Background: The impact of long epitopes on T-cell immunity remains unclear.
Results: We identified and characterized 15-mer epitopes restricted to HLA-A*02:01.
Conclusion: HLA-A*02:01, in addition to the HLA-B family, can bind long epitopes that represent new antigenic targets for CD8+ T-cells.
Significance: The characterization of 15-mer epitopes restricted to HLA-A*02:01 expands our knowledge of the HLA-ligandome.
Keywords: Crystallography, Immunology, Mass Spectrometry (MS), Peptide Conformation, Peptides, HLA-A*02:01, Human Leukocyte Antigen Class I, T-cell Immunity, Peptidomics
Abstract
Human leukocyte antigen (HLA) class I molecules generally present peptides (p) of 8 to 11 amino acids (aa) in length. Although an increasing number of examples with lengthy (>11 aa) peptides, presented mostly by HLA-B alleles, have been reported. Here we characterize HLA-A*02:01 restricted, in addition to the HLA-B*0702 and HLA-B*4402 restricted, lengthy peptides (>11 aa) arising from the B-cell ligandome. We analyzed a number of 15-mer peptides presented by HLA-A*02:01, and confirmed pHLA-I formation by HLA folding and thermal stability assays. Surprisingly the binding affinity and stability of the 15-mer epitopes in complex with HLA-A*02:01 were comparable with the values observed for canonical length (8 to 11 aa) HLA-A*02:01-restricted peptides. We solved the structures of two 15-mer epitopes in complex with HLA-A*02:01, within which the peptides adopted distinct super-bulged conformations. Moreover, we demonstrate that T-cells can recognize the 15-mer peptides in the context of HLA-A*02:01, indicating that these 15-mer peptides represent immunogenic ligands. Collectively, our data expand our understanding of longer epitopes in the context of HLA-I, highlighting that they are not limited to the HLA-B family, but can bind the ubiquitous HLA-A*02:01 molecule, and play an important role in T-cell immunity.
Introduction
Human leukocyte antigen (HLA)5 class I molecules are expressed on the surface of all nucleated cells presenting peptides for CD8+ T-cell recognition. The peptides presented in HLA class I molecules are protein fragments of intracellular origin, which are degraded by an array of proteases, the most prominent of which is the proteasome. The protein fragments are truncated to smaller peptides and translocated into the endoplasmic reticulum (ER). In the ER, the peptide-HLA class I molecule (pHLA) is assembled from a peptide, a polymorphic heavy chain, and the monomorphic light chain called β2-microglobulin (β2m). Both β2m and the peptide are required for the stability of the HLA class I molecule. A peptide with adequate binding motif residues will bind into the peptide-binding groove of the HLA class I molecule, allowing the assembled molecule to leave the ER and be transported via the Golgi complex to the cell surface to display the peptides to CD8+ T-cells (1).
It has long been reported that HLA class I molecules can accommodate 8–11-mer peptides, typically 9-mers (1–3). Over the last few years, different groups have reported the binding of 12-mer, 13-mer, 14-mer, and even a 16-mer peptides to HLA class I molecules (4–13). Crystallographic and biophysical studies showed the binding of a 13-mer viral epitope to the HLA-B*3508 molecule and T-cell recognition of the bulged peptide (12, 14–16). The synthetic elongation of previously defined T-cell epitopes by central amino acid insertion revealed binding of 8–25-mer peptides to HLA-B*3508, although central amino acid insertion was not generally tolerated well for all peptides (17).
Because some longer peptides are recognized by T-cells, such peptides may play an important role in T-cell-mediated therapies for cancer, and in vaccine design. So far, a rather limited number of naturally processed and presented longer peptides have been reported, and notably the majorities involve HLA-B alleles. Generally, previous reports on longer peptides have focused on a single or a few isolated peptides. A more general view on the contribution of longer peptides to the HLA-ligandome, in-depth analysis is required. One of our previous studies (8) provided an in-depth analysis, and allowed the selection of longer peptides for follow-up studies. Therefore, in the present study, we report on these longer peptides, i.e. 14–23-mers, binding to the HLA-B family members, namely HLA-B*4402 and HLA-B*0702, and more surprisingly to the HLA-A family molecule, HLA-A*02:01. Our analysis was focused on the common HLA-A*02:01 allele and its ability to bind 15-amino acid long epitopes. After elution and sequencing of the 15-mer peptides, bound to HLA class I molecules, we analyzed the pHLA-A*02:01 stability. We compare the binding affinity and stability of 15-mer·HLA-A*02:01 complexes with the canonical length 9- and 10-mer peptides bound to the same HLA molecule. We subsequently solved the structures of two distinct 15-mer epitopes in complex with the HLA-A*02:01 molecule, and show that they exhibited contrasting conformations of their central bulged region. Finally we formally establish that HLA-A*02:01 loaded with 15-mer peptides are antigenic targets for the T-cells, using tetramers loaded with the 15-mer epitopes to isolate reactive T-cells.
EXPERIMENTAL PROCEDURES
Cellular Sample Preparation
Sample preparation was as described in Ref. 8. Briefly, Epstein-Barr virus (EBV)-transformed B lymphoblastic cell lines (EBV-LCL) LCL-HHC (typing: HLA-A*02:01, B*0702, B*4402, Cw*0501, and Cw*0702) and LCL-JYpp65 (typing: HLA-A*02:01, B*0702, and Cw*0702) were expanded in roller bottles, using Iscove's modified Dulbecco's medium, supplemented with 10% heat-inactivated fetal bovine serum (FBS), penicillin/streptomycin, and l-glutamine, were collected, washed with ice-cold PBS, and stored at −80 °C until use. Antibodies were produced, purified, and stored as described in Ref. 8.
Isolation of HLA Class I-presented Peptides
Pellets of LCL-JYpp65 and LCL-HHC cell lines were lysed in 50 mm Tris-HCl, 150 mm NaCl, 5 mm EDTA, and 0.5% Nonidet-P40 (pH 8.0) and supplemented with Complete protease inhibitors (Roche Applied Science). The total concentration of cells in the lysis buffer was 0.1 × 109 cells/ml. After a 2-h incubation with tumbling of the cells in the lysis buffer at 4 °C, the preparation was centrifuged at 4 °C for 10 min at 2070 × g. The supernatant was transferred to a new tube and centrifuged at 4 °C for 35 min at 19,000 × g. The supernatant was pre-cleared with CL4B beads and subjected to the W6/32 immunoaffinity column with a flow rate of 2 ml/min. After washing, bound peptide-HLA class I complexes were eluted from the column, and dissociated, with 10% acetic acid. Peptides were separated from the HLA class I molecules by passage through a 10-kDa membrane (Pall macrosep centrifuge devices), and further purified by solid phase extraction (C18 Oasis, 100-μl bed volume, Waters), freeze dried, and resuspended in 95/3/0.1, water/ACN/FA (v/v/v).
Peptide Separation
The pools of peptides eluted form the two EBV-LCL lines were divided in three (LCL-JYpp65) or two portions (LCL-HHC). The LCL-JYpp65 pools were separated by peptide IEF, SCX, and C18 chromatography and the LCL-HHC pools were separated by peptide IEF and SCX chromatography, as described in Ref. 8 to achieve a high number of identified peptides. The fractions obtained from the three off line separation techniques were further fractionated and analyzed by nano-LC-MS/MS.
