Abstract
Formation of heterochromatin serves a critical role in organizing the genome and regulating gene expression. In most organisms, heterochromatin flanks centromeres and telomeres. To identify heterochromatic regions in the heavily studied model C. elegans, which possesses holocentric chromosomes with dispersed centromeres, we analyzed the genome-wide distribution of the heterochromatin protein 1 (HP1) ortholog HPL-2 and compared its distribution to other features commonly associated with heterochromatin. HPL-2 binding highly correlates with histone H3 mono- and dimethylated at lysine 9 (H3K9me1 and H3K9me2) and forms broad domains on autosomal arms. Although HPL-2, like other HP1 orthologs, binds H3K9me peptides in vitro, the distribution of HPL-2 in vivo appears relatively normal in mutant embryos that lack H3K9me, demonstrating that the chromosomal distribution of HPL-2 can be achieved in an H3K9me-independent manner. Consistent with HPL-2 serving roles independent of H3K9me, hpl-2 mutant worms display more severe defects than mutant worms lacking H3K9me. HPL-2 binding is enriched for repetitive sequences, and on chromosome arms is anticorrelated with centromeres. At the genic level, HPL-2 preferentially associates with well-expressed genes, and loss of HPL-2 results in up-regulation of some binding targets and down-regulation of others. Our work defines heterochromatin in an important model organism and uncovers both shared and distinctive properties of heterochromatin relative to other systems.
Eukaryotic genomes are packaged into two general types of chromatin: euchromatin and heterochromatin. This packaging is important for the regulation of gene expression and organization of the genome. Initially, heterochromatin was cytologically defined as the condensed, dark-staining chromatin that remains visible throughout the cell cycle (Heitz 1928). Since then, numerous molecular characteristics of heterochromatin have been identified. These include an enrichment of repetitive DNA elements, such as satellite DNA and sequences derived from transposable elements, and enrichment of histone H3 methylated at lysine 9 (H3K9me) (Grewal and Elgin 2002). Another hallmark of heterochromatin is the enrichment of heterochromatin protein 1 (HP1), a highly conserved, small nonhistone protein first identified in Drosophila (James and Elgin 1986). Heterochromatin is typically concentrated at pericentric and subtelomeric regions. How heterochromatin is distributed in an organism with numerous centromeres distributed along the length of each chromosome (i.e., holocentric) is not known. This paper defines the distribution of an HP1 protein and heterochromatin in the nematode C. elegans, which possesses holocentric chromosomes and is a valuable model for genome organization, chromosome segregation, gene expression, and development.
It has been shown through a variety of methods that the chromo domain (CD) of metazoan HP1 proteins specifically recognizes H3K9me2 and H3K9me3 (Bannister et al. 2001; Jacobs et al. 2001; Lachner et al. 2001). Evidence that this interaction is important for proper HP1 protein localization comes from the observation that loss or reduction of H3K9me results in loss or reduction of HP1 binding in vivo (Bannister et al. 2001; Lachner et al. 2001; Schotta et al. 2002; Seum et al. 2007; Tzeng et al. 2007). Other interactions besides the CD-H3K9me interaction are involved in HP1 localization as well, as Drosophila SU(VAR)205 (also known as HP1A) is able to associate with promoter regions of genes independently of H3K9me (Figueiredo et al. 2012), and HP1A lacking its CD is able to associate with heterochromatin (Smothers and Henikoff 2001). Furthermore, in vitro studies have shown that mouse CBX1, CBX3, and CBX5 (also known as HP1β, HP1γ, and HP1α, respectively) bind the histone-fold domain of histone H3 (Nielsen et al. 2001) and that fly HP1A binds DNA in a sequence-independent manner (Zhao et al. 2000). Interestingly, studies in fission yeast, flies, and mammals have demonstrated that the RNAi machinery and RNA itself contribute to HP1 protein localization (Pal-Bhadra et al. 2004; Verdel et al. 2004; Maison et al. 2011). Taken together, these studies implicate interactions between HP1 and methylated histone tails, histone cores, DNA, and RNA as contributing to the recruitment and retention of HP1 at particular DNA regions in vivo. In this study, we specifically tested whether H3K9me is required for proper HP1 localization in C. elegans.
The nematode C. elegans has two HP1 paralogous proteins: HP1 Like (heterochromatin protein) 1 and 2 (HPL-1 and HPL-2) (Couteau et al. 2002). HPL-2 serves more roles and/or more important roles than HPL-1, as hpl-2 mutants display diverse defects while hpl-1 mutants generally lack observable mutant phenotypes. HPL-2 is an important factor for germline health, as hpl-2 mutants display maternal-effect sterility at elevated temperature (25°C) (Coustham et al. 2006) and a reduced ability to silence exogenous “non-self” sequences in the germline (Couteau et al. 2002; Robert et al. 2005; Ashe et al. 2012; Shirayama et al. 2012). HPL-2 is also important in somatic development, as hpl-2 mutants show larval, somatic gonad, and vulval developmental defects (Schott et al. 2006). Comparisons of hpl-2 hpl-1 double mutants and hpl-2 single mutants suggest that HPL-2 and HPL-1 have some overlapping roles, as double mutant worms display more severe phenotypes than hpl-2 alone (Schott et al. 2006; Shirayama et al. 2012). Because HPL-2 is the more important of the two C. elegans HP1 homologs and is the only HP1 homolog in C. briggsae, a close relative of C. elegans (Vermaak and Malik 2009), we focused our current study on HPL-2.
Here, we show that HPL-2 binding to chromatin highly correlates with H3K9me1 and H3K9me2 throughout the genome and that HPL-2-enriched regions form domains that are also enriched for repetitive DNA elements. These observations suggest that HPL-2 indeed has functions associated with heterochromatin and that HPL-2-enriched domains represent the distribution of heterochromatin in C. elegans. Surprisingly, H3K9me is not necessary for the normal distribution of HPL-2, as the genome-wide pattern of HPL-2 is largely unchanged in met-2 set-25 mutant embryos, which Towbin et al. reported and we verified to lack H3K9me (Towbin et al. 2012). Consistent with HPL-2 having roles independent of H3K9me, met-2 set-25 mutants display less sterility at elevated temperature than hpl-2. Interestingly, worm heterochromatin has a unique distribution relative to other organisms: enrichment on the autosomal “arms” and on the leftmost region of the X chromosome. On autosomal arms, elevated HPL-2 levels flank centromeric chromatin, creating regions that resemble pericentric heterochromatin. HPL-2 shows a bias toward association with well-expressed genes, where it seems to repress the expression of some genes it binds and promote the expression of others. Our studies uncover both shared and unique properties of worm heterochromatin compared to other organisms, and reveal how heterochromatin is distributed in an organism with holocentric chromosomes.
Results
HPL-2 is concentrated along with H3K9 methylation on autosomal arms
To determine the distribution of the HP1 homolog HPL-2 in worms, we performed chromatin immunoprecipitation using a validated antibody (Supplemental Fig. S1) followed by microarray analysis (ChIP-chip) in early to mid-stage embryos. We observed large domains of HPL-2 enrichment on autosomal arms and the leftmost region of the X chromosome, and depletion of HPL-2 from the central regions of autosomes and most of the length of the X (Fig. 1A). Genomic coordinates of HPL-2-enriched arms and HPL-2-depleted centers are defined in Supplemental Figure S2. In agreement with low HPL-2 ChIP signal on the X, immunostaining of hermaphrodite germline nuclei for HPL-2 and H4K12ac, a histone modification enriched on autosomes in germ nuclei (Kelly et al. 2002), revealed that HPL-2 staining is lower on the X chromosomes than the autosomes (Fig. 1C).
Figure 1.
Features of heterochromatin are concentrated on autosomal arms and the leftmost region of the X chromosome in C. elegans. (A) Chromosomal heatmaps depicting median Z-scores of ChIP-chip signal over 2-kbp regions for HPL-2, H3K9me1, H3K9me2, H3K9me3, H3K27me3, and LEM-2, along with standardized repetitive element densities and gene densities over 10-kbp regions. Red indicates enrichment, blue indicates depletion. HPL-2-enriched arms are marked by black bars. Validations of anti-HPL-2 antibody and ChIP-chip of HPL-2 from hpl-2 mutant embryos are shown in Supplemental Figure S1. (B) Genome-wide Pearson correlation coefficients (PCCs) for median Z-scores of ChIP-chip signal between pairs of the chromatin marks and genomic features shown in panel A. PCCs for regions that lie either in HPL-2-enriched arms or in chromosome centers are shown in Supplemental Figure S3. (C) Immunofluorescence images showing DNA (blue), H4K12ac (green), and HPL-2 (red) staining of wild-type hermaphrodite germline nuclei. Arrows point to paired X chromosomes. Scale bar, 2 μm.
