Background: Knockdown of orosomucoid-like (ORMDL) proteins releases serine palmitoyltransferase (SPT) activity.
Results: Significant changes in SPT activity were detected when all three ORMDLs were overexpressed. Sphingolipids do not modify SPT-ORMDL interaction but rearrange ORMDLs. Macrophages suppress ORMDLs to induce de novo ceramide synthesis.
Conclusion: Coordinated ORMDL expression regulation strongly influences SPT activity.
Significance: SPT-ORMDL complex presents transcriptional and post-translational regulation.
Keywords: Ceramide, Enzyme Inhibitor, Macrophage, Serine Palmitoyltransferase, Sphingolipid
Abstract
The orosomucoid-like (ORMDL) protein family is involved in the regulation of de novo sphingolipid synthesis, calcium homeostasis, and unfolded protein response. Single nucleotide polymorphisms (SNPs) that increase ORMDL3 expression have been associated with various immune/inflammatory diseases, although the pathophysiological mechanisms underlying this association are poorly understood. ORMDL proteins are claimed to be inhibitors of the serine palmitoyltransferase (SPT). However, it is not clear whether individual ORMDL expression levels have an impact on ceramide synthesis. The present study addressed the interaction with and regulation of SPT activity by ORMDLs to clarify their pathophysiological relevance. We have measured ceramide production in HEK293 cells incubated with palmitate as a direct substrate for SPT reaction. Our results showed that a coordinated overexpression of the three isoforms inhibits the enzyme completely, whereas individual ORMDLs are not as effective. Immunoprecipitation and fluorescence resonance energy transfer (FRET) studies showed that mammalian ORMDLs form oligomeric complexes that change conformation depending on cellular sphingolipid levels. Finally, using macrophages as a model, we demonstrate that mammalian cells modify ORMDL genes expression levels coordinately to regulate the de novo ceramide synthesis pathway. In conclusion, we have shown a physiological modulation of SPT activity by general ORMDL expression level regulation. Moreover, because single ORMDL3 protein alteration produces an incomplete inhibition of SPT activity, this work argues against the idea that ORMDL3 pathophysiology could be explained by a simple on/off mechanism on SPT activity.
Introduction
Orosomucoid-like (ORMDL)3 proteins belong to a family of transmembrane proteins that contains three members located in the endoplasmic reticulum (1). The association of ORMDL3 with childhood asthma in a genome-wide association study (2) stimulated renewed interest in the study of these proteins. Despite the absence of the single nucleotide polymorphism (SNP) identified in the genome-wide association study (rs 7216389) in a coding region of the gene, this first study correlated increased expression of ORMDL3 with the risk allele. Since then, several SNPs around the ORMDL3 gene that are associated with pathologies like inflammatory bowel disease, type I diabetes, and rheumatoid arthritis have been described (3–6).
The genome-wide association study approach to the diagnosis of genetic risk factors is not focused on precandidate genes, making it an excellent tool to identify new genes involved in diseases. However, sometimes the identified genes have ill defined functions as in the case of ORMDL3 at the time it was detected. To elucidate the pathophysiology associated with ORMDL3, several laboratories have been trying to understand the role of ORMDLs in cell physiology. Our laboratory has focused on the effects of ORMDL3 expression levels in calcium homeostasis, a likely connection between an endoplasmic reticulum-resident protein and immune system dysfunction. We have found that the expression levels of this protein are inversely correlated with the calcium content of the endoplasmic reticulum due to an inhibition of sarco/endoplasmic reticulum Ca2+-ATPase pump activity (7). In addition, we have shown that the store-operated calcium entry, the main calcium entry pathway during T cell activation, is altered because ORMDL3 decreases the calcium buffering capacity of the mitochondria and the subsequent calcium-dependent inactivation of the calcium release-activated Ca2+ channel (8).
Conversely, it has been shown that the presence of ORMDLs acts as a break for the de novo sphingolipid synthesis pathway (9, 10). In yeast and mammalian cells, the complete knockdown of ORMDLs releases serine palmitoyltransferase (SPT) activity and generates an increase in long chain bases and ceramides. The expression of any of the isoforms in this knockdown condition rescues the normal functioning of the pathway (10). This fact, together with the interaction between the yeast ORMDL isoforms (Orms) and the SPT enzyme, has established the idea that ORMDLs are the endogenous inhibitors of SPT. In addition, the SPT-Orm interaction is dependent on a phosphorylation reaction that disrupts an oligomeric complex of Orms and interferes with SPT-Orm interaction (10). The regulation of the pathway implicated in Orm phosphorylation and its sensitivity to ceramide cell content have been described in yeast (11). However, the role of this phosphorylation in SPT-ORMDL interaction is not clear in mammals because the N-terminal regulatory region described in yeasts is absent in mammalian ORMDLs. More remarkable is the lack of evidence that different ORMDL3 expression levels in mammalian cells alter SPT activity; this is a critical gap in understanding the pathophysiology associated with this gene.
We herein evaluate the role of mammalian ORMDLs in the ceramide synthesis context with three specific aims: (i) to explore the effect of ORMDL3 overexpression on SPT activity, (ii) to study the ORMDL-SPT complex interaction and its dependence on ceramide cell content, and (iii) to find a physiological context in which cells modify ORMDL expression levels to modulate SPT activity. For this purpose, we used HEK293 cells as the heterologous expression system and palmitate treatment to stimulate SPT activity. We performed coimmunoprecipitation studies between SPT-ORMDL complex elements and FRET studies to confirm and explore conformational changes. Moreover, we used the RAW264.7 monocytic cell line to study the regulation of ORMDL expression during de novo sphingolipid generation under the activation process.
MATERIALS AND METHODS
Cell Culture and Transfection
HEK293 and RAW264.7 cells were grown in DMEM (Sigma-Aldrich) supplemented with 10% heat-inactivated fetal calf serum, 100 units/ml penicillin, and 100 units/ml streptomycin. The cells were maintained in a 5% CO2 environment at 37 °C. HEK293 cells were transiently transfected with the polycationic transfecting reagent polyethylenimine (PEI) (Polysciences), incubating cells with 6 eq of PEI/μg of DNA for 5 h before changing to normal growing medium. Myriocin (10 μm) and C6-ceramide (10 μm) were both obtained from Sigma, and dimethyl sulfoxide (DMSO) was used as vehicle.
