Background: The Pseudomonas aeruginosa cytotoxin ExoU localizes to the plasma membrane in eukaryotic cells.
Results: ExoU and related proteins utilize a conserved four-helical bundle to bind the lipid phosphatidylinositol 4,5-bisphosphate for localization.
Conclusion: The membrane localization domain of ExoU represents a novel phosphoinositide binding domain.
Significance: This is the first report of a four-helical bundle with specificity for phosphatidylinositol 4,5-bisphosphate.
Keywords: Bacterial Pathogenesis, Bacterial Toxin, Crystal Structure, Phosphatidylinositol Signaling, Phospholipase, Pseudomonas aeruginosa (P. aeruginosa)
Abstract
Bacterial toxins require localization to specific intracellular compartments following injection into host cells. In this study, we examined the membrane targeting of a broad family of bacterial proteins, the patatin-like phospholipases. The best characterized member of this family is ExoU, an effector of the Pseudomonas aeruginosa type III secretion system. Upon injection into host cells, ExoU localizes to the plasma membrane, where it uses its phospholipase A2 activity to lyse infected cells. The targeting mechanism of ExoU is poorly characterized, but it was recently found to bind to the phospholipid phosphatidylinositol 4,5-bisphosphate (PI(4,5)P2), a marker for the plasma membrane of eukaryotic cells. We confirmed that the membrane localization domain (MLD) of ExoU had a direct affinity for PI(4,5)P2, and we determined that this binding was required for ExoU localization. Previously uncharacterized ExoU homologs from Pseudomonas fluorescens and Photorhabdus asymbiotica also localized to the plasma membrane and required PI(4,5)P2 for this localization. A conserved arginine within the MLD was critical for interaction of each protein with PI(4,5)P2 and for localization. Furthermore, we determined the crystal structure of the full-length P. fluorescens ExoU and found that it was similar to that of P. aeruginosa ExoU. Each MLD contains a four-helical bundle, with the conserved arginine exposed at its cap to allow for interaction with the negatively charged PI(4,5)P2. Overall, these findings provide a structural explanation for the targeting of patatin-like phospholipases to the plasma membrane and define the MLD of ExoU as a member of a new class of PI(4,5)P2 binding domains.
Introduction
Bacteria use a number of mechanisms to inject toxins into eukaryotic cells (1). Once inside these cells, the toxins perform a variety of functions to disrupt host cell physiology (2–4). To adequately perform these functions, it is critical that toxins are localized to the appropriate intracellular compartment. A number of bacterial toxins have dedicated membrane localization domains (MLDs)4 that vary considerably in their targeting specificities and mechanisms (5). For instance, the botulinum neurotoxin A concentrates at the plasma membrane of neurons by direct affinity to the protein SNAP-25 (6). Other proteins localize to different membrane compartments by undergoing covalent lipid modification, such as the Salmonella effector SifA, which targets the plasma membrane following farnesylation (7). Still other proteins, such as the Pasteurella multocida toxin, have direct affinity for lipids (8). The many sophisticated and resourceful mechanisms by which bacterial toxins undergo intracellular targeting underscore the importance of proper localization to toxin function (7, 9). Although the examples listed above illustrate several well characterized mechanisms of toxin localization, it remains unclear how the majority of bacterial toxins are targeted to the appropriate intracellular compartment.
Patatin-like phospholipases are an important and intriguing family of microbial proteins. They are defined by the presence of a patatin domain, which encodes for phospholipase A2 (PLA2) activity that cleaves phospholipids at the sn-2 position (4). Patatin-like phospholipases are closely related to eukaryotic group IV cytosolic PLA2 (cPLA2) and group VI calcium-independent PLA2 enzymes, which share a defined serine-aspartate catalytic dyad (10, 11). While only recently identified, this family of proteins is quite large; ∼4400 potential proteins containing typical patatin domains are encoded in sequenced bacterial genomes (12, 13). Only a few of these proteins have been characterized, but these limited studies have found that patatin-like phospholipases can be delivered by type III, type IV, or type V secretion systems into host cells (11, 14, 15). Once in the eukaryotic intracellular environment, the PLA2 activities are associated with host cell death, disruption of signaling pathways, and bacterial internalization (15, 16, 18).
Arguably, the best characterized member of the family of patatin-like phospholipases is ExoU of Pseudomonas aeruginosa. This protein was initially discovered based upon its ability to confer cytotoxicity to P. aeruginosa strains and its secretion by the type III secretion system (19–21). Subsequent studies have defined it as one of the predominant virulence determinants of P. aeruginosa, as the secretion of ExoU is associated with poor clinical outcomes in human patients and with more severe infection in animal models (22, 23). ExoU does not act as a PLA2 enzyme on its own but instead requires eukaryotic host cofactors to be activated (24). Ubiquitin and ubiquitinated proteins have been identified as important for the activation of ExoU (25), as has the lipid phosphatidylinositol 4,5-bisphosphate (PI(4,5)P2) (26). Together, ubiquitin and PI(4,5)P2 activate ExoU synergistically, with each being important to the cytotoxic action of ExoU (26).
The C-terminal MLD of ExoU is necessary and sufficient for targeting of this effector protein to the plasma membrane (27). The crystal structure of ExoU in complex with its bacterial chaperone SpcU revealed that the MLD region encompasses residues 503–687 (28, 31). Furthermore, the C-terminal half (residues 604–687) of the MLD forms a four-helical bundle, a structural motif used by other toxins to target the plasma membrane (8, 29). Several key residues within the four-helical bundle have been identified as being critical to ExoU localization (27, 30), but the mechanism by which the MLD specifically recognizes the plasma membrane is unclear. Interestingly, it was recently shown that purified recombinant ExoU bound to PI(4,5)P2 immobilized on a solid support and that this binding required the MLD (31). Subsequent work has established that ExoU has a high affinity for PI(4,5)P2 and can use PI(4,5)P2 as a substrate (32). PI(4,5)P2 is a phospholipid found in the inner leaflet of the plasma membrane of eukaryotic cells (but not bacteria) (33), where it is recognized as a “signpost” to appropriately direct a number of endogenous proteins to this intracellular compartment (34). To dock at the plasma membrane, these proteins use a number of well characterized PI(4,5)P2 binding domains (e.g. pleckstrin homology, PSD-95/Dlg/ZO-1, and FERM domains), which play important roles in numerous cellular processes (35, 36). Although ExoU does not contain a known PI(4,5)P2 binding domain, its high affinity for PI(4,5)P2 has led others to hypothesize that it may localize to the plasma membrane by binding PI(4,5)P2 (31, 32). Thus, it is conceivable that the MLD of ExoU contains a novel PI(4,5)P2 binding domain.
In this study, we demonstrate that ExoU localizes to the plasma membrane through direct binding of the MLD to PI(4,5)P2. We extended these findings to patatin-like phospholipases of Pseudomonas fluorescens and Photorhabdus asymbiotica, demonstrating that the MLD of ExoU is a domain used by proteins of other bacterial genera and species to target the plasma membrane. To gain more information about this family of MLDs, we determined the crystal structure of the ExoU homolog of P. fluorescens. This structure was used along with the previously determined structure of P. aeruginosa ExoU to show that the four-helical bundle is conserved within the MLD and is positioned to expose a conserved positively charged arginine residue to interact with the negatively charged phosphate groups of PI(4,5)P2 in the plasma membrane. These results define a novel PI(4,5)P2 binding domain used by bacterial proteins to target the plasma membrane of host cells.
EXPERIMENTAL PROCEDURES
Cell Lines, Bacterial and Yeast Strains, and Media
Bacterial and yeast strains are listed in Tables 1 and 2, respectively. Escherichia coli strains were grown in Luria-Bertani (LB) broth, and when appropriate, media were supplemented with 100 μg/ml ampicillin. P. fluorescens strain A506 was a generous gift from Joyce Loper. P. asymbiotica strain ATCC 43949 was acquired from the ATCC. Saccharomyces cerevisiae strains SEY6210 (wild-type) and Δinp54/Δsac1, which were generous gifts from Peter Mayinger, were grown in yeast extract peptone dextrose (YPD) medium. Yeast strains expressing glucose-repressible, galactose-inducible pYC vectors were grown in synthetic complete medium lacking uracil and supplemented with 2% glucose (SC-ura + Glu) or 2% galactose, 1% raffinose (SC-ura + Gal) for varied expression of proteins (26). HeLa cells were cultured in modified Eagle's medium supplemented with 10% fetal bovine serum (Invitrogen).
TABLE 1.
Bacterial strains and plasmids
| Name | Relevant characteristics | Ref./Source |
|---|---|---|
| P. fluorescens A506 | Contains exoU homolog | Loper et al. (60) |
| P. asymbiotica ATCC 43949 | Contains exoU homolog | ATCC |
| E. coli strains | ||
| Top10 | F− mcrA Δ(mrr-hsdRMS-mcrBC) ϕ80lacZΔM15 ΔlacX74 recA1 araD139 Δ(ara-leu)7697 galU galK rpsL (StrR) endA1 nupG | Invitrogen |
| BL21 | F-ompT hsdSB (rB-mB-) gal dcm (DE3) | Invitrogen |
| E. coli Purification Plasmids | ||
| pEcoli-Cterm 6×HN | E. coli purification vector with C-terminal HN tag; Ampr | Clontech |
| pEcoli-Nterm 6×HN | E. coli purification vector with N-terminal HN tag; Ampr | Clontech |
| pEcoli-Cterm 6×HN-ExoUP.aer | For purification of full-length ExoUP.aer | 30 |
| pEcoli-Cterm 6×HN-ExoUP.flu | For purification of full-length ExoUP.flu | This study |
| pEcoli-Cterm 6×HN-ExoUP.asy | For purification of full-length ExoUP.asy | This study |
| pEcoli-Nterm 6×HN-ExoUP.aer MLD | For purification of ExoUP.aer MLD (503–687) | This study |
| pEcoli-Nterm 6×HN-ExoUP.aer MLD (LS608) | ExoUP.aer MLD construct with LS608 linker insertion | This study |
| pEcoli-Nterm 6×HN-ExoUP.aer MLD (I609N) | ExoUP.aer MLD construct with I609N substitution | This study |
| pEcoli-Nterm 6×HN-ExoUP.aer MLD (Q623R) | ExoUP.aer MLD construct with Q623R substitution | This study |
| pEcoli-Nterm 6×HN-ExoUP.aer MLD (N627I) | ExoUP.aer MLD construct with N627I substitution | This study |
| pEcoli-Nterm 6×HN-ExoUP.aer MLD (I654N) | ExoUP.aer MLD construct with I654N substitution | This study |
| pEcoli-Nterm 6×HN-ExoUP.aer MLD (R661L) | ExoUP.aer MLD construct with R661L substitution | This study |
| pEcoli-Nterm 6×HN-ExoUP.aer MLD (A678D) | ExoUP.aer MLD construct with A678D substitution | This study |
| pEcoli-Nterm 6×HN-ExoUP.flu MLD | For purification of ExoUP.flu MLD (453–639) | This study |
| pEcoli-Nterm 6×HN-ExoUP.flu MLD (R616L) | ExoUP.flu MLD construct with R616L substitution | This study |
| pEcoli-Nterm 6×HN-ExoUP.asy MLD | For purification of ExoUP.asy MLD (495–676) | This study |
| pEcoli-Nterm 6×HN-ExoUP.asy MLD (R652L) | ExoUP.asy MLD construct with R652L substitution | This study |
| E. coli transfection plasmids | ||
| pcDNA3.1 NT-GFP | Mammalian expression plasmid with N-terminal GFP tag; Ampr | Invitrogen |
| pcDNA3.1 ExoUP.aer MLD-GFP | Plasmid for expression of ExoUP.aer MLD-GFP | This study |
| pcDNA3.1 ExoUP.aer MLD (R661L)-GFP | ExoUP.aer MLD construct with R661L substitution | This study |
| pcDNA3.1 ExoUP.flu MLD-GFP | Plasmid for expression of ExoUP.flu MLD-GFP | This study |
| pcDNA3.1 ExoUP.flu MLD (R616L)-GFP | ExoUP.flu MLD construct with R616L substitution | This study |
| pcDNA3.1 ExoUP.asy MLD-GFP | Plasmid for expression of ExoUP.asy MLD-GFP | This study |
| pcDNA3.1 ExoUP.asy MLD (R652L)-GFP | ExoUP.asy MLD construct with R652L substitution | This study |
| pcDNA3.1 ExoUP.aer-GFP | Plasmid for expression of ExoUP.aer in mammalian cells | 65 |
| pcDNA3.1 ExoUP.aer-S142A-GFP | pcDNA3.1 ExoUP.aer-GFP with S142A substitution | 65 |
| pcDNA3.1 ExoUP.flu-GFP | For expression of ExoUP.flu-GFP in mammalian cells | This study |
| pcDNA3.1 ExoUP.flu-S92A-GFP | pcDNA3.1 ExoUP.flu-GFP with S92A substitution | This study |
| pcDNA3.1 ExoUP.asy-GFP | For expression of ExoUP.asy-GFP in mammalian cells | This study |
| pcDNA3.1 ExoUP.asy-S137A-GFP | pcDNA3.1 ExoUP.asy-GFP with S137A substitution | This study |
TABLE 2.
