Abstract
Background
The choice of a molecular test for first intention determination of paternal RHD zygosity, before entering into invasive diagnostics, is important for the management of pregnancies at risk of haemolytic disease of the foetus and newborn related to anti-RhD.
Materials and methods
RHD zygosity was evaluated in 370 RH:1 Tunisian donors by polymerase chain reaction - sequence-specific polymorphism (PCR-SSP) analysis and polymerase chain reaction - restriction fragment length polymorphism (PCR-RFLP) amplification of hybrid Rhesus box and by real time quantitative polymerase chain reaction (RQ-PCR) specific for RHD exon 5. To evaluate the accuracy of molecular tests in the cases of discordant results, the ten exons of RHD and Rhesus boxes were amplified by PCR and sequenced.
Results
Molecular investigations revealed that our 370 donors comprise 193 dizygous and 145 hemizygous individuals and 32 subjects whose zygosity remains unknown. Positive predictive values were higher than 99% for all the methods, reaching 100% for RQ-PCR. Negative predictive values were 83.24%, 87.27% and 98% for PCR-SSP, PCR-RFLP and RQ-PCR respectively. This study also revealed 19 novel Rhesus box polymorphisms and three novel RHD alleles: RHD(Trp185Stop), RHD(Ala176Thr) and RHD(Ile342Ile).
Discussion
RQ-PCR is the most convenient method for first intention determination of paternal RHD zygosity in Tunisians. However, taking into account positive and negative predictive values, PCR-RFLP could be an alternative despite the heterogeneity of Rhesus boxes and the complexity of RHD.
Keywords: zygosity, RHD alleles, Rhesus box polymorphisms
Introduction
RhD is a complex blood group antigen and anti-D has been implicated in haemolytic disease of the foetus and newborn (HDFN). Adoption of antenatal and postpartum use of Rh immune globulin in industrialised countries has resulted in a major decrease in the frequency of this disease1. In Tunisia, HDFN due to RhD immunisation is currently prevented in the vast majority of cases by administration of anti-D immunoglobulin to D-negative women within 72 hours of delivery of a RhD-positive neonate as well as in cases of abortion2. Systematic antenatal prophylaxis could enhance the prevention for all pregnant women but this strategy could create a shortage in anti-D immunoglobulin and carry significant potential costs. In order to develop an antenatal prophylaxis limited to foetuses that are potentially at risk of HDFN related to anti-D, it has been proposed that paternal zygosity could be determined as the first step in the management of the red cell alloimmunised pregnancy3. If the father is found to have a heterozygous genotype, genetic testing could be undertaken, through amniocentesis, to determine whether the foetus is at risk of foetal anaemia. The overall sensitivity and specificity of polymerase chain reaction (PCR) typing on amniotic fluid has been reported to be 99.5% and 98.6%, respectively, and both positive and negative predictive values were 99.1%4. However this invasive procedure is risky and might cause further sensitisation in RhD-negative women. RHD zygosity was once determined through serological testing using population statistics. Ethnicity played a major role in these calculations and recently molecular tests, which circumvent this issue, were shown to be more accurate5,6. A general problem concerning determination of the zygosity of the father is non-paternity. However, in a conservative society such as Tunisia, non-paternity is unlikely. The newest technology used in the determination of the foetal RhD type in the case of heterozygous paternity involves free foetal DNA in the maternal circulation. Based on the concept that cell-free tumour DNA could be found in the peripheral circulation of patients with cancer, Lo et al.7 were the first to report the presence of the RHD gene in the plasma of women pregnant with a RhD-positive foetus. Detection of foetal RHD in maternal plasma is used as a non-invasive method for assessing the risk of HDFN, but a remaining pitfall that hampers its use is the limited reliability of negative results since, without an internal control, true negative results cannot be distinguished from false negative results due to insufficient amounts of free foetal DNA. A recent report described the use of single nucleotide polymorphisms as internal controls, but a large study using this strategy is lacking8. Moreover, non-invasive antenatal diagnostic testing to target anti-D prophylaxis was shown to be unlikely to produce important clinical benefits and its reliability in different ethnic minority populations needs to be demonstrated rigorously9.
Since RHD is a very polymorphic gene attention should be paid to the specificity and sensitivity of molecular tests. In Caucasians, the most common mechanism of RH:-1 phenotype is deletion of the RHD gene10 occurring as a result of unequal crossing-over between the two Rhesus boxes flanking the RHD gene. This leaves a single hybrid Rhesus box as a target for RHD zygosity testing and detection of this hybrid Rhesus box has been used to demonstrate the presence of the RHD deletion, making it possible to distinguish RHD dizygous individuals from RHD hemizygous ones11. Different approaches to detect the hybrid Rhesus box have been described, but these molecular methods may lead to false results in Africans because of their large genetic diversity12.