In addition to enrichment for HP1 proteins, other hallmarks of heterochromatin are enrichment of H3K9me and repetitive DNA elements, as well as sparser gene density relative to euchromatin (Richards and Elgin 2002). To determine if HPL-2-enriched arms also possess these characteristics, we compared our HPL-2 ChIP-chip data to these features (Fig. 1A). HPL-2 has a similar overall distribution to that of previously published H3K9me ChIP signal (Liu et al. 2011). Like H3K9me, HPL-2 is more heavily enriched on pairing center arms compared to nonpairing center arms (Gu and Fire 2010; Liu et al. 2011). Interestingly, the pattern of HPL-2 most closely resembles H3K9me1 and H3K9me2 and less closely resembles H3K9me3, with genome-wide Pearson correlation coefficients (PCCs) of 0.72, 0.78, and 0.39, respectively (Fig. 1B). Within HPL-2-enriched arms, those PCCs are 0.68, 0.74, and 0.15, respectively (Supplemental Fig. S3). The distribution of HPL-2 does not match the previously published pattern of H3K27me3 (Liu et al. 2011) (PCC = 0.01) (Fig. 1A,B). Based on comparing the distribution of HPL-2 to standardized repetitive DNA element densities, HPL-2-enriched arms are also enriched for repetitive elements, with the genome-wide pattern of HPL-2 modestly matching that of repetitive sequences (PCC = 0.45) (Fig. 1A,B). This observation is consistent with previous work showing chromosome arms to be enriched for repetitive elements (The C. elegans Sequencing Consortium 1998). To explore the relationship between HPL-2 binding and gene density, we compared the distribution of HPL-2 to the number of protein-coding gene base pairs per unit length along each chromosome. Interestingly, there is no obvious relationship between HPL-2 and standardized gene densities (PCC = 0.01) (Fig. 1A,B). As HPL-2-enriched arms are also enriched for H3K9me and repetitive DNA elements, we conclude that these regions represent heterochromatin in C. elegans. However, differing from what has been observed in other organisms, worm heterochromatin does not show a depletion of genic DNA relative to the rest of the genome.
HPL-2 can associate with chromatin in an H3K9me-independent manner
The genomic distribution of HPL-2 correlates well with H3K9me1 and H3K9me2, and other studies, as well as our own, have shown that HPL-2 can bind H3K9me peptides in vitro (Supplemental Fig. S4; Wirth et al. 2009; Koester-Eiserfunke and Fischle 2011; Studencka et al. 2012). To test if the localization of HPL-2 and its persistence on chromatin depend on an interaction with H3K9me, we performed HPL-2 ChIP-chip using extracts prepared from mutant embryos lacking the two H3K9 histone methyltransferases MET-2 and SET-25. Towbin et al. previously reported that met-2 set-25 double-mutant embryos lack detectable H3K9me1, me2, and me3 (Towbin et al. 2012). Because a recent paper identified C. elegans SET-26 as another H3K9 histone methyltransferase (Greer et al. 2014), we repeated measurement of H3K9me1, me2, and me3 levels in met-2 set-25 double-mutant embryos by both immunostaining and mass spectrometry (Supplemental Fig. S5A,B). In agreement with Towbin et al., H3K9me was below the limit of detection in met-2 set-25 mutant embryos.
Surprisingly, the distribution of HPL-2 in met-2 set-25 mutant embryos was similar to that in wild type, although dampened. To generate the most comparable profiles of HPL-2 in wild-type and met-2 set-25 samples, ChIP signal was normalized relative to regions outside of HPL-2-enriched domains and not bound by HPL-2; those regions would not be expected to differ between wild type and mutant. At the genomic level, the distribution of HPL-2 over the length of each chromosome in met-2 set-25 embryos was very similar to wild type (genome-wide PCC = 0.79) but dampened (Fig. 2A). Importantly, different normalization methods yielded similar results (Supplemental Fig. S6A,B), indicating that the HPL-2 ChIP-chip signal present in met-2 set-25 embryos is indeed substantial. Consistent with these findings, HPL-2 showed similar chromosomal staining in wild-type and met-2 set-25 mutant nuclei (Fig. 2B). We conclude that, despite the extensive colocalization of HPL-2 and H3K9me in wild-type cells and the ability of HPL-2 to bind H3K9me peptides (Supplemental Fig. S4; Wirth et al. 2009; Koester-Eiserfunke and Fischle 2011; Studencka et al. 2012), H3K9me is not essential for HPL-2 to associate with chromatin. However, as the levels of HPL-2 bound to chromatin appear to be reduced in met-2 set-25 mutants relative to wild type, H3K9me may have roles in promoting the recruitment or retention of HPL-2.
Figure 2.
HPL-2 associates with chromatin independently of H3K9me. (A) Normalized mean Z-scores for HPL-2 ChIP-chip signal over the length of each chromosome in wild-type (WT) embryos and met-2 set-25 double mutant embryos, which lack H3K9me (Supplemental Fig. S5). The genome-wide PCC of HPL-2 ChIP-chip signal between WT and met-2 set-25 is 0.79. Chromosome arms are marked with black bars (see Fig. 1). Supplemental Figure S6 shows differently normalized HPL-2 ChIP-chip signal. (B) Immunofluorescence images of DNA, H3K9me2, and HPL-2 in WT and met-2 set-25 hermaphrodite germline nuclei. WT and met-2 set-25 samples were stained in parallel, and images were acquired using identical settings. Scale bar, 2 μm. (C) Immunofluorescence images of DNA (blue), H3K9me2 (green), and HPL-2 (red) in two WT male (XO) germline nuclei. Arrows point to single unpaired X chromosomes. Scale bar, 2 μm. (D) Percentage of hermaphrodites that were fertile (gray) or sterile (black) at 25°C in wild type (WT), hpl-2 M+Z-, hpl-2 M-Z-, met-2 set-25 M+Z-, and met-2 set-25 M-Z-. (M) Maternal supply of gene product, (Z) zygotic synthesis of gene product.
The ChIP analysis described above shows that H3K9me is not necessary for HPL-2 binding. To complement that analysis and test if in wild type HPL-2 preferentially associates with chromatin enriched for H3K9me, we took advantage of male germline nuclei, in which the single unpaired X chromosome in each pachytene nucleus is dramatically enriched for H3K9me2 relative to the autosomes (Kelly et al. 2002). If H3K9me2 recruits HPL-2 to chromatin, we would expect to see an enrichment of HPL-2 on unpaired X chromosomes as well. We observed that HPL-2 is not concentrated on and in fact appears to be absent from unpaired X chromosomes, despite their heavy enrichment for H3K9me2 (Fig. 2C). This observation demonstrates that H3K9me2 is not sufficient to recruit HPL-2 to chromatin.
Previous experiments demonstrated that hpl-2 mutants display maternal-effect sterility at elevated temperature (25°C) (Coustham et al. 2006). Even though the distribution of HPL-2 in met-2 set-25 mutants lacking H3K9me appears similar to wild type, mutant worms may have impaired HPL-2 function in the absence of H3K9me. To test this possibility, we compared the fertility of met-2 set-25 M+Z- and M-Z- mutants at elevated temperature with the fertility of hpl-2 M+Z- and M-Z- mutants, where M represents a maternal supply and Z represents zygotic synthesis of the gene product (Fig. 2D). Similar to the absence of detectable H3K9me in met-2 set-25 embryos (Supplemental Fig. S5), immunostaining of met-2 set-25 adult hermaphrodite germlines did not show any detectable H3K9me1 (data not shown), H3K9me2 (Fig. 2B), or H3K9me3 (Ho et al. 2014). As expected, 0% of hpl-2 M+Z- mutants and 100% of hpl-2 M-Z- mutants raised at 25°C were sterile. Interestingly, 4% of met-2 set-25 M+Z- mutants and only 32% of met-2 set-25 M-Z- mutants raised at 25°C were sterile, indicating that met-2 set-25 mutants display only weak maternal-effect sterility. Observing that met-2 set-25 mutants show significantly less sterility in the M-Z- generation than hpl-2 mutants suggests that HPL-2 can promote fertility in the absence of H3K9me. Conversely, the published finding that met-2 hpl-2 double mutants have more severe defects than either single mutant (Andersen and Horvitz 2007) suggests that H3K9me has HPL-2-independent roles and that reduction of H3K9me and loss of HPL-2 lead to additive defects.