S. cerevisiae strains and plasmids
| Name | Relevant characteristics | Ref./Source |
|---|---|---|
| S. cerevisiae strains | ||
| Wild-type (SEY6210) | MATα trp-Δ901 leu2-3, 112 his3-Δ200 ura3-52 lys2-801 suc2-Δ9 can1::hisG | Mayinger and co-workers (81) |
| Δinp54/Δsac1 | MATα trp-Δ901 leu2–3, 112 his3- Δ200 ura3–52 lys2–801 suc2-Δ9 can1::hisG sac1::TRP1, inp54::LEU2 | Mayinger and co-workers (61) |
| S. cerevisiae plasmids | ||
| pYC2/NT A | Glu-repressible, Gal-inducible yeast expression vector | Invitrogen |
| pYC-ExoUP.aer-S142A-GFP | For expression of ExoUP.aer-S142A-GFP | This study |
| pYC-ExoUP.flu-S92A-GFP | For expression of ExoUP.flu-S92A-GFP | This study |
| pYC-ExoUP.asy-S137A-GFP | For expression of ExoUP.asy-S137A-GFP | This study |
| pYC-ExoUP.asy MLD-GFP | For expression of ExoUP.asy MLD-GFP | This study |
| pYC-2x(GFP-PH) | For expression of GFP-tagged pleckstrin homology domain from PLCδ | Orth and co-workers (72) |
Purification of ExoU and Its Homologs
P. fluorescens and P. asymbiotica were grown overnight in LB broth, and genomic DNA was isolated with the DNeasy blood and tissue kit (Qiagen). Primers containing HindIII and NotI sites were used to amplify the exoU gene homolog from each species. (All primers used in this study are listed in Table 3.) PCR amplification products and the HN-C expression vector (Clontech) were digested with HindIII and NotI restriction enzymes, and the products were ligated and transformed into BL21 (DE3) Star competent cells. (All plasmids used in this study are listed in Tables 1 and 2.) ExoU proteins were purified as described previously using a HisTrap FF nickel column and a HiPrep 26/10 desalting column (GE Healthcare) (26). For purification of MLD proteins, corresponding primers were used to amplify the portion of the gene encoding the MLD, and amplification products were digested with HindIII and NotI. The digested products were ligated into a similarly digested HN-N vector (Clontech), and the ligated construct was transformed into BL21 (DE3) Star competent cells. Purification of these proteins was performed as described above.
TABLE 3.
Primers used in this study
| Name | Sequence (5′ → 3′)a |
|---|---|
| For purification | |
| 5′ ExoUP.aer MLD HindIII | AAAAAGCTTATCACAGACGGGGCGGTG |
| 3′ ExoUP.aer MLD NotI | AAAGCGGCCGCCTGTGAACTCCTTATTCCG |
| 5′ ExoUP.flu HindIII | AAAAAGCTTATGAAAGTCTCCAGTTCT |
| 3′ ExoUP.flu NotI | AAAGCGGCCGCCAACCGGAATACGCCAGGC |
| 5′ ExoUP.flu MLD HindIII | AAAAAGCTTGCCGGCGCCGAGGCGTTG |
| 5′ ExoUP.asy HindIII | AAAAAGCTTATGCAAATTCACATTAAT |
| 3′ ExoUP.asy NotI | AAAGCGGCCGCCTGCTGTTTTGATCATCCA |
| 5′ ExoUP.asy MLD HindIII | AAAAAGCTTTCAACCTCAAACATTGCT |
| For transfection | |
| 5′ ExoUP.flu AgeI | AAAACCGGTGATGAAAGTCTCCAGTTCT |
| 3′ ExoUP.flu NotI | AAAGCGGCCGCCCTAAACCGGAATACGCCA |
| 5′ ExoUP.asy AgeI | AAAACCGGTGATGCAAATTCACATTAAT |
| 3′ ExoUP.asy NotI | AAAGCGGCCGCCCTATGCTGTTTTGATCAT |
| 5′ ExoUP.aer MLD AgeI | AAAACCGGTGATCACAGACGGGGCGGTG |
| 3′ ExoUP.aer NotI | AAAGCGGCCGCCTCATGTGAACTCCTTATT |
| 5′ ExoUP.flu MLD AgeI | AAAACCGGTGGCCGGCGCCGAGGCGTTG |
| 5′ ExoUP.asy MLD AgeI | AAAACCGGTGTCAACCTCAAACATTGCT |
| For site-directed mutagenesis | |
| 5′ ExoUP.flu S92A | GTGGTTTCCGGCTCAGCGGCCGGTGCGATTTG |
| 3′ ExoUP.flu S92A | CAAATCGCACCGGCCGCTGAGCCGGAAACCAC |
| 5′ ExoUP.flu R616L | GTCAAACATTACCGAGCGCTCAATAAACCCTGGAGTAAAC |
| 3′ ExoUP.flu R616L | GTTTACTCCAGGGTTTATTGAGCGCTCGGTAATGTTTGAC |
| 5′ ExoUP.asy S137A | CACCATGTCAGGTTCTGCTGCGGGCGGGATCAC |
| 3′ ExoUP.asy S137A | GTGATCCCGCCCGCAGCAGAACCTGACATGGTG |
| 5′ ExoUP.asy R652L | GGCGGACAATTATGGCGCACTAATGAAACCTTGGAAAAAAC |
| 3′ ExoUP.asy R652L | GTTTTTTCCAAGGTTTCATTAGTGCGCCATAATTGTCCGCC |
| For yeast cloning | |
| 5′ GFP yeast | GACTCACTATAGGGAATATTAAGCTTACCATGGCCAGCAAAGGAGAAGAA |
| 3′ ExoUP.aer | GATAGGCTTACCTTCGAAGGGCCCTCTAGATCATGTGAACTCCTTATTCC |
| 3′ ExoUP.flu | GATAGGCTTACCTTCGAAGGGCCCTCTAGACTAAACCGGAATACGCCAGG |
| 3′ ExoUP.asy | GATAGGCTTACCTTCGAAGGGCCCTCTAGACTATGCTGTTTTGATCATCC |
a Restriction sites are in bold.
HeLa Cell Cytotoxicity Assays
ExoU proteins were expressed in HeLa cells by transfection of expression vectors. Constructs expressing wild-type ExoU from P. aeruginosa (hereafter referred to as ExoUP. aer) and variants containing amino acid substitutions had previously been generated using plasmid pcDNA3.1 NT-GFP (30). To generate similar constructs expressing ExoU homologs from P. fluorescens and P. asymbiotica (ExoUP. flu and ExoUP. asy, respectively) primers containing flanking AgeI and NotI sites were used to amplify the ExoU-encoding genes from the respective purification vectors. Then the pcDNA3.1 NT-GFP vector (Invitrogen) and the PCR products were digested with AgeI and NotI. The digested products were purified, ligated, and transformed into E. coli Top10 cells. The catalytic site substitutions were engineered by site-directed mutagenesis of exoUP. flu and exoUP. asy using the QuikChange site-directed mutagenesis kit (Agilent) (30) and the primers listed in Table 3. The constructs encoding the R661L substitution in ExoUP. aer and the corresponding substitutions in ExoUP. flu and ExoUP. asy were similarly engineered in the pcDNA plasmids by site-directed mutagenesis using primers listed in Table 3. Plasmids were purified using the QIAprep spin miniprep kit (Qiagen). X-treme gene transfection reagent (Roche Applied Science) was coupled with the DNA transfection constructs in serum-free medium to perform transfections with HeLa cells as described previously (30). Medium was collected after 24 h and measured for lactate dehydrogenase activity using the Cytotox 96 nonradioactive cytotoxicity assay (Promega).
Immunoblot Analysis for Detection of GFP-tagged Proteins
The same constructs used in the HeLa cell cytotoxicity assays were used for expression of GFP-tagged ExoU proteins from the pcDNA3.1 NT-GFP vector. Lipofectamine 2000 (Invitrogen) was coupled with DNA in serum-free medium and transfected onto 10-cm dishes of HeLa cells. After 24 h, cells were washed, collected, and lysed as described previously (26). Samples were run on a 10% SDS-polyacrylamide gel, transferred to a nitrocellulose membrane, and incubated in blocking buffer for 2 h at room temperature, as described previously (26). The membrane was incubated overnight at 4 °C with gentle shaking in the presence of Living Colors A.v. monoclonal antibody (Clontech) diluted 1:3000 in blocking buffer with 0.1% Tween 20. The membrane was washed and incubated with goat anti-mouse secondary antibody IR dye 800 (Li-Cor Biosciences) diluted 1:10,000 in blocking buffer supplemented with 0.1% Tween 20, for 1 h at room temperature with gentle shaking. The membrane was washed again, and blots were imaged using the Li-Cor Odyssey system.
HeLa Cell Fluorescence Microscopy
As described for the cytotoxicity assays, the MLD-encoding portions of exoU genes (both wild-type alleles and those encoding arginine-to-leucine substitutions) were amplified by PCR from the pcDNA3.1 NT-GFP ExoU constructs. The PCR product and the pcDNA3.1 NT-GFP vector were digested with AgeI and NotI, ligated, and transformed into Top10 cells. Expression constructs were purified using QIAprep spin miniprep kit. One day prior to transfection, HeLa cells were seeded into 24-well dishes containing coverslips. Cells were transfected as described for cytotoxicity assays and incubated for 20 h. Coverslips were washed and fixed in 3.7% formaldehyde and transferred to slides containing Prolong Gold anti-fade reagent (Molecular Probes). Coverslips were sealed with nail polish and visualized using a Nikon C2+ multispectral laser scanning confocal microscope at the Northwestern University Cell Imaging Facility. HeLa cell images were analyzed for fluorescence intensity distribution across individual cells using ImageJ version 1.43 software (National Institutes of Health).
Construction of Yeast Plasmids
Alleles encoding the catalytic variants of each of the ExoU homologs were PCR-amplified from the pcDNA3.1 NT-GFP vectors. The exoUP. asy MLD was similarly amplified. A pYC2/NT A vector (Invitrogen), which contains a glucose-repressible, galactose-inducible promoter (37), was digested with HindIII and XbaI. Using the LiAc heat shock method (37), 15 μl of PCR product and 1 μl of digested plasmid DNA were ligated within S. cerevisiae wild-type and Δinp54/Δsac1 strains, and appropriate colonies were selected on synthetic complete medium lacking uracil.
Visualization of Yeast Strains
Yeast strains were grown in SC-ura + Gal medium to induce expression of the GFP-tagged constructs. Yeast were immobilized in 1% agarose solution on coverslips and visualized using a Nikon C2+ multispectral laser scanning confocal microscope.
Liposome Binding Assays
Liposome binding assays were performed as described previously (38). Briefly, a neutral lipid backbone was prepared with 30% phosphatidylcholine (PC), 20% phosphatidylethanolamine (PE), and 20% cholesterol (Avanti Polar Lipids). The remaining lipids were 30% PC, 30% PE, 30% phosphatidylserine (PS), 30% phosphatidylinositol, or a combination of 25% PC with 5% phosphatidylinositol, PI(4)P, PI(5)P, PI(4,5)P2, or PI(3,4,5)P3. These lipids were mixed as chloroform stocks that were dried under nitrogen gas and then by vacuum. Lipids were resuspended in liposome buffer (100 mm KCl, 1 mm MgCl2, and 1 mm CaCl2 in 20 mm HEPES (pH 7.5)) to a final concentration of 3 mm and sonicated. ExoU protein was added to a final concentration of 3 μm (38). Protein and liposomes were coincubated at 37 °C for 5 min. Ultracentrifugation was then performed at 200,000 × g for 2 h at 25 °C. The supernatants were removed, and the pellets were resuspended in an equal volume of liposome buffer. Samples were run on 4–15% gradient SDS-polyacrylamide gels, and proteins were visualized by Coomassie staining. Band intensity was analyzed using ImageJ version 1.43 software.
PLA2 Assays
PLA2 assays were performed using the Cayman Chemical cPLA2 kit as described previously (26). Briefly, a total of 65 pmol of ExoU was added to 200 μl of 1.5 mm arachidonoyl thiophosphatidylcholine substrate for each assay condition. When indicated, 65 pmol of PI(4,5)P2 (Avanti Polar Lipids) or 1.3 nmol of 55% PC, 20% PE, 20% cholesterol, and 5% PI(4,5)P2-containing liposomes were added. In addition, 65 pmol or 65 fmol of ubiquitin (Sigma) were added when indicated. Absorbance was measured at 405 nm at the times indicated after the addition of 10 μl of 25 mm 5,5′-dithiobis(2-dinitrobenzoic acid). The PLA2 activity of ExoU was calculated using the following formula: A405/10.00 × 1/(nmol of ExoU), where 10.00 is the extinction coefficient for 5,5′-dithiobis(2-dinitrobenzoic acid).
Protein BLAST Analysis
Nonredundant protein sequences were analyzed for similarity to ExoUP. aer (UniProtKB accession number O34208) by performing protein-protein BLAST searches (39). MLD similarity was based upon ExoUP. aer residues 503–687, and homologs from different bacterial species were analyzed for alignment of key residues.
Surface Plasmon Resonance (SPR) Analysis
SPR analysis was performed on a BIAcore X instrument with a lipid-coated L1 chip as described previously (40). Lipids were prepared by mixing chloroform stocks and then drying them under nitrogen gas. Lipids were resuspended in 160 mm NaCl in 20 mm Tris (pH 7.4) to a final concentration of 400 μg/ml, sonicated, and extruded for uniform 100 nm liposome size. Kinetic and equilibrium SPR measurements were performed with the flow rate set at 30 and 10 μl/ml, respectively. Experiments determining Kd values were performed in triplicate. PC vesicles, to which ExoU has no detectable affinity, were used for the control surface. The active surface was coated with liposomes containing 77% PC, 20% PS, and 3% phosphoinositides or with 97% PC and 3% phosphoinositides. Assuming a Langmuir-type binding between the protein and protein-binding sites on vesicles, Req values were then plotted versus the total protein concentration ([P]o), and Kd values were determined by nonlinear least squares analysis of the binding isotherm using the equation Req = Rmax/(1 +Kd/[P]o).
Crystallization of ExoUP.flu
Crystallization of ExoUP. flu was performed at a protein concentration of 5.8 mg/ml in 10 mm Tris-HCl (pH 8.3) buffer containing 500 mm NaCl and 5 mm 2-mercaptoethanol by the sitting-drop vapor diffusion method at 295 K. Crystals suitable for data collection were obtained from the Classics II Suite (Qiagen Inc., Valencia, CA) condition consisting of 0.2 m ammonium acetate, 25% (w/v) PEG 3350 in 0.1 m BisTris (pH 5.5/6.5) buffer. Crystals were soaked in the well/crystallization solutions for cryoprotection and flash-frozen in liquid nitrogen for x-ray data collection.
