Abstract
Whole-exome sequencing studies have identified common mutations affecting genes encoding components of the RNA splicing machinery in hematological malignancies. Here, we sought to determine how mutations affecting the 3′ splice site recognition factor U2AF1 alter its normal role in RNA splicing. We find that U2AF1 mutations influence the similarity of splicing programs in leukemias, but do not give rise to widespread splicing failure. U2AF1 mutations cause differential splicing of hundreds of genes, affecting biological pathways such as DNA methylation (DNMT3B), X chromosome inactivation (H2AFY), the DNA damage response (ATR, FANCA), and apoptosis (CASP8). We show that U2AF1 mutations alter the preferred 3′ splice site motif in patients, in cell culture, and in vitro. Mutations affecting the first and second zinc fingers give rise to different alterations in splice site preference and largely distinct downstream splicing programs. These allele-specific effects are consistent with a computationally predicted model of U2AF1 in complex with RNA. Our findings suggest that U2AF1 mutations contribute to pathogenesis by causing quantitative changes in splicing that affect diverse cellular pathways, and give insight into the normal function of U2AF1’s zinc finger domains.
Myelodysplastic syndromes (MDS) represent a heterogeneous group of blood disorders characterized by dysplastic and ineffective hematopoiesis. Patients frequently suffer from cytopenias and are at increased risk for disease transformation to acute myeloid leukemia (AML) (Tefferi and Vardiman 2009). The only curative treatment is hematopoietic stem cell transplantation, for which most patients are ineligible due to advanced age at diagnosis. The development of new therapies has been slowed by our incomplete understanding of the molecular mechanisms underlying the disease.
Recent sequencing studies of MDS patient exomes identified common mutations affecting genes encoding components of the RNA splicing machinery, with ∼45%–85% of patients affected (Graubert et al. 2011; Papaemmanuil et al. 2011; Yoshida et al. 2011; Visconte et al. 2012). Spliceosomal genes are the most common targets of somatic point mutations in MDS, suggesting that dysregulated splicing may constitute a common theme linking the disparate disorders that comprise MDS. Just four genes—SF3B1, SRSF2, U2AF1, and ZRSR2—carry the bulk of the mutations, which are mutually exclusive and occur in heterozygous contexts (Yoshida et al. 2011). Targeted sequencing studies identified high-frequency mutations in these genes in other hematological malignancies as well, including chronic myelomonocytic leukemia and AML with myelodysplastic features (Yoshida et al. 2011). Of the four commonly mutated genes, SF3B1, U2AF1, and ZRSR2 encode proteins involved in 3′ splice site recognition (Shen et al. 2010; Cvitkovic and Jurica 2012), suggesting that altered 3′ splice site recognition is an important feature of the pathogenesis of MDS and related myeloid neoplasms.
U2AF1 (also known as U2AF35) may provide a useful model system to dissect the molecular consequences of MDS-associated spliceosomal gene mutations. U2AF1 mutations are highly specific—they uniformly affect the S34 and Q157 residues within the first and second CCCH zinc fingers of the protein—making comprehensive studies of all mutant alleles feasible (Fig. 1A). Furthermore, U2AF1’s biochemical role in binding the AG dinucleotide of the 3′ splice site is relatively well-defined (Merendino et al. 1999; Wu et al. 1999; Zorio and Blumenthal 1999). U2AF1 preferentially recognizes the core RNA sequence motif yAG|r (Fig. 1B), which matches the genomic consensus 3′ splice site and intron|exon boundary that crosslinks with U2AF1 (Wu et al. 1999). Nevertheless, our understanding of U2AF1:RNA interactions is incomplete. U2AF1’s U2AF homology motif (UHM) is known to mediate U2AF1:U2AF2 heterodimer formation (Kielkopf et al. 2001); however, both the specific protein domains that give rise to U2AF1’s RNA binding specificity and the normal function of U2AF1’s zinc fingers are unknown. Accordingly, the precise mechanistic consequences of U2AF1 mutations are difficult to predict.
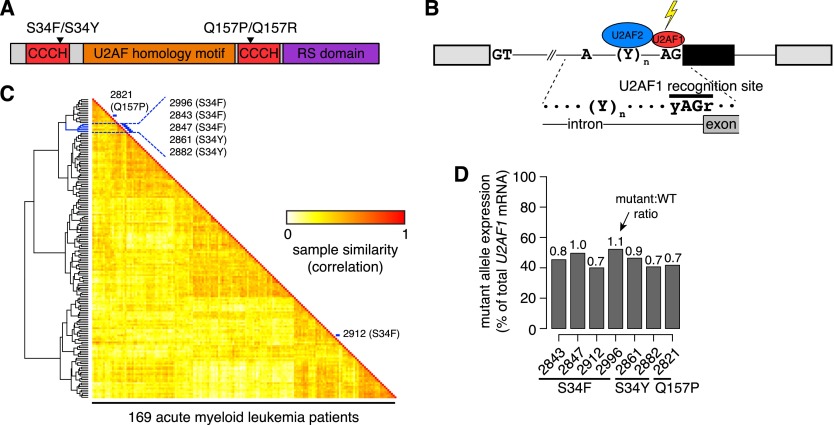
Figure 1.
U2AF1 mutations contribute to splicing programs in AML. (A) U2AF1 domain structure (Kielkopf et al. 2001; The UniProt Consortium 2012) and common mutations. (CCCH) CCCH zinc finger. (B) Schematic of U2AF1 interaction with the 3′ splice site of a cassette exon (black). (C) Heat map illustrating similarity of alternative splicing programs in AML transcriptomes. Dendrogram is from an unsupervised cluster analysis based on cassette exon inclusion levels. (Blue) Samples with U2AF1 mutations. (D) U2AF1 mutant allele expression as a percentage of total U2AF1 mRNA in AML transcriptomes. Numbers above bars indicate the ratio of mutant to WT allele expression.
Since the initial reports of common U2AF1 mutations in MDS, the molecular consequences of U2AF1 mutations have been controversial. An early study found that overexpression of mutant U2AF1 in HeLa cells resulted in dysfunctional splicing marked by frequent inclusion of premature termination codons and intron retention (Yoshida et al. 2011), while another early study reported increased exon skipping in a minigene assay following mutant U2AF1 expression in 293T cells, as well as increased cryptic splice site usage in the FMR1 gene in MDS samples (Graubert et al. 2011). U2AF1 mutations have been suggested to cause both alteration/gain of function (Graubert et al. 2011) and loss of function (Yoshida et al. 2011; Makishima et al. 2012). More recently, two studies analyzed acute myeloid leukemia transcriptomes from The Cancer Genome Atlas (TCGA) and found that exons with increased or decreased inclusion in samples with U2AF1 mutations exhibited different nucleotides prior to the AG of the 3′ splice site (Przychodzen et al. 2013; Brooks et al. 2014), suggesting that U2AF1 mutations may cause specific alterations to the RNA splicing process.
To determine how U2AF1 mutations alter RNA splicing in hematopoietic cells, we combined patient data, cell culture experiments, and biochemical studies. We found that U2AF1 mutations cause splicing alterations in biological pathways previously implicated in myeloid malignancies, including epigenetic regulation and the DNA damage response. U2AF1 mutations drive differential splicing by altering the preferred 3′ splice site motif in an allele-specific manner. Our results identify downstream targets of U2AF1 mutations that may contribute to pathogenesis, show that different U2AF1 mutations are not mechanistically equivalent, and give insight into the normal function of U2AF1’s zinc finger domains.
Results
U2AF1 mutations are associated with distinct splicing programs in AML
We first tested whether U2AF1 mutations were relevant to splicing programs in leukemias with an unbiased approach. We quantified genome-wide cassette exon splicing in the transcriptomes of 169 de novo adult acute myeloid leukemia (AML) samples that were sequenced as part of TCGA (The Cancer Genome Atlas Research Network 2013) and performed unsupervised cluster analysis. Five of the seven samples carrying a U2AF1 mutation clustered together (Fig. 1C). One of the samples that fell outside of this cluster had a Q157 rather than S34 U2AF1 mutation, and the other carried a mutation in the putative RNA splicing gene KHDRBS3 in addition to a U2AF1 mutation, potentially contributing to its placement in an outgroup. Both of the outgroup U2AF1 mutant samples additionally had low mutant allele expression relative to wild-type (WT) allele expression (Fig. 1D). These results suggest that U2AF1 mutations are associated with distinct splicing patterns in patients and are consistent with a recent report that spliceosomal mutations define a subgroup of myeloid malignancies based on gene expression and DNA methylation patterns (Taskesen et al. 2014).