HPL-2 and centromeric chromatin are generally anticorrelated
Heterochromatin is typically concentrated in pericentric regions (Hennig 1999). In Drosophila, heterochromatin boundaries have been shown to be hotspots for ectopic centromere formation (Olszak et al. 2011), suggesting that heterochromatin helps to define centromeres. C. elegans chromosomes are holocentric, with centromeres distributed along their lengths (Albertson and Thomson 1982), raising the interesting question of how the distribution of HPL-2 relates to dispersed centromeric regions. To explore this, we compared the distributions of HPL-2 and the C. elegans CENP-A homolog HCP-3 (also known as CeCENP-A), a highly conserved centromeric histone H3 variant (Buchwitz et al. 1999), in HPL-2-enriched arms and HPL-2-depleted centers. Using published CeCENP-A ChIP-chip data obtained from similar embryonic stages (Gassmann et al. 2012), we observed that, in arms, HPL-2 and CeCENP-A are anti-correlated: High HPL-2 signal is generally present in regions with low CeCENP-A signal, and vice versa (Fig. 3A). By examining HPL-2 levels at the borders of CeCENP-A domains (Gassmann et al. 2012), we found that, in arms, CeCENP-A domains are flanked by elevated HPL-2 levels (Fig. 3B,C), creating regions that resemble pericentric heterochromatin found in other organisms. However, in chromosome centers, CeCENP-A domains are not flanked by elevated HPL-2 (Fig. 3B,C), suggesting that HPL-2 is not critical for centromere formation.
Figure 3.
HPL-2 binding and CeCENP-A incorporation generally do not co-occur. (A–C) Comparisons of HPL-2 and CeCENP-A ChIP-chip signal in chromosome arms (left) versus centers (right). (A) Mean Z-scores of HPL-2 and CeCENP-A over 2-kbp regions. (B) Mean Z-scores with 95% confidence intervals of HPL-2 (blue) and CeCENP-A (red) over regions surrounding and across CeCENP-A domains. CeCENP-A domains were scaled to their average size (10 kbp). (C) Mean Z-scores of HPL-2 (blue) and CeCENP-A (red) distributed over representative 1.2-Mbp regions.
HPL-2 peaks are enriched for repetitive sequences
HP1 proteins in fission yeast, flies, and mammals have been shown to associate with regions containing or flanked by repetitive DNA elements (Partridge et al. 2000; Guenatri et al. 2004; de Wit et al. 2005). Consistent with HPL-2 binding sites overlapping with repetitive elements, we observed HPL-2-enriched arms to also be enriched for repetitive elements (Fig. 1A). Interestingly, after comparing HPL-2 peaks called using MA2C (Song et al. 2007) to repetitive regions, we found that HPL-2 peaks residing in chromosome arms as well as centers are enriched for repetitive elements (genome in arms = 18% repeats, HPL-2-bound regions = 20% repeats; genome in centers = 6% repeats, HPL-2-bound regions = 15% repeats) (Fig. 4A). To determine if specific classes of repetitive elements are enriched for HPL-2 binding, we identified the 10 species of repetitive elements with the highest numbers of individual repeats overlapping HPL-2 peaks (Fig. 4B). Strikingly, the repetitive elements CeRep5 and PALTTAA2 have 93% (1241/1339) and 62% (771/1236) of their individual repeats throughout the genome bound by HPL-2, respectively. Notably, most of the top 10 species of HPL-2-bound repeats have higher fractions of individual repeats bound by HPL-2 in arms compared to centers (Fig. 4B). These observations suggest that specific types of repetitive elements may serve an important role in helping to define regions where HPL-2 binds. However, as many HPL-2 peaks do not overlap with any identified repetitive elements (data not shown), possible repeat-centric mechanisms of HPL-2 binding cannot account for all observed HPL-2 peaks. Interestingly, four of the top 10 species of HPL-2-bound repeats are transposons or possible transposons (PALTTAA2, PALTA5, HelitronY1A, and CELE1). Perhaps binding of HPL-2 participates in preventing their expression.
Figure 4.
HPL-2 in arms versus centers is distributed differently over genomic features. (A–C) Analysis of HPL-2 in chromosome arms (left) versus centers (right). (A) Percentage of the genome in six genomic feature categories (repeat, promoter, genic, exon, intron, and intergenic) and percentage of the genome in those six categories covered by HPL-2 peaks. (B) Classes of repetitive elements that have the highest number of repeats bound by HPL-2. (C) Mean Z-scores of HPL-2 ChIP-chip signal over individual genes.
HPL-2 peaks in chromosome arms cover different genomic features than HPL-2 peaks in chromosome centers
To identify genic features on which HPL-2 may be enriched or depleted relative to the rest of the genome, we determined the total number of base pairs of all HPL-2 peaks that overlap defined genomic categories: promoter, genic, exon, intron, and intergenic. Interestingly, we found that the prevalence of genomic features covered by HPL-2 peaks residing in arms differs from those residing in chromosome centers. For example, HPL-2 peaks in arms are enriched for intron regions (genome in arms = 39% introns, HPL-2-bound regions = 49% introns), while HPL-2 peaks in centers are not (genome in centers = 30% introns, HPL-2-bound regions = 19% introns) (Fig. 4A). Conversely, HPL-2 peaks in centers are enriched for promoter regions (genome in centers = 10% promoter, HPL-2-bound regions = 28% promoter), while HPL-2 peaks in arms are not (genome in arms = 9% promoter, HPL-2-bound regions = 9% promoter) (Fig. 4A). Figure 4C shows individual genes with striking intron enrichment or promoter enrichment of HPL-2. As HPL-2 peaks are enriched for genic and promoter regions, we hypothesize that HPL-2 may serve direct roles in regulating at least some of the genes to which it binds.
HPL-2 preferentially associates with well-expressed genes
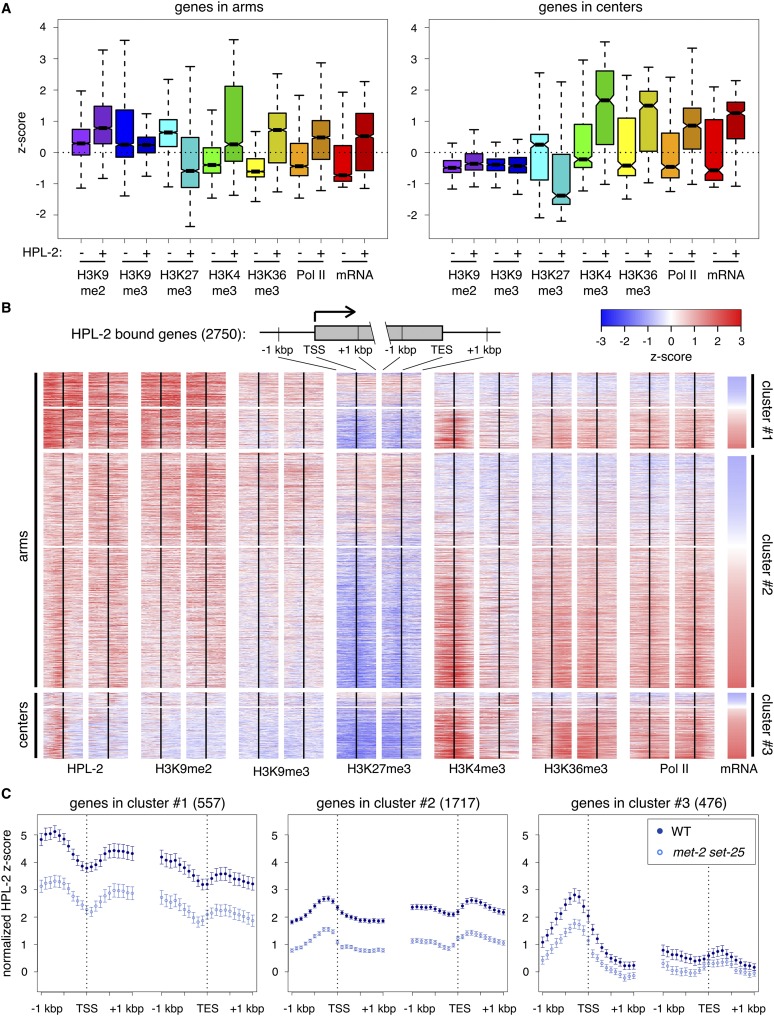
Although HP1 is best known for its role in generating repressed chromatin, HP1 proteins have been observed to associate with actively expressed genes. For example, in Drosophila, HP1A is enriched over well-expressed genes (de Wit et al. 2007), and HP1C is found in regions of active chromatin, where it promotes gene expression by interacting with the transcriptional machinery (Kwon et al. 2010). To determine whether HPL-2 preferentially associates with repressed or active genes, we compared the levels of histone modifications associated with repressed (H3K9me2, H3K9me3, H3K27me3) or active (H3K4me3, H3K36me3) transcriptional states, as well as RNA polymerase II (Pol II) and mRNA levels from previously published data (Rechtsteiner et al. 2010; Liu et al. 2011), on HPL-2-bound genes and genes not bound by HPL-2 (Fig. 5A). We found that HPL-2-bound genes, especially those located in arms, possess significantly higher levels of H3K9me2, but show no significant difference in H3K9me3 levels, compared to genes not bound by HPL-2. Interestingly, HPL-2-bound genes located either in arms or chromosome centers display significantly higher levels of H3K4me3, H3K36me3, Pol II, and mRNA, and significantly lower levels of H3K27me3 than genes not bound by HPL-2. These results show that, in worms, HPL-2 preferentially associates with well-expressed genes, consistent with HP1 associating with actively expressed genes in other organisms.