Data Collection and Structure Determination
A single-wavelength (λ = 0.97856) oscillation x-ray data set was collected on the Life Science Collaborative Access Team (LS-CAT) 21-ID-G. Diffraction images were indexed and scaled with HKL-2000 (41) to 2.5 Å resolution. Cell content analysis indicated the presence of two molecules of ExoUP. flu (Vm (Matthews coefficient) = 2.4; versus (solvent content) = 49.5%) within the P21 space group at 2.5 Å resolution. A partial molecular replacement solution of ExoUP. flu was obtained running Phaser (42) from the CCP4 package (43) and the ExoUP. aer structure (Protein Data Bank 3TU3 (28)) as a search model. The structure solution was rebuilt, and water molecules were added using ARP/wARP (44) from the CCP4 package. The structure was refined with REFMAC version 5.7 (45) from the CCP4 package with further manual building and alteration in Coot (46, 47). The quality of the structure was checked with the Protein Data Bank validation server (ADIT validation server) and MolProbity (48, 49). Total buried surface area of the ExoUP. flu dimer was determined by the “Protein interfaces, surfaces, and assemblies” service PISA at the European Bioinformatics Institute (50). Structure figures were generated with PyMOL (51). Data collection and structure-refinement statistics are given in Table 4. The final model was deposited in the Protein Data Bank under accession code 4QMK. Diffraction images for the deposited structure are available at the Center for Structural Genomics of Infectious Diseases (CSGID) website.
TABLE 4.
Data collection and structure-refinement statistics
Values in parentheses are for the highest resolution shell.
| Data collection | |
| Space group | P21 |
| Cell dimensions | |
| a, b, c (Å) | 67.2, 115.3, 88.4 |
| β (°) | 102.8 |
| No. of reflections | 43,226 (1,696) |
| Resolution (Å) | 30.00-2.50 (2.54-2.50) |
| Rmerge (%)a | 7.0 (44.1) |
| I/σ | 15.7 (2.1) |
| Completeness (%) | 95.2 (75.7) |
| Average redundancy | 3.7 (2.9) |
| Refinement | |
| Resolution (Å) | 29.86-2.50 (2.56-2.50) |
| Completeness (%) | 94.8 (74.1) |
| No. of reflections | 41,015 (2,344) |
| Rworkb/Rfreec (%) | 20.6/26.9 (32.4/42.1) |
| No. of atoms | |
| Protein (chain A/B) | 4,060/3,895 |
| Water (oxygen atoms) | 207 |
| B-factor (Å2) | |
| Overall | 56.7 |
| Protein (chain A/B) | 47.9/66.5 |
| Water | 47.5 |
| Coordinate deviation | |
| r.m.s.d.e bond lengths (Å) | 0.014 |
| r.m.s.d. bond angles (°) | 1.609 |
| Ramachandran statisticsd | |
| Most favorite (%) | 94.6 |
| Allowed (%) | 5.0 |
| Generously allowed (%) | 0.5 |
| Outside allowed (%) | 0.0 |
a Rmerge = Σhkl|I − 〈I〉|/ΣhklI.
b Rwork = Σ|Fobs − Fcalc|/Σ|Fobs|, where Fobs and Fcalc are the observed and the calculated structure factors, respectively.
c Rfree were calculated using 5% of total reflections randomly chosen and excluded from the refinement.
d Statistics are based on PROCHECK (17).
e r.m.s.d. is root mean square deviation.
Statistical Methods
Student's t test was used to compare means for cytotoxicity assays and yeast viability experiments. Significance was defined as a p value of <0.05. One-way analysis of variance was used for all other assays with multiple comparisons with the Bonferroni multiple comparisons test to determine significance.
RESULTS
MLD of ExoU Binds to PI(4,5)P2
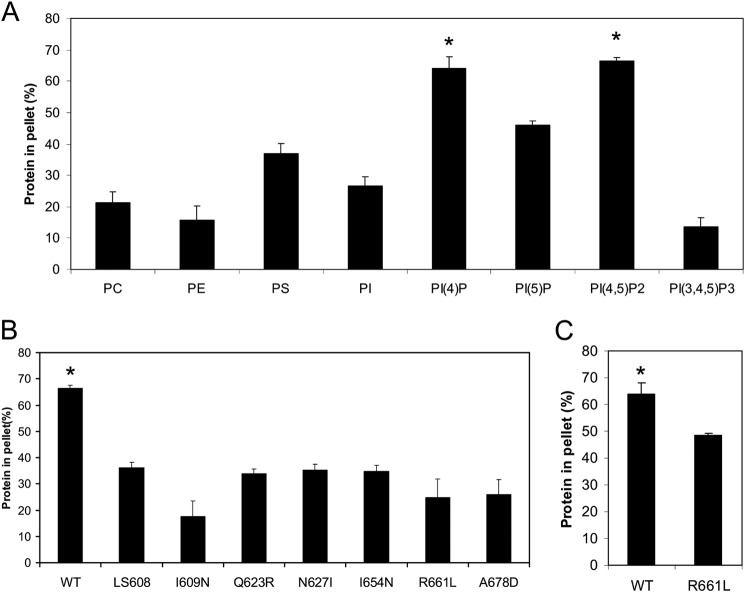
We sought to determine whether ExoU localization to the plasma membrane was mediated by direct affinity to phospholipids, as has been reported for several other toxins (8, 38). Although ExoU was known to bind to PI(4,5)P2, it was unclear whether this was required for localization or whether the MLD of ExoU was sufficient to mediate this binding (32). To investigate this, we purified the recombinant MLD of ExoU (residues 503–687) and tested it for co-sedimentation with neutral liposomes supplemented with various phospholipids (38, 52). We used a final lipid concentration of 3 mm, which has previously been used to effectively assess lipid-protein interactions (38, 52). We found that liposomes supplemented with either PI(4,5)P2 or PI(4)P bound to ExoU substantially more than other tested lipids (Fig. 1A). ExoU binding to PI(4,5)P2 is consistent with its plasma membrane targeting, as PI(4,5)P2 is found specifically in the plasma membrane of eukaryotic cells (27, 53). In contrast, PI(4)P is enriched in the Golgi, although a subpopulation of PI(4)P is in the plasma membrane and may contribute to plasma membrane targeting of some proteins (54). These findings are consistent with ExoU binding to PI(4,5)P2 (and potentially PI(4)P) to achieve plasma membrane localization.
FIGURE 1.
ExoU MLD requires specific residues to bind to PI(4,5)P2. A, recombinant ExoU MLD (residues 503–687) was tested for co-sedimentation in liposome binding assays with various lipids. Liposomes consisting of 30% PC, 20% PE, and 20% cholesterol were supplemented with 30% of the indicated lipids, except for PI(4)P, PI(5)P, PI(4,5)P2, and PI(3,4,5)P3, for which 5% was added (the remaining 25% was PC). Each assay was performed at least in triplicate; values are means, and error bars represent S.E. *, p < 0.05 compared with all other groups. The difference between PI(4)P and PI(4,5)P2 is not significant. PI, phosphatidylinositol. B, recombinant MLD proteins containing single amino acid substitutions (I609N, Q623R, N627I, I654N, R661L, and A678D) or a 5-amino acid insertion (LS608) were tested for binding to PI(4,5)P2-containing liposomes in co-sedimentation assays. Each assay was performed at least in triplicate; values are means, and error bars represent S.E. WT indicates the wild-type MLD of ExoU, using the PI(4,5)P2 binding data depicted in A. *, p < 0.001 compared with each variant. No difference between the variants is statistically significant. C, recombinant wild-type (WT) and R661L MLD protein were analyzed for binding to PI(4)P-containing liposomes. *, p < 0.05 relative to R661L variant.
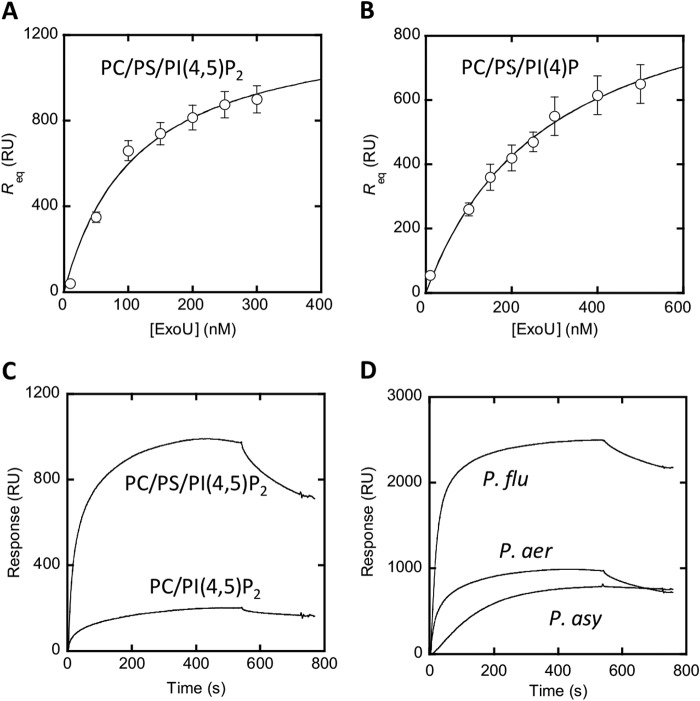
To obtain a more quantitative measure of ExoU lipid binding, we performed surface plasmon resonance (SPR) binding analysis using full-length recombinant ExoU protein and liposomes supported on a solid sensor surface (55). ExoU did not appreciably bind to phosphatidylcholine liposomes but did bind with high affinity to PI(4,5)P2 liposomes (Kd 110 ± 30 nm; Fig. 2A). ExoU also bound to PI(4)P but with lower affinity (Kd 290 ± 30 nm; Fig. 2B). These data confirm the liposome co-sedimentation assay results and demonstrate high affinity interactions between ExoU and PI(4,5)P2 and somewhat weaker interactions between ExoU and PI(4)P, which was not known previously. Liposome binding and SPR assays are more physiological methods of measuring protein-lipid binding than the lipid strips used previously by Gendrin et al. (56), potentially explaining why they did not observe ExoU-PI(4)P binding (31). Interestingly, although not required for PI(4,5)P2 binding, the presence of phosphatidylserine significantly increased ExoU-PI(4,5)P2 affinity (Fig. 2C). The addition of PS mimics its presence at the inner leaflet of the plasma membrane; this phospholipid contributes to the plasma membrane-targeting specificity of other phosphoinositide binding domains (57).
FIGURE 2.
Binding of ExoU to vesicles containing PI(4,5)P2 and PI(4)P measured by SPR analysis. A, determination of Kd for ExoU binding to PC/PS/PI(4,5)P2 (77:20:3) vesicles by equilibrium SPR analysis. The binding isotherm was generated from the Req (average of triplicate measurements) versus the concentration (P0) of ExoU plot. A solid line represents a theoretical curve constructed from Rmax (= 1300 ± 120) and Kd (= 110 ± 30 nm) values determined by nonlinear least squares analysis of the isotherm using the following equation: Req = Rmax/(1 + Kd/P0). B, determination of Kd values for ExoU binding to PC/PS/PI(4)P (77:20:3) vesicles by equilibrium SPR analysis. Rmax (= 1100 ± 60) and Kd (= 290 ± 30 nm) values were calculated as described for B. C, kinetic SPR sensorgrams for ExoUP. aer binding to PC/PS/PI(4,5)P2 (77:20:3) and PC/PI(4,5)P2 (97:3) vesicles. D, kinetic SPR sensorgrams for ExoUP. aer, ExoUP. flu, and ExoUP. asy binding to PC/PS/PI(4,5)P2 (77:20:3) vesicles. ExoUP. flu showed the highest degree of vesicle binding. Equilibrium SPR analysis confirmed that it has the highest affinity (Kd = 30 ± 6 nm). RU, resonance units. Error bars represent S.D.
In an earlier study, we had identified several residues of the MLD that were critical for the proper intracellular localization of ExoU but that did not disrupt the overall secondary structure of ExoU, as measured by circular dichroism (30). We reasoned that if MLD binding to PI(4,5)P2 truly was responsible for MLD localization to the plasma membrane, then these mislocalized ExoU variants should fail to bind PI(4,5)P2. We therefore purified recombinant MLD proteins with one of six single amino acid substitutions or with one five-amino acid insertion that had resulted in mislocalization inside host cells (30). These purified proteins were tested for their ability to bind liposomes containing PI(4,5)P2. Each was deficient in PI(4,5)P2 binding (Fig. 1B), demonstrating that ExoU residues necessary for localization to the plasma membrane of host cells were also critical for binding to PI(4,5)P2. One variant, R661L, was also tested for binding to PI(4)P. As was the case with PI(4,5)P2, it was deficient in PI(4)P binding, suggesting that the same mechanism may be used to bind both phosphoinositides (Fig. 1C).