RHD deletion was shown to be the main background of the RH:-1 phenotype in the Tunisian population13 suggesting that paternal RHD zygosity in Tunisians could be determined using molecular methods that are accurate in Caucasians. To identify the most accurate and reliable method for RHD zygosity assignment in Tunisians, RHD zygosity was determined in 405 Tunisian donors. Three molecular methods, based on polymerase chain reaction - sequence-specific polymorphism (PCR-SSP) analysis, polymerase chain reaction - restriction fragment length polymorphism (PCR-RFLP) amplification and real-time, quantitative polymerase chain reaction (RQ-PCR) specific for the RHD exon 5, were compared.
Materials and methods
Blood sampling, serological typing and DNA extraction
A total of 405 random EDTA blood samples were collected from blood donors recruited with informed consent according to the approved protocol of the Regional Blood Transfusion Centre of Sousse (Tunisia) to determine RHD zygosity. RhD phenotyping was routinely done as described in our previous study6 and the indirect antiglobulin test was performed systematically for apparently RH:-1 results. Genomic DNA was isolated from buffy coats by a salting-out method according to a standard protocol14.
Polymerase chain reaction - sequence-specific polymorphism analysis for the hybrid Rhesus box
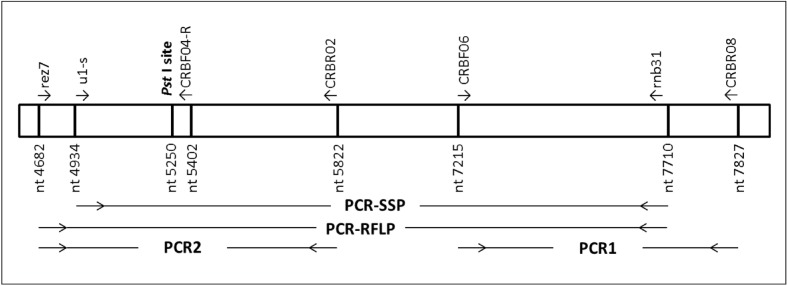
A 2778 bp product was amplified using forward primer u1-s15, which is specific for the hybrid and upstream Rhesus boxes, and reverse primer rnb31, which is specific for the hybrid and downstream Rhesus boxes (Figure 1). The PCR was performed as previously described6.
Figure 1.
Localisation of primers in the hybrid Rhesus box. Nucleotide 5250 represents the location of the PstI site used in the PCR-RFLP. Nucleotide positions are given corresponding to the standard downstream Rhesus box where position 1 represents the start of the homology between upstream and downstream Rhesus boxes and corresponds to nucleotide number 23 in GenBank accession number AJ252312.
Polymerase chain reaction - restriction fragment length polymorphism analysis for hybrid and downstream Rhesus boxes
The PCR-RFLP method was performed as described by Wagner and Flegel11. A 3,029 bp product was amplified using primers rez7 (universal primer for all Rhesus boxes) and rnb31 for amplification of the downstream and/or hybrid Rhesus boxes (Figure 1). The PCR amplicon was digested with 1.5 U/μL of Pst I (Fermentas, St. Leon-Rot, Germany) overnight at 37 °C and fragments were resolved using a 2% (w/v) agarose gel.
Real-time quantitative polymerase chain reaction analysis for RHD exon 5
Primers and probes for RHD exon 5 and CCR5 genes were from Finning16. The probes for RHD exon 5 and CCR5 were labelled with 6-FAM and Yakima yellow, respectively. Each primers was used at a final concentration of 300 nM and probes specific for RHD exon 5 and CCR5 were used at a final concentration of 200 nM and 100 nM, respectively. After 10 minutes at 95 °C, the reaction consisted of 40 cycles (95 °C for 10 sec and 60 °C for 30 sec). The MagNA Pure LC system (Roche Diagnostics, Indianapolis, IN, USA) was used to dispense master mix (TaqMan®, Applied Biosystems, Courtabœuf, France) and DNA (100 ng) in triplicate. Relative quantification (ΔCq) was done with CCR5 as the control gene and compared to results of control samples with one or two copies of the RHD gene17.
Genomic analysis of the RHD gene
To determine zygosity in discordant samples the ten exons of RHD were amplified by PCR and sequenced. Each exon amplification was performed using the primer sets described by Touinssi et al.18.
