Abstract
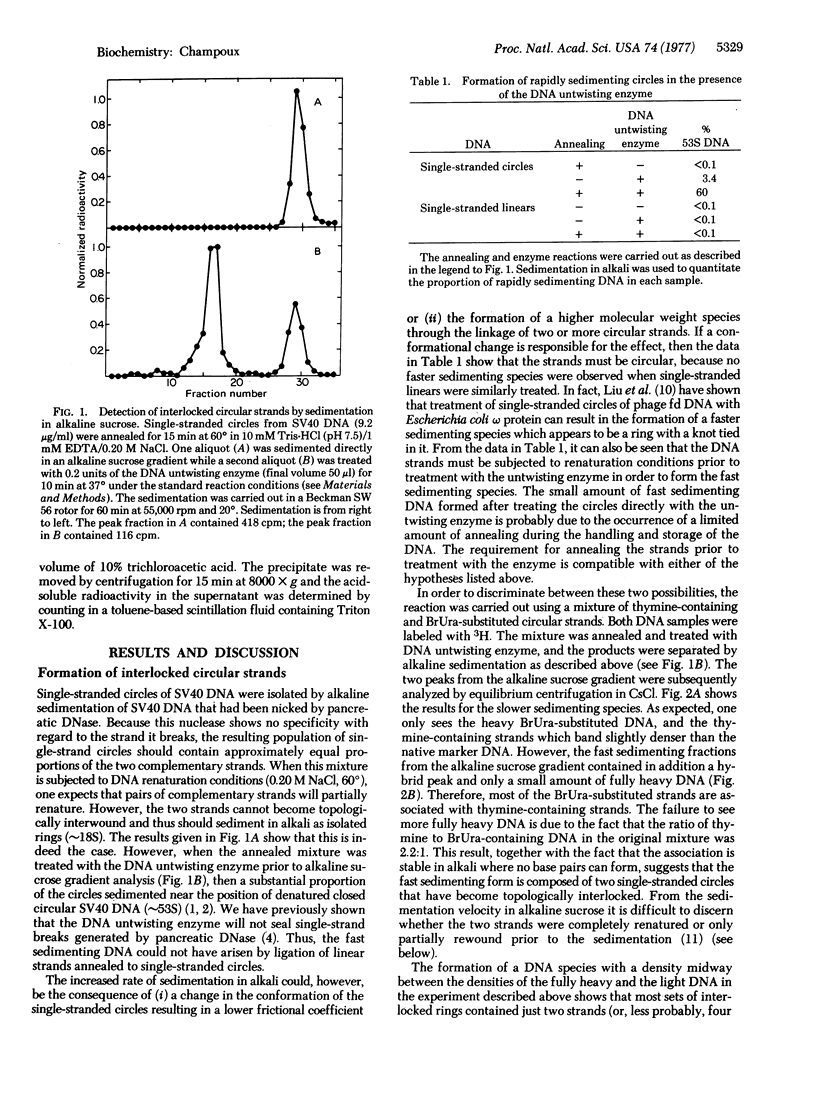
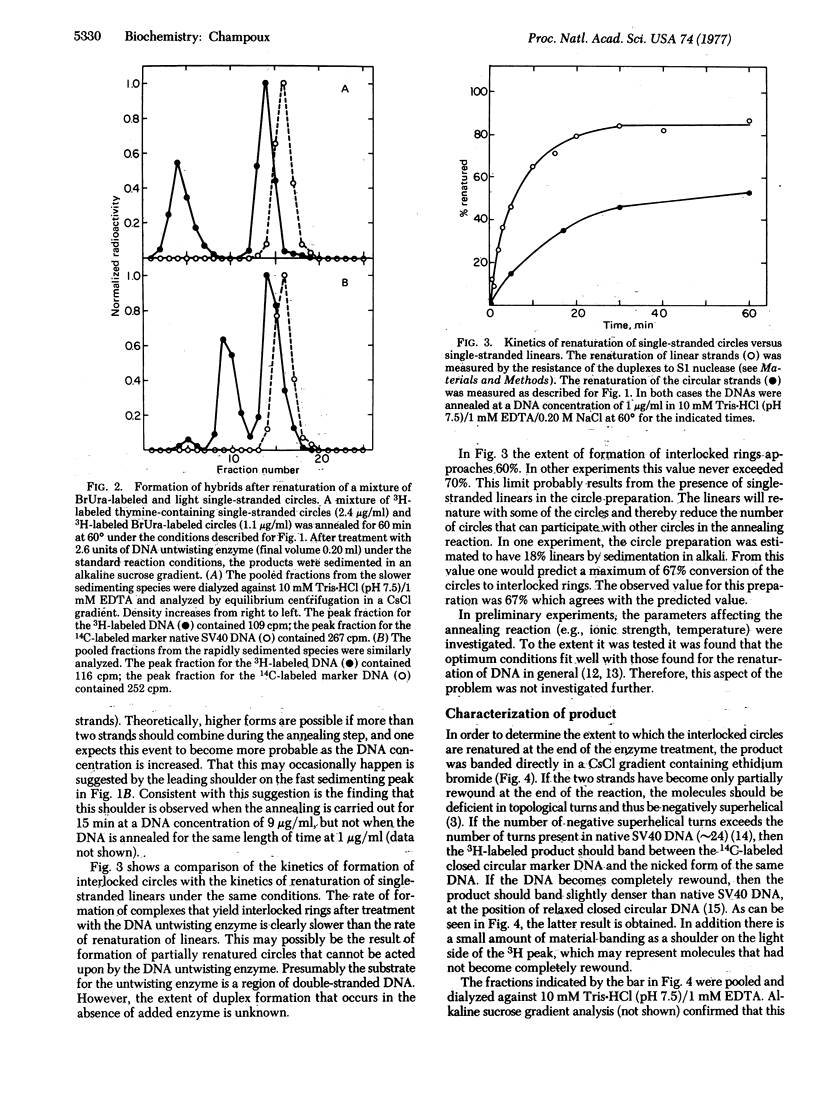
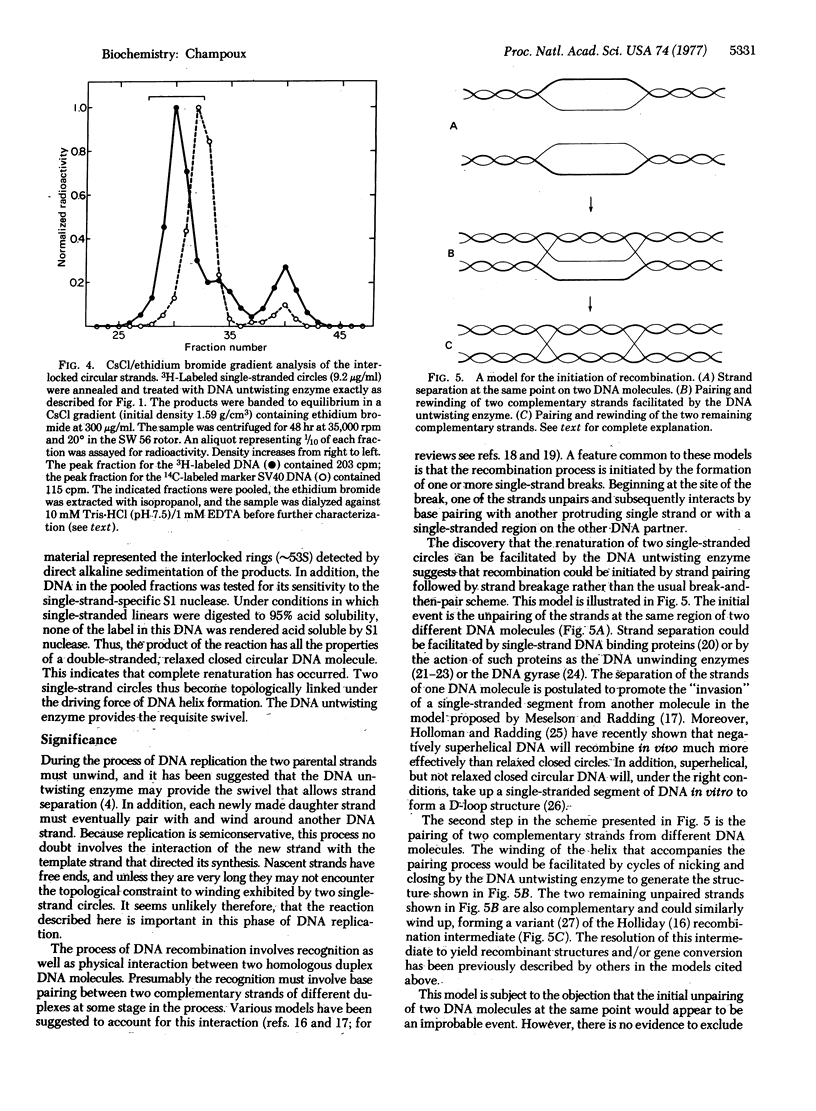
Renaturation of two complementary single-stranded circles should be limited by topological constraints against the rewinding of the DNA helix. If a mixture of complementary single-stranded rings is annealed and then treated with the DNA untwisting enzyme, the DNA circles completely renature as judged by (i) the presence of interlocked rings that sediment at 53 S in alkali, (ii) the buoyant density of the renatured DNA in CsCl gradients containing ethidium bromide, and (iii) the resistance of the product to the single-strand-specific S1 nuclease. Therefore, the DNA untwisting enzyme is able to provide a transient single-strand break that is sufficient to allow the two strands to completely rewind. The possibility that the untwisting enzyme might facilitate the initiation of the process of genetic recombination is discussed.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abdel-Monem M., Dürwald H., Hoffmann-Berling H. Enzymic unwinding of DNA. 2. Chain separation by an ATP-dependent DNA unwinding enzyme. Eur J Biochem. 1976 Jun 1;65(2):441–449. doi: 10.1111/j.1432-1033.1976.tb10359.x. [DOI] [PubMed] [Google Scholar]
- Alberts B. M., Frey L. T4 bacteriophage gene 32: a structural protein in the replication and recombination of DNA. Nature. 1970 Sep 26;227(5265):1313–1318. doi: 10.1038/2271313a0. [DOI] [PubMed] [Google Scholar]
- Broker T. R., Doermann A. H. Molecular and genetic recombination of bacteriophage T4. Annu Rev Genet. 1975;9:213–244. doi: 10.1146/annurev.ge.09.120175.001241. [DOI] [PubMed] [Google Scholar]