U2AF1 mutations alter RNA splicing in blood cells
To determine how U2AF1 mutations affect RNA splicing in an experimentally tractable system, we generated K562 erythroleukemic cell lines that stably expressed transgenic FLAG-tagged U2AF1 protein (WT, S34F, S34Y, Q157P, or Q157R mutations) at modest levels in the presence of the endogenous protein (Fig. 2A). This expression strategy, in which the transgene was modestly overexpressed at levels of 1.8–4.7× endogenous U2AF1 (Fig. 2B), is consistent with the coexpression of WT and mutant alleles at approximately equal levels that we observed in AML transcriptomes. Similar coexpression of WT and mutant alleles has been previously reported in MDS patients carrying U2AF1 mutations (Graubert et al. 2011). We separately knocked down (KD) endogenous U2AF1 to ∼13% of normal U2AF1 protein levels in the absence of transgenic expression to test whether the mutations cause gain or loss of function (Fig. 2A).
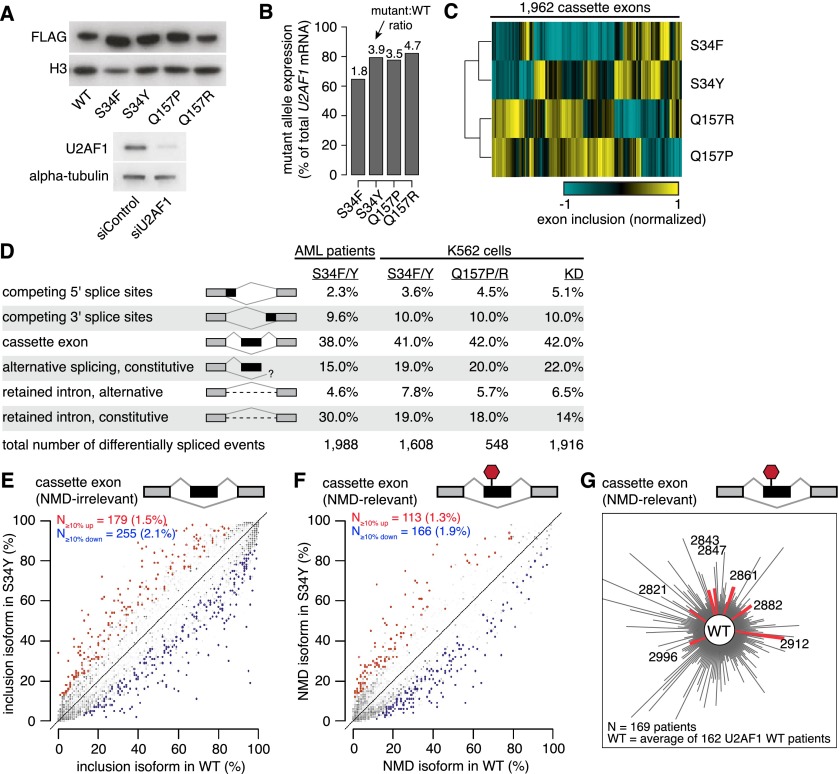
Figure 2.
U2AF1 mutations alter splicing, but do not cause splicing failure. (A) Western blots showing levels of FLAG-tagged U2AF1 in K562 cells stably expressing the indicated alleles (top) and levels of endogenous U2AF1 in K562 cells following transfection with a nontargeting siRNA or a siRNA pool against U2AF1 (bottom). (B) U2AF1 mutant allele expression as a percentage of total U2AF1 mRNA in K562 cells. (C) Heat map of K562 cells expressing mutant U2AF1. Dendrogram is from an unsupervised cluster analysis based on cassette exon inclusion levels. (D) U2AF1 mutation-dependent changes in splicing for AML S34 versus WT patients, K562 S34F or S34Y versus WT expression, K562 Q157P or Q157R versus WT expression, and K562 U2AF1 KD versus control KD. Percentages indicate the fraction of mutation-dependent splicing changes falling into each category of splicing event. (E) Levels of cassette exon inclusion in K562 cells expressing WT or S34Y U2AF1. (N) Numbers of alternatively spliced cassette exons with increased/decreased inclusion; (percentages) fraction of alternatively spliced cassette exons that are affected by S34Y expression. Events that do not change significantly are rendered transparent. Plot restricted to cassette exon events that are predicted to not induce nonsense-mediated decay (NMD). (F) Levels of NMD-inducing isoforms of cassette exon events in K562 cells expressing WT or S34Y U2AF1. (G) Levels of NMD-inducing isoforms of cassette exon events in AML transcriptomes. Distance from the center measures the splicing dissimilarity between each AML transcriptome and the average of all U2AF1 WT samples, defined as the sum of absolute differences in expression of NMD-inducing isoforms.
To identify mutation-dependent changes in splicing, we performed deep RNA-seq on these K562 cell lines stably expressing each mutant protein and on the control and U2AF1 KD cells (∼100M 2 × 49 bp reads per cell line). This provided sufficient read coverage to measure quantitative inclusion of ∼20,000 cassette exons that were alternatively spliced in K562 cells. Unsupervised cluster analysis of global cassette exon inclusion in these cell lines placed S34F/Y and Q157P/R as distinct groups and revealed that mutations within the first and second zinc fingers are associated with largely distinct patterns of exon inclusion (Fig. 2C). This is consistent with our cluster analysis of AML transcriptomes, in which the one sample with a Q157 mutation was placed as an outgroup to samples with S34 mutations.
We next assembled comprehensive maps of splicing changes driven by U2AF1 mutations in AML transcriptomes, K562 cells expressing mutant U2AF1, and K562 cells following U2AF1 KD. We tested ∼125,000 annotated alternative splicing events for differential splicing and assayed ∼160,000 constitutive splice junctions for evidence of novel alternative splicing or intron retention. We required a minimum change in isoform ratio of 10% to call an event differentially spliced. As our cluster analysis of K562 cells indicated that S34 and Q157 mutations generated distinct splicing patterns, we compared the six S34 AML samples to all U2AF1 WT AML samples. We separately identified splicing changes caused by both S34F and S34Y or both Q157P and Q157R in K562 cells relative to the WT control cells. The resulting catalogs of differentially spliced events revealed that all major classes of alternative splicing events, including cassette exons, competing splice sites, and retained introns, were affected by U2AF1 mutations (Fig. 2D; Supplemental Files S1–S5). Cassette exons constituted the majority of affected splicing events, followed by alternative splicing or intron retention of splice junctions annotated as constitutive in the UCSC Genome Browser (Meyer et al. 2013).
Thousands of splicing events were affected by each U2AF1 mutation, but the fraction of differentially spliced events was relatively low. For example, >400 frame-preserving cassette exons were differentially spliced in association with S34Y versus WT U2AF1 mutations; however, those >400 cassette exons constituted only ∼3.6% of frame-preserving cassette exons that are alternatively spliced in K562 cells (Fig. 2E). Expression of any mutant allele caused differential splicing of 2%–5% of frame-preserving cassette exons, with a bias toward exon skipping (Supplemental Fig. S1A). We did not observe increased levels of retained introns or isoforms that are predicted substrates for degradation by nonsense-mediated decay (NMD) in association with any U2AF1 mutation. Instead, constitutive intron removal appeared slightly more efficient in cells expressing mutant versus WT U2AF1 (Fig. 2F; Supplemental Fig. S1B,C). In contrast, we did observe increased levels of predicted NMD substrates and mRNAs with unspliced introns following U2AF1 KD (Supplemental Fig. S1B,C). Consistent with these findings in K562 cells, AML samples carrying U2AF1 mutations did not exhibit increased levels of NMD substrates or intron retention (Fig. 2G; Supplemental Figs. S2–S4). We conclude that S34 and Q157 U2AF1 mutations cause splicing changes affecting hundreds of exons, but do not give rise to widespread splicing failure.
These results contrast with a previous report that the U2AF1 S34F mutation causes overproduction of mRNAs slated for degradation and genome-wide intron retention (Yoshida et al. 2011). The discrepancy between those results and our observations are likely due to differing experimental designs. This previous study acutely expressed the S34F mutation at ∼50× WT levels in HeLa cells, whereas we stably expressed each mutant protein at 1.8–4.7× WT levels in blood cells (Fig. 1D). Maintaining a balance between WT and mutant protein expression—like that observed in AML and MDS patients—may be important to maintain efficient splicing.
U2AF1 mutations cause differential splicing of cancer-relevant genes
We next sought to identify downstream targets of U2AF1 mutations that might contribute to myeloid pathogenesis. We took a conservative approach of requiring differential splicing in AML transcriptomes as well as in K562 cells to help identify disease-relevant events that are likely direct consequences of U2AF1 mutations. We intersected differentially spliced events identified in three distinct comparisons: AML S34 versus WT samples, K562 S34 versus WT expression, and K562 Q157 versus WT expression. Of AML S34-associated differential splicing, 16.8% was phenocopied in K562 S34 cells versus 4.6% for K562 Q157 cells, consistent with allele-specific effects of U2AF1 mutations (Fig. 3A). The relatively low overlap of ∼17% between AML and K562 S34-associated differential splicing is likely due to differences in gene expression patterns between K562 and AML cells, the modest nature of splicing changes caused by U2AF1 mutations (such that many changes fall near the border of our statistical thresholds for differential splicing), and our stringent restriction to events that are differentially spliced in association with both S34F and S34Y mutations in K562 cells. This analysis revealed 54 splicing events that were affected by both S34 and Q157 mutations in AML transcriptomes and K562 cells. When we instead intersected genes containing differentially spliced events—not requiring that identical exons or splice sites be affected—we found a substantially increased intersection of 140 genes (Table 1).