ExoU-like MLDs from Other Patatin-like Proteins Also Bind to PI(4,5)P2
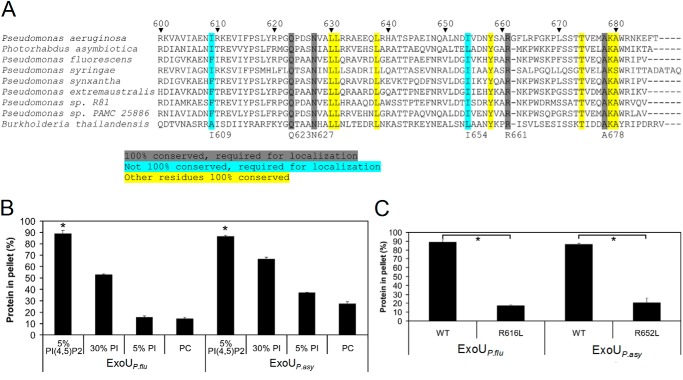
PI(4,5)P2 binding domains play important roles in intracellular trafficking of eukaryotic proteins, and each distinct binding domain is often utilized by a number of different proteins (33). For this reason, we sought to determine whether other bacterial proteins also used an ExoU MLD-like motif to bind PI(4,5)P2. Accordingly, BLAST analysis with the MLD of ExoU was used to identify several proteins containing similar MLD sequences (Fig. 3A). Genes encoding these proteins were found in the genomes of other Pseudomonas species such as P. fluorescens and Pseudomonas syringae but also in other genera, including Photorhabdus asymbiotica and Burkholderia thailandensis. Alignments revealed that a number of residues within these sequences were 100% conserved among this group of proteins, including some residues that had previously been shown to be important for ExoU localization, such as Gln-623, Asn-627, Arg-661, and Ala-678 (Fig. 3A) (30). Of special interest was Arg-661 of ExoU, a highly conserved and positively charged residue in a loop at the exposed cap of the four-helical bundle in the MLD (28). This residue could potentially directly interact with the negatively charged phosphate groups of PI(4,5)P2. In support of this model, substitution of Arg-661 with a leucine (R661L) had previously been shown to cause loss of activation of P. aeruginosa ExoU by PI(4,5)P2 (26). Likewise, recombinant MLD protein with this substitution did not bind PI(4,5)P2 in liposome binding assays (Fig. 1B). These results suggest that this conserved arginine residue might be critical for direct MLD binding to PI(4,5)P2.
FIGURE 3.
Several residues necessary for the binding of ExoU to PI(4,5)P2 are conserved in other proteins containing ExoU MLD-like domains. A, alignments of proteins containing ExoU MLD-like domains. The highlighted sequences identify highly conserved residues. Those residues highlighted in gray and blue were critical for ExoU localization in mammalian cells (30) and for binding to PI(4,5)P2-supplemented liposomes (Fig. 1B). Numbering system refers to ExoU of P. aeruginosa. Accession numbers for the indicated proteins in order are as follows: O34208, YP_003039880.1, YP_006326405.1, ZP_07262921.1, ZP_10142741.1, ZP_10438440.1, ZP_11185695.1, ZP_10426928.1, and ZP_18329767.1. B, recombinant putative MLD proteins from ExoUP. flu and ExoUP. asy (residues 453–639 and 495–676, respectively) were purified and assayed for liposome binding. Liposomes consisted of the same composition as depicted in Fig. 1, except that 5% PI/25% PC-supplemented liposomes were also tested. C, arginine-to-leucine substitutions corresponding to R661L in ExoUP. aer were generated in ExoUP. flu and ExoUP. asy recombinant MLD proteins. These proteins were purified and were tested for binding to liposomes supplemented with 5% PI(4,5)P2. The results for “WT,” which refers to wild-type protein, are the same as depicted in B. Each assay was performed in triplicate; values are means, and error bars represent S.E. *, p < 0.001 for PI(4,5)P2 compared with other lipids and wild-type compared with arginine-to-leucine substitution proteins.
To investigate whether ExoU MLD-like domains from other bacteria also bound PI(4,5)P2, we tested the recombinant MLDs from P. fluorescens and P. asymbiotica in the liposome binding assay. P. fluorescens is an environmental bacterium (58), and P. asymbiotica infects insects and is an emerging human pathogen (59). The MLD-containing proteins from these bacteria had not been previously studied (59, 60), but their homologies to ExoU extended beyond the MLDs. For this reason, we will refer to them as ExoUP. flu and ExoUP. asy, respectively. We found that the recombinant MLD proteins from ExoUP. flu and ExoUP. asy bound to PI(4,5)P2 significantly more than to phosphatidylcholine (Fig. 3B). Each protein also bound to phosphatidylinositol, albeit to a lesser degree than to PI(4,5)P2. These results were recapitulated with SPR analysis, which found that ExoUP. flu bound to PI(4,5)P2 with high affinity (Kd = 30 ± 6 nm) (Fig. 2D). SPR also detected ExoUP. asy binding to PI(4,5)P2, but this binding was of lower affinity (Kd value not calculated).
We then investigated whether substitution of the conserved arginine residue corresponding to Arg-661 in ExoUP. aer altered the ability of these homologs to bind PI(4,5)P2. Indeed, ExoUP. flu-R616L and ExoUP. asy-R652L were each substantially reduced in PI(4,5)P2 binding compared with their wild-type counterparts (Fig. 3C). These results indicate that the MLD of ExoU represents a new PI(4,5)P2 binding domain used by multiple bacterial species and that a conserved arginine residue is important for this binding.
ExoU and Other Patatin-like Proteins Localize in a PI(4,5)P2-dependent Manner
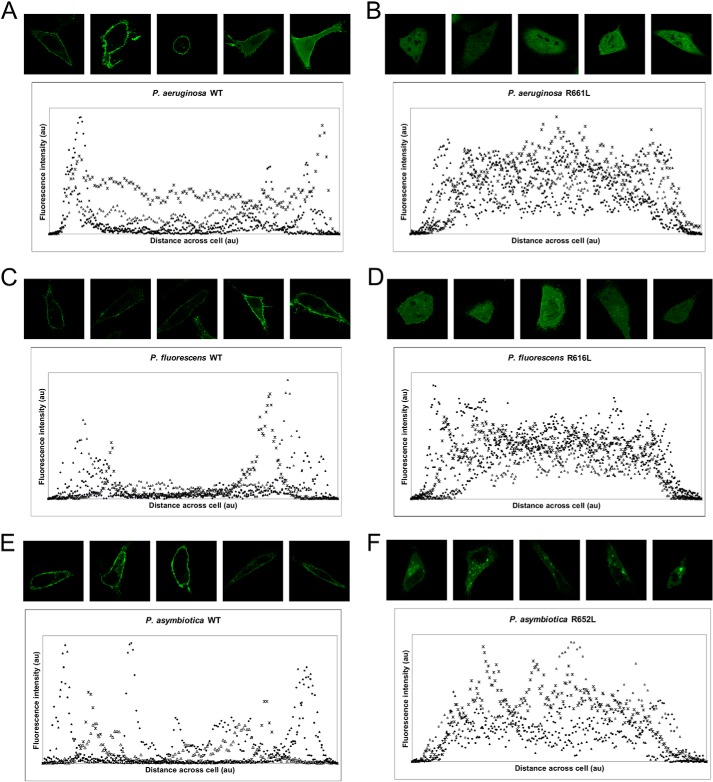
Having characterized the in vitro PI(4,5)P2 binding of the MLDs of several ExoU proteins, we sought to determine whether this binding contributed to localization. To this end, the intracellular localization of the MLD of each of the P. aeruginosa, P. fluorescens, and P. asymbiotica proteins was investigated by transfection of HeLa cells with constructs expressing GFP-tagged MLD proteins. Consistent with their ability to bind PI(4,5)P2 in vitro, each of the MLD proteins localized to the plasma membrane (Fig. 4, A, C, and E). As demonstrated previously, the R661L substitution in the ExoUP. aer MLD abolished membrane localization, with the protein becoming diffusely localized (Fig. 4B) (30). Similar results were observed with the corresponding R616L substitution in ExoUP. flu and the R652L substitution in ExoUP. asy, although the latter protein was associated with a less homogeneous intracellular distribution (Fig. 4, D and F). Overall, these results indicate that loss of PI(4,5)P2 binding in vitro is correlated with decreased localization to the plasma membrane and demonstrate the importance of the conserved MLD arginine residue to this process.
FIGURE 4.
MLDs from ExoUP. aer, ExoUP. flu, and ExoUP. asy localize to the plasma membrane of HeLa cells. HeLa cells were transfected with constructs expressing GFP-tagged MLD proteins from the ExoU homologs of P. aeruginosa, P. fluorescens, and P. asymbiotica. Representative images were visualized by fluorescence microscopy. Both wild-type (WT) (A, C, and E) and arginine-to-leucine substitutions (B, D, and F) of each protein were tested. Images were analyzed for fluorescence intensity distribution across cells using ImageJ. The distance and fluorescence intensity are in arbitrary units (au).
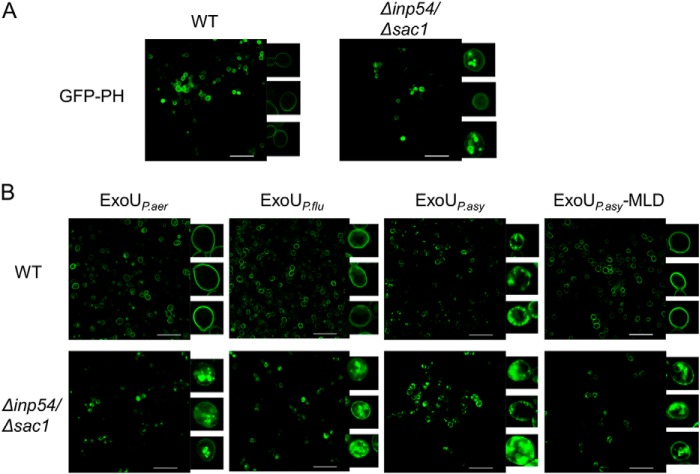
To solidify the importance of PI(4,5)P2 in mediating targeting of ExoU to the plasma membrane, we used a yeast model that was amenable to manipulation of PI(4,5)P2 localization. Yeast are an established model system for the study of ExoU activity and localization and have yielded results similar to those observed in mammalian cells (26, 37). We reasoned that if ExoU localization was truly dependent on binding to PI(4,5)P2, then mislocalization of PI(4,5)P2 should cause a corresponding mislocalization of ExoU. In these experiments, PI(4,5)P2 was visualized in yeast using a construct that expressed a GFP-tagged pleckstrin homology (PH) domain from mammalian phospholipase Cδ, which is a natural ligand of PI(4,5)P2 (Fig. 5A) (61). In wild-type yeast, PI(4,5)P2 exhibited a peripheral distribution, consistent with plasma membrane localization. However, in a Δinp54/Δsac1 yeast mutant, which lacks two phosphatases important for appropriate PI(4,5)P2 distribution, this phospholipid was mislocalized to the vacuole and cytosol (Fig. 5A), as described previously (61). Prior reports demonstrated that PI(4)P localization is unchanged in these yeast and that proteins that localize to the plasma membrane independently of PI(4,5)P2 are also unaffected (61). Full-length ExoUP. aer tagged with GFP localized to the plasma membrane in wild-type yeast but was largely associated with intracellular structures in Δinp54/Δsac1 mutant yeast (Fig. 5B). Thus, mislocalization of PI(4,5)P2 resulted in a similar mislocalization of ExoUP. aer, consistent with the model that ExoUP. aer binds to PI(4,5)P2 to target the plasma membrane.
FIGURE 5.
ExoUP. aer, ExoUP. flu, and ExoUP. asy localize to the plasma membrane of yeast in a PI(4,5)P2-dependent fashion. A, PI(4,5)P2 localization was visualized in wild-type (WT) and Δinp54/Δsac1 yeast by the expression of GFP-tagged pleckstrin homology domain from phospholipase C. Yeast were visualized using fluorescence confocal microscopy. B, GFP-tagged catalytically inactive ExoU proteins were visualized by fluorescence confocal microscopy following expression in yeast. The insets show individual yeast cells magnified ×15. ExoUP. asy-MLD refers to the membrane localization domain of ExoUP. asy Scale bars, 25 μm.
We next assessed the localization of ExoUP. flu and ExoUP. asy in yeast. In wild-type yeast, full-length ExoUP. flu localized to the plasma membrane similarly to ExoUP. aer (Fig. 5B). In the Δinp54/Δsac1 mutant yeast, ExoUP. flu displayed mislocalization away from the plasma membrane, indicating PI(4,5)P2-dependent localization. ExoUP. asy also localized to the plasma membrane in wild-type yeast but formed more punctate structures at or adjacent to the membrane (Fig. 5B). In the Δinp54/Δsac1 mutant yeast, the distribution of ExoUP. asy shifted somewhat to the interior of the yeast, although substantial amounts remained at the periphery. Because the localization of full-length ExoUP. asy in yeast differed from that of the ExoUP. asy MLD in HeLa cells, we examined the localization of the ExoUP. asy MLD in yeast. As observed in HeLa cells, the MLD alone localized specifically to the plasma membrane of yeast (Fig. 5B). Importantly, the ExoUP. asy MLD was mislocalized in Δinp54/Δsac1 yeast, suggesting that localization of the ExoUP. asy MLD is dependent on PI(4,5)P2. These results suggest that the MLD of ExoUP. asy requires PI(4,5)P2 for localization but that full-length ExoUP. asy may have residues outside the MLD that contribute to an altered intracellular distribution. Together these data indicate that ExoU and its homologs localize to the plasma membrane in a PI(4,5)P2-dependent manner, either as full-length proteins or as isolated MLD domains.