Figure 5.
HPL-2 preferentially associates with well-expressed genes. (A) Box plots depicting median Z-scores of H3K9me2 (purple), H3K9me3 (blue), H3K27me3 (cyan), H3K4me3 (green), H3K36me3 (yellow), and Pol II (orange) ChIP-chip signal, as well as Z-scores of mRNA levels (red) of genes bound by HPL-2 compared to an identical number of randomly selected genes not bound by HPL-2 that reside either in chromosome arms or in centers. Levels of H3K9me2, H3K9me3, H3K27me3, H3K36me3, and Pol II are over the entire lengths of genes, while levels of H3K4me3 are over a 1-kbp region centered over transcription start sites (TSSs). (B) Heatmap depicting mean Z-scores of HPL-2, H3K9me2, H3K9me3, H3K27me3, H3K4me3, H3K36me3, and Pol II ChIP-chip signal centered around TSSs and transcription end sites (TESs) of individual HPL-2-bound genes, along with standardized wild-type mRNA levels. Red indicates enrichment, blue indicates depletion. HPL-2-bound genes in arms were divided into two groups based upon the distributions of HPL-2 around TSSs, using the base R functions hclust (method = “complete”), dist (method = “maximum”), and cutree (k = 2) (R Core Team 2014). (C) Normalized mean Z-scores, with 95% confidence intervals, of HPL-2 ChIP-chip signal from WT (dark blue) and met-2 set-25 (light blue) centered around TSSs and TESs for HPL-2-bound genes in the three clusters defined in panel B.
HPL-2-bound genes show either promoter enrichment or a broad genic distribution of HPL-2
In Drosophila, HP1A is distributed in the promoter regions and over the bodies of genes it binds (Figueiredo et al. 2012). To investigate if HPL-2 has a similar genic distribution, we examined HPL-2 ChIP-chip signal surrounding the transcription start sites (TSSs) and transcription end sites (TESs) of all 2750 HPL-2-bound genes individually, along with various histone modifications (H3K9me2, H3K9me3, H3K27me3, H3K4me3, H3K36me3), Pol II, and mRNA levels from wild-type embryos (Fig. 5B; Rechtsteiner et al. 2010; Liu et al. 2011). HPL-2-bound genes were first divided into those in chromosome arms versus centers. Genes in arms were subsequently grouped based on the distribution of HPL-2 surrounding their TSSs. Bound genes show two general classes of genic HPL-2 distribution: broadly distributed (Fig. 5B, cluster #1) and enriched in promoter regions (Fig. 5B, clusters #2 and #3). Consistent with our previous results showing HPL-2 binding in the absence of H3K9me, the genic distributions of HPL-2 in met-2 set-25 mutants were similar to those in wild type but diminished in level (Fig. 5C). HPL-2 is more dramatically concentrated in the promoter region of genes in chromosome centers compared to genes in arms (Fig. 5B,C). Notably, despite the high correlation between HPL-2 and H3K9me2, many HPL-2-bound genes in chromosome centers display low levels of H3K9me2, consistent with HPL-2 targeting and/or retention at these promoters occurring in an H3K9me-independent manner. Interestingly, the genic distributions of HPL-2 are not correlated with expression level, as both promoter enriched HPL-2 and broadly distributed HPL-2 are found over genes with below- and above-average mRNA levels. In agreement with HPL-2 binding showing a bias toward well-expressed genes, the majority of HPL-2-bound genes have above-average expression levels, as well as above-average levels of H3K4me3, H3K36me3, and Pol II, and below-average levels of H3K27me3. Consistent with previous findings (Liu et al. 2011), H3K27me3 levels are strongly anticorrelated with expression levels, while H3K9me levels show less anticorrelation. Observing HPL-2 enriched in the promoter region of some genes versus distributed broadly over other genes raised the possibility that differing distributions of HPL-2 influence gene expression in different ways.
HPL-2 may directly regulate the expression of some genes to which it binds
To investigate if HPL-2 directly regulates transcription of at least some of the genes to which it binds, we performed transcript profiling of hpl-2 mutant embryos staged similarly to those used for our ChIP-chip analysis and compared transcript levels to previously published wild-type data that used the same NimbleGen microarrays and experimental conditions (Rechtsteiner et al. 2010). The hpl-2 mutant allele used is thought to be a null allele; it does not produce detectable transcript (Coustham et al. 2006). We identified 1297 genes as down-regulated and 942 genes as up-regulated in hpl-2 mutant embryos compared to wild type (Fig. 6A). If HPL-2 directly regulates expression of genes to which it binds, we would expect to see a significant overrepresentation of genes bound by HPL-2 among genes misregulated in hpl-2 mutants. Indeed, we observed 17% (222/1297) of down-regulated genes and 27% (255/942) of up-regulated genes in hpl-2 mutants to be bound by HPL-2, compared to 14% of either down- or up-regulated genes predicted by chance (Fig. 6A,B). These findings show that misregulated genes, especially those up-regulated in hpl-2 mutant embryos, are enriched for genes bound by HPL-2 and suggest that HPL-2 directly regulates at least some of the genes it binds. Our finding that the majority of misregulated genes in hpl-2 mutants are not bound by HPL-2 suggests that HPL-2 also influences gene expression indirectly, perhaps by modulating higher-order chromatin structure and/or organization within the nucleus, or by directly regulating the expression of factors that regulate other genes.
Figure 6.
HPL-2 likely directly regulates the expression of some genes it binds. (A) Volcano plot depicting log2(fold-change) versus −log10(adjusted P-value) for mRNA levels of 19,668 genes from microarray transcript profiling of hpl-2 mutant embryos compared to wild-type (WT) embryos (gray), with genes that are bound by HPL-2 (red). Significantly misregulated genes were defined as those having mRNA levels that showed a fold-change of at least ±1.5 and an adjusted P-value < 0.01. (B) Expected versus observed numbers of genes that are bound by HPL-2 and significantly down- or up-regulated in hpl-2 mutants. Displayed P-values were calculated using the χ2 test. (C) Table showing how HPL-2-bound genes within the three clusters defined in Figure 5B displaying below- or equal-to-or-above-average expression levels in wild type change in hpl-2 mutants (P < 0.01). See Supplemental Figure S7 for histograms of the data.