Sequencing analyses of the Rhesus boxes
With the aim of explaining discordant results, Rhesus boxes were amplified by PCR and sequenced using two PCR sets (Figure 1): PCR1 was applied with primers CRBF06 and CRBR0812 and PCR products were sequenced with the same couple of primers; PCR2 was done with Rez7 and CRBR02 primers12 and PCR products were sequenced with Rez7 and CRBF04-R (5′cctctgccagggcagtgca3′) primers.
Statistical analysis
The positive predictive value (PPV) was calculated for samples with no hybrid Rhesus Box detected by PCR-SSP and PCR-RFLP and for samples with two expressed alleles by RQ-PCR using the following formula:
The negative predictive value (NPV) was calculated for samples with one hybrid Rhesus Box detected by PCR-SSP and PCR-RFLP and for samples with one expressed alleles by RQ-PCR based on the same formula as that for the calculation of the PPV.
Results
RhD phenotype
RhD phenotyping of 405 donors randomly recruited from the centre of Tunisia revealed that 370 had the RH:1 phenotype and 35 had the RH:-1 phenotype. All RH:-1 phenotypes were confirmed through indirect antiglobulin testing.
Analysis of the RH:-1 samples
The RH:-1 samples (n=35) were used as controls for the PCR-SSP, PCR-RFLP and RQ-PCR analyses. PCR-SSP detected the hybrid Rhesus box in all RH:-1 samples. PCR-RFLP showed homozygous RHD gene deletion in 32 RH:-1 samples and three samples containing a single copy of hybrid Rhesus box. Sequencing of these samples showed a DIIIa-CE(4-7)-D, a weak D type 4.0 (omitted by serological techniques), and a new allele with a nucleotide change in RHD exon 7 at position 1026C/T (RHD [Ile342Ile]). According to the RQ-PCR analysis, RHD exon 5 was absent in 33 samples and present in weak D type 4.0 and RHD (Ile342Ile).
Analysis of the RH:1 samples
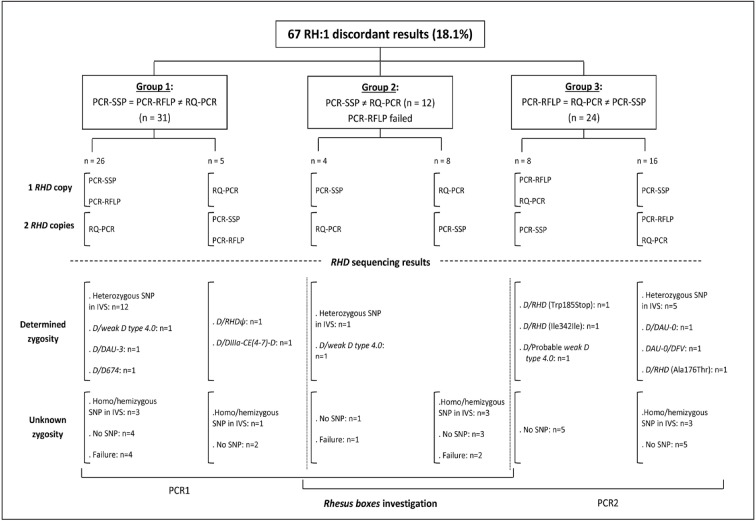
The comparison between the three methods for RHD zygosity assignment showed concordant results in 303 samples (81.9%). Discordant results (n=67, 18.1%) were classified into three groups (Figure 2). Group 1 (n=31) consisted of samples with results obtained by PCR-SSP similar to those obtained by PCR-RFLP but different from those obtained with RQ-PCR. Twenty-six samples had one copy of RHD gene by both PCR-SSP and PCR-RFLP and two copies by RQ-PCR, and five had two copies of RHD gene by both PCR-SSP and PCR-RFLP and one copy by RQ-PCR. Group 2 contained 12 samples with discordant PCR-SSP and RQ-PCR findings and without results in PCR-RFLP. We distinguished four samples in which the zygosity status differed in one copy of RHD gene by PCR-SSP and eight samples showing two copies. Group 3 consisted of 24 samples whose results obtained by PCR-RFLP were similar to those obtained by RQ-PCR but differed from those obtained by PCR-SSP.
Figure 2.
Classification and analysis of discordant results.
RHD sequencing results
RHD sequencing of the ten exons in 67 discordant samples are shown in Figure 2. All the RHD variants observed were heterozygous in trans to conventional RHD. We identified three new variants: (i) RHD(Trp185Stop): a G>A transition at position 555 leading to a stop codon; (ii) RHD(Ile342Ile): a 1026C>T transition in RHD exon 7, and (iii) RHD(Ala176Thr): a G>A transition at position 526 in RHD exon 4.