Mass Spectrometry Data Analysis
The tandem mass spectra were matched against the International Protein Index (IPI) human database version 3.87, using the Mascot search engine version 2.2.04 (Matrix Science, London, UK)), with a precursor mass tolerance of 2 ppm, with methionine oxidation as a variable modification, and a product ion tolerance of 0.5 Da. Scaffold software version 3 (Proteome software) was subsequently used to process the Mascot output files and generate spectrum reports. Duplicates were removed, and peptides longer than 11 amino acids with a Mascot ion score ≥35 were selected (supplemental Table S1). The selection of a Mascot ion score >35 has been thoroughly discussed in Hassan et al. (8).
Peptide Synthesis
Peptides were synthesized using standard fluorenylmethoxycarbonyl chemistry using a SyroII peptide synthesizer (MultiSynTech, Witten, Germany) (Table 1). The integrity of the peptides was checked using RP-HPLC and MS. The purity of the peptides was higher than 95%.
TABLE 1.
HLA-A*02:01 specific peptides
| Gene name | Protein name | Peptide sequence | Amino acid length | BMIa | IC50b |
|---|---|---|---|---|---|
| nm | |||||
| GYPC | Isoform Glycophorin-C of Glycophorin-C | ALQDAGDSSRKEYFI | 15 | 40 | 414 |
| GNB3 | Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit β-3 | ALWDIETGQQKTVFV | 15 | 39 | 15 |
| RAB9A | Ras-related protein Rab-9A | FLNKDLEVDGHFVTM | 15 | 76 | 83 |
| NUDCD2 | NudC domain-containing protein 2 | KLFDSTIADEGTWTL | 15 | 75 | 10 |
| NFKB1 | Isoform 2 of nuclear factor NF-κB p105 subunit | KLLEIPDPDKNWATL | 15 | 36 | 24 |
| ZNF828 | Zinc finger protein 828 | KLMEALEPPLEEQQI | 15 | 55 | 1366 |
| EEF2 | Elongation factor 2 | LLYEGPPDDEAAMGI | 15 | 92 | 55 |
| ARPC3 | Actin-related protein 2/3 complex subunit 3 | SLMDPDTKLIGNM*ALc | 15 | 57 | 51 |
| LB-SSR1–1S | Signal sequence receptor subunit α | VLFRGGPRGSLAVA | 14 | 29 | 78 |
| LB-NiSCH-1A | Imidazoline receptor antisera-selected protein2 | ALAPAPAEV | 9 | 42 | 52 |
| CMV | PP65 | NLVPMVATV | 9 | 28 | 45 |
| MART1-M-ELA | Melanoma antigen modified analogue | ELAGIGILTV | 10 | NAd | 46 |
| MART1-WT-AAG | Melanoma antigen wild type | AAGIGILTV | 9 | NA | 6955 |
a BMI, best Mascot ion score.
b IC50, and binding affinity of peptides.
c Oxidized methionine residue.
NA, not applicable.
Refolding of pHLA Monomers
Recombinant HLA-A*02:01 heavy chain and human β2m light chain were in-house produced in Escherichia coli. The refolding was performed by adding 1.8 mg of HLA-A*02:01 heavy chain solubilized in 8 m urea, 1.2 mg of β2m dialyzed to PBS and 2 mg of peptide dissolved in dimethyl sulfoxide, to 50 ml of cold refolding buffer (400 mm l-arginine HCl, 100 mm Tris-HCl, pH 8, 5 mm reduced glutathione, 0.5 mm oxidized glutatione-Na, 2 mm EDTA, 5% glycerol, Complete protease inhibitors (Roche Applied Science)), and vigorously mixed after each step. The mixture was incubated for 72 h at 10 °C. The refolded protein mixture was concentrated to a volume of 0.5 ml with an Amicon concentrator (membrane cutoff, 30 kDa), then purified by gel filtration using fast protein liquid chromatography on a Superdex 75 column (Amersham Biosciences) and PBS as eluent. Complexes were stored at −80 °C. The efficiency of the refolding (recovery) is determined by protein concentration measurement of the formed pHLA by the Bradford protein assay.
Preparation of pHLA Tetramers
Biotinylated pHLA complexes containing FLNKDLEVDGHFVTM (FLNKD) or ALQDAGDSSRKEYFI (ALQDA) peptides bound to HLA-A*02:01 (RAB9AFLNKD:HLA-A*02:01 or GYPCALQDA:HLA-A*02:01, respectively) were conjugated to streptavidin-coupled phycoerythrin (SA-PE, Invitrogen) or allophycocyanin (SA-APC, Invitrogen) to form pHLA-tetramers. Thereto, RAB9AFLNKD·HLA-A*02:01 and GYPCALQDA·HLA-A*02:01 complexes were incubated with SA-PE or SA-APC at empirically determined ratios of 12:1 and 10:1, respectively, based on biotinylation efficiency. Concentration of pHLA-tetramers was adjusted to 0.2 μg/μl with PBS. pHLA-tetramers were stored at 4 °C.
Isolation of Peptide-specific T-cell Clones
After having obtained informed consent, peripheral blood mononuclear cells of HLA-A*02:01-negative healthy individuals were isolated by Ficoll density gradient. To isolate RAB9A or GYPC-reactive T-cells by enrichment with pHLA-tetramers, a previously described protocol was used with minor modifications (18). Peripheral blood mononuclear cells were incubated with PE-labeled RAB9AFLNKD:HLA-A*02:01 and GYPCALQDA:HLA-A*02:01-tetramers for 1 h at 4 ºC. Cells were washed twice and incubated with anti-PE magnetic microbeads (Miltenyi Biotec, Bergisch Gladbach, Germany). PE-labeled cells were enriched via magnetic-associated cell sorting on a LS column (Miltenyi Biotec) according to the manufacturer's instructions (Miltenyi Biotec). Subsequently, positive fractions were incubated with an antibody against CD8 (Invitrogen) in combination with antibodies against CD4, CD14, and CD19 (BD Pharmingen) for 15 min at 4 °C. Cells were washed twice. pHLA-binding CD8+ T-cells were single-cell sorted on a FACSAria (BD Bioscience) into 96-well round-bottom culture plates containing 50,000 irradiated (35 gray) autologous peripheral blood mononuclear cells in 100 μl of culture medium composed of Iscove's modified Dulbecco's medium (Lonza, Basel, Switzerland) supplemented with 100 IU/ml of IL-2 (Proleukine; Novartis Pharma, Arnhem, The Netherlands), 5% FBS (Invitrogen), 5% human serum, and 0.8 μg/ml of phytohemagglutinin (Remel, Lenexa, KS).
FACS Analysis of Isolated T-cell Clones
20,000 T-cells of a particular clone were stained with 10 μl of pHLA-tetramers in a final concentration of 2 μg/ml/pHLA-tetramer for 15 min at 37 °C. Cells were washed once and analyzed on a LSRII (BD Biosciences) using Diva software (BD Biosciences).
Functional Analysis of T-cell Clones
2,000 T-cells of a particular clone were co-incubated with 30.000 T2 cells or EBV-transformed B lymphoblastic cell lines (B-LCLs). T2 cells were loaded with different concentrations of peptide for 30 min at 37 ºC prior to co-incubation with T-cell clones. Following 18 h of co-culture, the supernatant was harvested and GM-CSF secretion was assessed using standard enzyme-linked immunosorbent assay (ELISA, R&D Systems) following the manufacturer's instructions.
