blood 111, 1913–1923 [DOI] [PubMed] [Google Scholar]
- 39. Prieto-Remón I., Sánchez-Carrera D., López-Duarte M., Richard C., Pipaón C. (2013) Elevated levels of STAT1 in Fanconi anemia group A lymphoblasts correlate with the cells' sensitivity to DNA interstrand crosslinking drugs. Haematologica 98, 705–713 [DOI] [PMC free article] [PubMed] [Google Scholar]