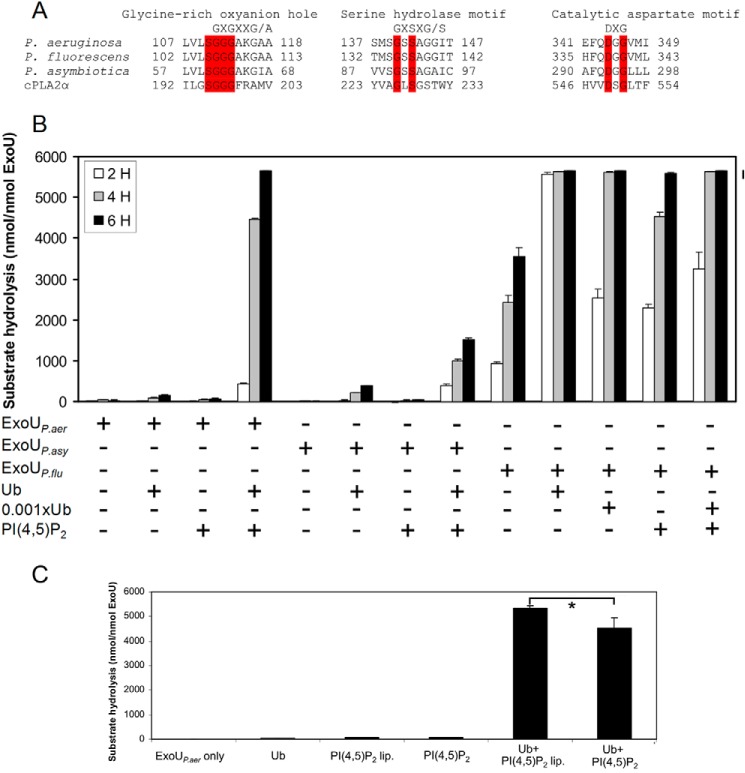
ExoUP. flu and ExoUP. asy Are Also Phospholipases
The proteins listed in Fig. 3A contained regions with similarity to not only the MLD of ExoUP. aer but also to its catalytic domain. In particular, both ExoUP. flu and ExoUP. asy have a putative PLA2 serine-aspartate catalytic dyad and glycine-rich oxyanion hole characteristic of patatin-like phospholipases (Fig. 6A). We therefore examined whether these proteins have PLA2 activity. We first confirmed that ExoUP. aer by itself does not have PLA2 activity but is synergistically activated by a combination of ubiquitin and PI(4,5)P2 (Fig. 6B) (25, 26). To test whether ExoUP. flu and ExoUP. asy act as phospholipases, recombinant versions of both proteins were purified and assessed in vitro for evidence of PLA2 activity. The enzymatic characteristics of ExoUP. asy were similar to those of ExoUP. aer, with no activity on its own and a synergistic activation with the addition of ubiquitin and PI(4,5)P2 (Fig. 6B). However, the overall activity of ExoUP. asy when combined with ubiquitin and PI(4,5)P2 was low, only 22% that of ExoUP. aer under the conditions of these assays. In contrast, ExoUP. flu alone had considerable PLA2 activity, and the addition of ubiquitin substantially increased this activity (Fig. 6B). In fact, addition of only 0.1% as much ubiquitin as ExoUP. flu resulted in significantly increased PLA2 activity. Supplementation with PI(4,5)P2, however, caused only a slight additional increase in catalytic activity, suggesting that ubiquitin plays a more predominant role in the activation of ExoUP. flu. These results demonstrate that MLD-containing patatin-like proteins from other bacterial species and genera also have PLA2 activity but differ from ExoUP. aer in how active they are by themselves or with ubiquitin and PI(4,5)P2. ExoUP. aer was also activated by PI(4,5)P2-containing liposomes, indicating that the liposome binding assay conditions are suitable for ExoU activation in the presence of ubiquitin (Fig. 6C).
FIGURE 6.
ExoUP. flu and ExoUP. asy have PLA2 activity. A, ExoUP. aer and its homologs from P. fluorescens and P. asymbiotica were aligned with cPLA2 (accession number P47712.2) at their putative catalytic sites. Residues highlighted in red are identical in each of these proteins. B, PLA2 activity of each of these proteins was measured in vitro using a synthetic phospholipid substrate. Each protein was analyzed for its ability to hydrolyze substrate by itself, with ubiquitin, with PI(4,5)P2, or with both ubiquitin and PI(4,5)P2. Ubiquitin (Ub) was added at a 1:1 ubiquitin/ExoU molar ratio or at a 1:1000 ratio (0.001×Ub). C, PLA2 activity of ExoUP. aer was measured with the addition of ubiquitin, PI(4,5)P2, or PI(4,5)P2-containing liposomes with the same overall PI(4,5)P2 concentration (PI(4,5)P2 lip.). PLA2 activity was measured at 5 h. Each assay was performed in triplicate; values are means, and error bars are S.E. *, p < 0.01.
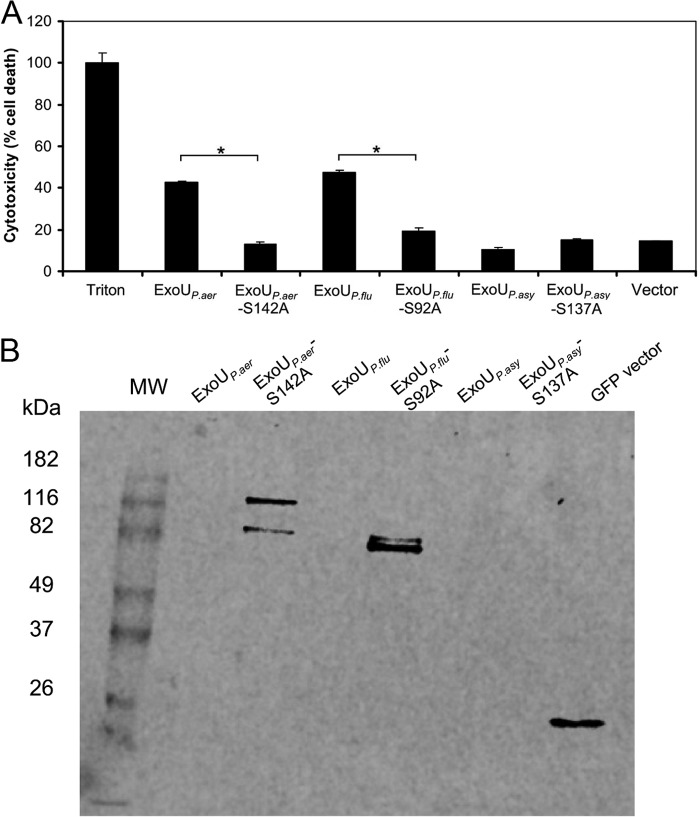
We next examined whether ExoUP. flu and ExoUP. asy, like ExoUP. aer, could kill eukaryotic cells. Each of these proteins was expressed in HeLa cells by transfection, and cytotoxicity was assessed by measuring lactate dehydrogenase release. ExoUP. aer, as shown previously, caused substantial cytotoxicity by 24 h, and this cytotoxicity was diminished by substituting an alanine for the catalytic serine at position 142 within the PLA2 domain (Fig. 7A) (30). Consistent with its high PLA2 activity, ExoUP. flu was also highly cytotoxic, and substitution of its putative catalytic serine also resulted in decreased cytotoxicity. Because of the rapid lysis of cells following expression of ExoUP. aer and ExoUP. flu, these proteins could not be detected within transfected cells, whereas expression of the proteins with catalytic site substitutions could be detected (Fig. 7B). Despite repeated attempts, we were not able to detect stable GFP-tagged protein in cells transfected with ExoUP. asy-expressing constructs and were therefore unable to assess the cytotoxicity of this protein (Fig. 7B). Detection following substitution of other tags in place of GFP was also not successful (data not shown). Interestingly, the MLD of ExoUP. asy was stable when expressed by itself in mammalian cells (Fig. 4), and the full-length GFP-tagged ExoUP. asy-S137A construct could be visualized in yeast (Fig. 5B). It is possible that the full-length ExoUP. asy protein may not fold properly in the mammalian cell cytosol or it may be destabilized or degraded. Overall, these experiments demonstrated that ExoUP. flu is cytotoxic to HeLa cells, but we could not assess the cytotoxicity of ExoUP. asy in this cell type.
FIGURE 7.
Cytotoxicity of ExoU homologs. A, HeLa cells were transfected with ExoU expression constructs and analyzed for release of lactate dehydrogenase as a measure of cell death. Results were normalized to treatment with Triton X-100 (100% cell death) and mock-transfected cells (0% cell death). Vector indicates transfection of pcDNA3.1 NT-GFP vector without insert. Each assay was performed in triplicate; values are means, and error bars represent S.E. *, p < 0.001. B, same GFP-tagged constructs shown in A were transfected into HeLa cells, which were subsequently analyzed for expression of the resulting proteins with anti-GFP antibody. MW denotes the molecular weight markers.
ExoUP. flu Crystal Structure and Its Comparison with the Structure of ExoUP. aer
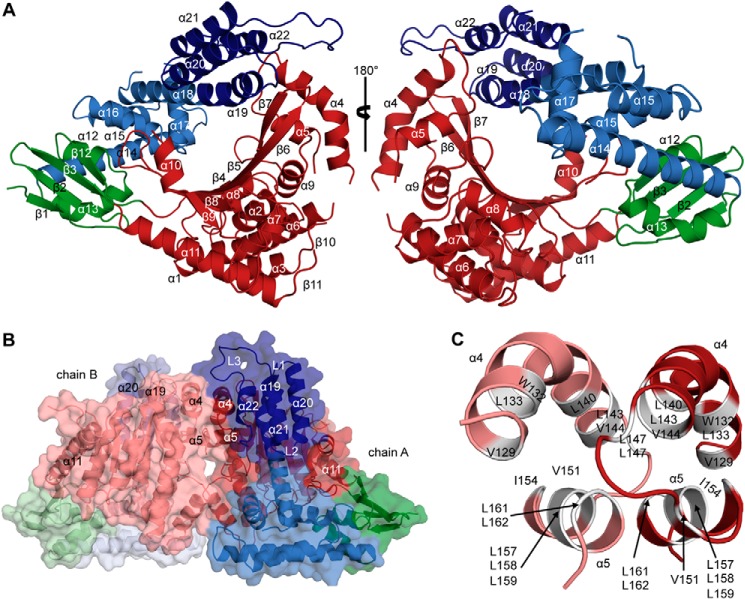
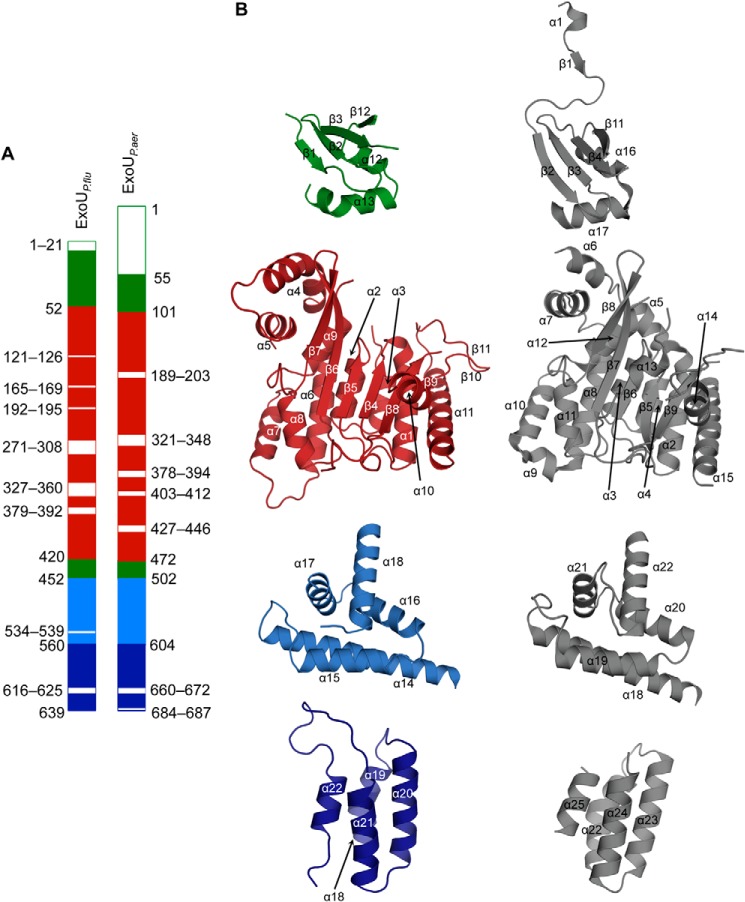
As mentioned, the crystal structure of ExoUP. aer was recently determined and suggested that the four-helical bundle region of the MLD might bind to the plasma membrane (28, 31), similar to the four-helical bundles of other toxins (8, 29). This structure suffered from two limitations. First, ExoUP. aer could only be crystallized in complex with its cognate chaperone SpcU, a bacterial protein that is not present inside eukaryotic cells. Thus, the relevance of this structure to ExoUP. aer interaction with eukaryotic membranes is unclear. Second, the loop containing the conserved Arg-661 was disordered in this structure, preventing an assessment of how it may promote membrane localization. In the hopes of overcoming these limitations and to further investigate MLD-PI(4,5)P2 interactions, we attempted to crystallize full-length ExoUP. flu. We successfully crystallized this protein in the absence of any chaperone proteins and determined its structure to 2.5 Å resolution (Fig. 8A). Surprisingly, ExoUP. flu crystallized as a dimer, with a total buried surface area of ∼2000 Å2 (Fig. 8B). Residues 130–169 (helices α4 and α5) of each patatin-like PLA2 domain constitute the primary interaction interface within the dimer. This dimeric structure is mainly stabilized by stretches of hydrophobic amino acids buried at the interface (Fig. 8C).
FIGURE 8.
Crystal structure of ExoU from P. fluorescens. A, ribbon diagram of a single ExoUP. flu molecule showing subunit domain 1 (the putative chaperone binding domain, green), domain 2 (the patatin PLA2 domain, red), domain 3 (light blue), and domain 4 (dark blue). Domains 3 and 4 together constitute the MLD. B, ribbon and surface representation of the asymmetric unit composition of the structure showing the dimeric arrangement of ExoUP. flu. L1, L2, and L3 are loops of domain 4 that connect the helices 19–20, 20–21, and 21–22, respectively, of the four-helical bundle. C, major dimerization interface generated by helices 4 and 5 of the patatin PLA2 domain. Buried hydrophobic residues (white and labeled with one-letter code) within the helices are displayed.