Ceramide Quantification
HEK293 cells were transfected with either pCDNA3 empty vector, pCDNA3-ORMDL1, pCDNA3-ORMDL2, pCDNA3-ORMDL3, pCDNA3-ORMDL3-Δ1–16,or the same final amount of all three ORMDL family members together. After 24 h of transfection, HEK293 cells were treated for 4 h with 500 μm palmitate (Sigma) complexed in 0.5% fatty acid-free BSA (Sigma), and BSA alone was used as a control. Cells were then washed twice with 1× PBS and centrifuged at 1800 rpm for 5 min at 4 °C, and the pellet was frozen in liquid nitrogen for ceramide quantification.
Lipid extraction and processing were performed as reported previously (12, 13). Lipid analysis was carried out by ultraperformance liquid chromatography coupled to time-of-flight (TOF) mass spectrometry in positive electrospray ionization mode. Instrument conditions were set as in previous studies (12, 13).
In Vitro LPS Stimulation
RAW264.7 cells were treated with 100 ng/ml Escherichia coli (055:B5) lipopolysaccharide (LPS) (Sigma-Aldrich) for the indicated time points. Total RNA or protein lysates were prepared from the cells and frozen at −80 °C until analysis. For ceramide quantification, RAW264.7 cells were washed twice in 1× PBS after LPS stimulation and centrifuged at 1800 rpm for 5 min at 4 °C, and the pellet was frozen in liquid nitrogen.
Western Blot and Immunoprecipitation Assays
HEK293 cells were transiently transfected with human ORMDL3-myc plus ORMDL1, ORMDL2, or ORMDL3 with different tags as indicated. In all conditions tested, cells were also transfected with a non-tagged ORMDL protein to ensure that all three proteins were present at the same ratio. After 24 h of transfection, cells were lysed with immunoprecipitation buffer (0.2% Triton X-100 plus protease inhibitor mixture in HEPES-buffered saline) and centrifuged at 100,000 × g to collect the total protein in supernatants. Then 1000 μg of total protein was incubated at 4 °C overnight with anti-myc antibody (Abcam) cross-linked with disuccinimidyl suberate (Pierce) to protein G-Sepharose beads. Immunocomplexes were washed with HEPES-buffered saline five times and eluted through a spin chromatography column with 0.2 m glycine, pH 2.5 before adding loading buffer and boiling for 5 min. Coimmunoprecipitation of ORMDL3-YFP, ORMDL1-YFP, or ORMDL2-YFP was detected by Western blot after separation by 4–12% gradient polyacrylamide gel electrophoresis and protein transfer to nitrocellulose membrane. Immunodetection was carried out using rabbit antibodies against ORMDL (1:300), SPTLC1 (1:300), and SPTLC2 (1:1000), all from Abcam. Secondary antibodies were horseradish peroxidase-conjugated anti-rabbit IgG (1:3000; GE Healthcare). The immunoreactive signal was detected by SuperSignal West Chemiluminescent substrate (Pierce) and visualized using the Molecular Imager Chemidoc XRS system (Bio-Rad).
In the case of RAW264.7 macrophages, after LPS stimulation, cells were washed with 1× PBS, lysed in 60 μl of lysis buffer (150 mm NaCl, 5 mm EDTA, 1% Triton X-100, 10 mm Tris-HCl, 1× Complete protease inhibitor) for 20 min on ice with agitation, and then centrifuged at 13,000 rpm at 4 °C for 30 min. Protein concentration in the supernatant was determined using the BCA Assay (Pierce). Equal amounts of protein (100 μg) were loaded into each lane and were separated on an SDS-polyacrylamide gel (12%).
Quantitative Real Time PCR Analysis
Total RNA of RAW264.7 cells was extracted using the Nucleospin RNA II kit (Macherey-Nagel) following the manufacturer's instructions. Total RNA (1 μg) was reverse transcribed to cDNA using the SuperScript-RT system (Invitrogen). Quantitative RT-PCR was performed on an ABI Prism 7900HT (Applied Biosystems) with SYBR Green (SYBR Green Power PCR Master Mix, Applied Biosystems). Gene-specific mouse primers used were: mORMDL1, 5′-GCATCCCCTTCTGCAGTGTT-3′ and 5′-CGGAGTCTCAAAAGGCGTTC-3′; mORMDL2, 5′-CGTCATCCATAACTTGGCAAT-3′ and 5′-AACTGTAGTCCATAGTCCATC-3′; mORMDL3, 5′-CTGCTGAGCATTCCCTTTGT-3′ and 5′-CACGGTGTGCAGAAAGATGT-3′; mβ-actin, 5′-TGGAATCCTGTGGCATCCATGAAAC-3′ and 5′-TAAAACGCAGCTCAGTAACAGTCCG-3′; mSPTLC1, 5′-GTCCCCTTCCAGAACTGGTTAAA-3′ and 5′-CCCATAGTGCTCGGTGACTC-3′; mSPTLC2, 5′-TGCAGCACTCGTCAGGAAAT-3′ and 5′-CAGGCAACCTTTGCCAACAA-3′. PCR conditions in all cases were 95 °C for 5 min, 95 °C for 30 s, 60 °C for 30 s, 72 °C for 30 s, and 72 °C for 5 min with 40 cycles of amplification.
Fluorescence Resonance Energy Transfer Experiments
Proximity of the different tails of ORMDLs was assessed by FRET experiments as described previously (14). Cells were seeded on 25-mm glass coverslips and transfected using the indicated combinations. In addition to the YFP/CFP protein pair studied, in each case, the rest of the ORMDLs were transfected without a tag to avoid artificial interactions. Acceptor photobleaching was carried out by gradual bleaching of the YFP acceptor on living cells using an SP2 confocal Leica microscope. The FRET value was expressed as the maximal CFP fluorescence increase compared with the initial value after bleaching of more than 90% of the YFP fluorescence signal. Images were analyzed using ImageJ software.
RESULTS
ORMDL Proteins Regulate de Novo Sphingolipid Synthesis
ORMDL proteins have been found to negatively regulate the activity of SPT, the enzyme that catalyzes the first rate-limiting step in de novo sphingolipid synthesis (10, 15). We used two different approaches, overexpression and silencing, to study how changes in ORMDL expression levels affect ceramide production and SPT activity. In overexpression experiments, monolayers of HEK293 cells were transiently transfected with either pCDNA3 empty vector (control), human pCDNA3-ORMDL1, -ORMDL2, -ORMDL3, or the combination of all three ORMDL proteins. For triple gene silencing, cells were treated with scrambled siRNA (control) and combined siRNA for ORMDL1, ORMDL2, and ORMDL3.