HLA Competition Refolding Assay
The competition refolding assay has been developed previously (19). Briefly, this assay employs unfolded recombinant HLA-A*02:01 heavy chain in combination with folded β2m and the fluorescent standard peptide (FLPSDCFlFPSV, a modified HBV epitope, available from the authors), and relies on protein folding during the assay. The peptide of interest competes with the labeled standard peptide for binding. After 24 h of incubation, protein complexes and free peptide are separated by size-exclusion chromatography, during which the fluorescence of protein and peptide fractions are monitored. Following peak integration of the fluorescent signals, the ratio of label in the protein and peptide fraction is calculated. The affinities of the peptides are expressed as IC50, the peptide concentration at which binding of the standard peptide is reduced to 50% (Table 1). In this assay we used three epitopes with high binding affinity to the HLA-A*02:01 molecules (LB-NiSCH-1A (ALAPAPAEV), CMV-pp65-NLV (NLVPMVATV), and MART1-M-ELA (ELAGIGILTV)). We used MART1-WT-AAG (AAGIGILTV) as a low affinity binder to the HLA-A*02:01 molecule (18).
Thermal Stability Assay
To assess the stability of each peptide in complex with the pHLA-A*02:01, a thermal shift assay was performed. The fluorescent dye Sypro Orange was used to monitor the protein unfolding. The thermal stability assay was performed in the Real Time Detection system (Corbett RotorGene 3000), originally designed for PCR. Each pHLA-A*02:01 complex in 10 mm Tris-HCl (pH 8), 150 mm NaCl, at two concentrations (5 and 10 μm) in duplicate, was heated from 25 to 95 °C with a heating rate of 1 °C/min. The fluorescence intensity was measured with excitation at 530 nm and emission at 555 nm. The Tm, or thermal melting point, represents the temperature required to unfold 50% of the protein (20) (Table 2).
TABLE 2.
Thermal stability of peptide·HLA-A*02:01 complexes
| Peptide-HLA-A*02:01 | Tma |
|---|---|
| °C | |
| FLNKDLEVDGHFVTM | 47.9 ± 0.5 |
| ALQDAGDSSRKEYFI | 48.0 ± 1.0 |
| KLLEIPDPDKNWATL | 66.5 ± 1.8 |
| ALWDIETGQQKTVFV | 58.0 ± 1.0 |
| NLVPMVATV | 63.9 ± 0.5 |
a Tm represents the temperature required to unfold 50% of the protein.
Crystallization, Data Collection, and Structure Determination
Crystals of the HLA-A*02:01·FLNKD and HLA-A*02:01·ALQDA complexes were grown by the hanging drop, vapor diffusion method at 20 °C with a protein/reservoir drop ratio of 1:1, at a concentration of 10 mg/ml of protein using 18–22% PEG 3350, 0.1 m HEPES, pH 7.5, and 0.1 m MgCl2. Crystals were soaked in a cryoprotectant solution containing mother liquor solution with the PEG concentration increase to 35% (w/v) and then flash frozen in liquid nitrogen. The data were collected on the MX1 beamline at the Australian Synchrotron (Clayton) using an ADSC-Quantum 210 CCD detector (at 100 K), processed using the XDS software (21) and scaled using SCALA software (22) from the CCP4 suite (23). The structures were determined by molecular replacement using the PHASER (24) program with the HLA-A*02:01 minus the peptide as the search model for the MHC (Protein Data Bank code 3GSO (25)). Manual model building was conducted using Coot software (26) followed by maximum-likelihood refinement with the PHENIX program (27). The final models have been validated using the Protein Data Base validation web site and the final refinement statistics are summarized in Table 3. Coordinates were submitted to the PDB database, HLA-A*02:01-FLNKD (code 4U6X) and HLA-A*02:01-ALQDA (code 4U6Y). All molecular graphics representations were created using PyMol (28). The interactions between the peptides and the HLA have been calculated using CONTACT in the CCP4 software suite (23).
TABLE 3.
Data collection and refinement statistics
Values in parentheses are for the highest resolution shell.
| HLA-A*02:01·FLNKD | HLA-A*02:01·ALQDA | |
|---|---|---|
| Data collection statistics | ||
| Resolution range (Å) | 29.44–1.46 (1.51–1.46) | 39.53–1.67 (1.73–1.67) |
| Space group | P 21 | P 21 |
| Cell dimensions (a,b,c) (Å) | 50.88, 79.76, 54.84 | 51.04, 79.06, 54.90 |
| β = 111.75° | β = 111.11° | |
| Total number of observations | 392,206 (37,550) | 264,156 (24,945) |
| Number of unique observations | 69,403 (6,859) | 46,434 (4,518) |
| Multiplicity | 5.6 (5.5) | 5.7 (5.5) |
| Data completeness (%) | 99.79 (98.69) | 99.71 (97.75) |
| I/σ(I) | 19.3 (2.5) | 17.8 (2.4) |
| Rpima (%) | 2.5 (29.7) | 3.1 (31.8) |
| Refinement Statistics | ||
| Rfactorb (%) | 16.12 (23.54) | 15.49 (21.89) |
| Rfreeb (%) | 19.04 (27.17) | 19.78 (28.18) |
| Number of non-hydrogen atoms | 4,037 | 3,859 |
| Macromolecules | 3,472 | 3,460 |
| Ligands | 15 | 27 |
| Water | 550 | 372 |
| Root mean square deviations from ideality | ||
| Bond lengths (Å) | 0.009 | 0.008 |
| Bond angles (°) | 1.26 | 1.13 |
| Ramachandran plot (%) | ||
| Allowed region (%) | 98 | 98 |
| Generously allowed region (%) | 2 | 2 |
| Ramachandran outliers (%) | 0 | 0 |
| Average B-factor | 21.5 | 23.7 |
| Macromolecules | 19.4 | 22.2 |
| Ligands | 42.4 | 59.7 |
| Water | 33.8 | 34.8 |
a Rp.i.m = Σhkl [1/(N-1)]1/2 Σi|Ihkl,i − 〈Ihkl〉|/Σhkl〈Ihkl〉.
b Rfactor = Σhkl‖Fo| − |Fc‖/Σhkl |Fo| for all data except ≈5%, which were used for Rfree calculation.
RESULTS
Non-canonical Peptides Presented in HLA Class I Molecules
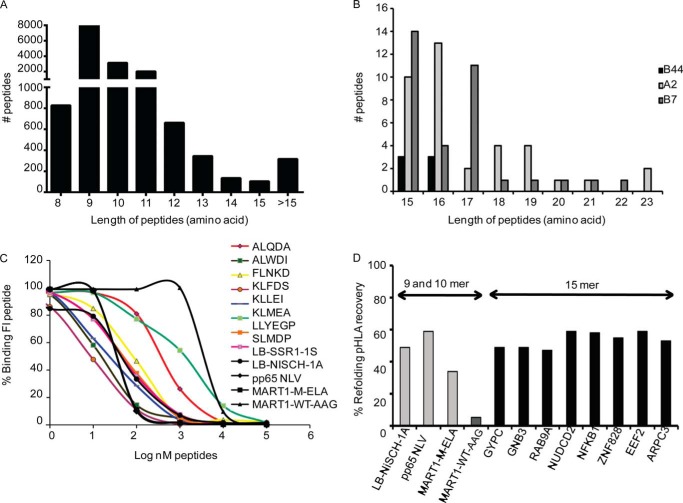
The list of eluted peptides from the two EBV-LCLs comprised 15,882 peptides, based on a length of 8–23 amino acids and a Mascot ion score >35. The list contained 1,568 peptides of 12–23 amino acids in length, of which 1,145 were 12–14-mers and 423 peptides are longer than 14 amino acids (supplemental Table S1 and Fig. 1).
FIGURE 1.
Peptide length distribution of the HLA-ligandome and binding affinity. Distribution of all peptides eluted from two EBV-LCL cell lines (A), and a focus on 15–23-mer peptides (B). A substantial percentage of HLA-ligands are longer than of canonical 8–11-mers. Binding affinity was as determined in an HLA competition refolding assay (C), and an HLA refolding efficiency assay (D).