We also identified a probable weak D type 4.0 (nucleotide changes: 667T>G in RHD exon 5, 819G>A in RHD exon 6 because RHD exon 2 and 4 amplifications failed). We found homo/hemizygous intronic single nucleotide polymorphisms (n=10) and heterozygous intronic single nucleotide polymorphisms (n=18). We were unable to identify seven samples, probably because of the quality of the DNA. In conclusion, zygosity remained unknown for 37 subjects: homo/hemizygous intronic single nucleotide polymorphisms (n=10), no single nucleotide polymorphisms (n=20), and genotyping failure (n=7). Therefore, 189 subjects were dizygous and 144 were hemizygous.
Rhesus boxes polymorphisms for RH:1 discordant samples
Since RHD genotyping failed in seven samples, only the remaining 60 discordant samples were analysed for Rhesus box polymorphisms by PCR1 and/or PCR2 (Figure 2). No single nucleotide polymorphism was observed at the 5′ part of Rhesus boxes in three samples (5%, with RHD/RHD(Ala176Thr), heterozygous intronic single nucleotide polymorphism in IVS1 and in IVS1+IVS8), whereas Rhesus boxes were mutated in 57 samples (95%). Table I summarises the different nucleotide positions of mutated Rhesus boxes in RH:1 discordant samples. In addition to single nucleotide polymorphisms reported by Wagner et al.19, we identified 19 new polymorphisms in Rhesus boxes, listed and underlined in Table I.
Table I.
Summary of mutated sequences of Rhesus boxes in discordant samples. Discrepant samples were compared with standard downstream and upstream Rhesus boxes.
| SNP or allele | Nucleotide positions | |||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| u1-s | rnb31 | |||||||||||||||||||||||||||
| 4929 | 4952 | 4978 | 5139 | 5265 | 5157 | 5177 | 7302 | 7315 | 7317 | 7320 | 7328 | 7331 | 7332 | 7359 | 7381 | 7394 | 7424 | 7454 | 7429 | 7555 | 7658 | 7689 | 7691 | 7695 | 7706 | 7712 | ||
| Standard DRB | n= | G | A | A | T | A | G | C | A | A | G | T | A | A | G | A | G | G | C | C | A | G | del | A | C | A | A | C |
| Standard URB | A | C | A | C | A | G | C | A | A | G | T | A | A | G | C | C | C | C | C | A | G | AT | A | C | C | A | C | |
| RHD/DIIIa-CE(4-7)-D | 1 | nd | nd | nd | nd | nd | nd | nd | - | - | - | - | R | R | R | C | C | - | - | - | - | - | AT | R | M | C | M | M |
| RHD/weak D type 4.0 | 1 | - | - | - | C | - | - | - | - | - | - | - | - | R | - | - | C | - | - | - | - | - | - | - | - | C | - | - |
| 1 | nd | nd | nd | nd | nd | nd | nd | - | - | - | - | - | R | - | - | C | - | - | - | - | - | - | - | - | - | - | - | |
| RHD/probable weak D type 4.0 | 1 | - | - | - | - | W | - | - | nd | nd | nd | nd | nd | nd | nd | nd | Nd | nd | nd | nd | nd | nd | nd | nd | nd | nd | nd | nd |
| RHD/RHDΨ | 1 | nd | nd | nd | nd | nd | nd | nd | - | - | - | - | - | R | R | - | C | - | - | - | - | - | - | - | - | - | - | - |
| RHD/DAU-0 and DAU-0/DFV | 2 | A | - | - | C | - | - | Y | nd | nd | nd | nd | nd | nd | nd | nd | Nd | nd | nd | nd | nd | nd | nd | nd | nd | nd | nd | nd |
| RHD/DAU-3 | 1 | A | - | - | - | - | - | Y | - | - | - | - | - | R | R | - | - | - | - | - | - | - | - | - | - | - | - | - |
| RHD/D674 | 1 | nd | nd | nd | nd | nd | nd | nd | - | - | - | - | - | R | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| RHD/RHD (Trp185Stop) | 1 | - | - | - | C | - | - | - | nd | nd | nd | nd | nd | nd | nd | nd | nd | nd | nd | nd | nd | nd | nd | nd | nd | nd | nd | nd |
| RHD/RHD (Ile342Ile) | 1 | - | - | - | C | - | R | - | nd | nd | nd | nd | nd | nd | nd | nd | nd | nd | nd | nd | nd | nd | nd | nd | nd | nd | nd | nd |
| Hetero in IVS1 | 1 | - | - | - | C | W | - | - | nd | nd | nd | nd | nd | nd | nd | nd | nd | nd | nd | nd | nd | nd | nd | nd | nd | nd | nd | nd |
| Homo/hemi in IVS1 | 1 | nd | nd | nd | nd | nd | nd | nd | - | - | - | - | - | R | R | - | C | - | - | - | - | - | - | - | - | - | - | - |