Knocking down ORMDL1–3 significantly increased the cellular ceramide content compared with scrambled control siRNA (Table 1) in agreement with a previous report in HeLa cells (15). All ceramide species contributed to the total increase with C16, C18, C22, and C24 the most abundant. On the contrary, there was no alteration of specific or total ceramide amount after overexpressing the individual ORMDL isoforms separately or overexpressing all three ORMDL proteins simultaneously compared with control (Table 1).
TABLE 1.
Ceramide content depending on ORMDL expression levels (pmol/1 × 106cells)
Ceramide content was quantified by mass spectrometry in HEK293 cells transiently transfected with empty vector (control), ORMDL1, ORMDL2, ORMDL3, or the same total amount of the three plasmids together (ORMDL123) for 24 h and with scrambled siRNA (siControl) or siRNA for the three members of the ORMDL family (si123) for 36 h (n = 3).
| Control | ORMDL1 | ORMDL2 | ORMDL3 | ORMDL123 | siControl | si123 | |
|---|---|---|---|---|---|---|---|
| C14 | 2.1 ± 0.3 | 2.8 ± 0.1 | 3.2 ± 0.5 | 3.2 ± 0.7 | 3.4 ± 0.5 | 0.8 ± 0.1 | 1.3 ± 0.0a |
| C16 | 60.3 ± 9.0 | 58.4 ± 0.6 | 75.8 ± 14 | 75.5 ± 18 | 77.4 ± 7.6 | 14.9 ± 1.7 | 29.9 ± 2.4a |
| C16:1 | 4.8 ± 0.8 | 5.5 ± 0.3 | 6.4 ± 1.6 | 6.5 ± 1.8 | 7.4 ± 0.9 | 1.4 ± 0.1 | 2.8 ± 0.2a |
| C18 | 9.9 ± 1.9 | 6.7 ± 3.2 | 12.5 ± 2.5 | 10.4 ± 1.4 | 11.0 ± 0.3 | 2.7 ± 0.5 | 5.7 ± 0.1a |
| C20 | 3.8 ± 1.0 | 2.5 ± 0.1 | 3.9 ± 0.8 | 3.6 ± 0.2 | 3.0 ± 0.7 | 0.9 ± 0.2 | 2.3 ± 0.4b |
| C22 | 28.2 ± 4.9 | 26.0 ± 0.6 | 33.0 ± 7.4 | 27.6 ± 2.9 | 29.4 ± 0.9 | 3.7 ± 1.8 | 15.3 ± 1.5a |
| C24 | 104.9 ± 16 | 98.4 ± 1.4 | 109.5 ± 22 | 89.1 ± 7.8 | 111.1 ± 6.8 | 25.0 ± 4.1 | 47.8 ± 3.3b |
| Total | 214.1 ± 34 | 201.3 ± 3.8 | 244.3 ± 48 | 215.8 ± 33 | 242.8 ± 15 | 49.4 ± 4.8 | 105 ± 7.5a |
a p < 0.01.
b p < 0.05.
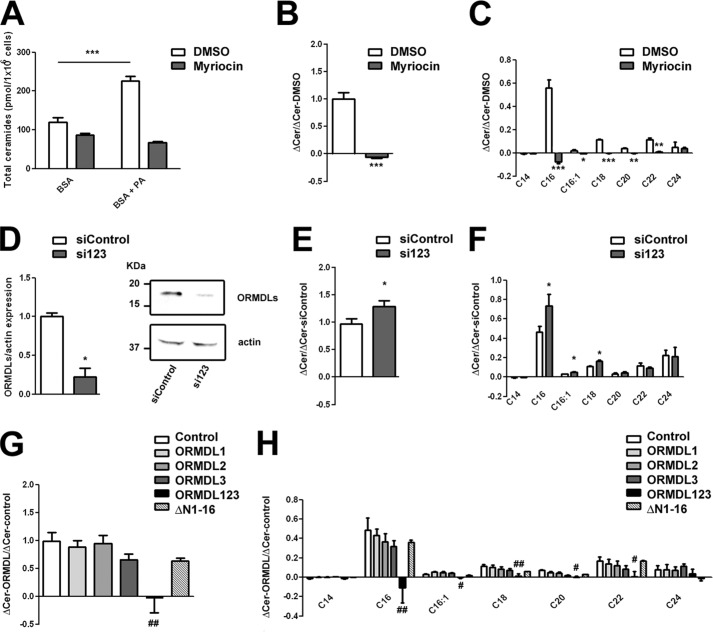
We also evaluated the production of ceramides induced by addition of palmitate (PA), the substrate of the SPT enzyme. The increase of ceramide levels observed after 4-h incubation with 500 μm PA conjugated with BSA compared with control cells incubated with BSA alone was blocked in the presence of myriocin, a specific SPT inhibitor. This blockage demonstrated that the ceramide production using this protocol came mainly from the de novo pathway as it has been described previously (16). We normalized the value of the difference in ceramide content between cells treated with PA and control cells in the presence of myriocin or DMSO (Fig. 1B) and studied the contribution of the different ceramide species to this increase. After a 4-h incubation with PA, the newly formed ceramides contained mainly the C16 species, an early ceramide species from the de novo synthesis pathway (Fig. 1C). We then decided to explore, following the same protocol and normalization, the production of ceramides in cells knocked down for the three ORMDL isoforms with specific siRNAs (Fig. 1D). We observed a significant increase in total ceramide production, especially the C16 species, after a 4-h PA treatment in cells knocked down for ORMDLs compared with control cells with scrambled siRNA (Fig. 1, E and F). These results further demonstrated, together with the higher ceramide content in cells knocked down for the ORMDLs (Table 1), that the absence of ORMDL members promotes SPT activity. Conversely, cells overexpressing individual ORMDL isoforms did not show altered ceramide production under PA incubation (Fig. 1, G and H). A similar lack of effect was observed in cells expressing an ORMDL3 mutant lacking the first 16 amino acids of the N-terminal tail (ΔN1–16) that are essential for ORMDL3 calcium signaling function (8) (Fig. 1, G and H). Remarkably, only the overexpression of all three ORMDL proteins provoked a complete blockage in ceramide production after PA treatment in a manner similar to myriocin (Fig. 1, G and H). These results indicate that all three ORMDL isoforms are needed to efficiently block SPT activity. To date, there is no clear mechanistic explanation for the regulation of SPT by ORMDL proteins in mammals. Our observation that coordinated expression changes in all ORMDL isoforms are required to modify SPT suggests the formation of a complex with close interaction among the three ORMDL members.