The 8–11-mer peptides have been reported by Hassan et al. (8) (Fig. 1A), and so we concentrated our study on the peptides of non-canonical length (>11 aa). It is important to note that in large scale proteomics experiments a certain false discovery rate is acceptable. For HLA-presented peptides 5% is accepted as the false discovery rate (8, 29). Therefore, it cannot be excluded that a few peptides might have been incorrectly assigned, but the large majority will have been correctly assigned. In addition, we performed our immunopurification experiments with a pan class I-antibody, w6/32, which might complicate assignment of peptides to a particular allele. However, in this study the A alleles and B alleles have clearly distinct motifs. The known HLA C allele present in our cells, as known from the SYFPEITHI database, do not fulfill our A and B motifs.
To estimate the number of potentially relevant non-canonical binders to the HLA molecules we used NetMHC, and initially used a simple definition of binders by definition of the P2 anchor: HLA-A*02:01 (P2, LMV), HLA-B*0702 (P2, P), HLA-B*4402 (P2, E). 922 of the 1,145 12–14-mers (81%) fulfilled this P2 anchor criterion, which compares well with the 75% of binders as found using NetMHC (with a score <1,000 nm).
Of the listed 15–23-mer peptides 214 of 423 (51%) fulfilled the P2 anchor criterion (Fig. 1B). Because NetMHC does not allow calculation of binding affinities beyond 14-mers, we defined potential binders more stringently, based on the following mandatory anchor residues, including the PΩ position: HLA-A*02:01 (P2, LMV; and PΩ, LMIVA), HLA-B*0702 (P2, Pro and PΩ, Leu), and HLA-B*4402 (P2, Glu; and PΩ, FY). This additional constraint resulted in the selection of 77 peptides with a length of 15–23 amino acids (supplemental Table S2). Surprisingly, our result indicated that whereas previous studies on longer epitopes were based on HLA-B molecules, it was evident from the elution assay and mass spectrometry analysis that HLA-A molecules, including HLA-A*02:01, also have the ability to bind longer epitopes (Fig. 1B).
Competition Refolding Assay
From the 10 15-mer peptides found fulfilling the P2 and PΩ criteria for HLA-A*02:01, eight with P2 = Leu were synthesized for further characterization (Table 1). To show the binding efficacy of these naturally processed 15-mer peptides, we performed refolding and competition assays (Table 1) (19). The two assays are complementary. The competition refolding assay shows the ability of the peptide to bind. The refolding assay shows the efficiency of formation (i.e. the yield of the HLA-monomer folding process). The yield is an additional important parameter for pHLA stability and a predictor of efficient tetramer formation. Several other peptides with known binding affinities were included in the assay to evaluate the relative binding affinity of the 15-mer peptides. LB-NISCH-1A (ALAPAPAEV), MART1-M-ELA (ELAGIGILTV), and CMV-pp65-NLV (NLVPMVATV) peptides are known high affinity binders to the HLA-A*02:01 molecule, and were included as control (18). The MART1-WT-AAG (AAGIGILTV) epitope was included as a low affinity binder to the HLA-A*02:01 molecule. In the competition assay the fluorescein (F1)-labeled reference peptide (FLPSDCFlFPSV), known to bind efficiently to the HLA-A*02:01 molecule, and the peptide of interest compete for binding in the HLA class I groove during folding. The affinities of the peptides are expressed as IC50 (Table 1). The calculated percentage of bound fluorescent reference peptide after competition with the 15-mer peptides, and the high and low affinity reference peptides are listed in Table 1, and are plotted in Fig. 1C.
The results showed that all eight synthesized 15-mer peptides, fulfilling the HLA A*0201 motif, have an IC50 between 10 and 1366 nm, most of which are in the high binding affinity range (19). For comparison, the low binding affinity peptide MART1-WT-AAG (AAGIGILTV) has a higher IC50 of ∼7,000 nm, whereas the high binding affinity peptide pp65-NLV has an IC50 of 45 nm. These results illustrate that the 15-mer peptides bind to the HLA-A*02:01 molecule with similar affinity as 8–11-mer peptides, and some even with higher affinity such as the KLFDS (IC50 of 10 nm, Table 1). In summary the 15-mer epitopes exhibited affinities comparable with that of 9–10-mers bound to HLA-A*02:01, showing that the length was not an obstacle for peptides to bind the common HLA-A*02:01 allele.
pHLA Complexes Refolding Efficiency Assay
We next applied an HLA refolding efficiency assay to assess the binding of 15-mer peptides to HLA-A*02:01 by measurement of the yield of formation of pHLA. This assay determined the capacity of the peptides to support stable refolding of the heavy chain and β2m recombinant subunits of the HLA-A*02:01 complex. The yield of folded pHLA-A*02:01 was determined for the classical length (9-mer) and longer peptides (15-mer) under the same refolding conditions. HLA recovery levels of 47–59% were obtained for the eight 15-mer peptides. The yields of the three known high affinity binders LB-NiSCH-1A, CMV-pp65-NLV, and MART1-M-ELA were 49, 58.8, and 34% respectively. The weak binder MART-1-WT-AAG showed a pHLA recovery yield of 5.4% (Fig. 1D). These results indicate that the 15-mer peptides have a similar binding efficiency as the classical high affinity 9-mer peptides, and so are able to stabilize the formation of the HLA-A*02:01·β2m complex as well as canonical peptides.
Stability of the 15-mer·HLA-A*02:01 Complexes
We then assessed the thermal stability, after refolding, of HLA-A*02:01 bound to four distinct 15-mer peptides and compared these values to HLA-A*02:01 bound to a canonical 9-mer epitope CMV-pp65-NLV (25). The thermal melting point, or Tm, observed for HLA-A*02:01 in complex with the CMV-pp65-NLV peptide was 63.9 °C (Table 2). We then performed the same assay with the four HLA-A*02:01·15-mer complexes, along with the HLA-A*02:01·NLV complex. The FLNKD and ALQDA peptides exhibited the lowest Tm, with a value of ∼48 °C, which was notably lower than the HLA-A*02:01·NLV complex. In contrast, the ALWDI and KLLEI Tm were 58 and 66.5 °C, respectively (Table 2). Interestingly the two 15-mer peptides with the lowest Tm have non-optimal HLA-A*02:01 anchor residue at PΩ, namely a valine residue (Table 1). In summary the HLA-A*02:01-restricted 15-mer epitopes can exhibit a range of Tm, with some of them highly stable in the cleft of HLA-A*02:01.
Crystal Structures of 15-mer·HLA-A*02:01 Complexes
To date only seven structures of HLA in complex with long epitopes (>11 aa) are available (4, 7, 12, 13, 31, 32) as well as one structure of a rat MHC in complex with a 13-mer peptide (33). The seven pHLA structures include: two 12-mer EBV epitopes bound to HLA-B*4403 (32) and to HLA-B*3508 (13); a 13-mer EBV epitope in complex with closely related allomorphs HLA-B*3501 and HLA-B*3508 (12); a 13-mer epitope bound to HLA-B*0702 (7), a self 14-mer peptide in complex with HLA-B*3501 (7, 31); and a self 16-mer peptide bound to HLA-B*4102 (4). These structures solved to date reveal that the N and C termini of the peptides bind in similar fashion to the one observed for the classical length peptides, and that the central part of the peptide bulges out of the binding cleft.