As HPL-2 is concentrated in the promoter regions of some genes it binds and is more broadly distributed over others, we examined whether the loss of HPL-2 has different effects on genes with different HPL-2 distributions. In each of the three clusters of HPL-2-bound genes defined in Figure 5B, we determined the numbers of genes up- and down-regulated in hpl-2 embryos that have below- and equal-to-or-above-average wild-type expression levels, and compared those numbers to the numbers expected by chance (Supplemental Fig. S7). The results, summarized in Figure 6C, suggest that HPL-2 does not regulate the transcription of low-expression genes. For high-expression genes in arms, HPL-2 participates in activating some and repressing others, regardless of the distribution of HPL-2 on those genes. For high-expression genes in chromosome centers, HPL-2 serves a predominantly repressive role.
Among genes that interact with the inner nuclear membrane, those bound by HPL-2 are expressed at significantly higher levels than those not bound by HPL-2
Based on ChIP-chip analysis of the inner nuclear membrane protein LEM-2, the autosomal arms and leftmost region of the X chromosome are associated with the nuclear periphery (Fig. 1A; Ikegami et al. 2010). Strikingly, despite the overlap of HPL-2-enriched and LEM-2-enriched domains and a modest correlation at the genomic level (PCC = 0.46) (Fig. 1B), the distributions of HPL-2 and LEM-2 in chromosome arms show little correlation (arm PCC = 0.03) (Supplemental Fig. S3). To explore the relationship between HPL-2 and LEM-2 in arms, we used previously published LEM-2 subdomains (Ikegami et al. 2010) and compared the numbers of genes bound by LEM-2 alone, HPL-2 alone, and both LEM-2 and HPL-2. Of the genes located in arms, the majority (93%; 8189/8794) is bound by LEM-2; 6049 are bound by LEM-2 alone, and 2140 are bound by both LEM-2 and HPL-2 (Fig. 7A). Only 134 genes are bound by HPL-2 alone (Fig. 7A). Thus, the overwhelming majority (94%) of arm genes bound by HPL-2 interact with the nuclear periphery.
Figure 7.
HPL-2-bound genes that interact with the inner nuclear membrane are well expressed. (A) Venn diagram depicting the overlap of LEM-2-bound and HPL-2-bound genes in chromosome arms. (B) Box plots depicting median Z-scores of H3K9me2 (purple), H3K9me3 (blue), H3K27me3 (cyan), H3K4me3 (green), H3K36me3 (yellow), and Pol II (orange) ChIP-chip signal, as well as Z-scores of mRNA levels (red) of genes in arms bound by LEM-2 alone, both LEM-2 and HPL-2, or HPL-2 alone. Heatmaps of individual genes are shown in Supplemental Figure S8.
Genes that interact with the inner nuclear membrane in flies and mammals are predominantly transcriptionally repressed (Guelen et al. 2008; Wen et al. 2009; Filion et al. 2010), and prior work in worms showed that LEM-2-associated genes are predominantly poorly expressed (Ikegami et al. 2010). Most HPL-2-bound genes are also bound by LEM-2 (Fig. 7A), and yet HPL-2 preferentially associates with well-expressed genes (Fig. 5A,B), raising a conundrum. To determine the expression levels of LEM-2-interacting genes that are also bound by HPL-2, we compared median levels of various histone modifications (H3K9me2, H3K9me3, H3K27me3, H3K4me3, and H3K36me3), along with Pol II and mRNA levels obtained from previously published data (Rechtsteiner et al. 2010; Liu et al. 2011), for arm genes bound by LEM-2 alone, both LEM-2 and HPL-2, or HPL-2 alone (Fig. 7B). Genes bound by LEM-2 alone show significantly lower expression levels compared to genes bound by both LEM-2 and HPL-2, which show lower expression levels relative to genes bound by HPL-2 alone. Consistent with these relative expression levels, genes bound by LEM-2 alone display significantly higher H3K27me3 and lower H3K4me3, H3K36me3, and Pol II relative to genes bound by both LEM-2 and HPL-2, which display higher H3K27me3 and lower H3K4me3, H3K36me3, and Pol II than genes bound by HPL-2 alone. Heatmaps showing histone modification and mRNA levels of individual genes bound by LEM-2 alone, LEM-2 and HPL-2, and HPL-2 alone are in Supplemental Figure S8. Taken together, our results show that there is widespread overlap between LEM-2-bound and HPL-2-bound genes, and that LEM-2-bound genes also bound by HPL-2 are expressed at significantly higher levels than those not bound by HPL-2. These observations suggest that HPL-2 may promote the expression of LEM-2-bound genes that would otherwise be repressed.
Discussion
Our studies reveal the genome-wide distribution of an HP1 ortholog (HPL-2) in C. elegans. HPL-2 is enriched in previously defined H3K9me domains on the autosomal arms and the leftmost region of the X chromosome (Liu et al. 2011). Surprisingly, we found that the distribution of HPL-2 can be achieved in an H3K9me-independent manner. The co-enrichment of three conserved features of heterochromatin—an HP1 protein, H3K9me, and repetitive elements—allows identification of autosomal arms and the left end of the X as C. elegans heterochromatin. While worm heterochromatin shares some features associated with heterochromatin in other systems, it displays some unique properties, most notably an apparent lack of involvement in defining centromeres and high rates of meiotic recombination, as discussed below.
Current models propose that HP1 proteins are initially recruited to chromatin independently of H3K9me, with subsequent spreading from initial binding sites in cis and retention on chromatin occurring in an H3K9me-dependent manner (Grewal and Moazed 2003; Dialynas et al. 2006; Maison et al. 2011; Figueiredo et al. 2012). We investigated the H3K9me dependence of HPL-2 by analyzing the genome-wide distribution of HPL-2 in met-2 set-25 double-mutant embryos in which H3K9me1, me2, and me3 were previously reported and which we verified are undetectable by mass spectrometry (Towbin et al. 2012). Our observation that HPL-2 persists on chromatin in mutant embryos lacking H3K9me demonstrates that HPL-2 can be recruited to and retained on chromatin in the absence of H3K9me. Strikingly, the distribution of HPL-2 in embryos lacking H3K9me closely resembles the pattern in wild-type embryos. These findings differ from those reported in other organisms, where ChIP-chip of HP1 proteins in mutants lacking H3K9me ranges from loss of HP1 on chromatin to an altered pattern of HP1, with selective persistence at promoter regions (Cam et al. 2005; Figueiredo et al. 2012). Our findings suggest that either the pattern of worm HPL-2 is dictated by recruitment and does not involve much spreading, or the pattern involves spreading that is H3K9me-independent. Candidate factors for recruiting HPL-2 to chromatin are the worm-specific zinc finger protein LIN-13 and the retinoblastoma-like pocket protein LIN-35 (Coustham et al. 2006; Kudron et al. 2013). Although the distribution of HPL-2 in worms lacking H3K9me resembles that in wild type, HPL-2 levels appear to be diminished in the H3K9me-lacking mutant, suggesting that recruitment and/or retention of HPL-2 is reduced in the absence of H3K9me. Our finding that hpl-2 mutants display fully penetrant (100%) maternal-effect sterility at elevated temperature, while H3K9me-lacking mutants display low penetrance (32%) maternal-effect sterility underscores the different requirements for H3K9 methylation and HPL-2 function in the germline. However, the fact that H3K9me-lacking mutants display any sterility may mean that HPL-2 function is compromised in the absence of H3K9me.
Most well-studied organisms are monocentric, with a single centromere on each chromosome, and in those systems heterochromatin is concentrated in pericentric regions (Grewal and Elgin 2002). Components of heterochromatin, such as HP1 proteins and repetitive DNA sequences, play important roles in centromere formation (Harrington et al. 1997; Ikeno et al. 1998; Folco et al. 2008). As discussed above, C. elegans heterochromatin has a unique distribution, with broad domains on autosomal arms and the leftmost region of the X chromosome, and is enriched for repetitive elements. Interestingly, CeCENP-A domains, which define potential centromeres, are evenly distributed throughout the genome with no apparent relationship to repetitive regions, suggesting that worm centromeres form in a repeat-independent manner (Gassmann et al. 2012; Subirana and Messeguer 2013). Within heterochromatic autosomal arms, HPL-2 enrichment flanks CeCENP-A domains, implying that there is a conserved relationship between HP1 proteins and centromeric regions in worms. However, outside of heterochromatic autosomal arms, CeCENP-A domains are not flanked by HPL-2 enrichment, suggesting that HPL-2 is not required to define worm centromeres. Indeed, both worm HP1 proteins (HPL-1 and HPL-2) are dispensable for de novo centromere formation (Yuen et al. 2011). In fact, new centromeres form more readily in the absence of worm HP1 proteins, suggesting that HP1 in worms has roles in restricting centromere formation (Yuen et al. 2011). Whether the distribution of CeCENP-A or of functional centromeres along C. elegans chromosomes changes in the absence of HP1 proteins remains to be determined.