FIGURE 1.
ORMDLs modulate ceramide production. Ceramide content from HEK293 cells was quantified by mass spectrometry after incubation with 500 μm PA conjugated with 0.5% BSA or with vehicle (0.5% BSA alone). A–C, effect of myriocin on ceramide production under PA treatment. A, absolute numbers of total ceramides detected. B, difference in total ceramide (Cer) levels between cells incubated with PA and vehicle in the presence of myriocin versus the difference in the DMSO control value. C, same as in B but for the different ceramide species (n = 3; *, p < 0.05; **, p < 0.01; ***, p < 0.001). D and E, ceramide content quantification of cells transiently transfected for 36 h with scrambled siRNA (siControl) or siRNA against the three members of the ORMDL family (si123). D, representative Western blot and total quantification of the ORMDL expression repression. E, difference in total ceramide levels between cells incubated with PA and vehicle (0.5% BSA) of cells treated with si123 versus the difference in the scrambled siRNA value. F, same as in E but for the different ceramide species (n = 6; *, p < 0.05). G and H, ceramide content of cells transiently transfected with empty vector (Control), ORMDL1, ORMDL2, ORMDL3, or the same total amount of the three plasmids together (ORMDL123). G, difference in total ceramide levels between cells incubated with PA and vehicle (0.5% BSA) after transfection with the indicated plasmids versus the difference in the control value. H, same as in G but for the different ceramide species (n = 6–8; analysis of variance test compared with control; #, p < 0.05; ##, p < 0.01). Error bars represent S.E.
ORMDL Complex Formation and Rearrangement Depending on Lipid Environment
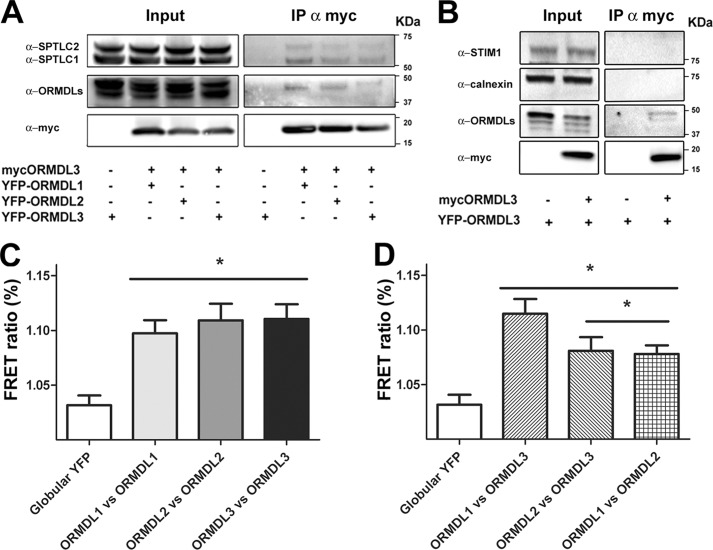
To evaluate possible interactions among ORMDL1, ORMDL2, and ORMDL3, immunoprecipitation assays were performed. HEK293 cells were transiently transfected with myc-ORMDL3 and either YFP-ORMDL1 or YFP-ORMDL2 to study hetero-oligomerization. For homo-oligomer formation studies of ORMDL3, cells were additionally transfected with YFP-ORMDL3. As a control, we used cells transfected with YFP-ORMDL3 in the absence of myc-ORMDL3. In all conditions tested, ORMDL isoforms without a tag were co-transfected at the same ratio to guarantee the presence of all members and to minimize artificial interactions due to the overexpression approach. Immunoprecipitation targeted the myc epitope in all cases, including the control condition. Coimmunoprecipitation was studied using an anti-ORMDL antibody, and the bands obtained migrated around 42 kDa, the expected size for the YFP-ORMDL constructs. As shown in Fig. 2A, mammalian ORMDL1 and ORMDL2 are able to directly interact with ORMDL3, forming heteromeric structures. These heteromeric structures showed the same extent of interaction (around 10%) as the homomeric structures of ORMDL3 as measured by band intensity analysis.
FIGURE 2.
ORMDLs form a complex with SPT. HEK293 cells were transfected with ORMDL1, ORMDL2, or ORMDL1 + ORMDL2 and the indicated tagged constructs to ensure every condition had all three members of the family. A and B, Western blot analysis of immunoprecipitations (IP) with an anti-myc antibody for the different conditions as indicated. C and D, FRET acceptor photobleaching studies of homomeric pairs, ORMDL1 versus ORMDL1 (ORMDL1-CFP + ORMDL1-YFP), ORMDL2 versus ORMDL2 (ORMDL2-CFP + ORMDL2-YFP), ORMDL3 versus ORMDL3 (ORMDL3-CFP + ORMDL3-YFP), and heteromeric pairs, ORMDL1 versus ORMDL3 (ORMDL1-CFP + ORMDL3-YFP), ORMDL2 versus ORMDL3 (ORMDL2-CFP + ORMDL3-YFP), and ORMDL1 versus ORMDL2 (ORMDL2-CFP + ORMDL1-YFP). Globular YFP, ORMDL3-CFP + PCDNA3-YFP. The data were obtained from three independent experiments. Error bars represent S.E. (n = 24; *, p < 0.05).
Regarding the formation of a complex between ORMDLs and SPT enzyme, we confirmed previous studies showing that ORMDL3 was able to interact with one subunit of the SPT complex, namely SPTLC1 (10). Moreover, we detected interaction between ORMDL3 and the catalytic subunit of the SPT complex, SPTLC2. The fraction of coimmunoprecipitation was around 4% with both subunits, demonstrating the existence of a functional ORMDL-SPT complex. Besides, we did not obtain coimmunoprecipitation with other endoplasmic reticulum membrane proteins such as STIM1 and calnexin (Fig. 2B). This result confirmed that the interactions we observed were not unspecific.