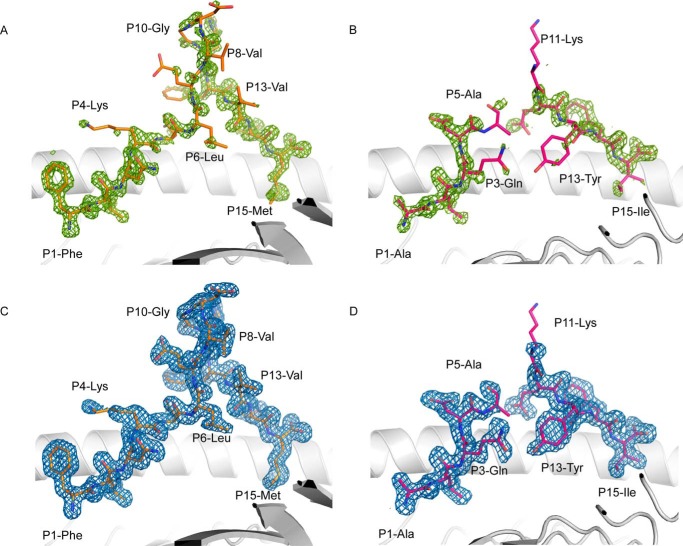
Interestingly of the long epitopes characterized none of them were in complex with the HLA-A molecule. To understand how the HLA-A*02:01 molecule can present long epitopes of 15 residues in length, we determined the structure of the HLA-A*02:01·FLNKD and HLA-A*02:01·ALQDA complexes at high resolution (Table 3). The two peptide·HLA complexes were crystallized in the same space group with the same unit cell dimension. Therefore, the difference in peptide structures was attributable to the peptide sequence. The two peptides bind with the canonical P2-Leu into the B pocket and with non-canonical PΩ-Val residues in the F-pocket for HLA-A*02:01, a methionine for the FNLKD peptide, and an isoleucine for the ALQDA peptide (Fig. 2, A and B). The FNLKD peptide density was clear and unambiguous in the cleft of the HLA-A*02:01 molecule (Fig. 2, A and C), whereas the central part of the ALQDA was poorly defined (Fig. 2, B and D). Despite the two 15-mer peptides exhibiting a similar Tm value to the same HLA-A*02:01 (Table 2), conformation of the two peptides were notably different (Fig. 3).
FIGURE 2.
Electron density map of 15-mer epitopes bound to HLA-A*02:01. The A and B panels show the omit map (Fo − Fc) at 3σ in green for the FLNKD and ALQDA peptides in complex with HLA-A*02:01, respectively. The C and D panels show the 2Fo − Fc map contoured at 1σ in blue after the final refinement for the FLNKD and ALQDA peptides, respectively. The HLA-A*02:01 is represented as a white schematic; the peptides are represented in stick and colored in orange for FLNKD and pink for ALQDA.
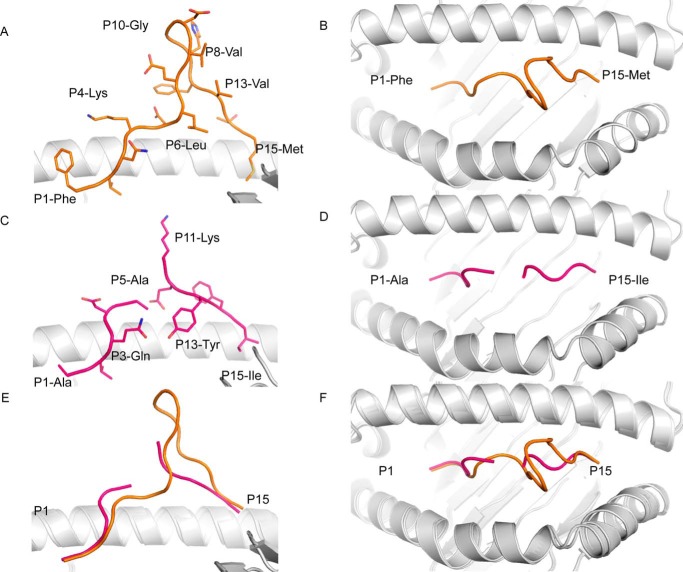
FIGURE 3.
Crystal structures of 15-mer peptides in complex with HLA-A*02:01 molecule. Side view (A, C, and E panels) and top view (B, D, and F panels) of the HLA-A*02:01 cleft (white schematic) bind to the FLNKD peptide (orange stick and loop) or the ALQDA peptide (pink stick and loop). The bottom panels (E and F) show a superimposition of the two peptides in the cleft of the HLA-A*02:01 molecule in the same orientation as the upper panels.
The ALQDA was mobile in the cleft of the HLA-A*02:01 molecule (Fig. 2D), and as a result the central region from P6 to P10 was not built in the final model of the pHLA complex. Flexibility is often associated with a long peptide presentation by HLA class I molecules, as exemplified by the 16-mer AEMY self-peptide presented by the HLA-B*41:03 molecule (4). The ALQDA binds the HLA-A*02:01 molecule via 9 of its residues and forms 188 contacts with the HLA (9 salt-bridges and 16 hydrogen bonds). The number of bonds formed by the 15-mer ALQDA was similar to the 9-mer NLV peptide (185 contacts, 2 salt bridges, and 14 hydrogen bonds), despite the extra 6 residues. A small amino acid such as valine is optimal at the C-terminal part of the peptide sequence as it fits well in the F pocket of the HLA-A*0201 cleft. As observed in the NLV peptide structure (PDB code 3GSO (25)), whereby the P9-Val sat on top of tyrosine 116 of the HLA-A*0201 molecule. The change to larger amino acids, such as methionine or isoleucine, at the C-terminal position of the peptide leads to rotation of tyrosine 116 to avoid steric clashes that pushes arginine 97. This rearrangement of buried amino acids within the antigen binding cleft appears to be less favorable to the overall stability of the pHLA-A*0201 complex (Table 2).
Contrasting the flexible ALQDA 15-mer, the FNLKD peptide was well defined in the cleft of the HLA-A*02:01 molecule (Fig. 2C), and is the longest well defined epitope observed in complex with a HLA class I molecule to date. The FNLKD bulges out of the HLA-A*02:01 cleft forming a β-sheet hairpin structure from P7 to P12 residues (Fig. 3A). The secondary structure formation in the bulged part of the peptide made intra-molecular contacts, constraining and rigidifying the peptide (34), and probably explains how the FNLKD can be such a long peptide and being so rigid in HLA-A*02:01 cleft. Interestingly the β-sheet hairpin formation is higher than the hinge of the α2-helix of the HLA-A*02:01 and would represent an immediate contact point for the T-cell receptor. The stable conformation of the FLNKD was also associated with a higher number of contacts with the HLA-A*02:01 molecule, with the peptide engaging 11 of its residues to interact with the HLA, and making a total of 215 contacts (6 salt bridges and 14 hydrogen bonds). This β-sheet hairpin structure is the first reported for an epitope bound to class I HLA. An α-helix has been previously reported in the 12-mer CPS bound to the HLA-B*3508 complex (13). The crystal structures of the two 15-mers in complex with HLA-A*02:01 show that, like the HLA-B molecule, HLA-A can present long peptides in a diverse array of conformations from mobile to highly stable, and could represent some new antigen for T-cells.