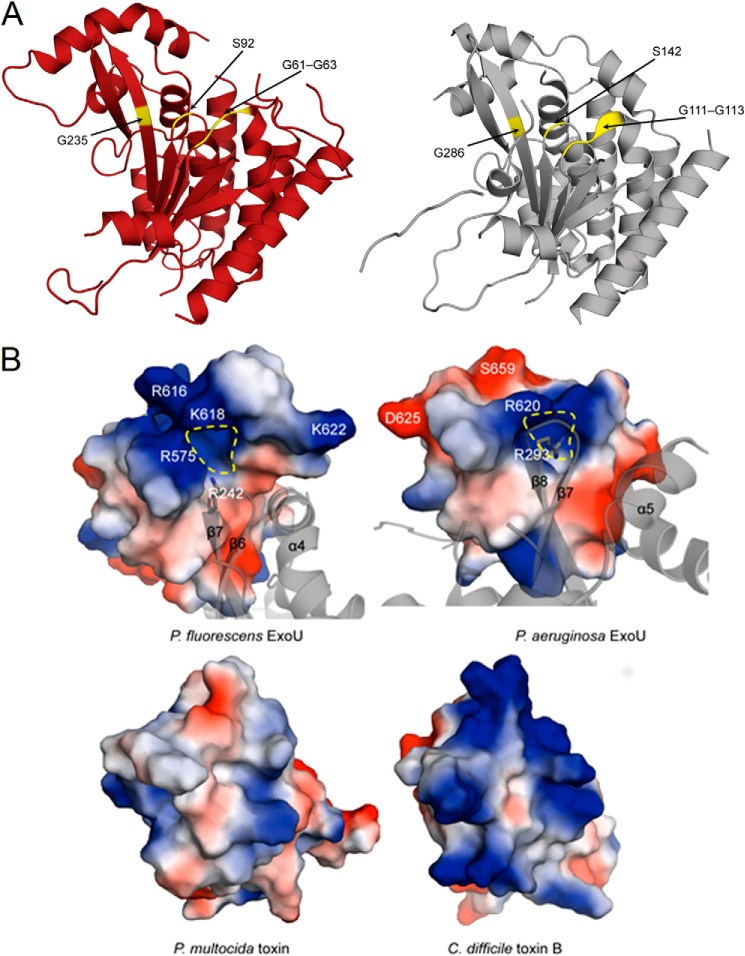
Comparison with the domain architecture of ExoUP. aer (28) allowed the ExoUP. flu structure to be broadly divided into four domains as follows: a putative chaperone binding domain (residues 21–51 and 421–451), a patatin-like PLA2 domain (residues 52–420), and an MLD region, which encompasses domains three and four (residues 452–559 and 560–639, respectively) (Fig. 8A). ExoUP. aer and ExoUP. flu are quite similar in their overall tertiary structures, overlapping with root-mean-square deviation of 2.2 Å (based on the DaliLite server (62–64)) over 473 Cα atoms (Fig. 9). The two ExoU structures also have comparable disordered regions (Fig. 9). The catalytic region of the two proteins are similar as well, with the glycine-rich oxyanion hole (Gly-61–63 in ExoUP. flu and Gly-111–113 in ExoUP. aer) and the serine hydrolase motif containing catalytic Ser-92 (Ser-142 in ExoUP. aer) each aligning well structurally (Fig. 10A) (65). Gly-286, an additional glycine important for ExoUP. aer activity, is also conserved structurally (Fig. 10A, Gly-235 in ExoUP. flu) (65). The catalytic Asp-294 was not modeled due to disorder, as was previously the case for ExoUP. aer (28). Notably, the chaperone binding domains of these two proteins are less similar, suggesting that ExoUP. flu has a structurally distinct chaperone, lacks a chaperone, or that crystal packing has affected the domain's orientation in ExoUP. flu (Fig. 9). It is unclear whether this may explain potential PLA2 activity differences between the two proteins.
FIGURE 9.
Comparison of the ExoUP. flu and ExoUP. aer structures. A, linear depiction of ExoUP. flu and ExoUP. aer domains. The putative chaperone binding domain (domain 1) is shown in green, the patatin PLA2 domain (domain 2) in red, and the MLD in blue. The MLD is composed of domain 3 (light blue) and domain 4 (dark blue). Disordered regions in both structures are shown in white. B, domain-to-domain comparisons of the least squares superimposed ExoUP. flu (colored as in A) and ExoUP. aer (gray) structures.
FIGURE 10.
Comparisons of the catalytic regions and surface charge distributions of ExoU proteins. A, comparison of the catalytic patatin PLA2 domain of ExoUP. flu (red, chain A used) and ExoUP. aer (silver). The glycine-rich oxyanion hole region (Gly-61–63 in ExoUP. flu and Gly-111–113 in ExoUP. aer), catalytic serine residue (Ser-92 in ExoUP. flu and Ser-142 in ExoUP. aer), and an additional conserved glycine residue important in activity (Gly-235 in ExoUP. flu and Gly-286 in ExoUP. aer) are highlighted. B, charged surface and ribbon representations of the domain four portions of ExoUP. flu and ExoUP. aer and the corresponding portions of the PMT and C. difficile toxin B (TcdB). Dashed yellow line indicates a potential PI(4,5)P2-binding/recognition site.
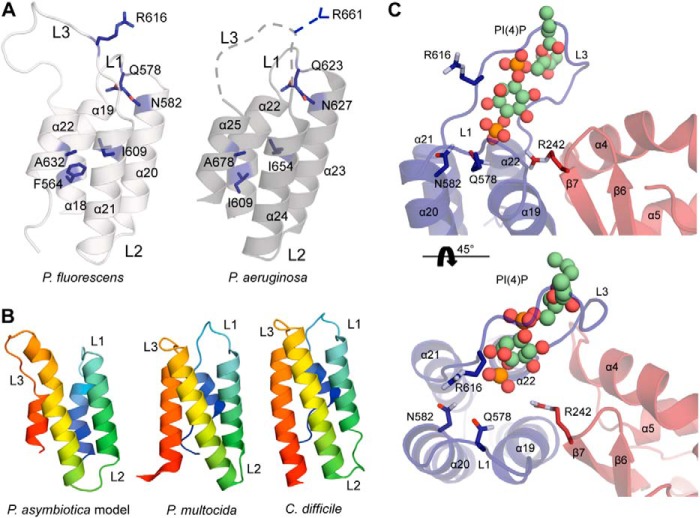
The MLD regions (domains three and four) of ExoUP. flu and ExoUP. aer are very similar structurally, with domain four of each protein forming a four-helical bundle that overlaps with root-mean-square deviation of just 0.7 Å (Fig. 11A). The conserved Arg-616 of ExoUP. flu is part of the L3 loop that protrudes from the four-helical bundle, similarly located to Arg-661 of ExoUP. aer. As mentioned, the conserved nature of this arginine, its location at the cap of the four-helical bundle, its critical role in membrane localization (Fig. 4), and its positive charge makes it an attractive candidate for binding to the negatively charged phosphates of PI(4,5)P2. Whereas the portion of the ExoUP. aer L3 loop containing Arg-661 was disordered, this loop was modeled in the ExoUP. flu structure (Fig. 11A). One of the ExoUP. flu monomers showed the L3 loop protruding and well positioned to interact with membranes (Fig. 8B). Importantly, the ExoUP. flu monomers have both the four-helical bundle and the PLA2 catalytic residues oriented to the same face of the protein (Fig. 8B). This arrangement is consistent with the four-helical bundle positioning ExoU at the plasma membrane in an orientation that facilitates placement of substrate membrane phospholipids into the catalytic pocket. The caps of the four-helical bundles of both ExoUP. flu and ExoUP. aer form positively charged surface-exposed pockets, suggesting potential additional contacts with negatively charged PI(4,5)P2 molecules (Fig. 10B). Interestingly, this surface charge distribution differs from structurally similar four-helical bundle domains of other toxins such as P. multocida toxin (PMT) and Clostridium difficile toxin B (TcdB) that do not have specificity for PI(4,5)P2 (Fig. 10B). Additional conserved residues important for the localization of ExoUP. aer (30) are also structurally conserved in ExoUP. flu (Figs. 3A and 11A). Furthermore, the conserved polar or charged residues Arg-616, Gln-578, and Asn-582 of ExoUP. flu (Arg-661, Gln-623, and Asn-627 in ExoUP. aer) were all located in the cap of the four-helical bundle, suggesting a role in binding to PI(4,5)P2. In contrast, the conserved hydrophobic residues Phe-564, Ile-609, and Ala-632 of ExoUP. flu (Ile-609, Ile-654, and Ala-678 in ExoUP. aer) were located within the α-helices themselves, signifying a role in maintaining the overall four-helical bundle configuration (Fig. 11A). In summary, structural data suggest a model whereby the four-helical bundle structural motif is used by many bacterial proteins to interact with a variety of membranes, but the caps of the four-helical bundles of patatin-like proteins have been “customized” to bind PI(4,5)P2 and target these proteins to the plasma membrane of eukaryotic cells.
FIGURE 11.
Four-helical bundle structures of ExoU MLDs. A, domain 4 structures of ExoUP. flu and ExoUP. aer. Residues critical to PI(4,5)P2 binding by ExoUP. aer (Fig. 1B) are indicated, as are the homologous residues present in ExoUP. flu. The L3 of ExoUP. aer, which contains Arg-661 (blue dashed line), was disordered, but the corresponding L3 of ExoUP. flu was ordered. B, depictions of the four-helical bundle structures of ExoUP. asy, PMT, and TcdB. The ExoUP. asy structure is a model derived from the ExoUP. aer MLD (rainbow colors; blue, N terminus and red, C terminus). C, ExoUP. flu (colored as in Fig. 8B) with modeled PI(4)P molecule (ball-and-stick model with carbon atoms in green) based on the L. pneumophila effector DrrA/SidM structure (DrrA/SidM omitted for clarity; Protein Data Bank code 4MXP).
DISCUSSION
We describe the mechanism by which three members of the patatin-like phospholipases bind PI(4,5)P2 to localize to the plasma membrane of host cells. Using liposome binding assays, we found that the MLD of ExoUP. aer has specific affinity for PI(4,5)P2 and that MLD residues important for plasma membrane localization are also critical for PI(4,5)P2 binding. Although the ability of ExoU to bind PI(4,5)P2 was already known (31, 32), we found that PI(4,5)P2 mislocalization disrupted targeting of ExoUP. aer to the plasma membrane. The use of this novel PI(4,5)P2 binding domain is not restricted to ExoUP. aer but was also utilized by patatin-like proteins from P. fluorescens and P. asymbiotica. The crystal structures of the full-length ExoUP. flu and ExoUP. aer demonstrated that their MLDs likely use a four-helical bundle structural motif to present a positively charged pocket containing a conserved arginine residue important for PI(4,5)P2 binding. Binding of PI(4,5)P2 by this motif may orient the ExoU patatin-like catalytic site toward phospholipid substrates in the plasma membrane. Together, these findings define a new PI(4,5)P2-binding motif used by bacterial proteins to target the plasma membrane of eukaryotic cells, where they are well positioned to access phospholipid substrates within the plasma membrane and to cause cell lysis.
Phosphoinositides (phosphatidylinositol lipids with phosphates attached to the 3-, 4-, or 5-positions of the inositol ring) such as PI(4,5)P2 are critical for the function of eukaryotic cells, often serving as signposts for defined membrane compartments (34). PI(4,5)P2 is predominantly found in the plasma membrane, PI(4)P in the Golgi (66), and PI(3)P in endosomes (67). This allows proteins with particular lipid-binding specificities to be targeted to the corresponding membrane compartments. PI(4,5)P2 binding domains in particular are critical for the function of eukaryotic cells. For example, PH domains, epsin N-terminal homology domains, and FERM domains each target endogenous proteins to the plasma membrane (35, 36, 68). These and other domains play critical roles in cell processes such as endocytosis, motility, and cytoskeletal anchoring (35, 69, 70). They use different mechanisms for PI(4,5)P2 recognition, but a common theme is the presence of basic residues that bind to the negatively charged PI(4,5)P2 (71). Our results add further evidence to the notion that bacteria have evolved mechanisms to co-opt this elaborate localization system.
Although PI(4,5)P2 binding domains are common among eukaryotic proteins, the ExoU MLD is only the second report of a bacterial PI(4,5)P2 binding domain (72). Orth and co-workers (72) recently identified a bacterial phosphoinositide binding domain (BPD) in four effectors of the type III secretion systems of Vibrio parahaemolyticus, Yersinia spp., and P. syringae. The BPD localized these effectors to the plasma membrane of eukaryotic cells following injection. Unlike the MLD of ExoU, however, BPDs are at the N termini and overlap with the chaperone binding domains of these proteins. Structurally, BPDs have two β-strands and two α-helices instead of a four-helical bundle. These differences indicate that the ExoU MLD is quite distinct from the BPD and acts by a different mechanism. Perhaps this is because the PLA2 activities of ExoU and other patatin-like proteins impose more stringent constraints, requiring that the MLDs not only localize to membranes but also provide access to phospholipid substrates in the membrane. In support of this, ExoU phospholipases appear to more closely mimic the strategy employed by some eukaryotic phospholipases, which use PI(4,5)P2 to regulate not only their localization but also their activity. For instance, phospholipase D contains both a dedicated PH domain and an additional PI(4,5)P2-binding site responsible for activation of its phospholipase activity (73). cPLA2 is also localized to and activated by PI(4,5)P2, further illustrating the important role of PI(4,5)P2 in both intracellular targeting and activation of eukaryotic phospholipases (74). ExoU strongly binds to PI(4,5)P2, with a Kd of just 110 nm, comparable with the PH domain of phospholipase C, a prototype mammalian PI(4,5)P2 binding domain (75). It will be interesting to see whether eukaryotic PI(4,5)P2 binding domains can substitute for the MLD of ExoU or whether the structural uniqueness of the MLD confers specific PI(4,5)P2-binding properties that facilitate cell lysis by ExoU. In this regard, it should be noted that artificial targeting to eukaryotic membranes by farnesylation was insufficient to restore cell lysis to an ExoU variant with a disrupted MLD (30).