To further verify the observed interactions and extend the studies to all combinations of the three ORMDLs, the C terminus of ORMDL proteins was tagged with CFP/YFP FRET pairs. As in the previous experiments, the additional ORMDL isoforms were co-transfected at the same ratio without a tag to avoid artificial interactions. All tested combinations showed a positive FRET signal compared with the globular YFP control. Regarding the homomeric structures (ORMDL1-ORMDL1, ORMDL2-ORMDL2, and ORMDL3-ORMDL3), a similar FRET signal was obtained between the individual ORMDL isoforms (Fig. 2B). In the heteromerization studies, FRET signals for ORMDL1-ORMDL2 and ORMDL2-ORMDL3 were lower than those for ORMDL1-ORMDL3 (Fig. 2C). Considering that we observed no differences between homomeric and heteromeric interactions in the coimmunoprecipitation experiments, a possible explanation for the lower FRET signals in the ORMDL1-ORMDL2 and ORMDL2-ORMDL3 combinations is that the final conformation places the C terminus of ORMDL2 more distant from ORMDL1 and ORMDL3.
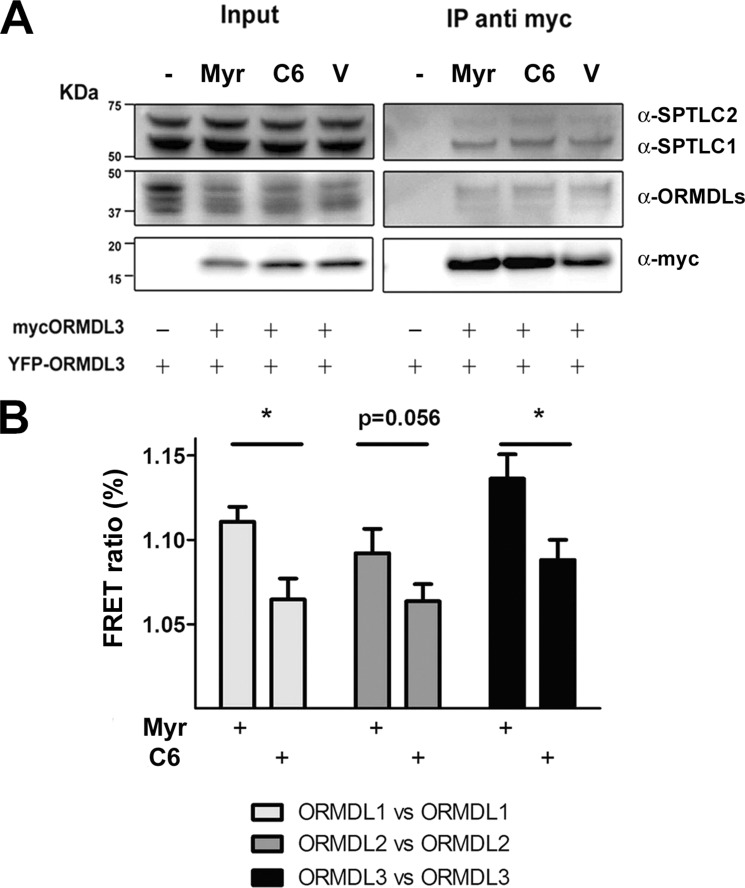
Previous studies revealed that complex formation by yeast ORMDL orthologs is influenced by the amount of cellular sphingolipid composition. A decreased sphingolipid content not only reduces Orm oligomerization but also the Orm-Lcb1 binding (10). This release is supposed to allow for new sphingolipid generation. To explore the influence of sphingolipid content of mammalian ORMDL-SPTLC complex formation, we studied this process in both low and high sphingolipid-content conditions. For a low sphingolipid environment, cells were treated with myriocin, a specific inhibitor of SPT; for a high sphingolipid environment, cells were incubated with C6-ceramide, a cell-permeable sphingolipid that has been shown to modulate SPT in an ORMDL-dependent manner (15). Our coimmunoprecipitation studies using a myc-ORMDL3 antibody showed that neither the ORMDL3-ORMDL3 interaction nor its binding to SPTLC1 or SPTLC2 was altered by changing the sphingolipidic environment (myriocin or C6-ceramide treatment) (Fig. 3A). These data suggest that the interaction of ORMDL-SPT complexes is independent of the sphingolipidic environment. To evaluate putative conformational changes of ORMDL homo-oligomers due to the sphingolipid composition, FRET experiments were performed in both low and high sphingolipid content conditions. Interestingly, FRET efficiency was significantly reduced in a high sphingolipid environment (C6-ceramide treatment) for all three ORMDL isoforms compared with low sphingolipid content (myriocin treatment) (Fig. 3B). Taken together, these results indicate the existence of a complex rearrangement between the ORMDL-SPTLC structures rather than a binding release mechanism.
FIGURE 3.
ORMDLs rearrange under different sphingolipid environments. HEK293 cells were transfected with ORMDL1, ORMDL2, or ORMDL1 + ORMDL2 and the indicated tagged constructs to ensure every condition had all three members of the family. Myriocin (Myr), C6-ceramide (C6), or DMSO (V) was applied to cells for 4 h. A, Western blot analysis of immunoprecipitations (IP) with an anti-myc antibody for the different conditions as indicated. B, FRET acceptor photobleaching studies between ORMDL members: pairs ORMDL1 versus ORMDL1 (ORMDL1-CFP + ORMDL1-YFP), ORMDL2 versus ORMDL2 (ORMDL2-CFP + ORMDL2-YFP), and ORMDL3 versus ORMDL3 (ORMDL3-CFP + ORMDL3-YFP). Globular YFP, ORMDL3-CFP + PCDNA3-YFP. The data were obtained from three independent experiments. Error bars represent S.E. (n = 24; *, p < 0.05).