15-mer Epitopes Presented by HLA-A*02:01 Can Activate CD8+ T-cells
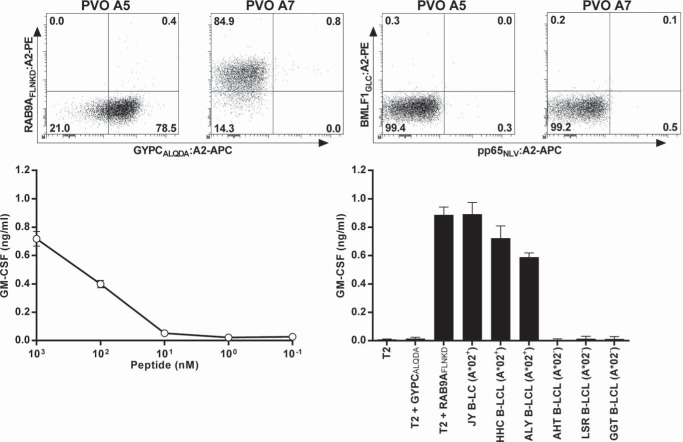
We demonstrate that HLA-A*02:01 can bind longer peptides with high binding affinity, forming stable pHLA complexes, and determined how HLA-A*02:01 can present 15-mer peptides. To establish if 15-mer·HLA*0201 complexes were antigenic and recognized by T-cells, we used pHLA-tetramers with the two structurally characterized 15-mer peptides FLNKD and ALQDA (RAB9AFLNKD:HLA-A*02:01 and GYPCALQDA:HLA-A*02:01, respectively). Because negative selection during thymic development depletes T-cells recognizing such self-antigens bound to self-HLA, T-cells were isolated from HLA-A*02:01-negative healthy individuals, which contain a naive T-cell repertoire capable of recognizing such self-antigens presented in HLA-A*02:01. From peripheral blood mononuclear cells of HLA-A*02:01-negative individuals, CD8+ T-cells clones were expanded that bind pHLA-tetramers RAB9AFLNKD:HLA-A*02:01 and GYPCALQDA:HLA-A*02:01 by first enriching pHLA-tetramer binding cells by magnetic-associated cell sorting followed by immediate single-cell FACS sorting. Among the isolated T-cells, clone PVO A5 showed specific binding of GYPCALQDA:HLA-A*02:01-tetramer, whereas binding to RAB9AFLNKD:HLA-A*02:01-tetramer was absent (Fig. 4A). In contrast, T-cell clone PVO A7 specifically bound to tetramer RAB9AFLNKD:HLA-A*02:01, whereas binding to GYPCALQDA:HLA-A*02:01 was absent (Fig. 4A). In addition, both T-cell clones did not bind to two control tetramers composed of HLA-A*02:01 displaying either EBV-derived peptide GLCTLVAML or human cytomegalovirus (CMV)-derived peptide NLVPMVATV, further indicating specific binding of both T-cell clones to their respective pHLA-tetramer displaying a 15-mer peptide (Fig. 4B). Next, peptide-dependent recognition for both T-cell clones was assessed by pulsing HLA-A*02:01-positive T2 cells with the two 15-mer peptides. GYPCALQDA-specific T-cell clone PVO A5 did not recognize peptide pulsed T2-cells indicating insufficient sensitivity for HLA-bound ALQDA (data not shown). In contrast, T-cell clone PVO A7 recognized T2-cells pulsed with peptide FLNKD (Fig. 4C). This recognition was peptide-specific because no recognition of T2-cells pulsed with ALQDA was observed (Fig. 4D). In addition, the T-cell clone PVO A7 recognized three HLA-A*02:01-positive B-LCLs, which naturally express RAB9A and were used to elute the 15-mer peptide FLNKD (Fig. 4D). Lack of recognition of three HLA-A*02:01-negative B-LCLs indicates that the observed reactivity of T-cell clone PVO A7 was HLA-A*02:01-dependent (Fig. 4D). These data indicate that the 15-mer peptide FLNKD presented in HLA-A*02:01 on the cell surface can be recognized by T-cells in a peptide-dependent manner.
FIGURE 4.
Specific T-cell clone recognition of 15-mer peptide FLNKD presented by HLA-A*02:01. T-cell clones PVO A5 and PVO A7 were isolated using pHLA-tetramers composed of GYPC-derived peptide ALQDA- or RAB9A-derived peptide FLNKD bound to HLA-A*02:01 (GYPCALQDA:HLA-A*02:01 or RAB9AFLNKD:HLA-A*02:01) from an HLA-A*02:01-negative individual. A, T-cell clones PVO A5 (left) and PVO A7 (right) specifically bound to GYPCALQDA:HLA-A*02:01-tetramer and RAB9AFLNKD:HLA-A*02:01-tetramer, respectively, after staining with PE-labeled RAB9AFLNKD:HLA-A*02:01 and APC-labeled GYPCALQDA:HLA-A*02:01-tetramer. B, the panel shows the lack of staining with two control tetramers composed of HLA-A*02:01 displaying two virus-derived epitopes for T-cell clones PVO A5 (left) and PVO A7 (right). All plots are shown with bi-exponential axis. Numbers in corners indicate percent cells in each quadrant. C, GM-CSF production was measured after co-culturing T-cell clone PVO A7 with HLA-A*02:01-positive T2 cells loaded with decreasing concentration of FLNKD peptide. D, GM-CSF production was measured after co-culturing T-cell clone PVO A7 with T2 cells, T2-cells loaded with 500 nm peptide (+GYPCALQDA or +RAB9AFLNKD), three HLA-A*02:01-positive B-LCLs (A2+), which naturally express and present RAB9A, or three HLA-A*02:01-negative B-LCLs (A2−). Experiments were performed in triplicate. Shown is one representative experiment of two independent experiments. Error bars indicate S.D.
DISCUSSION
Classically, HLA class I molecules present 8–11-mer peptides, although, an expanding list of lengthy (>11 aa) HLA-restricted peptides have emerged (6). Crystallographic studies have reported on seven pHLAs structures involving 12- to 16-mer epitopes (4, 7, 12, 13, 31, 32). These previous studies were all focused on HLA-B molecules, and here we describe the ability of the HLA-A*02:01 molecule to bind long epitopes too, with 538 12–14-mers being defined. Furthermore, 77 peptides are listed of 15–23 amino acids long that fulfill both the P2 and PΩ anchors criteria in either HLA-A*02:01, HLA-B*0702, or HLA-B*44. A comparable percentage of longer peptides was found in the reprocessed data of Mommen et al. (9), in particular in HLA-A*0301 and HLA-B*0702, and to a lesser extent in HLA-A*0101 and HLA-B*2705. The listing of peptides shows that HLA-A molecules appear to be just as suitable for presenting longer peptides as the HLA-B alleles. Both the intensity and the hydrophobicity of the longer peptides resemble that of the canonical 8- to 11-mer peptides. There was a steady decline in the number of longer peptides for every additional amino acid, which probably represents the probability of a peptide to survive in the cellular proteolytic environment. Longer peptides have an increasing chance of being cut by a protease. Of note, the amino acids between the P2 and PΩ anchors are not generally enriched for specific amino acid residues, so, the amino acid stretch between the anchors does not seem to be specifically resistant to proteolytic degradation on the basis of its primary structure. Longer peptides can be translocated by TAP into the ER, although generally somewhat less efficiently (35). In the ER, peptides are protected from being trimmed to short peptides for presentation in HLA by the nature of ERAAP (30). The fact that there seems to be no clear-cut length limitation (on the long side) imposed by the HLA class I binding groove can be explained by the phenomenon of (super)bulging of the peptide, with the P2 and PΩ anchor residues position fixed in the peptide binding groove, but with freedom to “leave” the binding groove for (part) of the peptide between these anchors residues. The two refolding assays we employed, both showed that the behavior of the 15-mer peptides resembled that of the canonical 8–11-mer peptides, i.e. the 15-mer peptides just as easily formed pHLA complexes and competed to the same degree as known good binders of 8–11-mer length. From the two 15-mer peptides solved in complex with the HLA-A*02:01, we observed two different conformations of the long epitopes in the cleft of HLA-A*02:01. First the ALQDA peptide was highly mobile, and its central region was poorly defined, reminiscent of the 16-mer self-peptide observed in complex with the HLA-B*4103 (4). Contrasting with the high flexibility of the ALQDA, the FLNKD was well defined and adopted one single rigid conformation when bound to HLA-A*02:01, similar to the 13-mer EBV epitope in complex with HLA-B*3508 (12). The FLNKD peptide central region formed a β-hairpin secondary structure that bulged out of the HLA-A*02:01 cleft, and could be a potential contact point for FLNKD-specific T-cells, and it will be of high interest to know how T-cells can engage a highly rigid bulge peptide like the FLNKD epitope. The TCR could potentially “struggle” to bind it or it will mostly focus on the peptide (like SB27,(15)) or might bind on the side of the peptide.