| Hetero in IVS2 | 1 | nd | nd | nd | nd | nd | nd | nd | R | - | S | K | R | - | - | - | - | - | - | - | R | - | - | - | - | C | - | - |
| 1 | nd | nd | nd | nd | nd | nd | nd | - | - | - | - | - | R | R | C | C | C | M | Y | - | - | - | R | M | C | M | M | |
| 1 | nd | nd | nd | nd | nd | nd | nd | - | - | - | - | - | R | R | - | C | - | - | Y | - | - | - | R | M | C | M | M | |
| Homo/hemi in IVS2 | 1 | nd | nd | nd | nd | nd | nd | nd | - | - | - | - | - | R | R | - | C | - | - | - | - | - | - | R | M | C | M | M |
| 1 | - | - | - | - | W | - | - | - | - | - | - | - | R | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| Homo/hemi in IVS5 | 1 | - | - | - | C | - | R | - | nd | nd | nd | nd | nd | nd | nd | nd | nd | nd | nd | nd | nd | nd | nd | nd | nd | nd | nd | nd |
| Hetero in IVS8 | 1 | nd | nd | nd | nd | nd | nd | nd | - | - | - | K | - | R | - | - | - | - | - | - | - | - | - | - | - | C | - | - |
| 1 | - | - | - | - | - | R | - | - | - | - | - | - | R | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| Homo/hemi in IVS8 | 1 | - | - | - | - | - | - | - | - | - | - | - | - | R | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| 1 | A | - | - | - | - | - | - | - | - | - | - | - | R | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| Hetero in IVS1, Homo/hemi in IVS1, Hetero in IVS2 and Homo/hemi in IVS4 | 4 | nd | nd | nd | nd | nd | nd | nd | - | - | - | - | - | R | - | - | C | - | - | - | - | - | - | - | - | C | - | - |
| Homo/hemi in IVS1 and in IVS4 | 2 | - | - | - | C | - | - | - | nd | nd | nd | nd | nd | nd | nd | nd | nd | nd | nd | nd | nd | nd | nd | nd | nd | nd | nd | nd |
| Hetero in IVS0+IVS8 | 1 | nd | nd | nd | nd | nd | nd | nd | - | R | - | K | - | R | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| Hetero in IVS1+IVS8 | 1 | nd | nd | nd | nd | nd | nd | nd | - | - | - | - | - | R | - | - | - | - | - | - | - | - | - | - | - | C | - | - |
| Homo/hemi in IVS1+IVS8, Hetero in IVS2 and in IVS5+IVS8 | 3 | nd | nd | nd | nd | nd | nd | nd | - | - | - | - | - | R | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| Hetero inIVS2+IVS8 | 1 | - | A | - | - | - | - | Y | - | - | - | - | - | R | - | - | - | - | - | - | - | - | - | - | - | C | - | - |
| Hetero inIVS3+IVS8 and in IVS8 | 2 | A | - | - | - | - | - | - | nd | nd | nd | nd | nd | nd | nd | nd | nd | nd | nd | nd | nd | nd | nd | nd | nd | nd | nd | nd |
| No SNP | 1 | A | - | - | C | - | - | - | - | - | - | - | - | R | R | C | C | C | - | Y | - | - | - | R | M | C | M | M |
| 1 | nd | nd | nd | nd | nd | nd | nd | - | - | - | - | - | R | R | - | C | C | - | C | - | - | - | R | M | C | M | M | |
| 1 | - | - | W | C | - | - | - | nd | nd | nd | nd | nd | nd | nd | nd | nd | nd | nd | nd | nd | nd | nd | nd | nd | nd | nd | nd | |
| 1 | - | - | - | C | - | - | - | - | - | - | - | - | R | - | - | - | - | - | - | - | R | - | - | - | - | - | - | |
| 2 | - | - | - | C | - | - | - | - | - | - | - | - | R | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| 2 | nd | nd | nd | nd | nd | nd | nd | - | - | - | - | - | R | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| 3 | nd | nd | nd | nd | nd | nd | nd | - | - | - | - | - | R | - | - | C | - | - | - | - | - | - | - | - | - | - | - | |
| 9 | - | - | - | C | - | - | - | nd | nd | nd | nd | nd | nd | nd | nd | nd | nd | nd | nd | nd | nd | nd | nd | nd | nd | nd | nd | |
Positions of no described nucleotides changes of mutated Rhesus boxes are underlined; grey columns represent nucleotide positions with mutations under rnb31 and/or u1-s primers. Homo/hemi: homozygous or hemizygous; Hetero: heterozygous. DRB: downstream Rhesus box; URB: upstream Rhesus box; SNP; single nucleotide polymorphism; “del” indicates a two-nucleotide deletion; “−” indicates that no mutation was noted; “nd” not determined.