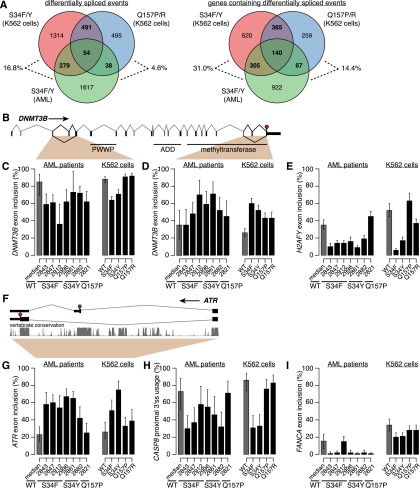
Figure 3.
U2AF1 mutations affect genes involved in disease-relevant cellular processes. (A) Overlap between mutation-dependent differential splicing in AML S34F/Y patients, K562 S34F/Y cells, and K562 Q157P/R cells. Overlap taken at the level of specific events (left) or genes containing differentially spliced events (right). (Percentages) The fraction of differentially spliced events (left) or genes containing differentially spliced events (right) in S34F/Y AML transcriptomes that are similarly differentially spliced in K562 cells expressing S34F/Y or Q157P/R U2AF1. (B) DNMT3B gene structure and protein domains (The UniProt Consortium 2012). Upstream 5′ UTR not shown. (PWWP) Pro-Trp-Trp-Pro domain; (ADD) ATRX-DNMT3-DNMT3L domain; (red stop sign) stop codon. (C,D) Inclusion of DNMT3B cassette exons. (Error bars) 95% confidence intervals as estimated from read coverage levels by MISO (Katz et al. 2010). (E) Inclusion of H2AFY cassette exon. (F) Cassette exon at 3′ end of ATR. Conservation is phastCons (Siepel et al. 2005) track from UCSC (Meyer et al. 2013). (G) Inclusion of cassette exon in ATR. (H) Usage of intron-proximal 3′ splice site of CASP8. (I) Inclusion of cassette exon in FANCA.
Table 1.
Genes that are differentially spliced in association with U2AF1 mutations
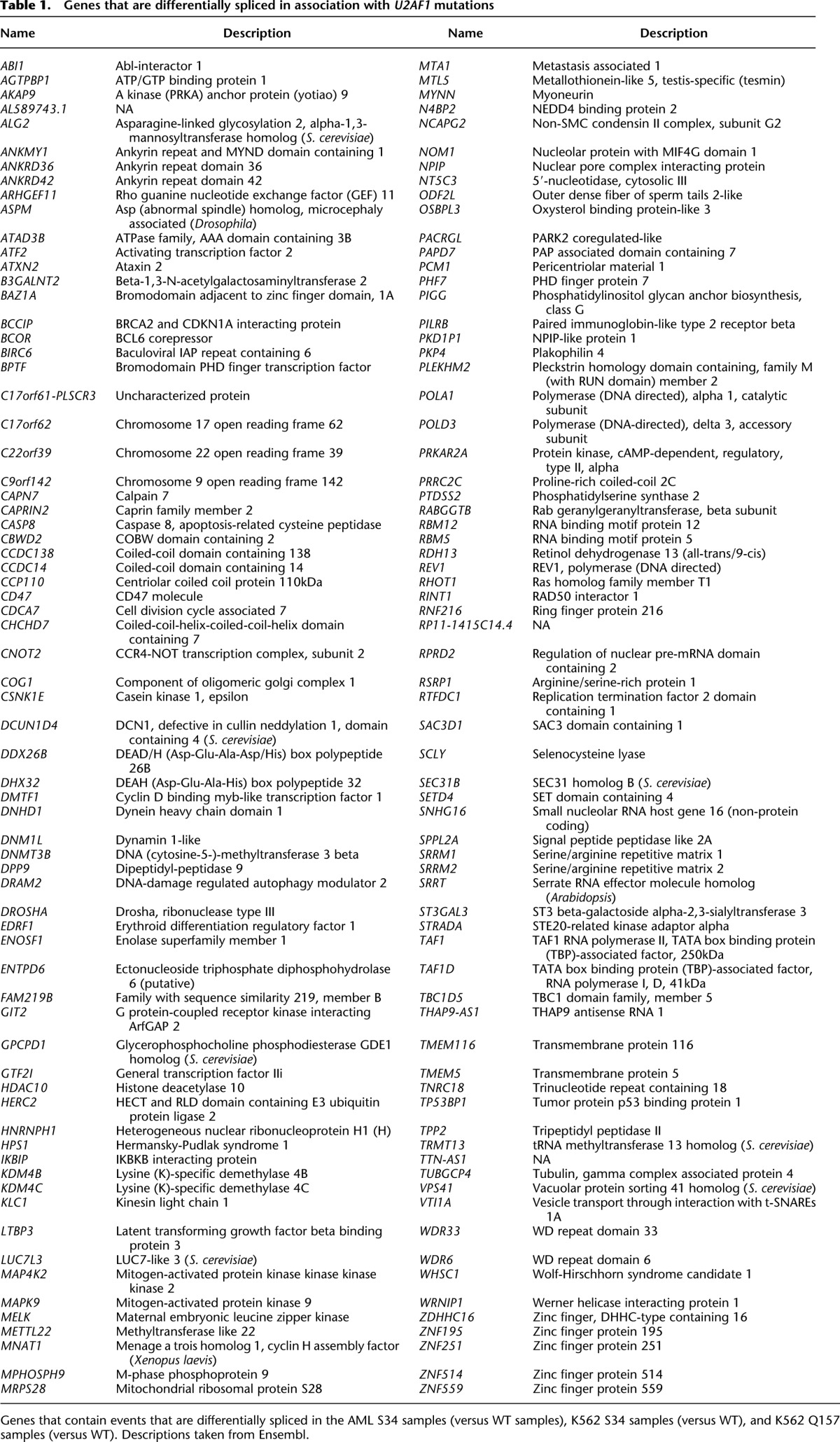
Many genes that were differentially spliced in association with U2AF1 mutations participate in biological pathways previously implicated in myeloid malignancies. For example, DNMT3A encodes a de novo DNA methyltransferase and is a common mutational target in myelodysplastic syndromes and acute myeloid leukemia (Ley et al. 2010; Walter et al. 2011). Multiple exons of its paralog DNMT3B, including an exon encoding part of the methyltransferase domain, are differentially spliced in AML patients carrying U2AF1 mutations as well as in K562 cells expressing U2AF1 mutant protein (Fig. 3B–D; Supplemental Fig. S5A,B). Similarly, different exons of ASXL1 are alternatively spliced in association with S34 mutations in AML transcriptomes and K562 cells, although the same exons are not consistently affected (Supplemental Files S1–S5). ASXL1 is a common mutational target in myelodysplastic syndromes and related disorders (Gelsi-Boyer et al. 2009), and U2AF1 and ASXL1 mutations co-occur more frequently than expected by chance (Thol et al. 2012). Other genes participating in epigenetic processes are differentially spliced as well, such as H2AFY (Fig. 3E; Supplemental Fig. S5C). H2AFY encodes the core histone macro-H2A.1, which is important for X chromosome inactivation (Hernández-Muñoz et al. 2005). As loss of X chromosome inactivation causes an MDS-like disease in mice (Yildirim et al. 2013), differential splicing of macro-H2A.1 could potentially be relevant to disease processes.
Isoform switches, wherein a previously minor isoform becomes the major isoform, were relatively rare but did occur. For example, a cassette exon at the 3′ end of the ATR gene, which encodes a PI3K-related kinase that activates the DNA damage checkpoint, is included at high rates in association with S34, but not Q157, mutations. This cassette exon alters the C terminus of the ATR protein, may render the mRNA susceptible to nonsense-mediated decay, and is highly conserved (Fig. 3F,G; Supplemental Fig. S5D). S34 mutations similarly cause an isoform switch from an intron-proximal to an intron-distal 3′ splice site of CASP8 that is predicted to shorten the N terminus of the protein (Fig. 3H; Supplemental Fig. S5E).