- Champoux J. J., Dulbecco R. An activity from mammalian cells that untwists superhelical DNA--a possible swivel for DNA replication (polyoma-ethidium bromide-mouse-embryo cells-dye binding assay). Proc Natl Acad Sci U S A. 1972 Jan;69(1):143–146. doi: 10.1073/pnas.69.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champoux J. J. Evidence for an intermediate with a single-strand break in the reaction catalyzed by the DNA untwisting enzyme. Proc Natl Acad Sci U S A. 1976 Oct;73(10):3488–3491. doi: 10.1073/pnas.73.10.3488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champoux J. J., McConaughy B. L. Priming of superhelical SV40 DNA by Escherichia coli RNA polymerase for in vitro DNA synthesis. Biochemistry. 1975 Jan 28;14(2):307–316. doi: 10.1021/bi00673a017. [DOI] [PubMed] [Google Scholar]
- Champoux J. J., McConaughy B. L. Purification and characterization of the DNA untwisting enzyme from rat liver. Biochemistry. 1976 Oct 19;15(21):4638–4642. doi: 10.1021/bi00666a014. [DOI] [PubMed] [Google Scholar]
- DULBECCO R., VOGT M. EVIDENCE FOR A RING STRUCTURE OF POLYOMA VIRUS DNA. Proc Natl Acad Sci U S A. 1963 Aug;50:236–243. doi: 10.1073/pnas.50.2.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gellert M., Mizuuchi K., O'Dea M. H., Nash H. A. DNA gyrase: an enzyme that introduces superhelical turns into DNA. Proc Natl Acad Sci U S A. 1976 Nov;73(11):3872–3876. doi: 10.1073/pnas.73.11.3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray H. B., Jr, Upholt W. B., Vinograd J. A buoyant method for the determination of the superhelix density of closed circular DNA. J Mol Biol. 1971 Nov 28;62(1):1–19. doi: 10.1016/0022-2836(71)90127-6. [DOI] [PubMed] [Google Scholar]
- Hirt B. Selective extraction of polyoma DNA from infected mouse cell cultures. J Mol Biol. 1967 Jun 14;26(2):365–369. doi: 10.1016/0022-2836(67)90307-5. [DOI] [PubMed] [Google Scholar]