T-cells could be isolated from HLA-A*02:01-negative healthy individuals that contain a naive T-cell repertoire capable of recognizing self-antigens presented in HLA-A*02:01. T-cell clones demonstrated specific binding of pHLA-tetramer and furthermore, peptide-dependent recognition was observed for selected T-cell clones with HLA-A*02:01-positive T2-cells pulsed with the peptides as well as recognition of endogenously processed peptide on HLA-A*02:01-positive B-LCLs. Clone PVO A5 lacks functional reactivity against peptide-loaded target cells although there is specific staining of that clone with GYPCALQDA:A2 pHLA-tetramer. We have previously demonstrated that pHLA-tetramer staining alone is a poor indicator of functional avidity of a T-cell clone (18). Therefore, it is most likely that the clone avidity of the PVO A5 for the HLA-bound peptide GYPCALQDA is insufficient to trigger T-cell activity, whereas binding of pHLA-tetramer is possible. To circumvent the depletion of high avidity T-cells targeting self-peptides presented in self-HLA during thymic development, pHLA-tetramer binding T-cells were isolated from a healthy HLA-A2-negative individual. Based on previous results we estimated to isolate both high as well as low affinity T-cells (18). The results demonstrate that PVO A5 represents a low avidity T-cell clone for GYPCALQDA:A2, whereas clone PVO A7 represents a high avidity T-cell clone specific for RAB9AFLNKD:A2.
Clone PVO A7 demonstrated peptide-dependent recognition of a peptide-loaded T2 stimulator cells in the nanomolar range. Virus-specific T-cells demonstrate peptide sensitivity as low as in the picomolar range. However, caution must be exerted when comparing sensitivity between T-cell clones recognizing different epitopes based solely on recognition of peptide-loaded stimulator cells. Not only affinity of the TCR for its cognate peptide is important but also binding properties of the peptide to its respective HLA-molecule is critical, because exogenously loaded peptide need to compete with already HLA-bound peptide. These properties can differ between peptides. Furthermore, PVO A7 is able to recognize the endogenously processed and presented peptide on three HLA-A2-positive B-LCL indicating high functional avidity comparable with virus-specific T cells.
From these findings, T-cells appear to be capable to specifically recognize longer peptides. So far, there seems to be no clear limitation on peptide length for T-cell recognition of HLA class I presented peptides.
Altogether our data show that HLA class I restricted presentation and recognition is less restrictive than previously anticipated. Our data expand our understanding of HLA class I ligand presentation, and show that longer peptides are regular members of the HLA-ligandome, and should not be discarded in epitope discovery experiments, because these peptides might be useful in immunotherapy. Furthermore, the non-canonical peptide sequences presented here provide insight in antigen presentation and antigen processing.
Supplementary Material
Acknowledgments
We thank N. Dolezal, R. Cordfunke, and W. Benckhuijsen for peptide synthesis. We thank Kristy Campbell and Nathan Croft for their technical assistance and the staff at the Australian Synchrotron for assistance with data collection.
This work was supported by Landsteiner Foundation for Blood Transfusion Research Grant LSBR0713 and the Australian Research Council and the National Health and Medical Research Council of Australia.

This article contains supplemental Tables S1 and S2.
The atomic coordinates and structure factors (codes 4U6Y and 4U6X) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- HLA
- human leukocyte antigen
- pHLA
- peptide-human leukocyte antigen
- aa
- amino acid
- B-LCL
- B lymphoblastic cell line
- ER
- endoplasmic reticulum
- PE
- phycoerythrin
- β2m
- β2-microglobulin
- SA
- streptavidin.
REFERENCES
- 1. Neefjes J., Jongsma M. L., Paul P., Bakke O. (2011) Towards a systems understanding of MHC class I and MHC class II antigen presentation. Nat. Rev. Immunol. 11, 823–836 [DOI] [PubMed] [Google Scholar]
- 2. Rammensee H. G., Falk K., Rötzschke O. (1993) Peptides naturally presented by MHC class I molecules. Annu. Rev. Immunol. 11, 213–244 [DOI] [PubMed] [Google Scholar]
- 3. Yewdell J. W., Reits E., Neefjes J. (2003) Making sense of mass destruction: quantitating MHC class I antigen presentation. Nat. Rev. Immunol. 3, 952–961 [DOI] [PubMed] [Google Scholar]
- 4. Bade-Döding C., Theodossis A., Gras S., Kjer-Nielsen L., Eiz-Vesper B., Seltsam A., Huyton T., Rossjohn J., McCluskey J., Blasczyk R. (2011) The impact of human leukocyte antigen (HLA) micropolymorphism on ligand specificity within the HLA-B*41 allotypic family. Haematologica 96, 110–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ben Dror L., Barnea E., Beer I., Mann M., Admon A. (2010) The HLA-B*2705 peptidome. Arthritis Rheum. 62, 420–429 [DOI] [PubMed] [Google Scholar]
- 6. Burrows S. R., Rossjohn J., McCluskey J. (2006) Have we cut ourselves too short in mapping CTL epitopes? Trends Immunol. 27, 11–16 [DOI] [PubMed] [Google Scholar]
- 7. Ebert L. M., Liu Y. C., Clements C. S., Robson N. C., Jackson H. M., Markby J. L., Dimopoulos N., Tan B. S., Luescher I. F., Davis I. D., Rossjohn J., Cebon J., Purcell A. W., Chen W. (2009) A long, naturally presented immunodominant epitope from NY-ESO-1 tumor antigen: implications for cancer vaccine design. Cancer Res. 69, 1046–1054 [DOI] [PubMed] [Google Scholar]
- 8. Hassan C., Kester M. G., de Ru A. H., Hombrink P., Drijfhout J. W., Nijveen H., Leunissen J. A., Heemskerk M. H., Falkenburg J. H., van Veelen P. A. (2013) The human leukocyte antigen-presented ligandome of B lymphocytes. Mol. Cell. Proteomics 12, 1829–1843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mommen G. P., Frese C. K., Meiring H. D., van Gaans-van den Brink J., de Jong A. P., van Els C. A., Heck A. J. (2014) Expanding the detectable HLA peptide repertoire using electron-transfer/higher-energy collision dissociation (EThcD). Proc. Natl. Acad. Sci. U.S.A. 111, 4507–4512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Probst-Kepper M., Stroobant V., Kridel R., Gaugler B., Landry C., Brasseur F., Cosyns J. P., Weynand B., Boon T., Van Den Eynde B. J. (2001) An alternative open reading frame of the human macrophage colony-stimulating factor gene is independently translated and codes for an antigenic peptide of 14 amino acids recognized by tumor-infiltrating CD8 T lymphocytes. J. Exp. Med. 193, 1189–1198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Scull K. E., Dudek N. L., Corbett A. J., Ramarathinam S. H., Gorasia D. G., Williamson N. A., Purcell A. W. (2012) Secreted HLA recapitulates the immunopeptidome and allows in-depth coverage of HLA A*02:01 ligands. Mol. Immunol. 51, 136–142 [DOI] [PubMed] [Google Scholar]