We noticed that splicing changes frequently affected multiple genes relevant to a specific biological process, such as DNA damage (ATR and FANCA) (Fig. 3G,I; Supplemental Fig. S5D,F). Consistent with this observation, Gene Ontology analysis indicated that genes involved in the cell cycle, chromatin modification, DNA methylation, DNA repair, and RNA processing pathways, among others, are enriched for differential splicing in both AML transcriptomes and K562 cells in association with U2AF1 mutations. This enrichment could be due to high basal rates of alternative splicing within these genes, which frequently are composed of many exons, or instead caused by specific targeting by mutant U2AF1. Upon correcting for gene-specific variation in the number of possible alternatively spliced isoforms, these pathways were no longer enriched in Gene Ontology analyses. We conclude that U2AF1 mutations preferentially affect specific biological pathways, but that this enrichment is due to frequent alternative splicing within such genes rather than specific targeting by U2AF1 mutant protein.
U2AF1 mutations cause allele-specific alterations in the 3′ splice site consensus
Previous biochemical studies showed that U2AF1 recognizes the core sequence motif yAG|r of the 3′ splice site (Merendino et al. 1999; Wu et al. 1999; Zorio and Blumenthal 1999). Accordingly, we hypothesized that the splicing changes caused by U2AF1 mutations might be due to preferential activation or repression of 3′ splice sites in a sequence-specific manner. To test this hypothesis, we identified consensus 3′ splice sites of cassette exons that were promoted or repressed in AML transcriptomes carrying U2AF1 mutations relative to WT patients. For each mutant U2AF1 sample, we enumerated all cassette exons that were differentially spliced between the sample and an average U2AF1 WT sample, requiring a minimum change in isoform ratio of 10%. Exons whose inclusion was increased or decreased in U2AF1 mutant samples exhibited different consensus nucleotides at the −3 and +1 positions flanking the AG of the 3′ splice site. As these positions correspond to the yAG|r motif bound by U2AF1, this data supports our hypothesis that U2AF1 mutations alter 3′ splice site recognition activity in a sequence-specific manner (Fig. 4A).
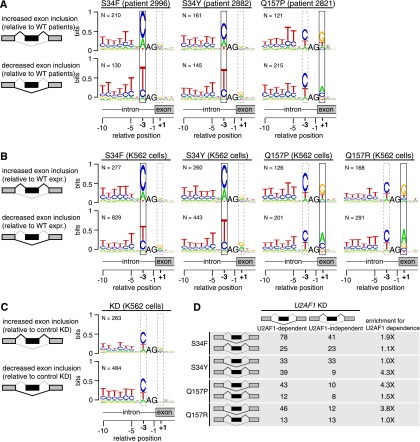
Figure 4.
U2AF1 mutations alter 3′ splice site consensus sequences. (A) Consensus 3′ splice sites of cassette exons with increased or decreased inclusion in U2AF1 mutant relative to WT AML transcriptomes. Boxes highlight sequence preferences at the −3 and +1 positions that differ from the normal 3′ splice site consensus. (Vertical axis) Information content in bits; (N) number of cassette exons with increased or decreased inclusion in each sample. Data for all U2AF1 mutant samples is shown in Supplemental Figure S6. (B) As in A, but for K562 cells expressing the indicated mutation versus WT. (C) As in A, but for K562 cells following U2AF1 KD or control KD. (D) Overlap between cassette exons that are promoted or repressed by mutant versus WT expression (rows) and U2AF1 versus control KD (columns) in K562 cells. The third column indicates the enrichment for U2AF1 dependence, defined as the overlap between exons affected by mutant U2AF1 expression and exons repressed versus promoted by U2AF1 KD.
Mutations affecting different residues of U2AF1 were associated with distinct alterations in the consensus 3′ splice site motif yAG|r of differentially spliced exons. The S34F and S34Y mutations, affecting the first zinc finger, were associated with nearly identical alterations at the −3 position in all six S34 mutant samples, whereas the Q157P mutation, affecting the second zinc finger, was associated with alterations at the +1 position (Supplemental Fig. S6). In contrast, cassette exons that were differentially spliced in randomly chosen U2AF1 WT samples relative to an average AML sample did not exhibit altered consensus sequences at the −3 or +1 positions (Supplemental Fig. S7). These results confirm the findings of two recent studies of this cohort of AML patients—which reported a frequent preference for C instead of T at the −3 position of differentially spliced cassette exons in U2AF1 mutant samples (Przychodzen et al. 2013; Brooks et al. 2014)—and extend their observations of altered splice site preference to show allele-specific effects of U2AF1 mutations, which have not been previously identified.
U2AF1 mutation-dependent sequence preferences (C/A >> T at the −3 position for S34F/Y and G >> A at the +1 position for Q157P) differ from the genomic consensus for cassette exons. C/T and G/A appear at similar frequencies at the −3 and +1 positions of 3′ splice sites of cassette exons (Supplemental Fig. S6,7), and minigene and genomic studies of competing 3′ splice sites indicate that C and T are approximately equally effective at the −3 position (Smith et al. 1993; Bradley et al. 2012). The consensus 3′ splice sites associated with promoted/repressed cassette exons in U2AF1 mutant transcriptomes also differ from U2AF1’s known RNA binding specificity. A previous study reported a core tAG|g motif in the majority of RNA sequences bound by U2AF1 in a SELEX experiment (Wu et al. 1999). Comparing that motif with preferences observed in U2AF1 mutant transcriptomes, we hypothesize that S34 U2AF1 promotes unusual recognition of C instead of T at the −3 position, while Q157 U2AF1 reinforces preferential recognition of G instead of A at the +1 position.
We next tested whether these alterations in 3′ splice site preference are a direct consequence of U2AF1 mutations. Comparing K562 cells expressing mutant versus WT U2AF1, we found that cassette exons that were promoted or repressed by each mutation exhibited sequence preferences at the −3 and +1 positions that were highly similar to those observed in AML patient samples (Fig. 4B). Mutations affecting identical residues (S34F/Y and Q157P/R) caused similar alterations in 3′ splice site preference, whereas mutations affecting different residues did not, confirming the allele-specific consequences of U2AF1 mutations. In contrast, cassette exons that were differentially spliced following KD of endogenous U2AF1 did not exhibit sequence-specific changes at the −3 or +1 positions of the 3′ splice site (Fig. 4C). We therefore conclude that S34 and Q157 mutations cause alteration or gain of function, consistent with the empirical absence of inactivating (nonsense or frameshift) U2AF1 mutations observed in patients.
U2AF1 mutations preferentially affect U2AF1-dependent 3′ splice sites
U2AF1 mutations are associated with altered 3′ splice site consensus sequences, yet only a relatively small fraction of cassette exons are affected by expression of U2AF1 mutant protein. Previous biochemical studies found that only a subset of exons have “AG-dependent” 3′ splice sites that require U2AF1 binding for proper splice site recognition (Reed 1989; Wu et al. 1999). We therefore speculated that exons that are sensitive to U2AF1 mutations might also rely upon U2AF1 recruitment for normal splicing. We empirically defined U2AF1-dependent exons as those with decreased inclusion following U2AF1 KD and computed the overlap between U2AF1-dependent exons and exons that were affected by U2AF1 mutant protein expression. For every mutation, we observed an enrichment for overlap with U2AF1-dependent exons, suggesting that U2AF1 mutations preferentially affect exons with AG-dependent splice sites (Fig. 4D).