Role of ORMDLs in Macrophage Activation
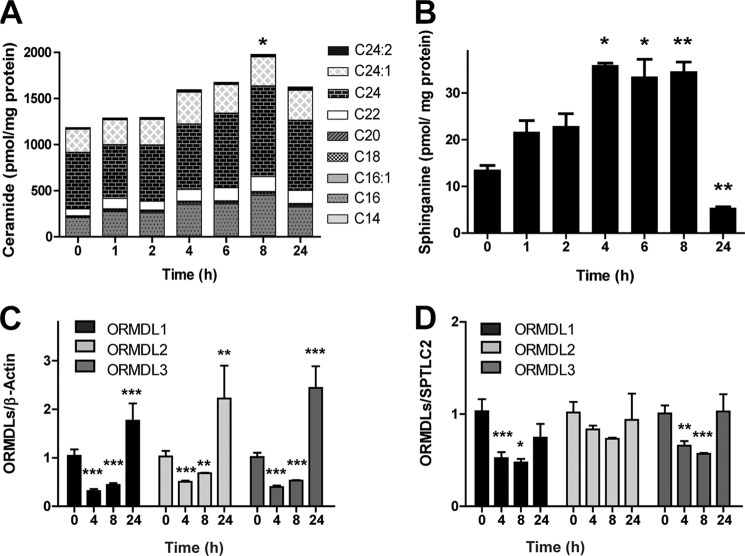
The importance of the ORMDL complex formation was further studied in cells of the innate immune response, a more physiological system. Previous studies have shown an increase in ceramide levels after activation of macrophages with bacterial LPS (16–19). In this process, the contribution of the de novo synthesis pathway at early time points is necessary for proper activation (19, 20), making it an interesting process for use in evaluating the role of the ORMDL complex in the regulation of ceramide levels. Therefore, RAW264.7 cells were treated with LPS, and the resulting changes in the sphingolipid content were monitored. Ceramide content increased steadily during the time of LPS activation, showing a peak after 8 h that decreases after long time activation (Fig. 4A). A similar trend was exhibited by sphinganine, another intermediate metabolite in the de novo ceramide synthesis pathway (Fig. 4B). These results agree with an early activation of the de novo pathway.
FIGURE 4.
Reduced ORMDL/SPT ratio promotes de novo ceramide synthesis in activated macrophages. RAW267.4 cells were treated with 100 ng/ml LPS for 24 h. A, time course of total ceramide content in activated RAW cells. Bars are divided into the different ceramide species detected. Statistical analysis considers the total ceramide content. B, time course of sphinganine content in activated RAW267.4 cells. C and D, gene expression analyzed by RT-PCR of ORMDLs during activation. Data are normalized to β-actin (C) and SPTLC2 (D). Error bars represent S.E. (n = 5; *, p < 0.05; **, p < 0.01; ***, p < 0.001).
The endogenous expression of the ORMDLs after macrophage activation revealed a time-dependent regulation at both transcriptional and translational levels of all three ORMDL isoforms with a significant decrease at early time points (4 and 8 h) (Fig. 4C) followed by an increase at later times (24 h). To correlate the reduction of ORMDL expression with an increase in ceramide production, we compared the expression levels of ORMDLs with SPTLC2, the main SPT present in these cells. We observed a reduced ORMDL/SPTLC2 ratio during higher de novo ceramide production (Fig. 4D).
DISCUSSION
Complete absence of ORMDL protein expression in mammalian and yeast cells leads to an overproduction of ceramides through the de novo synthesis pathway (9, 10, 15). This fact, together with the positive coimmunoprecipitation of ORMDLs and SPT, supports the scenario in which ORMDLs are endogenous inhibitors of the first enzyme of de novo sphingolipid synthesis. Conversely, the rescue of normal growth on the Orm1/Orm2 double KO yeast strain by the expression of mammalian ORMDL3 isoform has generated the idea that all the ORMDL members have redundant functions and that this is a regulatory mechanism conserved throughout evolution (10). In addition to the different regulatory elements described in yeast, it has been shown that ORMDLs participate in the cascade that senses and inhibits SPT depending on the intracellular ceramide content (11, 15). However, the current knowledge regarding ORMDL function in controlling sphingolipid synthesis still has many blanks that hamper the understanding of the pathophysiological role of ORMDL3, a gene implicated as a risk factor in several diseases (2–6). The present study tries to cast light on the relationship between mammalian ORMDLs and SPT.
Until now it was not known whether single variations in ORMDL3 expression could cause changes in SPT function and cellular sphingolipid content. This information is important to understand whether SNPs that exert a cis regulation on ORMDL3 gene expression, claimed to be risk factors of several pathologies (2, 21), could be linked to the function of this protein in sphingolipid synthesis. Our results show that increased expression levels of each different ORMDL or the three isoforms together do not affect the ceramide content of the cell. Interestingly, triple overexpression blocks SPT function when stimulating the de novo ceramide synthesis with palmitate. These findings can only be explained by additional regulatory mechanisms besides changes in expression. Thus, ORMDLs would not be allowed to lower the homeostatic content of ceramides in the cell, but their presence suffices to block SPT when ceramides increase to putatively deleterious levels. This effect is observed only in triple overexpression, meaning that all three members are needed for an effective sensing and blockage of the de novo pathway. Nevertheless, our results on the effect of ORMDL expression on SPT activity have to be cautiously interpreted as they may be partially dependent on the experimental settings, i.e. cell type and palmitate treatment. In this sense, we have modified exclusively ORMDL expression levels to mimic, in our in vitro model, as much as possible the pathophysiological context related to the diseases in which ORMDL3 has been implicated. Genetic analysis of ORMDL3-related inflammatory diseases only pointed to changes in ORMDL3 expression without information about possible changes in other elements participating in the SPT-ORMDL complex. In this context and based on the fact that overexpression of ORMDL3 alone did not modify ceramide content and palmitate-induced sphingolipid synthesis, our results do not support a pathophysiological mechanism based on the complete SPT inhibition. Besides, deletion of the first 16 amino acids of the ORMDL3 N-terminal tail has been shown to abolish its impact on calcium signaling (8). However, the lack of differences regarding ceramide content and production observed in the present work for overexpression of this mutant and wild type ORMDL3 argues against an explanation for the effect of ORMDL3 on calcium homeostasis based on its role in sphingolipid synthesis.
Another aspect we have addressed in this work is the dynamic regulation of ORMDL-SPTLC complexes that has been proposed in light of the experiments performed in yeasts (10). Our results demonstrate that the regulation based on interaction of the yeast ORMDL isoforms is lost in mammalian ORMDLs. The immunoprecipitation experiments did not support the occurrence of changes in the ORMDL-ORMDL or ORMDL-SPTLC interaction when the ceramide content of the cell was manipulated. Interestingly, another regulatory mechanism based on ORMDL phosphorylation via the TORC1 pathway that does not modify the interaction of ORMDL oligomers necessary for complex sphingolipid formation has been found in yeast (22). Whether this modulation is similar to the conformational changes observed in our experiments is a question that requires further investigation. It is also important to note that the regulation described in yeast is based on the phosphorylation of the N-terminal part of yeast Orms (10, 11). This domain is only present in yeast and plants, whereas vertebrate proteins cluster differently (1). Regarding the stoichiometry of the complex, we propose that ORMDLs are able to form homo- and hetero-oligomers with similar affinity because in our experiments we always overexpressed all the ORMDL members to avoid artificial interactions. Hetero-oligomerization likely results in a slightly different conformation based on the different FRET observed. Finally, our results also suggest that ORMDL regulation of SPT might be achieved by structural rearrangements depending on the intracellular ceramide content that do not involve changes in ORMDL-SPTLC binding affinity.