By obtaining the full-length crystal structure of a second ExoU patatin-like phospholipase, we were able to compare these two members of this protein family (28). In particular, the MLDs of ExoUP. aer and ExoUP. flu were quite similar, illustrating a conserved four-helical bundle fold that is important for PI(4,5)P2 binding. PI(4,5)P2 binding domains frequently contain positively charged residues that interact with the negatively charged phosphates of PI(4,5)P2 (71). Our findings suggest that Arg-661 of ExoUP. aer is such a residue. In a previous examination of ExoUP. aer for important MLD residues, a substitution in Arg-661 significantly attenuated ExoU localization and cytotoxicity (30). In this study, the homologous Arg-616 residue of ExoUP. flu was also important for localization, and both residues were contained within the L3 loop of the four-helical bundle. Additionally, modeling of the ExoUP. asy MLD structure based on the previously characterized ExoUP. aer structure indicated that it too could form a four-helical bundle containing the conserved arginine in the L3 loop (Fig. 11B). This is consistent with its similar localization pattern, as well as the importance of its conserved arginine residue for localization. Interestingly, other unrelated bacterial toxins, such as P. multocida PMT and C. difficile TcdB, also contain four-helical bundles (Fig. 11B) (8, 76), but these proteins have different lipid-binding specificities. Unlike ExoUP.aer and ExoUP. flu, PMT and TcdB bind with high affinity to a variety of negatively charged phospholipids such as phosphatidylserine and phosphatidylinositol (8, 38). Substitutions of positively charged amino acids in both the L1 and L3 loops of PMT and TcdB resulted in disruption of localization to membranes rich in these phospholipids (38). These other proteins also have conserved hydrophobic residues in the L3 loops that help establish membrane binding (38). ExoU does have hydrophobic residues in its L3 loop (residues 660–672), but they are not well conserved, suggesting they may not play a major role in localization (Fig. 3A).
The similarities between the four-helical bundles of the ExoU homologs and the other bacterial toxins suggest that four-helical bundles are a conserved structural platform for binding phospholipids in membranes but that alterations within these domains (and in particular in their L1 and L3 loops) “customize” their lipid-binding specificities. It will be interesting to see whether other four-helical bundle proteins are tailored to recognize phosphoinositides characteristic of other intracellular membrane compartments.
A more detailed mechanism of MLD binding will require a crystal structure of ExoU in complex with PI(4,5)P2. Although such a structure is not yet available, the structure of the Legionella pneumophila toxin DrrA in complex with PI(4)P was recently solved (77). The membrane-binding portion of DrrA forms a three-helical bundle rather than a four-helical bundle, and the structure shows binding to PI(4)P rather than PI(4,5)P2, but we reasoned that the comparison may nevertheless be informative. For this reason, we used this structure to model ExoUP. flu binding to PI(4)P. Superposition of the helical bundle domains positioned PI(4)P within the immediate vicinity of the L3 loop with the conserved Arg-616 of ExoUP. flu, as we had postulated (Fig. 11C). This supports the importance of the L3 loop and Arg-616 of ExoUP. flu in direct PI(4,5)P2 binding and plasma membrane localization. Additional contacts outside the MLD may contribute to this binding, as Arg-242 (Arg-293 of ExoUP. aer) is also well positioned to bind PI(4,5)P2 (Fig. 10B). However, the MLD alone is sufficient for PI(4,5)P2 binding (Fig. 1A) and for localization (Fig. 4). A more comprehensive screen is necessary to determine other residues in the L3 loop and the four-helical bundle of ExoU necessary for PI(4,5)P2 binding and the potential contribution of residues within the MLD but outside the four-helical bundle or outside the MLD (27). These studies could help inform the differences in apparent affinity for PI(4,5)P2 by the ExoU homologs, despite their maintaining a conserved mechanism (Fig. 2D). Of the several other residues already demonstrated to be important for PI(4,5)P2 binding by ExoU (Fig. 1B), most were buried within the four-helical bundle and not located near the postulated membrane-binding interface (28). It is possible that Arg-661 (of ExoUP. aer) makes initial contact with PI(4,5)P2, causing a conformational change in the four-helical bundle, which allows these buried residues to contact the membrane. Alternatively, these other residues may play a structural role in helical packing and maintenance of the overall four-helical conformation, as has been postulated for critical residues in other four-helical bundle MLDs (29).
ExoUP. aer has been extensively investigated for its importance both clinically and in animal models of infection (23, 78). In contrast, the homologous ExoU proteins from the emerging human and insect pathogen P. asymbiotica and the plant commensal P. fluorescens have not been previously studied. Although these proteins need to be examined more thoroughly in the context of the bacteria that naturally produce them, several features suggest that they may be important for interaction with eukaryotic cells. First, they are postulated to be effector proteins of type III secretion systems, suggesting that they are injected into eukaryotic cells. Indeed, ExoUP. asy is the sole identified putative effector in the P. asymbiotica type III secretion system locus (59). Second, they localize by binding to PI(4,5)P2, which is only found in eukaryotic cells. Third, they have PLA2 activities that are modulated by eukaryotic factors, which, at least in the case of ExoUP. aer, is used to subvert a broad spectrum of eukaryotic cells (79, 80). Interestingly, the enzymatic properties of these ExoU homologs differed substantially despite quite similar structures (Fig. 6). For example, compared with ExoUP. aer, ExoUP. flu was active in the absence of co-activators and highly active in the presence of ubiquitin. A more detailed and expanded comparative structural analysis of similar MLD-containing patatin-like proteins may identify mechanistic explanations for the different enzymatic properties of this family of proteins.
In summary, we have characterized a PI(4,5)P2 binding domain used by proteins from several different bacteria, including human and insect pathogens and a plant saprophyte. The four-helical bundle of the ExoU MLD binds PI(4,5)P2 through a mechanism that requires a conserved arginine in an exposed loop. It will be interesting to determine whether proteins other than patatin-like phospholipases use similar MLD domains to bind PI(4,5)P2.
Acknowledgments
We thank Peter Mayinger for the yeast strains, Kim Orth for the GFP-PH yeast expression construct, and Joyce Loper for the P. fluorescens strain. We thank members of the Hauser Laboratory for critical reading of the manuscript and the Northwestern University Cell Imaging Facility for assistance with microscopy. We thank the staff of the LS-CAT 21-ID-G station for help during x-ray data collection. Microscopy work utilized the Northwestern University Cell Imaging Facility, which is generously supported by National Institutes of Health Grant CCSG P30 CA060553 from NCI awarded to the Robert H. Lurie Comprehensive Cancer Center. The Center for Structural Genomics of Infectious Diseases project has been funded in whole or in part by National Institutes of Health through Contracts HHSN272200700058C and HHSN272201200026C from the NIAID and the Department of Health and Human Services (to W. F. A.).
Note Added in Proof
During the revision of this manuscript, Anderson et al. published an article (Anderson, D. M., Sato, H., Dirck, A. T., Feix, J. B., and Frank, D. W. (2014) J. Bacteriol. 10.1128/JB.02402-14) also demonstrating that some ExoU homologs bind PI(4,5)P2 and are cytotoxic and that ubiquitin and PI(4,5)P2 differentially activate these homologs.
This work was supported, in whole or in part, by National Institutes of Health Grants AI053674, AI075191, AI099269, AI04831, and AI088286 (to A. R. H.) and AI092825 and AI051490 (to K. J. S.). This work was also supported by American Heart Association Grant 12PRE8660003 (to G. H. T.).
The atomic coordinates and structure factors (code 4QMK) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- MLD
- membrane localization domain
- PC
- phosphatidylcholine
- PE
- phosphatidylethanolamine
- PS
- phosphatidylserine
- PI
- phosphatidylinositol
- PI(4)P
- phosphatidylinositol 4-phosphate
- PI(5)P
- phosphatidylinositol 5-phosphate
- PI(4,5)P2
- phosphatidylinositol 4,5-bisphosphate
- PI(3,4,5)P3
- phosphatidylinositol 3,4,5-triphosphate
- PLA2
- phospholipase A2
- SPR
- surface plasmon resonance
- PMT
- P. multocida toxin
- FERM
- 4.1/ezrin/radixin/moesin
- BPD
- bacterial phosphoinositide binding domain
- PH
- pleckstrin homology
- cPLA2
- cytosolic PLA2
- ExoUP. aer
- ExoU from P. aeruginosa
- ExoUP. flu
- ExoU from P. fluorescens
- ExoUP. asy
- ExoU from P. asymbiotica
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol.
REFERENCES
- 1. Tseng T. T., Tyler B. M., Setubal J. C. (2009) Protein secretion systems in bacterial-host associations, and their description in the Gene Ontology. BMC Microbiol. 9, Suppl. 1, S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Aktories K. (2011) Bacterial protein toxins that modify host regulatory GTPases. Nat. Rev. Microbiol. 9, 487–498 [DOI] [PubMed] [Google Scholar]
- 3. Satchell K. J. (2009) Actin crosslinking toxins of Gram-negative bacteria. Toxins 1, 123–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sitkiewicz I., Stockbauer K. E., Musser J. M. (2007) Secreted bacterial phospholipase A2 enzymes: better living through phospholipolysis. Trends Microbiol. 15, 63–69 [DOI] [PubMed] [Google Scholar]
- 5. Geissler B. (2012) Bacterial toxin effector-membrane targeting: outside in, then back again. Front. Cell. Infect. Microbiol. 2, 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chen S., Barbieri J. T. (2011) Association of botulinum neurotoxin serotype A light chain with plasma membrane-bound SNAP-25. J. Biol. Chem. 286, 15067–15072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Boucrot E., Beuzón C. R., Holden D. W., Gorvel J. P., Méresse S. (2003) Salmonella typhimurium SifA effector protein requires its membrane-anchoring C-terminal hexapeptide for its biological function. J. Biol. Chem. 278, 14196–14202 [DOI] [PubMed] [Google Scholar]
- 8. Kamitani S., Kitadokoro K., Miyazawa M., Toshima H., Fukui A., Abe H., Miyake M., Horiguchi Y. (2010) Characterization of the membrane-targeting C1 domain in Pasteurella multocida toxin. J. Biol. Chem. 285, 25467–25475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhang Y., Deng Q., Porath J. A., Williams C. L., Pederson-Gulrud K. J., Barbieri J. T. (2007) Plasma membrane localization affects the RhoGAP specificity of Pseudomonas ExoS. Cell. Microbiol. 9, 2192–2201 [DOI] [PubMed] [Google Scholar]
- 10. Hirschberg H. J., Simons J. W., Dekker N., Egmond M. R. (2001) Cloning, expression, purification and characterization of patatin, a novel phospholipase A. Eur. J. Biochem. 268, 5037–5044 [DOI] [PubMed] [Google Scholar]
- 11. Sato H., Frank D. W., Hillard C. J., Feix J. B., Pankhaniya R. R., Moriyama K., Finck-Barbançon V., Buchaklian A., Lei M., Long R. M., Wiener-Kronish J., Sawa T. (2003) The mechanism of action of the Pseudomonas aeruginosa-encoded type III cytotoxin, ExoU. EMBO J. 22, 2959–2969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lang C., Flieger A. (2011) Characterisation of Legionella pneumophila phospholipases and their impact on host cells. Eur. J. Cell Biol. 90, 903–912 [DOI] [PubMed] [Google Scholar]
- 13. Banerji S., Flieger A. (2004) Patatin-like proteins: a new family of lipolytic enzymes present in bacteria? Microbiology 150, 522–525 [DOI] [PubMed] [Google Scholar]
- 14. Salacha R., Kovacipć F., Brochier-Armanet C., Wilhelm S., Tommassen J., Filloux A., Voulhoux R., Bleves S. (2010) The Pseudomonas aeruginosa patatin-like protein PlpD is the archetype of a novel type V secretion system. Environ. Microbiol. 12, 1498–1512 [DOI] [PubMed] [Google Scholar]
- 15. VanRheenen S. M., Luo Z. Q., O'Connor T., Isberg R. R. (2006) Members of a Legionella pneumophila family of proteins with ExoU (phospholipase A) active sites are translocated to target cells. Infect. Immun. 74, 3597–3606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Aurass P., Pless B., Rydzewski K., Holland G., Bannert N., Flieger A. (2009) bdhA-patD operon as a virulence determinant, revealed by a novel large-scale approach for identification of Legionella pneumophila mutants defective for amoeba infection. Appl. Environ. Microbiol. 75, 4506–4515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Laskowski R. A., MacArthur M. W., Moss D. S., Thornton J. M. (1993) PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26, 283–291 [Google Scholar]
- 18. Zhu W., Hammad L. A., Hsu F., Mao Y., Luo Z. Q. (2013) Induction of caspase 3 activation by multiple Legionella pneumophila Dot/Icm substrates. Cell. Microbiol. 15, 1783–1795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kang P. J., Hauser A. R., Apodaca G., Fleiszig S. M., Wiener-Kronish J., Mostov K., Engel J. N. (1997) Identification of Pseudomonas aeruginosa genes required for epithelial cell injury. Mol. Microbiol. 24, 1249–1262 [DOI] [PubMed] [Google Scholar]
- 20. Hauser A. R., Kang P. J., Engel J. (1998) PepA, a novel secreted protein of Pseudomonas aeruginosa, is necessary for cytotoxicity and virulence. Mol. Microbiol. 27, 807–818 [DOI] [PubMed] [Google Scholar]
- 21. Finck-Barbançon V., Goranson J., Zhu L., Sawa T., Wiener-Kronish J. P., Fleiszig S. M., Wu C., Mende-Mueller L., Frank D. W. (1997) ExoU expression by Pseudomonas aeruginosa correlates with acute cytotoxicity and epithelial injury. Mol. Microbiol. 25, 547–557 [DOI] [PubMed] [Google Scholar]