Meiotic recombination is usually suppressed within heterochromatin, preventing genomic expansions and contractions due to erroneous recombination events between identical but differently positioned repetitive elements (Grewal and Jia 2007). Intriguingly, the heterochromatic autosomal arms in C. elegans display high recombination rates, and chromosome centers show low recombination rates (Barnes et al. 1995; Rockman and Kruglyak 2009). One possibility to account for this seemingly contradictory observation is that within worm heterochromatin, meiotic recombination may be suppressed within HPL-2-bound and/or repetitive regions, with hotspots of recombination occurring between these regions. However, recent high-resolution mapping of recombination frequencies over a segment of chromosome II that spans a center-to-arm transition revealed that meiotic recombination rates within these center and arm regions are relatively uniform and show little relation to the distribution of H3K9me2 (Kaur and Rockman 2014). It will be interesting to explore whether loss of HPL-2 influences recombination rates or leads to genomic instability due to aberrant recombination.
Here we define heterochromatin in the important model organism C. elegans. We uncover interesting relationships between worm heterochromatin and centromeric regions defined by CeCENP-A, the inner nuclear membrane defined by LEM-2, genes and their expression levels, and meiotic recombination rates. Strikingly, the chromosomal distribution of the worm HP1 ortholog HPL-2 is mostly unchanged in the absence of H3K9me. This work uncovers both unique and shared properties of worm heterochromatin relative to other organisms.
Methods
Worm strains and culture
C. elegans strains were maintained as previously described (Brenner 1974) unless otherwise noted. Strains used in this study include: N2 (Bristol) as wild type (WT); PFR40 hpl-2(tm1489) III polq-1(?); SS1183 hpl-2(tm1489) III; SS1184 hpl-2(tm1489) III/hT2G[bli-4(e937) let-?(q782) qIs48] (I:III); GW638 met-2(n4256) set-25(n5021) III; and SS1148 met-2(n4256) set-25(n5021) III/hT2G[bli-4(e937) let-?(q782) qIs48] (I:III). Recently, the hpl-2(tm1489) strain PFR40 was discovered to contain a linked deletion in polq-1 (J Ahringer, pers. comm.). We generated hpl-2 mutant strains lacking the polq-1 deletion (SS1183 and SS1184).
Anti-HPL-2 antibodies
Affinity-purified rabbit antibodies raised against residues 12-111 of HPL-2 were generated by Strategic Diagnostic Inc. Antibodies obtained from rabbit Q2324 were used in this study.
Preparation of embryo extracts and chromatin
Growth and harvest of wild-type (N2) and hpl-2 mutant (PFR40) early embryos were performed as previously described (Rechtsteiner et al. 2010). met-2 set-25 mixed-stage embryo growth and harvest were performed in an identical manner, except that worms were allowed to become fully gravid before harvesting embryos. Chromatin extracts were prepared as previously described for L3s (Liu et al. 2011).
Chromatin immunoprecipitation (ChIP) from embryos
ChIPs were performed as previously described (Rechtsteiner et al. 2010), with 0.5 mg of chromatin extract and 2.5 μg of anti-HPL-2 antibody (SDQ2324) used for each reaction. For HPL-2 ChIPs, three biological replicates were performed for wild-type embryos, two biological replicates for met-2 set-25 embryos, and one replicate for hpl-2 (PFR40) embryos.
ChIP DNA hybridizations to microarrays and data analysis
LM-PCR amplified ChIP DNA was prepared and hybridized to arrays as previously described (Rechtsteiner et al. 2010), with input samples labeled with Cy3 and ChIP samples with Cy5. WS170 (ce4) array probe coordinates were mapped to the WS220 (ce10) genome build using the default parameters of the UCSC Genome Browser liftOver tool (Kuhn et al. 2013). After removing probes that align to repetitive regions, log2(IP/Input) ratios were scaled to obtain Z-scores, and replicate Z-scores were averaged. To compare HPL-2 ChIP-chip signal between wild type and met-2 set-25, normalization was performed as previously described (Rechtsteiner et al. 2010), except using regions outside of HPL-2-enriched arms after removal of HPL-2 peaks (called from both wild type and met-2 set-25).
Peak calling and genomic feature analysis
HPL-2 ChIP-chip peaks were called using MA2C (Song et al. 2007), using an FDR of 1%. All genomic coordinates were obtained from genome build WS220 (ce10) (http://www.wormbase.org/); for repetitive elements, nonrepetitive low complexity regions were removed from the UCSC Genome Browser RepeatMasker track (Smit et al. 1996–2010; Jurka 2000; Karolchik et al. 2014). To determine proportions of genomic features bound by HPL-2 (promoters, genes, introns, exons, intergenic regions, repeats), the number of base pairs from each category that overlapped with HPL-2 peaks was determined and compared to the total number of base pairs in each category. Only protein-coding genes were considered; promoters were defined as regions within 500 bp upstream of TSSs. To account for splicing variants, introns and exons for all isoforms were merged; consequently, exonic and intronic regions overlap at several regions. Genes were defined to be bound by HPL-2 if a HPL-2 peak overlapped at least 50 bp of its gene body or promoter region.
Transcript profiling from hpl-2 mutant embryos and comparison to wild type
Early embryo RNA from hpl-2 mutant (PFR40) was collected in quadruplicate, processed as previously described (Rechtsteiner et al. 2010), and compared to wild-type early embryo expression data obtained from the same publication, which utilized identical NimbleGen microarrays. Similar growth conditions and stages were used to generate hpl-2 embryos as had been used for wild-type embryos. Using R (R Core Team 2014), expression data for both wild type and hpl-2 were quantile-normalized together using the affy package (Bolstad et al. 2003), and the Limma package (Smyth et al. 2005) was used to determine the log2(fold-change) for each gene, along with adjusted P-values using empirical Bayes methods. In cases where multiple isoforms of the same gene were represented on the arrays, the fold-change value for the isoform with the highest significance was used.
Immunocytochemistry
All samples were fixed using methanol/acetone as previously described (Strome and Wood 1983), except samples were incubated in methanol for 10 min, and PBS included 0.1% Tween-20. Final concentrations of primary antibodies were: anti-HPL-2 (SDQ2324) at 0.25 μg/mL, anti-H3K9me1 (Abcam 9045) at 0.025 μg/mL, anti-H3K9me2 (Kimura 6D11) at 0.18 μg/mL, and anti-H3K9me3 (Kimura 2F3) at 0.36 μg/mL. Secondary antibodies conjugated to either Alexa Fluor 488 or 594 (Molecular Probes) were used at 1:500 for 2 h at room temperature. Images were acquired using a Volocity spinning disk confocal system (Perkin-Elmer/Improvision) coupled with a Nikon Eclipse TE2000-E inverted microscope.
Fertility assays
Parent worms were reared under standard growth conditions at 20°C and up-shifted as L4s to 25°C ± 0.5°C. Subsequent generations were maintained at 25°C. F1 larvae were plated individually, and 2–3 d later scored for fertility/sterility by looking for the presence of F2 progeny. For M-Z- worms, homozygous parents were upshifted to 25°C; for M+Z- worms, heterozygous parents were up-shifted, and homozygous F1 progeny were analyzed.
Histone tail binding assays
Nuclear extracts were prepared from mixed-stage wild-type embryos grown as described above and incubated with synthetic histone H3 peptides. After washing and eluting from the peptides, western blotting was performed to assay for binding of HPL-2. For details, see Supplemental Methods.
Mass spectrometry of histone H3 peptides
Bulk histones were acid-extracted from nuclei obtained from mixed-stage wild-type and met-2 set-25 mutant embryos grown as described above, derivatized, desalted, and analyzed by liquid chromatography coupled to mass spectrometry. For details, see Supplemental Methods.
Data access
Microarray data (ChIP-chip and transcript profiling) from this study are available at the NCBI Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE58764. Mass spectrometry data are available at the Chorus repository (http://chorusproject.org/) under accession number 457.