U2AF1 mutations alter the preferred 3′ splice site motif yAG|r
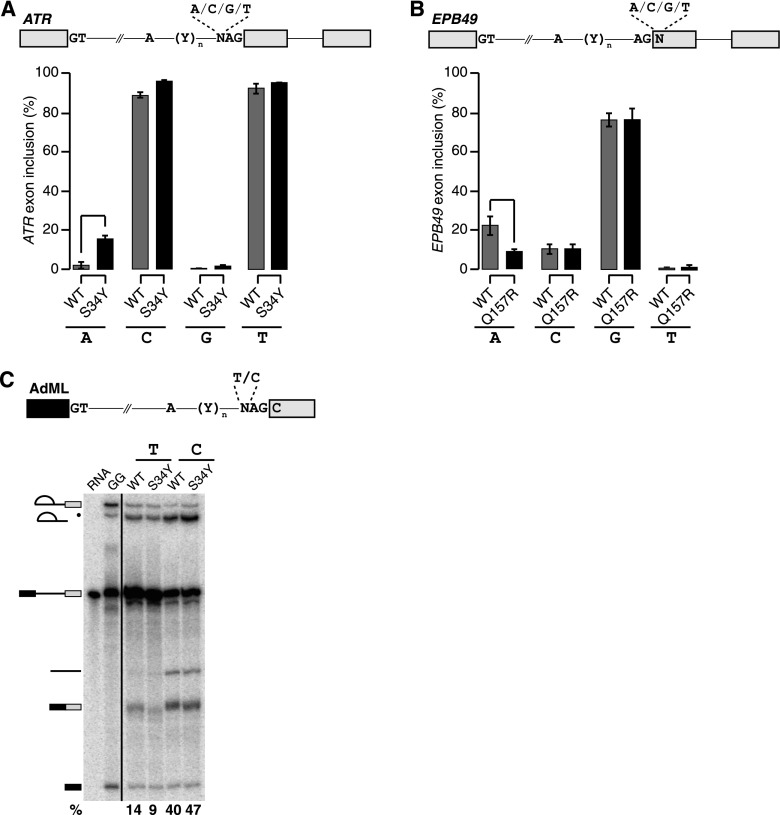
Our genomics data shows that cassette exons promoted/repressed by U2AF1 mutations have 3′ splice sites differing from the consensus. We therefore tested whether altering the core 3′ splice site motif of an exon influenced its recognition in the presence of WT versus mutant U2AF1. We created a minigene encoding a cassette exon of ATR, which responds robustly to S34 mutations in AML transcriptomes and K562 cells (Fig. 3F,G), by cloning the genomic locus containing the ATR cassette exon and flanking constitutive introns and exons into a plasmid. The minigene exhibited mutation-dependent splicing of the cassette exon, as expected, although cassette exon recognition was less efficient than from the endogenous locus. We then mutated the −3 position of the cassette exon’s 3′ splice site to A/C/G/T and measured cassette exon inclusion in WT and S34Y K562 cells. Robust mutation-dependent increases in splicing required the A at the −3 position found in the endogenous locus, consistent with the unusual preference for A observed in our analyses of AML and K562 transcriptomes. We additionally observed a small but reproducible increase for C (Fig. 5A). We next performed similar experiments for Q157-dependent splicing changes. We created a minigene encoding a cassette exon of EPB49 (encoding the erythrocyte membrane protein band 4.9), mutated the +1 position of the 3′ splice site to A/C/G/T, and measured cassette exon inclusion in WT and Q157R K562 cells. Cassette exon recognition was suppressed by Q157R expression when the +1 position was an A, consistent with our genomic prediction, and was not affected by Q157R when the +1 position was mutated to another nucleotide. Therefore, for both ATR and EPB49, robust S34 and Q157-dependent changes in splicing required the endogenous nucleotides at the −3 and +1 positions.
Figure 5.
U2AF1 mutations cause sequence-dependent changes in 3′ splice site recognition. (A) Schematic of ATR minigene (top) and inclusion of ATR cassette exon transcribed from minigenes with A/C/G/T at the −3 position of the 3′ splice site in K562 cells expressing WT or S34Y U2AF1 (bottom). (Error bars) Standard deviation from biological triplicates. (B) Schematic of EPB49 minigene (top) and inclusion of EPB49 cassette exon transcribed from minigenes with A/C/G/T at the +1 position of the 3′ splice site in K562 cells expressing WT or Q157R U2AF1 (bottom). (C) Schematic of AdML pre-mRNA substrate used for in vitro splicing (top) and in vitro splicing of AdML substrate incubated with nuclear extract from K562 cells expressing WT or S34Y U2AF1 (bottom). Percentages are the fraction of second step products (spliced mRNA and lariat intron) relative to all RNA species after 60 min of incubation. (RNA) Input radiolabeled RNA; (GG) pre-mRNA with the AG dinucleotide replaced by GG to illustrate the first step product of splicing; (black dot) exonucleolytic “chew back” product of the lariat intermediate.
We next tested how U2AF1 mutations influence constitutive, rather than alternative, splicing in an in vitro context. We used the adenovirus major late (AdML) substrate, a standard model of constitutive splicing, and mutated the −3 position of the 3′ splice site to C/T. We measured AdML splicing efficiency following in vitro transcription and incubation with nuclear extract of K562 cells expressing WT or S34Y U2AF1. The AdML substrate exhibited sequence-specific changes in splicing efficiency in association with U2AF1 mutations. Consistent with our genomic analyses, AdML with C/T at the −3 position was more/less efficiently spliced in S34Y versus WT cells (Fig. 5C). Taken together, our data demonstrate that U2AF1 mutations cause sequence-specific alterations in the preferred 3′ splice site motif in patients, in cell culture, and in vitro.
U2AF1 mutations may modify U2AF1:RNA interactions
Because U2AF1 mutations alter the preferred 3′ splice site motif yAG|r—the same motif that is recognized and bound by U2AF1 (Wu et al. 1999)—we next investigated whether U2AF1 mutations could potentially modify U2AF1’s RNA binding activity. U2AF1’s RNA binding specificity could originate from its U2AF homology motif (UHM) and/or its two CCCH zinc fingers. The UHM domain mediates U2AF heterodimer formation and is sufficient to promote splicing of an AG-dependent pre-mRNA substrate (Guth et al. 2001; Kielkopf et al. 2001). However, this domain binds a consensus 3′ splice site sequence with low affinity (Kielkopf et al. 2001), suggesting that it may be insufficient to generate U2AF1’s sequence specificity. Because U2AF1’s zinc fingers are independently required for U2AF RNA binding (Webb and Wise 2004), and our data indicate that zinc finger mutations alter splice site preferences, we hypothesized that U2AF1’s zinc fingers might directly interact with the 3′ splice site.
To evaluate whether this hypothesis is sterically possible, we started from the experimentally determined structure of the UHM domain in complex with a peptide from U2AF2 (Kielkopf et al. 2001), modeled the conformations of the zinc finger domains bound to RNA by aligning them to the CCCH zinc finger domains in the TIS11d:RNA complex structure (Hudson et al. 2004), and sampled the conformations of the two short linker regions using fragment assembly techniques (Leaver-Fay et al. 2011). The RNA was built in two segments taken from the TIS11d complex, one anchored in the N-terminal zinc finger and one in the C-terminal finger. We modeled multiple 3′ splice site sequences (primarily variants of uuAG|ruu) and explored a range of possible alignments of the 3′ splice site within the complex. The final register was selected on the basis of energetic analysis and manual inspection using known features of the specificity pattern of the 3′ splice site (in particular, the lack of a significant genomic consensus at the −4 and +3 positions, consistent with the experimental absence of a crosslink between U2AF1 and the −4 position) (Wu et al. 1999).
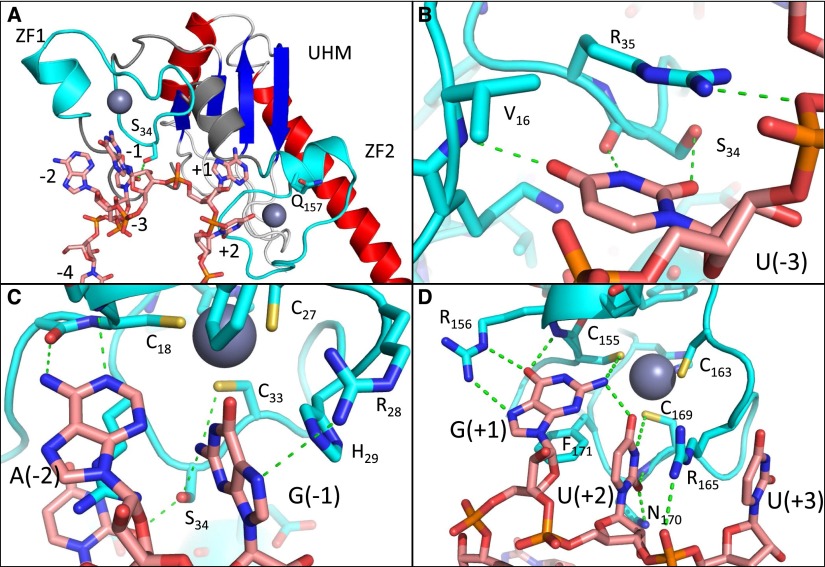
Based on these simulations, we propose a theoretical model of U2AF1 in complex with RNA wherein the zinc finger domains guide recognition of the yAG|r motif, consistent with the predictions of our mutational data. The model has the following features (Fig. 6A; Supplemental File S6). The first zinc finger contacts the bases immediately preceding the splice site, including the AG dinucleotide (Fig. 6B,C), while the second zinc finger binds immediately downstream (Fig. 6D). The RNA is kinked at the splice site and bent overall throughout the complex so that both the 5′ and 3′ ends of the motif are oriented toward the UHM domain and U2AF2 peptide. Contacts compatible with the 3′ splice site consensus are observed at the sequence-constrained RNA positions. The mutated positions S34 and Q157 are near the bases at which perturbed splice site preferences are observed for their respective mutations. Moreover, the modified preferences can, to some extent, be rationalized by contacts seen in our simulations. S34 forms a hydrogen bond with U(−1), and preference for U at −1 appears to decrease upon mutation; the Q157P mutation would improve electrostatic complementarity with G at +1 by removing a backbone NH group, in agreement with increased G preference in this mutant.
Figure 6.