- 12. Tynan F. E., Borg N. A., Miles J. J., Beddoe T., El-Hassen D., Silins S. L., van Zuylen W. J., Purcell A. W., Kjer-Nielsen L., McCluskey J., Burrows S. R., Rossjohn J. (2005) High resolution structures of highly bulged viral epitopes bound to major histocompatibility complex class I. Implications for T-cell receptor engagement and T-cell immunodominance. J. Biol. Chem. 280, 23900–23909 [DOI] [PubMed] [Google Scholar]
- 13. Wynn K. K., Fulton Z., Cooper L., Silins S. L., Gras S., Archbold J. K., Tynan F. E., Miles J. J., McCluskey J., Burrows S. R., Rossjohn J., Khanna R. (2008) Impact of clonal competition for peptide-MHC complexes on the CD8+ T-cell repertoire selection in a persistent viral infection. Blood 111, 4283–4292 [DOI] [PubMed] [Google Scholar]
- 14. Liu Y. C., Miles J. J., Neller M. A., Gostick E., Price D. A., Purcell A. W., McCluskey J., Burrows S. R., Rossjohn J., Gras S. (2013) Highly divergent T-cell receptor binding modes underlie specific recognition of a bulged viral peptide bound to a human leukocyte antigen class I molecule. J. Biol. Chem. 288, 15442–15454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tynan F. E., Burrows S. R., Buckle A. M., Clements C. S., Borg N. A., Miles J. J., Beddoe T., Whisstock J. C., Wilce M. C., Silins S. L., Burrows J. M., Kjer-Nielsen L., Kostenko L., Purcell A. W., McCluskey J., Rossjohn J. (2005) T cell receptor recognition of a “super-bulged” major histocompatibility complex class I-bound peptide. Nat. Immunol. 6, 1114–1122 [DOI] [PubMed] [Google Scholar]
- 16. Liu Y. C., Chen Z., Burrows S. R., Purcell A. W., McCluskey J., Rossjohn J., Gras S. (2012) The energetic basis underpinning T-cell teceptor tecognition of a super-bulged peptide bound to a major histocompatibility complex class I molecule. J. Biol. Chem. 287, 12267–12276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bell M. J., Burrows J. M., Brennan R., Miles J. J., Tellam J., McCluskey J., Rossjohn J., Khanna R., Burrows S. R. (2009) The peptide length specificity of some HLA class I alleles is very broad and includes peptides of up to 25 amino acids in length. Mol. Immunol. 46, 1911–1917 [DOI] [PubMed] [Google Scholar]
- 18. Hombrink P., Hassan C., Kester M. G., de Ru A. H., van Bergen C. A., Nijveen H., Drijfhout J. W., Falkenburg J. H., Heemskerk M. H., van Veelen P. A. (2013) Discovery of T cell epitopes implementing HLA-peptidomics into a reverse immunology approach. J. Immunol. 190, 3869–3877 [DOI] [PubMed] [Google Scholar]
- 19. Tan T. L., Geluk A., Toebes M., Ottenhoff T. H., Drijfhout J. W. (1997) A novel, highly efficient peptide-HLA class I binding assay using unfolded heavy chain molecules: identification of HIV-1 derived peptides that bind to HLA-A*0201 and HLA-A*0301. J. Immunol. Methods 205, 201–209 [DOI] [PubMed] [Google Scholar]
- 20. Gras S., Wilmann P. G., Chen Z., Halim H., Liu Y. C., Kjer-Nielsen L., Purcell A. W., Burrows S. R., McCluskey J., Rossjohn J. (2012) A structural basis for varied alphabeta TCR usage against an immunodominant EBV antigen restricted to a HLA-B8 molecule. J. Immunol. 188, 311–321 [DOI] [PubMed] [Google Scholar]
- 21. Kabsch W. (2010) Xds. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Evans P. (2006) Scaling and assessment of data quality. Acta Crystallogr. D Biol. Crystallogr. 62, 72–82 [DOI] [PubMed] [Google Scholar]
- 23. Collaborative Computational Project, Number 4. (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 [DOI] [PubMed] [Google Scholar]
- 24. Read R. J. (2001) Pushing the boundaries of molecular replacement with maximum likelihood. Acta Crystallogr. D Biol. Crystallogr. 57, 1373–1382 [DOI] [PubMed] [Google Scholar]
- 25. Gras S., Saulquin X., Reiser J. B., Debeaupuis E., Echasserieau K., Kissenpfennig A., Legoux F., Chouquet A., Le Gorrec M., Machillot P., Neveu B., Thielens N., Malissen B., Bonneville M., Housset D. (2009) Structural bases for the affinity-driven selection of a public TCR against a dominant human cytomegalovirus epitope. J. Immunol. 183, 430–437 [DOI] [PubMed] [Google Scholar]
- 26. Emsley P., Lohkamp B., Scott W. G., Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. DeLano W. L. (2002) The PyMOL Molecular Graphic System. Schrödinger, LLC, New York [Google Scholar]
- 29. Bourdetsky D., Schmelzer C. E., Admon A. (2014) The nature and extent of contributions by defective ribosome products to the HLA peptidome. Proc. Natl. Acad. Sci. U.S.A. 111, E1591–E1599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Serwold T., Gaw S., Shastri N. (2001) ER aminopeptidases generate a unique pool of peptides for MHC class I molecules. Nat. Immunol. 2, 644–651 [DOI] [PubMed] [Google Scholar]
- 31. Probst-Kepper M., Hecht H. J., Herrmann H., Janke V., Ocklenburg F., Klempnauer J., van den Eynde B. J., Weiss S. (2004) Conformational restraints and flexibility of 14-meric peptides in complex with HLA-B*3501. J. Immunol. 173, 5610–5616 [DOI] [PubMed] [Google Scholar]
- 32. Rist M. J., Theodossis A., Croft N. P., Neller M. A., Welland A., Chen Z., Sullivan L. C., Burrows J. M., Miles J. J., Brennan R. M., Gras S., Khanna R., Brooks A. G., McCluskey J., Purcell A. W., Rossjohn J., Burrows S. R. (2013) HLA peptide length preferences control CD8+ T cell responses. J. Immunol. 191, 561–571 [DOI] [PubMed] [Google Scholar]
- 33. Speir J. A., Stevens J., Joly E., Butcher G. W., Wilson I. A. (2001) Two different, highly exposed, bulged structures for an unusually long peptide bound to rat MHC class I RT1-Aa. Immunity 14, 81–92 [DOI] [PubMed] [Google Scholar]
- 34. Theodossis A., Guillonneau C., Welland A., Ely L. K., Clements C. S., Williamson N. A., Webb A. I., Wilce J. A., Mulder R. J., Dunstone M. A., Doherty P. C., McCluskey J., Purcell A. W., Turner S. J., Rossjohn J. (2010) Constraints within major histocompatibility complex class I restricted peptides: presentation and consequences for T-cell recognition. Proc. Natl. Acad. Sci. U.S.A. 107, 5534–5539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Koopmann J. O., Post M., Neefjes J. J., Hämmerling G. J., Momburg F. (1996) Translocation of long peptides by transporters associated with antigen processing (TAP). Eur. J. Immunol. 26, 1720–1728 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.