- 22. Hauser A. R., Cobb E., Bodi M., Mariscal D., Vallés J., Engel J. N., Rello J. (2002) Type III protein secretion is associated with poor clinical outcomes in patients with ventilator-associated pneumonia caused by Pseudomonas aeruginosa. Crit. Care Med. 30, 521–528 [DOI] [PubMed] [Google Scholar]
- 23. Schulert G. S., Feltman H., Rabin S. D., Martin C. G., Battle S. E., Rello J., Hauser A. R. (2003) Secretion of the toxin ExoU is a marker for highly virulent Pseudomonas aeruginosa isolates obtained from patients with hospital-acquired pneumonia. J. Infect. Dis. 188, 1695–1706 [DOI] [PubMed] [Google Scholar]
- 24. Sato H., Feix J. B., Hillard C. J., Frank D. W. (2005) Characterization of phospholipase activity of the Pseudomonas aeruginosa type III cytotoxin, ExoU. J. Bacteriol. 187, 1192–1195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Anderson D. M., Schmalzer K. M., Sato H., Casey M., Terhune S. S., Haas A. L., Feix J. B., Frank D. W. (2011) Ubiquitin and ubiquitin-modified proteins activate the Pseudomonas aeruginosa T3SS cytotoxin, ExoU. Mol. Microbiol. 82, 1454–1467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tyson G. H., Hauser A. R. (2013) Phosphatidylinositol 4,5-bisphosphate is a novel co-activator of the Pseudomonas aeruginosa cytotoxin ExoU. Infect. Immun. 81, 2873–2881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rabin S. D., Veesenmeyer J. L., Bieging K. T., Hauser A. R. (2006) A C-terminal domain targets the Pseudomonas aeruginosa cytotoxin ExoU to the plasma membrane of host cells. Infect. Immun. 74, 2552–2561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Halavaty A. S., Borek D., Tyson G. H., Veesenmeyer J. L., Shuvalova L., Minasov G., Otwinowski Z., Hauser A. R., Anderson W. F. (2012) Structure of the type III secretion effector protein ExoU in complex with its chaperone SpcU. PLoS One 7, e49388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Geissler B., Tungekar R., Satchell K. J. (2010) Identification of a conserved membrane localization domain within numerous large bacterial protein toxins. Proc. Natl. Acad. Sci. U.S.A. 107, 5581–5586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Veesenmeyer J. L., Howell H., Halavaty A. S., Ahrens S., Anderson W. F., Hauser A. R. (2010) Role of the membrane localization domain of the Pseudomonas aeruginosa effector protein ExoU in cytotoxicity. Infect. Immun. 78, 3346–3357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gendrin C., Contreras-Martel C., Bouillot S., Elsen S., Lemaire D., Skoufias D. A., Huber P., Attree I., Dessen A. (2012) Structural basis of cytotoxicity mediated by the type III secretion toxin ExoU from Pseudomonas aeruginosa. PLoS Pathog. 8, e1002637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sato H., Frank D. W. (2014) Intoxication of host cells by the T3SS phospholipase ExoU: PI(4,5)P2-associated, cytoskeletal collapse and late phase membrane blebbing. PLoS One 9, e103127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. McLaughlin S., Wang J., Gambhir A., Murray D. (2002) PIP(2) and proteins: interactions, organization, and information flow. Annu. Rev. Biophys. Biomol. Struct. 31, 151–175 [DOI] [PubMed] [Google Scholar]
- 34. Behnia R., Munro S. (2005) Organelle identity and the signposts for membrane traffic. Nature 438, 597–604 [DOI] [PubMed] [Google Scholar]
- 35. Hamada K., Shimizu T., Matsui T., Tsukita S., Hakoshima T. (2000) Structural basis of the membrane-targeting and unmasking mechanisms of the radixin FERM domain. EMBO J. 19, 4449–4462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lemmon M. A., Ferguson K. M. (2000) Signal-dependent membrane targeting by pleckstrin homology (PH) domains. Biochem. J. 350, 1–18 [PMC free article] [PubMed] [Google Scholar]
- 37. Rabin S. D., Hauser A. R. (2003) Pseudomonas aeruginosa ExoU, a toxin transported by the type III secretion system, kills Saccharomyces cerevisiae. Infect. Immun. 71, 4144–4150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Geissler B., Ahrens S., Satchell K. J. (2012) Plasma membrane association of three classes of bacterial toxins is mediated by a basic-hydrophobic motif. Cell. Microbiol. 14, 286–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. (1990) Basic local alignment search tool. J. Mol. Biol. 215, 403–410 [DOI] [PubMed] [Google Scholar]
- 40. Kim H., Afsari H. S., Cho W. (2013) High-throughput fluorescence assay for membrane-protein interaction. J. Lipid Res. 54, 3531–3538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Otwinowski Z., Minor W. (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 42. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Winn M. D., Ballard C. C., Cowtan K. D., Dodson E. J., Emsley P., Evans P. R., Keegan R. M., Krissenel E. B., Leslie A. G., McCoy A., McNicholas S. J., Murshudov G. N., Pannu N. S., Potterton E. A., Powell H. R., Read R. J., Vagin A., Wilson K. S. (2011) Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67, 235–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Morris R. J., Perrakis A., Lamzin V. S. (2003) ARP/warp and automatic interpretation of protein electron density maps. Methods Enzymol. 374, 229–244 [DOI] [PubMed] [Google Scholar]
- 45. Murshudov G. N., Skubák P., Lebedev A. A., Pannu N. S., Steiner R. A., Nicholls R. A., Winn M. D., Long F., Vagin A. A. (2011) REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D Biol. Crystallogr. 67, 355–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Emsley P., Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 47. Emsley P., Lohkamp B., Scott W. G., Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Chen V. B., Arendall W. B., 3rd, Headd J. J., Keedy D. A., Immormino R. M., Kapral G. J., Murray L. W., Richardson J. S., Richardson D. C. (2010) MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Davis I. W., Leaver-Fay A., Chen V. B., Block J. N., Kapral G. J., Wang X., Murray L. W., Arendall W. B., 3rd, Snoeyink J., Richardson J. S., Richardson D. C. (2007) MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 35, W375–W383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Krissinel E., Henrick K. (2007) Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797 [DOI] [PubMed] [Google Scholar]
- 51. DeLano W. L. (2012) The PyMOL Molecular Graphics System, Version 1.5.0.4, Schrödinger, LLC, New York [Google Scholar]
- 52. Mesmin B., Robbe K., Geny B., Luton F., Brandolin G., Popoff M. R., Antonny B. (2004) A phosphatidylserine-binding site in the cytosolic fragment of Clostridium sordellii lethal toxin facilitates glucosylation of membrane-bound Rac and is required for cytotoxicity. J. Biol. Chem. 279, 49876–49882 [DOI] [PubMed] [Google Scholar]
- 53. Szentpetery Z., Balla A., Kim Y. J., Lemmon M. A., Balla T. (2009) Live cell imaging with protein domains capable of recognizing phosphatidylinositol 4,5-bisphosphate; a comparative study. BMC Cell Biol. 10, 67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Hammond G. R., Fischer M. J., Anderson K. E., Holdich J., Koteci A., Balla T., Irvine R. F. (2012) PI4P and PI(4,5)P2 are essential but independent lipid determinants of membrane identity. Science 337, 727–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Chen Y., Sheng R., Källberg M., Silkov A., Tun M. P., Bhardwaj N., Kurilova S., Hall R. A., Honig B., Lu H., Cho W. (2012) Genome-wide functional annotation of dual-specificity protein- and lipid-binding modules that regulate protein interactions. Mol. Cell 46, 226–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Várnai P., Balla T. (2008) Live cell imaging of phosphoinositides with expressed inositide binding protein domains. Methods 46, 167–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Manna D., Bhardwaj N., Vora M. S., Stahelin R. V., Lu H., Cho W. (2008) Differential roles of phosphatidylserine, PtdIns(4,5)P2, and PtdIns(3,4,5)P3 in plasma membrane targeting of C2 domains. Molecular dynamics simulation, membrane binding, and cell translocation studies of the PKCα C2 domain. J. Biol. Chem. 283, 26047–26058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Varivarn K., Champa L. A., Silby M. W., Robleto E. A. (2013) Colonization strategies of Pseudomonas fluorescens Pf0–1: activation of soil-specific genes important for diverse and specific environments. BMC Microbiol. 13, 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Costa S. C., Girard P. A., Brehélin M., Zumbihl R. (2009) The emerging human pathogen Photorhabdus asymbiotica is a facultative intracellular bacterium and induces apoptosis of macrophage-like cells. Infect. Immun. 77, 1022–1030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Loper J. E., Hassan K. A., Mavrodi D. V., Davis E. W., 2nd, Lim C. K., Shaffer B. T., Elbourne L. D., Stockwell V. O., Hartney S. L., Breakwell K., Henkels M. D., Tetu S. G., Rangel L. I., Kidarsa T. A., Wilson N. L., van de Mortel J. E., Song C., Blumhagen R., Radune D., Hostetler J. B., Brinkac L. M., Durkin A. S., Kluepfel D. A., Wechter W. P., Anderson A. J., Kim Y. C., Pierson L. S., 3rd, Pierson E. A., Lindow S. E., Kobayashi D. Y., Raaijmakers J. M., Weller D. M., Thomashow L. S., Allen A. E., Paulsen I. T. (2012) Comparative genomics of plant-associated Pseudomonas spp.: insights into diversity and inheritance of traits involved in multitrophic interactions. PLoS Genet. 8, e1002784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Wiradjaja F., Ooms L. M., Tahirovic S., Kuhne E., Devenish R. J., Munn A. L., Piper R. C., Mayinger P., Mitchell C. A. (2007) Inactivation of the phosphoinositide phosphatases Sac1p and Inp54p leads to accumulation of phosphatidylinositol 4,5-bisphosphate on vacuole membranes and vacuolar fusion defects. J. Biol. Chem. 282, 16295–16307 [DOI] [PubMed] [Google Scholar]
- 62. Goujon M., McWilliam H., Li W., Valentin F., Squizzato S., Paern J., Lopez R. (2010) A new bioinformatics analysis tools framework at EMBL-EBI. Nucleic Acids Res. 38, W695–W699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Holm L., Park J. (2000) DaliLite workbench for protein structure comparison. Bioinformatics 16, 566–567 [DOI] [PubMed] [Google Scholar]
- 64. McWilliam H., Li W., Uludag M., Squizzato S., Park Y. M., Buso N., Cowley A. P., Lopez R. (2013) Analysis tool web services from the EMBL-EBI. Nucleic Acids Res. 41, W597–W600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Rabin S. D., Hauser A. R. (2005) Functional regions of the Pseudomonas aeruginosa cytotoxin ExoU. Infect Immun. 73, 573–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Bishé B., Syed G. H., Field S. J., Siddiqui A. (2012) Role of phosphatidylinositol 4-phosphate (PI4P) and its binding protein GOLPH3 in hepatitis C virus secretion. J. Biol. Chem. 287, 27637–27647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Gillooly D. J., Morrow I. C., Lindsay M., Gould R., Bryant N. J., Gaullier J. M., Parton R. G., Stenmark H. (2000) Localization of phosphatidylinositol 3-phosphate in yeast and mammalian cells. EMBO J. 19, 4577–4588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Itoh T., Koshiba S., Kigawa T., Kikuchi A., Yokoyama S., Takenawa T. (2001) Role of the ENTH domain in phosphatidylinositol-4,5-bisphosphate binding and endocytosis. Science 291, 1047–1051 [DOI] [PubMed] [Google Scholar]
- 69. Chen X., Rotenberg S. A. (2010) PhosphoMARCKS drives motility of mouse melanoma cells. Cell. Signal. 22, 1097–1103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Martin T. F. (2001) PI(4,5)P(2) regulation of surface membrane traffic. Curr. Opin. Cell Biol. 13, 493–499 [DOI] [PubMed] [Google Scholar]
- 71. Li L., Shi X., Guo X., Li H., Xu C. (2014) Ionic protein-lipid interaction at the plasma membrane: what can the charge do? Trends Biochem. Sci. 39, 130–140 [DOI] [PubMed] [Google Scholar]
- 72. Salomon D., Guo Y., Kinch L. N., Grishin N. V., Gardner K. H., Orth K. (2013) Effectors of animal and plant pathogens use a common domain to bind host phosphoinositides. Nat. Commun. 4, 2973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Sciorra V. A., Rudge S. A., Prestwich G. D., Frohman M. A., Engebrecht J., Morris A. J. (1999) Identification of a phosphoinositide binding motif that mediates activation of mammalian and yeast phospholipase D isoenzymes. EMBO J. 18, 5911–5921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Le Berre L., Takano T., Papillon J., Lemay S., Cybulsky A. V. (2006) Role of phosphatidylinositol 4,5-bisphosphate in the activation of cytosolic phospholipase A2-α. Prostaglandins Other Lipid Mediat. 81, 113–125 [DOI] [PubMed] [Google Scholar]
- 75. Yoon Y., Lee P. J., Kurilova S., Cho W. (2011) In situ quantitative imaging of cellular lipids using molecular sensors. Nat. Chem. 3, 868–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Reinert D. J., Jank T., Aktories K., Schulz G. E. (2005) Structural basis for the function of Clostridium difficile toxin B. J. Mol. Biol. 351, 973–981 [DOI] [PubMed] [Google Scholar]
- 77. Del Campo C. M., Mishra A. K., Wang Y. H., Roy C. R., Janmey P. A., Lambright D. G. (2014) Structural basis for PI(4)P-specific membrane recruitment of the legionella pneumophila effector DrrA/SidM. Structure 22, 397–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Shaver C. M., Hauser A. R. (2004) Relative contributions of Pseudomonas aeruginosa ExoU, ExoS, and ExoT to virulence in the lung. Infect Immun. 72, 6969–6977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Matz C., Moreno A. M., Alhede M., Manefield M., Hauser A. R., Givskov M., Kjelleberg S. (2008) Pseudomonas aeruginosa uses type III secretion system to kill biofilm-associated amoebae. ISME J. 2, 843–852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Miyata S., Casey M., Frank D. W., Ausubel F. M., Drenkard E. (2003) Use of the Galleria mellonella caterpillar as a model host to study the role of the type III secretion system in Pseudomonas aeruginosa pathogenesis. Infect. Immun. 71, 2404–2413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Schorr M., Then A., Tahirovic S., Hug N., Mayinger P. (2001) The phosphoinositide phosphatase Sac1p controls trafficking of the yeast Chs3p chitin synthase. Curr. Biol. 11, 1421–1426 [DOI] [PubMed] [Google Scholar]