Theoretical model of the U2AF1:RNA complex. (A) Overview, with the zinc finger domains colored cyan, the RNA in salmon, the UHM beta sheet in blue, and alpha helices in red. The frequently mutated positions S34 and Q157 are shown in stick representation. (ZF) Zinc finger. (B–D) Interactions with individual bases characteristic of the 3′ splice site consensus. Green dotted lines indicate hydrogen bonds and favorable electrostatic interactions; RNA and selected side chains are shown in stick representation.
Discussion
Here, we have described the mechanistic consequences of U2AF1 mutations in hematopoietic cells and provided a catalog of splicing changes driven by each common U2AF1 mutation. U2AF1 mutations cause highly specific alterations in 3′ splice site recognition in myeloid neoplasms. Taken together with the high frequency of mutations targeting U2AF1 and genes encoding other 3′ splice site recognition factors, our results support the hypothesis that specific alterations in 3′ splice site recognition are important contributors to the molecular pathology of MDS and related hematological disorders.
We observed consistent differential splicing of multiple genes, such as DNMT3B and FANCA, that participate in molecular pathways previously implicated in blood disease. It is tempting to speculate that differential splicing of a few such genes in well-characterized pathways explain how U2AF1 mutations drive disease. However, we instead hypothesize that spliceosomal mutations contribute to dysplastic hematopoiesis and tumorigenesis by dysregulating a multitude of genes involved in many aspects of cell physiology. This hypothesis is consistent with two notable features of our data. First, hundreds of exons are differentially spliced in response to U2AF1 mutations. Second, many of the splicing changes are relatively modest. In both the AML and K562 data, we observed relatively few isoform switches, with the ATR and CASP8 examples illustrated in Figure 3 being notable exceptions. Therefore, we expect that specific targets such as DNMT3B probably contribute to, but do not wholly explain, U2AF1 mutation-induced pathophysiology. As additional data from tumor transcriptome sequencing become available—for example, as more patient transcriptomes carrying Q157 mutations are sequenced—precisely identifying disease-relevant changes in splicing will become increasingly reliable.
Our understanding of the molecular consequences of U2AF1 mutations will also benefit from further experiments conducted during the differentiation process. Both the AML and K562 data arose from relatively “static” systems, in the sense that the bulk of the assayed cells were not actively undergoing lineage specification. U2AF1 mutations likely cause similar changes in splice site recognition in both precursor and more differentiated cells, but altered splice site recognition could have additional consequences in specific cell types. A recent study reported that regulated intron retention is important for granulopoiesis (Wong et al. 2013), consistent with the idea that as-yet-unrecognized shifts in RNA processing may occur during hematopoiesis. By disrupting such global processes, altered splice site recognition could contribute to the ineffective hematopoiesis that characterizes MDS.
Relevance to future studies of spliceosomal mutations
Both mechanistic and phenotypic studies of cancer-associated somatic mutations frequently focus on single mutations, even when multiple distinct mutations affecting that gene occur at high rates. Similarly, distinct mutations affecting the same gene are frequently grouped together in prognostic and other clinical studies, thereby implicitly assuming that different mutations have similar physiological consequences. Our finding that different U2AF1 mutations are not mechanistically equivalent illustrates the value of studying all high-frequency mutations when feasible. The distinctiveness of S34 and Q157 mutation-induced alterations in 3′ splice site preference suggests that they could theoretically constitute clinically relevant disease subtypes, potentially contributing to the heterogeneity of MDS. Mutations affecting other spliceosomal genes may likewise have allele-specific consequences. For example, mutations at codons 625 versus 700 of SF3B1 are most commonly associated with uveal melanoma (Harbour et al. 2013; Martin et al. 2013) versus MDS (Graubert et al. 2011; Papaemmanuil et al. 2011; Yoshida et al. 2011; Visconte et al. 2012) and chronic lymphocytic leukemia (Quesada et al. 2011). Accordingly, we speculate that stratifying patients by mutation could prove fruitful for future studies of spliceosomal gene mutations.
Our study additionally illustrates how investigating disease-associated somatic mutations can give insight into the normal function of proteins. With a fairly restricted set of assumptions, we computationally predicted a family of models in which the first zinc finger of U2AF1 recognizes the AG dinucleotide of the 3′ splice site. As a computational prediction, the model must be tested with future experiments. Nonetheless, given the concordance between our theoretical model of U2AF1:RNA interactions and our mutational data, this model may provide a useful framework for future studies of U2AF1 function in both healthy and diseased cells.
Methods
Vector construction and cell culture
Inserts encoding bicistronic constructs of the form U2AF1 + Gly Gly + FLAG + T2A + mCherry were created by standard methods (details in Supplemental Methods). These inserts were cloned into the self-inactivating lentiviral vector pRRLSIN.cPPT.PGK-GFP.WPRE (Addgene Plasmid 12252). The resulting plasmids coexpress U2AF1 and mCherry under control of the PGK promoter. K562 erythroleukemia cells were grown in RPMI-1640 supplemented with 10% FCS. To generate stable cell lines, K562 cells were infected with concentrated lentiviral supernatants at a MOI of ∼5 in growth media supplemented with 8 μg/mL protamine sulfate. Cells were then expanded, and transduced cells expressing mCherry were isolated by fluorescence activated cell sorting (FACS) using a Becton Dickinson FACSAria II equipped with a 561-nm laser. For RNAi studies, K562 cells were transfected with a control (nontargeting) siRNA (Dharmacon D-001810-03-20) or a siRNA pool against U2AF1 (Dharmacon ON-TARGETplus SMARTpool L-012325-01-0005) using the Nucleofector II device from Lonza with the Cell Line Nucleofector Kit V (program T16), and RNA and protein were collected 48 h after transfection.
mRNA sequencing
Total RNA was obtained by lysing 10 million K562 cells for each sample in TRIzol, and RNA was extracted using Qiagen RNeasy columns. Using 4 μg of total RNA, we prepared poly(A)-selected, unstranded libraries for Illumina sequencing using a modified version of the TruSeq protocol (details in Supplemental Methods). RNA-seq libraries were then sequenced on the Illumina HiSeq 2000 to a depth of ∼100 million 2 × 49 bp reads per sample.
Accession numbers
For the AML analysis, BAM files were downloaded from CGHub (“LAML” project) and converted to FASTQ files of unaligned reads for subsequent read mapping. For the HeLa cell analysis, FASTQ files were downloaded from DDBJ series DRA000503 (http://trace.ddbj.nig.ac.jp/DRASearch/), and the reads were trimmed to 50 bp (after removing the first five bp) to restrict to the high-quality portion of the reads. A similar trimming procedure was performed in the original manuscript (Yoshida et al. 2011).
Genome annotations and read mapping
MISO v2.0 annotations were used for cassette exon, competing 5′ and 3′ splice sites, and retained intron events (Katz et al. 2010). Constitutive junctions were defined as splice junctions that were not alternatively spliced in any isoform of the UCSC knownGene track (Meyer et al. 2013). For read mapping purposes, a gene annotation file was created by combining isoforms from the MISO v2.0 (Katz et al. 2010), UCSC knownGene (Meyer et al. 2013), and Ensembl 71 (Flicek et al. 2013) annotations for the UCSC hg19 (NCBI GRCh37) human genome assembly, and a splice junction annotation file was created by enumerating all possible combinations of annotated splice sites as previously described (Hubert et al. 2013). RSEM (Li and Dewey 2011) and Bowtie (Langmead et al. 2009) were used to map reads to the gene annotation file, and TopHat (Trapnell et al. 2009) was used to align remaining unaligned reads to the genome and splice junctions (full details in Supplemental Methods).
Isoform expression measurements
MISO (Katz et al. 2010) and v2.0 of its annotations were used to quantify isoform ratios for annotated alternative splicing events, and alternative splicing of constitutive junctions and retention of constitutive introns was quantified with junction reads as previously described (Hubert et al. 2013). All analyses were restricted to splicing events with at least 20 relevant reads (reads supporting either or both isoforms) that were alternatively spliced in our data. Events were defined as differentially spliced between two samples if they satisfied the following criteria: (1) at least 20 relevant reads in both samples; (2) a change in isoform ratio of at least 10%; and (3) a Bayes factor ≥ 2.5 (AML data) or 5 (K562 data). Because the AML data had approximately twofold lower read coverage than the K562 data, we reduced the Bayes factor by a factor of two to compensate for the loss in statistical power. Wagenmakers’ framework (Wagenmakers et al. 2010) was used to compute Bayes factors for differences in isoform ratios between samples. A description of the isoform-specific PCR used for Supplemental Figure S5 is given in Supplemental Methods. To identify splicing events that were differentially spliced in AML S34 samples versus WT samples (Supplemental File S1), we used the Mann-Whitney U test and required P < 0.01. To identify splicing events that were differentially spliced in each AML sample with a U2AF1 mutation (Fig. 4A; Supplemental Fig. S6), each U2AF1 mutant sample was compared to an average U2AF1 WT sample. The average U2AF1 WT sample was created by averaging isoform ratios over all 162 U2AF1 WT samples.