Supplementary Material
Acknowledgments
We thank Andreas Rechtsteiner for assistance and advice on bioinformatic analysis, members of the Strome laboratory for helpful discussions, Julie Ahringer for helpful discussions and sharing unpublished data, Hiroshi Kimura for antibodies, Susan Gasser for providing the met-2 set-25 double-mutant strain (GW638), and Ole Nørregaard Jensen for help on the preliminary quantification of H3K9me by mass spectrometry. This research was funded by NIH grant GM34059 to S. Strome, NIH NHGRI grant U01 HG004270 to The modENCODE Consortium headed by J.D. Lieb, and a Eugene Cota-Robles Fellowship to J.M.G. B.A.G. gratefully acknowledges support from an NSF Early Faculty CAREER award and an NIH Innovator award (DP2OD007447) from the Office of the Director, NIH. Some strains were provided by the CGC, which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440).
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.180489.114.
References
- Albertson DG, Thomson JN. 1982. The kinetochores of Caenorhabditis elegans. Chromosoma 86: 409–428. [DOI] [PubMed] [Google Scholar]
- Andersen EC, Horvitz HR. 2007. Two C. elegans histone methyltransferases repress lin-3 EGF transcription to inhibit vulval development. Development 134: 2991–2999. [DOI] [PubMed] [Google Scholar]
- Ashe A, Sapetschnig A, Weick E, Mitchell J, Bagijn MP, Cording AC, Doebley A, Goldstein LD, Lehrbach NJ, Le Pen J, et al. 2012. piRNAs can trigger a multigenerational epigenetic memory in the germline of C. elegans. Cell 150: 88–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannister AJ, Zegerman P, Partridge JF, Miska EA, Thomas JO, Allshire RC, Kouzarides T. 2001. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 410: 120–124. [DOI] [PubMed] [Google Scholar]
- Barnes TM, Kohara Y, Coulson A, Hekimi S. 1995. Meiotic recombination, noncoding DNA and genomic organization in Caenorhabditis elegans. Genetics 141: 159–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolstad BM, Irizarry RA, Astrand M, Speed TP. 2003. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 19: 185–193. [DOI] [PubMed] [Google Scholar]
- Brenner S. 1974. The genetics of Caenorhabditis elegans. Genetics 77: 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchwitz BJ, Ahmad K, Moore LL, Roth MB, Henikoff S. 1999. A histone-H3-like protein in C. elegans. Nature 401: 547–548. [DOI] [PubMed] [Google Scholar]
- The C. elegans Sequencing Consortium 1998. Genome sequence of the nematode C. elegans: a platform for investigating biology. Science 282: 2012–2018. [DOI] [PubMed] [Google Scholar]
- Cam HP, Sugiyama T, Chen ES, Chen X, FitzGerald PC, Grewal SIS. 2005. Comprehensive analysis of heterochromatin- and RNAi-mediated epigenetic control of the fission yeast genome. Nat Genet 37: 809–819. [DOI] [PubMed] [Google Scholar]
- Coustham V, Bedet C, Monier K, Schott S, Karali M, Palladino F. 2006. The C. elegans HP1 homologue HPL-2 and the LIN-13 zinc finger protein form a complex implicated in vulval development. Dev Biol 297: 308–322. [DOI] [PubMed] [Google Scholar]
- Couteau F, Guerry F, Muller F, Palladino F. 2002. A heterochromatin protein 1 homologue in Caenorhabditis elegans acts in germline and vulval development. EMBO Rep 3: 235–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit E, Greil F, van Steensel B. 2005. Genome-wide HP1 binding in Drosophila: developmental plasticity and genomic targeting signals. Genome Res 15: 1265–1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit E, Greil F, van Steensel B. 2007. High-resolution mapping reveals links of HP1 with active and inactive chromatin components. PLoS Genet 3: e38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dialynas GK, Makatsori D, Kourmouli N, Theodoropoulos PA, McLean K, Terjung S, Singh PB, Georgatos SD. 2006. Methylation-independent binding to histone H3 and cell cycle-dependent incorporation of HP1β into heterochromatin. J Biol Chem 281: 14350–14360. [DOI] [PubMed] [Google Scholar]
- Figueiredo MLA, Philip P, Stenberg P, Larsson J. 2012. HP1a recruitment to promoters is independent of H3K9 methylation in Drosophila melanogaster. PLoS Genet 8: e1003061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filion GJ, van Bemmel JG, Braunschweig U, Talhout W, Kind J, Ward LD, Brugman W, de Castro IJ, Kerkhoven RM, Bussemaker HJ, et al. 2010. Systematic protein location mapping reveals five principal chromatin types in Drosophila cells. Cell 143: 212–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folco HD, Pidoux AL, Urano T, Allshire RC. 2008. Heterochromatin and RNAi are required to establish CENP-A chromatin at centromeres. Science 319: 94–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gassmann R, Rechtsteiner A, Yuen KW, Muroyama A, Egelhofer T, Gaydos L, Barron F, Maddox P, Essex A, Monen J, et al. 2012. An inverse relationship to germline transcription defines centromeric chromatin in C. elegans. Nature 484: 534–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer EL, Beese-Sims SE, Brookes E, Spadafora R, Zhu Y, Rothbart SB, Aristizábal-Corraleset D, Chen S, Badeauz AI, Jin Q, et al. 2014. A histone methylation network regulates transgenerational epigenetic memory in C. elegans. Cell Rep 7: 113–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grewal SIS, Elgin SCR. 2002. Heterochromatin: new possibilities for the inheritance of structure. Curr Opin Genet Dev 12: 178–187. [DOI] [PubMed] [Google Scholar]
- Grewal SIS, Jia S. 2007. Heterochromatin revisited. Nat Rev Genet 8: 35–46. [DOI] [PubMed] [Google Scholar]
- Grewal SIS, Moazed D. 2003. Heterochromatin and epigenetic control of gene expression. Science 301: 798–802. [DOI] [PubMed] [Google Scholar]
- Gu SG, Fire A. 2010. Partitioning the C. elegans genome by nucleosome modification, occupancy, and positioning. Chromosoma 119: 73–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guelen L, Pagie L, Brasset E, Meuleman W, Faza MB, Talhout W, Eussen BH, de Klein A, Wessels L, de Laat W, et al. 2008. Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature 453: 948–951. [DOI] [PubMed] [Google Scholar]
- Guenatri M, Bailly D, Maison C, Almouzni G. 2004. Mouse centric and pericentric satellite repeats form distinct functional heterochromatin. J Cell Biol 166: 493–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrington JJ, Van Bokkelen G, Mays RW, Gustashaw K, Willard HF. 1997. Formation of de novo centromeres and construction of first-generation human artificial microchromosomes. Nat Genet 15: 345–355. [DOI] [PubMed] [Google Scholar]
- Heitz E. 1928. Das heterochromatin der Moose I. Jahrb Wiss Botanik 69: 762–818. [Google Scholar]
- Hennig W. 1999. Heterochromatin. Chromosoma 108: 1–9. [DOI] [PubMed] [Google Scholar]
- Ho JWK, Jung YL, Liu T, Alver BH, Lee S, Ikegami K, Sohn K, Minoda A, Tolstorukov MY, Appert A, et al. 2014. Comparative analysis of metazoan chromatin organization. Nature 512: 449–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikegami K, Egelhofer TA, Strome S, Lieb JD. 2010. Caenorhabditis elegans chromosome arms are anchored to the nuclear membrane via discontinuous association with LEM-2. Genome Biol 11: R120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeno M, Grimes B, Okazaki T, Nakano M, Saitoh K, Hoshino H, McGill NI, Cooke H, Masumoto H. 1998. Construction of YAC-based mammalian artificial chromosomes. Nat Biotechnol 16: 431–439. [DOI] [PubMed] [Google Scholar]
- Jacobs SA, Taverna SD, Zhang Y, Briggs SD, Li J, Eissenberg JC, Allis CD, Khorasanizadeh S. 2001. Specificity of the HP1 chromo domain for the methylated N-terminus of histone H3. EMBO J 20: 5232–5241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James TC, Elgin SC. 1986. Identification of a nonhistone chromosomal protein associated with heterochromatin in Drosophila melanogaster and its gene. Mol Cell Biol 6: 3862–3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurka J. 2000. Repbase Update: a database and an electronic journal of repetitive elements. Trends Genet 16: 418–420. [DOI] [PubMed] [Google Scholar]