Observation of polymorphisms under rnb31 led us to assign a hemizygous status to one subject and a dizygous status to four subjects whose zygosity was previously unknown. Thus, zygosity remained unknown for 32 subjects because of homo/hemizygous intronic single nucleotide polymorphisms (n=7), genotyping failure (n=7) and no single nucleotide polymorphism in the RHD gene (n=18). Of the remaining 338 samples, 193 were dizygous and 145 hemizygous.
Positive and negative predictive values of the three methods
The PPV and NPV were calculated for each technique using the 338 samples with known zygosity (Table II). The PPV were higher than 99% for all methods, reaching 100% for RQ-PCR. The NPV were 83.24%, 87.27% and 98% for PCR-SSP, PCR-RFLP and RQ-PCR, respectively.
Table II.
Positive and negative predictive values and Youden index for the three molecular methods.
| HDFN risk | Two non-silent RHD alleles | One non-silent RHD allele | PPV | NPV | Youden index | |
|---|---|---|---|---|---|---|
|
| ||||||
| High (100%) | Moderate (50%) | |||||
| Need for amniocentesis | No | Yes | ||||
| PCR-SSP | No hybrid Rhesus box detected | 164 | 1 | 99.39 | 83.24 | 0.84 |
| One hybrid Rhesus box detected | 29 | 144 | ||||
|
| ||||||
| PCR-RFLP | No hybrid Rhesus box detected | 167 | 1 | 99.4 | 87.27 | 0.88 |
| One hybrid Rhesus box detected | 21 | 144 | ||||
|
| ||||||
| RQ-PCR | Two expressed alleles | 188 | 0 | 100 | 98 | 0.98 |
| One expressed alleles | 3 | 147 | ||||
Positive predictive value (PPV), negative predictive value (NPV) and Youden index were calculated using: http://www.aly-abbara.com/utilitaires/statistiques/sensibilite_specificite_vpp_vpn.html.
Discussion
Antibodies to the RhD antigen can be produced during pregnancy in a RhD-negative mother carrying a RhD-positive foetus, in particular following foetal-maternal haemorrhage at birth. While the first baby is usually not harmed, these antibodies may cause HDFN in subsequent RhD-positive babies. RhD incompatibility is a major cause of HDFN. Establishing paternal RHD zygosity to determine the risk of HDFN related to anti-D has the advantage of predicting the risk of HDFN in future pregnancies. Indeed, if the father is homozygous for the RHD gene (dizygous), the chance of inheriting the RHD gene is 100% for each pregnancy. Conversely, in the case of a hemizygous father (RHD/d) the risk is 50%. The aim of the present study was to assess the most convenient molecular method for determination of RHD zygosity in the Tunisian population as a method of first intention for the evaluation of HDFN risk.
Analysis of 370 random RhD-positive Tunisians using three molecular tests showed 81.9% concordant results and through investigation of RHD alleles and Rhesus box polymorphisms in the 67 discordant samples we concluded that our cohort consisted of 193 dizygous subjects, 145 hemizygous individuals and 32 subjects whose zygosity remains unknown because of lack of heterozygous single nucleotide polymorphisms or technical failure. Based on the 338 samples with known zygosity, the PPV, NPV and Youden index were calculated for each molecular test. The highest Youden index was observed for RQ-PCR (0.98) showing that this method was the most reliable and convenient for the Tunisian population. However, taking into account the high PPV (99.4%) and despite the lower NPV (87.27%) of PCR-RFLP, this method could be an alternative to the use of RQ-PCR for determining RHD zygosity in Tunisia. If the father is found to be hemizygous, second intention invasive methods would then be used to evaluate the risk of HDFN. Thus, in cases of falsely labelled hemizygous fathers, invasive investigations will be performed uselessly correcting the false zygosity assignment of the first intention test. Since some RHD variant alleles in the Tunisian population were shown to be related to some in Caucasians, we suggest that the false negative results using RQ-PCR (n=3) could be linked to presence in trans to a conventional RHD allele of a hybrid RHD-CE-D gene, such as DVI type 1 or 2 which are the most frequent partial alleles encountered in Caucasians20,21. In the heterozygous state such a hybrid gene cannot be distinguish by either haemagglutination tests or DNA analysis. False positive and false negative results using PCR-SSP and PCR-RFLP were the result of: (i) mutations in Rhesus boxes leading to lack of amplification of hybrid Rhesus boxes, and (ii) the presence of non-functional RHD alleles from African origin leading to an incorrect estimation of the risk of HDFN.