Cluster analysis, sequence logos, and gene ontology enrichment
To perform the cluster analysis of AML transcriptomes (Fig. 1C) and K562 cells (Fig. 2C), we identified cassette exons that displayed changes in isoform ratios ≥ 10% across the samples and then further restricted to cassette exons with at least 100 informative reads across all samples. An informative read is defined as a RNA-seq read that supports either isoform, but not both. We created a similarity matrix using the Pearson correlation computed from the z-score normalized cassette exon inclusion values and clustered the samples using Ward’s method. Sequence logos were created with v1.26.0 of the seqLogo package in Bioconductor (Gentleman et al. 2004). Gene ontology analysis was performed with GOseq (Young et al. 2010) and is described further in Supplemental Methods.
Western blotting
Protein lysates from K562 cells pellets were generated by resuspension in RIPA buffer and protease inhibitor along with sonication. Protein concentrations were determined using the Bradford protein assay. Ten micrograms of protein was then subjected to SDS-PAGE and subsequently transferred to nitrocellulose membranes. Membranes were blocked with 5% milk in Tris-buffered saline (TBS) for 1 h at room temperature and then incubated with primary antibody 1:1000 anti-U2AF1 (Bethyl Laboratories, catalog no. A302-080A), anti-FLAG (Thermo, catalog no. MA1-91878), anti-Histone H3 (Abcam, catalog no. ab1791), or anti-alpha-tubulin (Sigma, catalog no. T9026) for 1 h at room temperature. Blots were washed with TBS containing 0.005% Tween 20 and then incubated with the appropriate secondary antibody for 1 h at room temperature.
Minigenes
An insert containing the ATR genomic locus (Chr 3: 142168344–142172070) or EPB49 genomic locus (Chr 8: 21938036–21938724) was cloned into the EcoRV site of pUB6/V5-HisA vector (Invitrogen) by Gibson assembly cloning (NEB). Site-directed mutagenesis was used to generate different nucleotides at the −3 position of the 3′ splice site. Details of minigene transfection and real-time PCR are specified in Supplemental Methods.
In vitro splicing
A pre-mRNA substrate transcribed from the AdML derivative HMS388 was used in all splicing reactions (Jurica et al. 2002; Reichert et al. 2002). T7 runoff transcription was used to generate G(5′)ppp(5′)G-capped radiolabeled pre-mRNA using UTP [α-32P], and K562 nuclear extracts were isolated following a published protocol (Folco et al. 2012) with a minor modification. Pre-mRNA substrates were incubated in standard splicing conditions, and RNA species were separated in a 12% denaturing polyacrylamide gel and visualized using a phosphoimager (full details in Supplemental Methods). For quantification in Figure 5, each species was normalized by subtracting the background and then dividing by the number of uracil nucleotides in that species. The percentage of the second step products was calculated by dividing the second step species (spliced mRNA and lariat intron) by the total of all species in the lane.
Protein structure prediction
Models of U2AF1 (residues 9–174) in complex with a RNA fragment extending from the 3′ splice site positions −4 to +3 were built by combining template-based modeling, fragment assembly methods, and all-atom refinement. Models were built using the software package Rosetta (Leaver-Fay et al. 2011) with template coordinate data taken from the UHM:ULM complex structure (Kielkopf et al. 2001) (PDB ID 1jmt: residues A/46–143) and the TIS11d:RNA complex structure (Hudson et al. 2004) (PDB ID 1rgo: U2AF1 residues 16–37 mapped to A/195–216; residues 155–174 mapped to A/159–179; RNA positions −4 to −1 mapped to D/1–4; RNA positions +1 to +3 mapped to D/7–9). The remainder of the modeled region (residues 9–15, 38–45, and 144–154) was built using fragment assembly (with templated regions held internally fixed) in a low-resolution representation (backbone heavy atoms and side chain centroids) and force field. The fragment assembly simulation consisted of 6000 fragment-replacement trials, for which fragments of size 6 (trials 1–3000), 3 (trials 3001–5000), and 1 (trials 5001–6000) were used. The RNA was modeled in two pieces, one anchored in the N-terminal zinc finger and the other in the C-terminal zinc finger, with docking geometries taken from the TIS11d:RNA complex. A pseudo-energy term favoring chain closure was added to the potential function to reward closure of the chain break between the RNA fragments. The fragment assembly simulation was followed by all-atom refinement during which all side chains as well as the nontemplated protein backbone and the RNA were flexible. Roughly 100,000 independent model building simulations were conducted, each with a different random number seed and using a randomly selected member of the 1rgo NMR ensemble as a template. Low-energy final models were clustered to identify frequently sampled conformations (the model depicted in Fig. 5A was the center of the largest cluster). We explored a range of possible alignments of the splice site RNA within the complex, with the final model selected on the basis of all-atom energies, RNA chain closure, manual inspection, and known sequence features of the 3′ splice site motif.
Data access
The RNA-seq data from K562 cells have been submitted to the NCBI Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE58871.
Supplementary Material
Acknowledgments
We thank Beverly Torok-Storb for project assistance and advice, and Sue Biggins, Toshi Tsukiyama, and members of the Bradley laboratory for comments on the manuscript. This research was supported by the Hartwell Innovation Fund (R.K.B., A.R.), Damon Runyon Cancer Research Foundation DFS 04-12 (R.K.B.), Ellison Medical Foundation AG-NS-1030-13 (R.K.B.), NIH/NCI P30 CA015704 recruitment support (R.K.B.), Fred Hutchinson Cancer Research Center institutional funds (R.K.B.), NIH/NCI training grant T32 CA009657 (J.O.I.), NIH/NIDDK P30 DK056465 pilot study (J.O.I.), NIH/NHLBI U01 HL099993 (A.R.), NIH/NIDDK K08 DK082783 (A.R.), the J.P. McCarthy Foundation (A.R.), the Storb Foundation (A.R.), and NIH/NIGMS R01 GM088277 (P.B.).
Author contributions: J.O.I. designed the molecular genetics and biochemistry experiments. A.R. designed the cell culture and U2AF1 expression strategies. J.O.I., A.R., B.H., M.E.M., and A.S.Z. performed experimental work, including cloning, cell culture, and flow cytometry. R.K.B. and P.B. performed computational analyses and wrote the manuscript with contributions from other authors. R.K.B. and A.R. initiated the study.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.181016.114.
Freely available online through the Genome Research Open Access option.