- Karolchik D, Barber GP, Casper J, Clawson H, Cline MS, Diekhans M, Dreszer TR, Fujita PA, Guruvadoo L, Haeussler M, et al. 2014. The UCSC Genome Browser database: 2014 update. Nucleic Acids Res 42: D764–D770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur T, Rockman MV. 2014. Crossover heterogeneity in the absence of hotspots in Caenorhabditis elegans. Genetics 196: 137–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly WG, Schaner CE, Dernburg AF, Lee M, Kim SK, Villeneuve AM, Reinke V. 2002. X-chromosome silencing in the germline of C. elegans. Development 129: 479–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koester-Eiserfunke N, Fischle W. 2011. H3K9me2/3 binding of the MBT domain protein LIN-61 is essential for Caenorhabditis elegans vulva development. PLoS Genet 7: e1002017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudron M, Niu W, Lu Z, Wang G, Gerstein M, Snyder M, Reinke V. 2013. Tissue-specific direct targets of Caenorhabditis elegans Rb/E2F dictate distinct somatic and germline programs. Genome Biol 14: R5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn RM, Haussler D, Kent WJ. 2013. The UCSC genome browser and associated tools. Brief Bioinform 14: 144–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon SH, Florens L, Swanson SK, Washburn MP, Abmayr SM, Workman JL. 2010. Heterochromatin protein 1 (HP1) connects the FACT histone chaperone complex to the phosphorylated CTD of RNA polymerase II. Genes Dev 24: 2133–2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachner M, O’Carroll D, Rea S, Mechtler K, Jenuwein T. 2001. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature 410: 116–120. [DOI] [PubMed] [Google Scholar]
- Liu T, Rechtsteiner A, Egelhofer TA, Vielle A, Latorre I, Cheung M, Ercan S, Ikegami K, Jensen M, Kolasinska-Zwierz P, et al. 2011. Broad chromosomal domains of histone modification patterns in C. elegans. Genome Res 21: 227–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maison C, Bailly D, Roche D, de Oca RM, Probst AV, Vassias I, Dingli F, Lombard B, Loew D, Quivy J, et al. 2011. SUMOylation promotes de novo targeting of HP1α to pericentric heterochromatin. Nat Genet 43: 220–227. [DOI] [PubMed] [Google Scholar]
- Nielsen AL, Oulad-Abdelghani M, Ortiz JA, Remboutsika E, Chambon P, Losson R. 2001. Heterochromatin formation in mammalian cells: interaction between histones and HP1 proteins. Mol Cell 7: 729–739. [DOI] [PubMed] [Google Scholar]
- Olszak AM, van Essen D, Pereira AJ, Diehl S, Manke T, Maiato H, Saccani S, Heun P. 2011. Heterochromatin boundaries are hotspots for de novo kinetochore formation. Nat Cell Biol 13: 799–808. [DOI] [PubMed] [Google Scholar]
- Pal-Bhadra M, Leibovitch BA, Gandhi SG, Rao M, Bhadra U, Birchler JA, Elgin SCR. 2004. Heterochromatic silencing and HP1 localization in Drosophila are dependent on the RNAi machinery. Science 303: 669–672. [DOI] [PubMed] [Google Scholar]
- Partridge JF, Borgstrøm B, Allshire RC. 2000. Distinct protein interaction domains and protein spreading in a complex centromere. Genes Dev 14: 783–791. [PMC free article] [PubMed] [Google Scholar]
- R Core Team 2014. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria http://www.R-project.org/.
- Rechtsteiner A, Ercan S, Takasaki T, Phippen TM, Egelhofer TA, Wang W, Kimura H, Lieb JD, Strome S. 2010. The histone H3K36 methyltransferase MES-4 acts epigenetically to transmit the memory of germline gene expression to progeny. PLoS Genet 6: e1001091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards EJ, Elgin SCR. 2002. Epigenetic codes for heterochromatin formation and silencing: rounding up the usual suspects. Cell 108: 489–500. [DOI] [PubMed] [Google Scholar]
- Robert VJP, Sijen T, van Wolfswinkel J, Plasterk RHA. 2005. Chromatin and RNAi factors protect the C. elegans germline against repetitive sequences. Genes Dev 19: 782–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockman MV, Kruglyak L. 2009. Recombinational landscape and population genomics of Caenorhabditis elegans. PLoS Genet 5: e1000419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schott S, Coustham V, Simonet T, Bedet C, Palladino F. 2006. Unique and redundant functions of C. elegans HP1 proteins in post-embryonic development. Dev Biol 298: 176–187. [DOI] [PubMed] [Google Scholar]
- Schotta G, Ebert A, Krauss V, Fischer A, Hoffmann J, Rea S, Jenuwein T, Dorn R, Reuter G. 2002. Central role of Drosophila SU(VAR)3-9 in histone H3-K9 methylation and heterochromatic gene silencing. EMBO J 21: 1121–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seum C, Reo E, Peng H, Rauscher FJ III, Spierer P, Bontron S. 2007. Drosophila SETDB1 is required for chromosome 4 silencing. PLoS Genet 3: e76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirayama M, Seth M, Lee H, Gu W, Ishidate T, Conte D Jr, Mello CC. 2012. piRNAs initiate an epigenetic memory of nonself RNA in the C. elegans germline. Cell 150: 65–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit AFA, Hubley R, Green P 1996–2010. RepeatMasker Open-3.0 http://www.repeatmasker.org/.
- Smothers JF, Henikoff S. 2001. The hinge and chromo shadow domain impart distinct targeting of HP1-like proteins. Mol Cell Biol 21: 2555–2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth GK, Michaud J, Scott HS. 2005. Use of within-array replicate spots for assessing differential expression in microarray experiments. Bioinformatics 21: 2067–2075. [DOI] [PubMed] [Google Scholar]
- Song JS, Johnson WE, Zhu X, Zhang X, Li W, Manrai AK, Liu JS, Chen R, Liu XS. 2007. Model-based analysis of two-color arrays (MA2C). Genome Biol 8: R178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strome S, Wood WB. 1983. Generation of asymmetry and segregation of germ-line granules in early C. elegans embryos. Cell 35: 15–25. [DOI] [PubMed] [Google Scholar]
- Studencka M, Wesołowski R, Opitz L, Salinas-Riester G, Wisniewski JR, Jedrusik-Bode M. 2012. Transcriptional repression of Hox genes by C. elegans HP1/HPL and H1/HIS-24. PLoS Genet 8: e1002940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subirana JA, Messeguer X. 2013. A satellite explosion in the genome of holocentric nematodes. PLoS ONE 8: e62221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towbin BD, González-Aguilera C, Sack R, Gaidatzis D, Kalck V, Meister P, Askjaer P, Gasser SM. 2012. Step-wise methylation of histone H3K9 positions heterochromatin at the nuclear periphery. Cell 150: 934–947. [DOI] [PubMed] [Google Scholar]
- Tzeng T, Lee C, Chan L, Shen CJ. 2007. Epigenetic regulation of the Drosophila chromosome 4 by the histone H3K9 methyltransferase dSETDB1. Proc Natl Acad Sci 104: 12691–12696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdel A, Jia S, Gerber S, Sugiyama T, Gygi S, Grewal SIS, Moazed D. 2004. RNAi-mediated targeting of heterochromatin by the RITS complex. Science 303: 672–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermaak D, Malik HS. 2009. Multiple roles for heterochromatin protein 1 genes in Drosophila. Annu Rev Genet 43: 467–492. [DOI] [PubMed] [Google Scholar]
- Wen B, Wu H, Shinkai Y, Irizarry RA, Feinberg AP. 2009. Large histone H3 lysine 9 dimethylated chromatin blocks distinguish differentiated from embryonic stem cells. Nat Genet 41: 246–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirth M, Paap F, Fischle W, Wenzel D, Agafonov D, Samatov T, Wisniewski J, Jedrusik-Bode M. 2009. Linker histone HIS-24 and SIR-2.1 deacetylase induce H3K27me3 in C. elegans germline. Mol Cell Biol 29: 3700–3709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuen KWY, Nabeshima K, Oegema K, Desai A. 2011. Rapid de novo centromere formation occurs independently of heterochromatin protein 1 in C. elegans embryos. Curr Biol 21: 1800–1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao T, Heyduk T, Allis CD, Eissenberg JC. 2000. Heterochromatin protein 1 binds to nucleosomes and DNA in vitro. J Biol Chem 275: 28332–28338. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.