Finally, we demonstrate for the first time using physiological stimuli that mammalian cells modulate ceramide content by modifying expression levels of the ORMDL family. We have focused on macrophages where de novo ceramide synthesis has been characterized during LPS activation (16–18). The increase in ceramides through the SPT pathway is necessary for correct activation, and its blockage affects several processes such as autophagy and interleukin production (19, 20). In this context, we show that expression of the three ORMDL members is repressed at early time points in correlation with ceramide synthesis induction. The reduction in the ORMDL/SPTLC ratio suggests a regulatory scenario in which ceramide synthesis is induced by reducing the expression of the inhibitory subunits of the SPT complex. These results agree with the heterologous expression analysis where the triple knockdown of ORMDLs is necessary to release SPT activity (15).
Our work reinforces the idea that ORMDLs are negative modulators of SPT activity, providing physiological evidence for this role. However, increased individual expression of these proteins does not reduce the ceramide content in the cell, which conflicts with the idea that the pathophysiological link between disease-associated SNPs that change ORMDL3 expression is related to changes in cellular sphingolipid content due to a complete inhibition of the de novo pathway. This also implies that alternative mechanisms should be considered in determining the role of ORMDLs in immune/inflammatory diseases, i.e. the control of calcium homeostasis (7, 8) and the unfolded protein response (23, 24).
This work was supported by Spanish Ministry of Economy and Competitiveness Grants SAF2010-16725 and SAF2011-22444, Fondo de Investigación Sanitaria Red HERACLES Grant RD12/0042/0014, Fondos Europeos de Desarrollo Regional, Generalitat de Catalunya Grant SGR2014, and Fundació la Marató de TV3 Grant 20134030.
- ORMDL
- orosomucoid-like
- SPT
- serine palmitoyltransferase
- CFP
- cyan fluorescent protein
- PA
- palmitate.
REFERENCES
- 1. Hjelmqvist L., Tuson M., Marfany G., Herrero E., Balcells S., Gonzàlez-Duarte R. (2002) ORMDL proteins are a conserved new family of endoplasmic reticulum membrane proteins. Genome Biol. 3, RESEARCH0027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Moffatt M. F., Kabesch M., Liang L., Dixon A. L., Strachan D., Heath S., Depner M., von Berg A., Bufe A., Rietschel E., Heinzmann A., Simma B., Frischer T., Willis-Owen S. A., Wong K. C., Illig T., Vogelberg C., Weiland S. K., von Mutius E., Abecasis G. R., Farrall M., Gut I. G., Lathrop G. M., Cookson W. O. (2007) Genetic variants regulating ORMDL3 expression contribute to the risk of childhood asthma. Nature 448, 470–473 [DOI] [PubMed] [Google Scholar]
- 3. McGovern D. P., Gardet A., Törkvist L., Goyette P., Essers J., Taylor K. D., Neale B. M., Ong R. T., Lagacé C., Li C., Green T., Stevens C. R., Beauchamp C., Fleshner P. R., Carlson M., D'Amato M., Halfvarson J., Hibberd M. L., Lördal M., Padyukov L., Andriulli A., Colombo E., Latiano A., Palmieri O., Bernard E. J., Deslandres C., Hommes D. W., de Jong D. J., Stokkers P. C., Weersma R. K., NIDDK IBD Genetics Consortium, Sharma Y., Silverberg M. S., Cho J. H., Wu J., Roeder K., Brant S. R., Schumm L. P., Duerr R. H., Dubinsky M. C., Glazer N. L., Haritunians T., Ippoliti A., Melmed G. Y., Siscovick D. S., Vasiliauskas E. A., Targan S. R., Annese V., Wijmenga C., Pettersson S., Rotter J. I., Xavier R. J., Daly M. J., Rioux J. D., Seielstad M. (2010) Genome-wide association identifies multiple ulcerative colitis susceptibility loci. Nat. Genet. 42, 332–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Barrett J. C., Hansoul S., Nicolae D. L., Cho J. H., Duerr R. H., Rioux J. D., Brant S. R., Silverberg M. S., Taylor K. D., Barmada M. M., Bitton A., Dassopoulos T., Datta L. W., Green T., Griffiths A. M., Kistner E. O., Murtha M. T., Regueiro M. D., Rotter J. I., Schumm L. P., Steinhart A. H., Targan S. R., Xavier R. J., NIDDK IBD Genetics Consortium, Libioulle C., Sandor C., Lathrop M., Belaiche J., Dewit O., Gut I., Heath S., Laukens D., Mni M., Rutgeerts P., Van Gossum A., Zelenika D., Franchimont D., Hugot J. P., de Vos M., Vermeire S., Louis E., Belgian-French IBD Consortium, Wellcome Trust Case Control Consortium, Cardon L. R., Anderson C. A., Drummond H., Nimmo E., Ahmad T., Prescott N. J., Onnie C. M., Fisher S. A., Marchini J., Ghori J., Bumpstead S., Gwilliam R., Tremelling M., Deloukas P., Mansfield J., Jewell D., Satsangi J., Mathew C. G., Parkes M., Georges M., Daly M. J. (2008) Genome-wide association defines more than 30 distinct susceptibility loci for Crohn's disease. Nat. Genet. 40, 955–962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Barrett J. C., Clayton D. G., Concannon P., Akolkar B., Cooper J. D., Erlich H. A., Julier C., Morahan G., Nerup J., Nierras C., Plagnol V., Pociot F., Schuilenburg H., Smyth D. J., Stevens H., Todd J. A., Walker N. M., Rich S. S. (2009) Genome-wide association study and meta-analysis find that over 40 loci affect risk of type 1 diabetes. Nat. Genet. 41, 703–707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kurreeman F. A., Stahl E. A., Okada Y., Liao K., Diogo D., Raychaudhuri S., Freudenberg J., Kochi Y., Patsopoulos N. A., Gupta N., CLEAR investigators, Sandor C., Bang S. Y., Lee H. S., Padyukov L., Suzuki A., Siminovitch K., Worthington J., Gregersen P. K., Hughes L. B., Reynolds R. J., Bridges S. L., Jr., Bae S. C., Yamamoto K., Plenge R. M. (2012) Use of a multiethnic approach to identify rheumatoid-arthritis-susceptibility loci, 1p36 and 17q12. Am. J. Hum. Genet. 90, 524–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cantero-Recasens G., Fandos C., Rubio-Moscardo F., Valverde M. A., Vicente R. (2010) The asthma-associated ORMDL3 gene product regulates endoplasmic reticulum-mediated calcium signaling and cellular stress. Hum. Mol. Genet. 19, 111–121 [DOI] [PubMed] [Google Scholar]