References
- Bradley RK, Merkin J, Lambert NJ, Burge CB. 2012. Alternative splicing of RNA triplets is often regulated and accelerates proteome evolution. PLoS Biol 10: e1001229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks AN, Choi PS, de Waal L, Sharifnia T, Imielinski M, Saksena G, Pedamallu CS, Sivachenko A, Rosenberg M, Chmielecki J, et al. . 2014. A pan-cancer analysis of transcriptome changes associated with somatic mutations in U2AF1 reveals commonly altered splicing events. PLoS ONE 9: e87361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Cancer Genome Atlas Research Network 2013. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med 368: 2059–2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cvitkovic I, Jurica MS. 2012. Spliceosome Database: a tool for tracking components of the spliceosome. Nucleic Acids Res 41: D132–D141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flicek P, Ahmed I, Amode MR, Barrell D, Beal K, Brent S, Carvalho-Silva D, Clapham P, Coates G, Fairley S, et al. . 2013. Ensembl 2013. Nucleic Acids Res 41: D48–D55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folco EG, Lei H, Hsu JL, Reed R. 2012. Small-scale nuclear extracts for functional assays of gene-expression machineries. J Vis Exp 64: e4140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelsi-Boyer V, Trouplin V, Adélaïde J, Bonansea J, Cervera N, Carbuccia N, Lagarde A, Prébet T, Nezri M, Sainty D, et al. . 2009. Mutations of polycomb-associated gene ASXL1 in myelodysplastic syndromes and chronic myelomonocytic leukaemia. Br J Haematol 145: 788–800. [DOI] [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, et al. . 2004. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 5: R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graubert TA, Shen D, Ding L, Okeyo-Owuor T, Lunn CL, Shao J, Krysiak K, Harris CC, Koboldt DC, Larson DE, et al. . 2011. Recurrent mutations in the U2AF1 splicing factor in myelodysplastic syndromes. Nat Genet 44: 53–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guth S, Tange TØ, Kellenberger E, Valcárcel J. 2001. Dual function for U2AF35 in AG-dependent pre-mRNA splicing. Mol Cell Biol 21: 7673–7681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harbour JW, Roberson EDO, Anbunathan H, Onken MD, Worley LA, Bowcock AM. 2013. Recurrent mutations at codon 625 of the splicing factor SF3B1 in uveal melanoma. Nat Genet 45: 133–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernández-Muñoz I, Lund AH, van der Stoop P, Boutsma E, Muijrers I, Verhoeven E, Nusinow DA, Panning B, Marahrens Y, van Lohuizen M. 2005. Stable X chromosome inactivation involves the PRC1 Polycomb complex and requires histone MACROH2A1 and the CULLIN3/SPOP ubiquitin E3 ligase. Proc Natl Acad Sci 102: 7635–7640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubert CG, Bradley RK, Ding Y, Toledo CM, Herman J, Skutt-Kakaria K, Girard EJ, Davison J, Berndt J, Corrin P, et al. . 2013. Genome-wide RNAi screens in human brain tumor isolates reveal a novel viability requirement for PHF5A. Genes Dev 27: 1032–1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson BP, Martinez-Yamout MA, Dyson HJ, Wright PE. 2004. Recognition of the mRNA AU-rich element by the zinc finger domain of TIS11d. Nat Struct Mol Biol 11: 257–264. [DOI] [PubMed] [Google Scholar]
- Jurica MS, Licklider LJ, Gygi SR, Grigorieff N, Moore MJ. 2002. Purification and characterization of native spliceosomes suitable for three-dimensional structural analysis. RNA 8: 426–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz Y, Wang ET, Airoldi EM, Burge CB. 2010. Analysis and design of RNA sequencing experiments for identifying isoform regulation. Nat Methods 7: 1009–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kielkopf CL, Rodionova NA, Green MR, Burley SK. 2001. A novel peptide recognition mode revealed by the x-ray structure of a core U2AF35/U2AF65 heterodimer. Cell 106: 595–605. [DOI] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leaver-Fay A, Tyka M, Lewis SM, Lange OF, Thompson J, Jacak R, Kaufman K, Renfrew PD, Smith CA, Sheffler W, et al. . 2011. ROSETTA3: an object-oriented software suite for the simulation and design of macromolecules. Methods Enzymol 487: 545–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley TJ, Ding L, Walter MJ, McLellan MD, Lamprecht T, Larson DE, Kandoth C, Payton JE, Baty J, Welch J, et al. . 2010. DNMT3A mutations in acute myeloid leukemia. N Engl J Med 363: 2424–2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Dewey CN. 2011. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12: 323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makishima H, Visconte V, Sakaguchi H, Jankowska AM, Abu Kar S, Jerez A, Przychodzen B, Bupathi M, Guinta K, Afable MG, et al. . 2012. Mutations in the spliceosome machinery, a novel and ubiquitous pathway in leukemogenesis. Blood 119: 3203–3210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M, Maßhöfer L, Temming P, Rahmann S, Metz C, Bornfeld N, van de Nes J, Klein-Hitpass L, Hinnebusch AG, Horsthemke B, et al. . 2013. Exome sequencing identifies recurrent somatic mutations in EIF1AX and SF3B1 in uveal melanoma with disomy 3. Nat Genet 45: 933–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merendino L, Guth S, Bilbao D, Martínez C, Valcárcel J. 1999. Inhibition of msl-2 splicing by sex-lethal reveals interaction between U2AF35 and the 3′ splice site AG. Nature 402: 838–841. [DOI] [PubMed] [Google Scholar]
- Meyer LR, Zweig AS, Hinrichs AS, Karolchik D, Kuhn RM, Wong M, Sloan CA, Rosenbloom KR, Roe G, Rhead B, et al. . 2013. The UCSC Genome Browser database: extensions and updates 2013. Nucleic Acids Res 41: D64–D69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papaemmanuil E, Cazzola M, Boultwood J, Malcovati L, Vyas P, Bowen D, Pellagatti A, Wainscoat JS, Hellstrom-Lindberg E, Gambacorti-Passerini C, et al. . 2011. Somatic SF3B1 mutation in myelodysplasia with ring sideroblasts. N Engl J Med 365: 1384–1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Przychodzen B, Jerez A, Guinta K, Sekeres MA, Padgett R, Maciejewski JP, Makishima H. 2013. Patterns of missplicing due to somatic U2AF1 mutations in myeloid neoplasms. Blood 122: 999–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quesada V, Conde L, Villamor N, Ordóñez GR, Jares P, Bassaganyas L, Ramsay AJ, Beà S, Pinyol M, Martínez-Trillos A, et al. . 2011. Exome sequencing identifies recurrent mutations of the splicing factor SF3B1 gene in chronic lymphocytic leukemia. Nat Genet 44: 47–52. [DOI] [PubMed] [Google Scholar]
- Reed R. 1989. The organization of 3′ splice-site sequences in mammalian introns. Genes Dev 3: 2113–2123. [DOI] [PubMed] [Google Scholar]
- Reichert VL, Le Hir H, Jurica MS, Moore MJ. 2002. 5′ exon interactions within the human spliceosome establish a framework for exon junction complex structure and assembly. Genes Dev 16: 2778–2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H, Zheng X, Luecke S, Green MR. 2010. The U2AF35-related protein Urp contacts the 3′ splice site to promote U12-type intron splicing and the second step of U2-type intron splicing. Genes Dev 24: 2389–2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siepel A, Bejerano G, Pedersen JS, Hinrichs AS, Hou M, Rosenbloom K, Clawson H, Spieth J, Hillier LW, Richards S, et al. . 2005. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res 15: 1034–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CW, Chu TT, Nadal-Ginard B. 1993. Scanning and competition between AGs are involved in 3′ splice site selection in mammalian introns. Mol Cell Biol 13: 4939–4952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taskesen E, Havermans M, van Lom K, Sanders MA, van Norden Y, Bindels E, Hoogenboezem R, Reinders MJT, Figueroa ME, Valk PJM, et al. . 2014. Two splice factor mutant leukemia subgroups uncovered at the boundaries of MDS and AML using combined gene expression and DNA-methylation profiling. Blood 123: 3327–3335. [DOI] [PubMed] [Google Scholar]
- Tefferi A, Vardiman JW. 2009. Myelodysplastic syndromes. N Engl J Med 361: 1872–1885. [DOI] [PubMed] [Google Scholar]
- Thol F, Kade S, Schlarmann C, Löffeld P, Morgan M, Krauter J, Wlodarski MW, Kölking B, Wichmann M, Görlich K, et al. . 2012. Frequency and prognostic impact of mutations in SRSF2, U2AF1, and ZRSR2 in patients with myelodysplastic syndromes. Blood 119: 3578–3584. [DOI] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL. 2009. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25: 1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The UniProt Consortium 2012. Reorganizing the protein space at the Universal Protein Resource (UniProt). Nucleic Acids Res 40: D71–D75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visconte V, Makishima H, Jankowska A, Szpurka H, Traina F, Jerez A, O’Keefe C, Rogers HJ, Sekeres MA, Maciejewski JP, et al. . 2012. SF3B1, a splicing factor is frequently mutated in refractory anemia with ring sideroblasts. Leukemia 26: 542–545. [DOI] [PubMed] [Google Scholar]
- Wagenmakers EJ, Lodewyckx T, Kuriyal H, Grasman R. 2010. Bayesian hypothesis testing for psychologists: a tutorial on the Savage-Dickey method. Cognit Psychol 60: 158–189. [DOI] [PubMed] [Google Scholar]
- Walter MJ, Ding L, Shen D, Shao J, Grillot M, McLellan M, Fulton R, Schmidt H, Kalicki-Veizer J, O’Laughlin M, et al. . 2011. Recurrent DNMT3A mutations in patients with myelodysplastic syndromes. Leukemia 25: 1153–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb CJ, Wise JA. 2004. The splicing factor U2AF small subunit is functionally conserved between fission yeast and humans. Mol Cell Biol 24: 4229–4240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong JJL, Ritchie W, Ebner OA, Selbach M, Wong JWH, Huang Y, Gao D, Pinello N, Gonzalez M, Baidya K, et al. . 2013. Orchestrated intron retention regulates normal granulocyte differentiation. Cell 154: 583–595. [DOI] [PubMed] [Google Scholar]
- Wu S, Romfo CM, Nilsen TW, Green MR. 1999. Functional recognition of the 3′ splice site AG by the splicing factor U2AF35. Nature 402: 832–835. [DOI] [PubMed] [Google Scholar]
- Yildirim E, Kirby JE, Brown DE, Mercier FE, Sadreyev RI, Scadden DT, Lee JT. 2013. Xist RNA is a potent suppressor of hematologic cancer in mice. Cell 152: 727–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida K, Sanada M, Shiraishi Y, Nowak D, Nagata Y, Yamamoto R, Sato Y, Sato-Otsubo A, Kon A, Nagasaki M, et al. . 2011. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature 478: 64–69. [DOI] [PubMed] [Google Scholar]
- Young MD, Wakefield MJ, Smyth GK, Oshlack A. 2010. Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol 11: R14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zorio DA, Blumenthal T. 1999. Both subunits of U2AF recognize the 3′ splice site in Caenorhabditis elegans. Nature 402: 835–838. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.