The present study also led to the identification of three novel RHD alleles: RHD(Trp185Stop), RHD(Ala176Thr) and RHD(Ile342Ile). Because of the premature stop, RHD(Trp185Stop) is predicted to be a silent allele which is associated with over-estimation of HDFN risk in the context of zygosity determination. RHD(Ala176Thr) is predicted to encode a weak phenotype since amino-acid 176 is located in the fifth transmembrane helix22. However, since these alleles were in trans to conventional RHD, no serological investigations were performed and we have no certainty about the RhD phenotype associated with these alleles. Allele encoding RHD(Ile342Ile) was found in two samples. Despite the silent nature of the polymorphism one of the donors bearing this allele typed as RhD-negative. Unfortunately, no blood sample was available for verification of serological results and no conclusion can be made based only on the molecular analysis. This study also demonstrated the complexity of Rhesus boxes with the description of 19 novel polymorphisms. The heterogeneity of Rhesus boxes highlights the limitations of Rhesus box approaches to the determination of RHD zygosity.
Altogether, the results of our analysis of 338 random samples with known zygosity demonstrated that RQ-PCR can accurately assign zygosity status of samples in Tunisians. In the case of determination of paternal zygosity, RQ-PCR enables prediction of the risk that a foetus will inherit RHD and will be useful in identifying HDFN cases which may require further genotyping.
Acknowledgements
We thank all blood donors at the Regional Blood Transfusion Centre in Sousse (Tunisia) who made this work for a doctoral thesis possible. We acknowledge the team from EFS-AM (the French Blood Institute), especially Sophie Beley and Thomas Granier for their technical help.
Footnotes
Authorship contribution
Narjes Kacem and Monique Silvy performed the research study and wrote the paper. Narjes Kacem, Monique Silvy and Pascal Bailly analysed the data. Monique Silvy, Pascal Bailly, Saloua Jemni-Yacoub and Jacques Chiaroni contributed essential reagents and tools.
The Authors declare no conflict of interest.
References
- 1.Moise KJ, Jr, Argoti PS. Management and prevention of red cell alloimmunization in pregnancy: a systematic review. Obstet Gynecol. 2012;120:1132–9. doi: 10.1097/aog.0b013e31826d7dc1. [DOI] [PubMed] [Google Scholar]
- 2.Moussa H, Tsochandaridis M, Jemni-Yacoub S, et al. Fetal RhD genotyping by real time quantitative PCR in maternal plasma of RhD-negative pregnant women from the Sahel of Tunisia. Ann Biol Clin. 2012;70:683–8. doi: 10.1684/abc.2012.0769. [DOI] [PubMed] [Google Scholar]
- 3.Moise KJ., Jr Management of rhesus alloimmunization in pregnancy. Obstet Gynecol. 2008;112:164–76. doi: 10.1097/AOG.0b013e31817d453c. [DOI] [PubMed] [Google Scholar]
- 4.Goebel JC, Soergel P, Pruggmayer M, et al. Prenatal diagnosis of the Rhesus D fetal blood type on amniotic fluid in daily practice. Arch Gynecol Obstet. 2008;277:155–60. doi: 10.1007/s00404-007-0437-y. [DOI] [PubMed] [Google Scholar]
- 5.Pirelli KJ, Pietz BC, Johnson ST, et al. Molecular determination of RHD zygosity: predicting risk of hemolytic disease of the fetus and newborn related to anti-D. Prenat Diagn. 2010;30:1207–12. doi: 10.1002/pd.2652. [DOI] [PubMed] [Google Scholar]
- 6.Kacem N, Chakroun T, Moussa H, et al. RHD zygosity assignments based on most probable genotype and hybrid Rhesus box detection in Tunisia. Transfus Med. 2012;22:362–6. doi: 10.1111/j.1365-3148.2012.01172.x. [DOI] [PubMed] [Google Scholar]