- Holloman W. K., Radding C. M. Recombination promoted by superhelical DNA and the recA gene of Escherichia coli. Proc Natl Acad Sci U S A. 1976 Nov;73(11):3910–3914. doi: 10.1073/pnas.73.11.3910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holloman W. K., Wiegand R., Hoessli C., Radding C. M. Uptake of homologous single-stranded fragments by superhelical DNA: a possible mechanism for initiation of genetic recombination. Proc Natl Acad Sci U S A. 1975 Jun;72(6):2394–2398. doi: 10.1073/pnas.72.6.2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotchkiss R. D. Models of genetic recombination. Annu Rev Microbiol. 1974;28(0):445–468. doi: 10.1146/annurev.mi.28.100174.002305. [DOI] [PubMed] [Google Scholar]
- Keller W. Determination of the number of superhelical turns in simian virus 40 DNA by gel electrophoresis. Proc Natl Acad Sci U S A. 1975 Dec;72(12):4876–4880. doi: 10.1073/pnas.72.12.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L. F., Depew R. E., Wang J. C. Knotted single-stranded DNA rings: a novel topological isomer of circular single-stranded DNA formed by treatment with Escherichia coli omega protein. J Mol Biol. 1976 Sep 15;106(2):439–452. doi: 10.1016/0022-2836(76)90095-4. [DOI] [PubMed] [Google Scholar]
- Mackay V., Linn S. Selective inhibition of the dnase activity of the recBC enzyme by the DNA binding protein from Escherichia coli. J Biol Chem. 1976 Jun 25;251(12):3716–3719. [PubMed] [Google Scholar]
- McMilin K. D., Stahl M. M., Stahl F. W. Rec-mediated recombinational hot spot activity in bacteriophage lambda. I. Hot spot activity associated with spi-deletions and bio substitutions. Genetics. 1974 Jul;77(3):409–423. doi: 10.1093/genetics/77.3.