- 8. Carreras-Sureda A., Cantero-Recasens G., Rubio-Moscardo F., Kiefer K., Peinelt C., Niemeyer B. A., Valverde M. A., Vicente R. (2013) ORMDL3 modulates store-operated calcium entry and lymphocyte activation. Hum. Mol. Genet. 22, 519–530 [DOI] [PubMed] [Google Scholar]
- 9. Han S., Lone M. A., Schneiter R., Chang A. (2010) Orm1 and Orm2 are conserved endoplasmic reticulum membrane proteins regulating lipid homeostasis and protein quality control. Proc. Natl. Acad. Sci. U.S.A. 107, 5851–5856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Breslow D. K., Collins S. R., Bodenmiller B., Aebersold R., Simons K., Shevchenko A., Ejsing C. S., Weissman J. S. (2010) Orm family proteins mediate sphingolipid homeostasis. Nature 463, 1048–1053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Roelants F. M., Breslow D. K., Muir A., Weissman J. S., Thorner J. (2011) Protein kinase Ypk1 phosphorylates regulatory proteins Orm1 and Orm2 to control sphingolipid homeostasis in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 108, 19222–19227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Munoz-Olaya J. M., Matabosch X., Bedia C., Egido-Gabás M., Casas J., Llebaria A., Delgado A., Fabriàs G. (2008) Synthesis and biological activity of a novel inhibitor of dihydroceramide desaturase. ChemMedChem 3, 946–953 [DOI] [PubMed] [Google Scholar]
- 13. Canals D., Mormeneo D., Fabriàs G., Llebaria A., Casas J., Delgado A. (2009) Synthesis and biological properties of pachastrissamine (jaspine B) and diastereoisomeric jaspines. Bioorg. Med. Chem. 17, 235–241 [DOI] [PubMed] [Google Scholar]
- 14. Garcia-Elias A., Mrkonjic S., Pardo-Pastor C., Inada H., Hellmich U. A., Rubio-Moscardó F., Plata C., Gaudet R., Vicente R., Valverde M. A. (2013) Phosphatidylinositol-4,5-biphosphate-dependent rearrangement of TRPV4 cytosolic tails enables channel activation by physiological stimuli. Proc. Natl. Acad. Sci. U.S.A. 110, 9553–9558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Siow D. L., Wattenberg B. W. (2012) Mammalian ORMDL proteins mediate the feedback response in ceramide biosynthesis. J. Biol. Chem. 287, 40198–40204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Schilling J. D., Machkovech H. M., He L., Sidhu R., Fujiwara H., Weber K., Ory D. S., Schaffer J. E. (2013) Palmitate and lipopolysaccharide trigger synergistic ceramide production in primary macrophages. J. Biol. Chem. 288, 2923–2932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Andreyev A. Y., Fahy E., Guan Z., Kelly S., Li X., McDonald J. G., Milne S., Myers D., Park H., Ryan A., Thompson B. M., Wang E., Zhao Y., Brown H. A., Merrill A. H., Raetz C. R., Russell D. W., Subramaniam S., Dennis E. A. (2010) Subcellular organelle lipidomics in TLR-4-activated macrophages. J. Lipid Res. 51, 2785–2797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chang Z. Q., Lee S. Y., Kim H. J., Kim J. R., Kim S. J., Hong I. K., Oh B. C., Choi C. S., Goldberg I. J., Park T. S. (2011) Endotoxin activates de novo sphingolipid biosynthesis via nuclear factor κB-mediated upregulation of Sptlc2. Prostaglandins Other Lipid Mediat. 94, 44–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sims K., Haynes C. A., Kelly S., Allegood J. C., Wang E., Momin A., Leipelt M., Reichart D., Glass C. K., Sullards M. C., Merrill A. H., Jr. (2010) Kdo2-lipid A, a TLR4-specific agonist, induces de novo sphingolipid biosynthesis in RAW264.7 macrophages, which is essential for induction of autophagy. J. Biol. Chem. 285, 38568–38579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yu H., Valerio M., Bielawski J. (2013) Fenretinide inhibited de novo ceramide synthesis and proinflammatory cytokines induced by Aggregatibacter actinomycetemcomitans. J. Lipid Res. 54, 189–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Verlaan D. J., Berlivet S., Hunninghake G. M., Madore A. M., Larivière M., Moussette S., Grundberg E., Kwan T., Ouimet M., Ge B., Hoberman R., Swiatek M., Dias J., Lam K. C., Koka V., Harmsen E., Soto-Quiros M., Avila L., Celedón J. C., Weiss S. T., Dewar K., Sinnett D., Laprise C., Raby B. A., Pastinen T., Naumova A. K. (2009) Allele-specific chromatin remodeling in the ZPBP2/GSDMB/ORMDL3 locus associated with the risk of asthma and autoimmune disease. Am. J. Hum. Genet. 85, 377–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Shimobayashi M., Oppliger W., Moes S., Jenö P., Hall M. N. (2013) TORC1-regulated protein kinase Npr1 phosphorylates Orm to stimulate complex sphingolipid synthesis. Mol. Biol. Cell 24, 870–881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Miller M., Tam A. B., Cho J. Y., Doherty T. A., Pham A., Khorram N., Rosenthal P., Mueller J. L., Hoffman H. M., Suzukawa M., Niwa M., Broide D. H. (2012) ORMDL3 is an inducible lung epithelial gene regulating metalloproteases, chemokines, OAS, and ATF6. Proc. Natl. Acad. Sci. U.S.A. 109, 16648–16653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Miller M., Rosenthal P., Beppu A., Mueller J. L., Hoffman H. M., Tam A. B., Doherty T. A., McGeough M. D., Pena C. A., Suzukawa M., Niwa M., Broide D. H. (2014) ORMDL3 transgenic mice have increased airway remodeling and airway responsiveness characteristic of asthma. J. Immunol. 192, 3475–3487 [DOI] [PMC free article] [PubMed] [Google Scholar]