- 7.Lo YM, Corbetta N, Chamberlain PF, et al. Presence of fetal DNA in maternal plasma and serum. Lancet. 1997;350:485–7. doi: 10.1016/S0140-6736(97)02174-0. [DOI] [PubMed] [Google Scholar]
- 8.Doescher A, Petershofen EK, Wagner FF, et al. Evaluation of single-nucleotide polymorphisms as internal controls in prenatal diagnosis of fetal blood groups. Transfusion. 2013;53:353–62. doi: 10.1111/j.1537-2995.2012.03738.x. [DOI] [PubMed] [Google Scholar]
- 9.Szczepura A, Osipenko L, Freeman K. A new fetal RHD genotyping test: costs and benefits of mass testing to target antenatal anti-D prophylaxis in England and Wales. BMC Pregnancy Childbirth. 2011;11:5. doi: 10.1186/1471-2393-11-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Colin Y, Chérif-Zahar B, Le Van Kim C, et al. Genetic basis of the RhD-positive and RhD-negative blood group polymorphism as determined by Southern analysis. Blood. 1991;78:2747–52. [PubMed] [Google Scholar]
- 11.Wagner FF, Flegel WA. RHD gene deletion occurred in the Rhesus box. Blood. 2000;95:3662–8. [PubMed] [Google Scholar]
- 12.Grootkerk-Tax MG, Maaskant-van Wijk PA, van Drunen J, et al. The highly variable RH locus in nonwhite persons hampers RHD zygosity determination but yields more insight into RH-related evolutionary events. Transfusion. 2005;45:327–37. doi: 10.1111/j.1537-2995.2005.04199.x. [DOI] [PubMed] [Google Scholar]
- 13.Moussa H, Tsochandaridis M, Chakroun T, et al. Molecular background of D-negative phenotype in the Tunisian population. Transfus Med. 2012;22:192–8. doi: 10.1111/j.1365-3148.2012.01142.x. [DOI] [PubMed] [Google Scholar]
- 14.Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988;16:1215. doi: 10.1093/nar/16.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Perco P, Shao CP, Mayr WR, et al. Testing for the D zygosity with three different methods revealed altered Rhesus boxes and a new weak D type. Transfusion. 2003;43:335–9. doi: 10.1046/j.1537-2995.2003.00313.x. [DOI] [PubMed] [Google Scholar]
- 16.Finning KM, Martin PG, Soothill PW, et al. Prediction of fetal D status from maternal plasma: introduction of a new non-invasive fetal RHD genotyping service. Transfusion. 2002;42:1079–85. doi: 10.1046/j.1537-2995.2002.00165.x. [DOI] [PubMed] [Google Scholar]
- 17.Silvy M, Chapel-Fernandes S, Beley S, et al. Molecular characterization of a new D- - haplotype in a Comorian man. Vox Sang. 2012;103:352–5. doi: 10.1111/j.1423-0410.2012.01620.x. [DOI] [PubMed] [Google Scholar]
- 18.Touinssi M, Chapel-Fernandes S, Granier T, et al. Molecular analysis of inactive and active RHD alleles in native Congolese cohorts. Transfusion. 2009;49:1353–60. doi: 10.1111/j.1537-2995.2009.02161.x. [DOI] [PubMed] [Google Scholar]
- 19.Wagner FF, Moulds JM, Flegel WA. Genetic mechanisms of Rhesus box variation. Transfusion. 2005;45:338–44. doi: 10.1111/j.1537-2995.2005.04339.x. [DOI] [PubMed] [Google Scholar]
- 20.Muller TH, Wagner FF, Trockenbacher A, et al. PCR screening for common weak D types shows different distributions in three Central European populations. Transfusion. 2001;41:45–52. doi: 10.1046/j.1537-2995.2001.41010045.x. [DOI] [PubMed] [Google Scholar]
- 21.Roubinet F, Apoil PA, Blancher A. Frequency of partial D phenotypes in the south western region of France. Transfus Clin Biol. 1996;3:247–55. doi: 10.1016/s1246-7820(96)80004-8. [DOI] [PubMed] [Google Scholar]
- 22.Silvy M, Barrault A, Velliquette RW, et al. RHCE*cE734C allele encodes an altered c antigen and a suppressed E antigen not detected with standard reagents. Transfusion. 2013;53:955–61. doi: 10.1111/j.1537-2995.2012.03860.x. [DOI] [PubMed] [Google Scholar]