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meselson M. S., Radding C. M. A general model for genetic recombination. Proc Natl Acad Sci U S A. 1975 Jan;72(1):358–361. doi: 10.1073/pnas.72.1.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmir M., Révet B. M., Vinograd J. Dependence of the sedimentation coefficient of denatured closed circular DNA in alkali on the degree of strand interwinding. The absolute sense of supercoils. J Mol Biol. 1974 Feb 15;83(1):35–45. doi: 10.1016/0022-2836(74)90422-7. [DOI] [PubMed] [Google Scholar]
- Scott J. F., Eisenberg S., Bertsch L. L., Kornberg A. A mechanism of duplex DNA replication revealed by enzymatic studies of phage phi X174: catalytic strand separation in advance of replication. Proc Natl Acad Sci U S A. 1977 Jan;74(1):193–197. doi: 10.1073/pnas.74.1.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobell H. M. Symmetry in protein-nucleic acid interaction and its genetic implications. Adv Genet. 1973;17:411–490. doi: 10.1016/s0065-2660(08)60175-3. [DOI] [PubMed] [Google Scholar]
- Studier F. W. Effects of the conformation of single-stranded DNA on renaturation and aggregation. J Mol Biol. 1969 Apr;41(2):199–209. doi: 10.1016/0022-2836(69)90385-4. [DOI] [PubMed] [Google Scholar]
- Vinograd J., Lebowitz J., Watson R. Early and late helix-coil transitions in closed circular DNA. The number of superhelical turns in polyoma DNA. J Mol Biol. 1968 Apr 14;33(1):173–197. doi: 10.1016/0022-2836(68)90287-8. [DOI] [PubMed] [Google Scholar]
- WEIL R., VINOGRAD J. THE CYCLIC HELIX AND CYCLIC COIL FORMS OF POLYOMA VIRAL DNA. Proc Natl Acad Sci U S A. 1963 Oct;50:730–738. doi: 10.1073/pnas.50.4.730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wetmur J. G., Davidson N. Kinetics of renaturation of DNA. J Mol Biol. 1968 Feb 14;31(3):349–370. doi: 10.1016/0022-2836(68)90414-2. [DOI] [PubMed] [Google Scholar]


