Abstract
The eyes of the horseshoe crab Limulus polyphemus have long been used for studies of basic mechanisms of vision, and the structure and physiology of Limulus photoreceptors have been examined in detail. Less is known about the opsins Limulus photoreceptors express. We previously characterized a UV opsin (LpUVOps1) that is expressed in all three types of Limulus eyes (lateral compound eyes, median ocelli and larval eyes) and three visible light-sensitive rhabdomeric opsins (LpOps1, -2 and -5) that are expressed in Limulus lateral compound and larval eyes. Physiological studies showed that visible light-sensitive photoreceptors are also present in median ocelli, but the visible light-sensitive opsins they express were unknown. In the current study we characterize three newly identified, visible light-sensitive rhabdomeric opsins (LpOps6, -7 and -8) that are expressed in median ocelli. We show that they are ocellar specific and that all three are co-expressed in photoreceptors distinct from those expressing LpUVOps1. Our current findings show that the pattern of opsin expression in Limulus eyes is much more complex than previously thought and extend our previous observations of opsin co-expression in visible light-sensitive Limulus photoreceptors. We also characterize a Limulus peropsin/RGR (LpPerOps1). We examine the phylogenetic relationship of LpPerOps1 with other peropsins and RGRs, demonstrate that LpPerOps1 transcripts are expressed in each of the three types of Limulus eyes and show that the encoded protein is expressed in membranes of cells closely associated with photoreceptors in each eye type. These finding suggest that peropsin was in the opsin repertoire of euchelicerates.
KEY WORDS: Limulus photoreceptor, Ocellar specific opsin, Opsin co-expression, Peropsin
INTRODUCTION
The eyes of the American horseshoe crab Limulus polyphemus Linnaeus 1758 have long served as models for studies of phototransduction, light and dark adaptation, retinal integration and circadian changes in visual function. Limulus polyphemus (hereafter referred to as Limulus) is one of only four extant species of xiphosauran chelicerates, an early branching sister group to arachnids (Regier et al., 2010; Edgecombe and Legg, 2014). Therefore, studies of the structure and function of Limulus eyes may provide insights into the structure and function of the eyes of Euarthropoda (Nilsson and Kelber, 2007).
Limulus has three different types of eyes: a pair of median ocelli or median eyes (MEs), a pair of lateral compound eyes (LEs) and three pairs of larval eyes – lateral, median and ventral. The locations of these eyes are illustrated in Fig. 1A. Horseshoe crabs are the only extant chelicerates with compound eyes. All other chelicerates with eyes have camera-type eyes. Much is known about the physiology and structure of photoreceptors in each eye type; less is known about the opsins they express. Opsin, the protein component of photopigments, largely determines the spectral sensitivity and function of the photopigment (Yokoyama, 2000). In the present study we examine the expression of four newly identified Limulus opsins in each eye type; therefore the retina and photoreceptors in each are described here briefly.
Fig. 1.
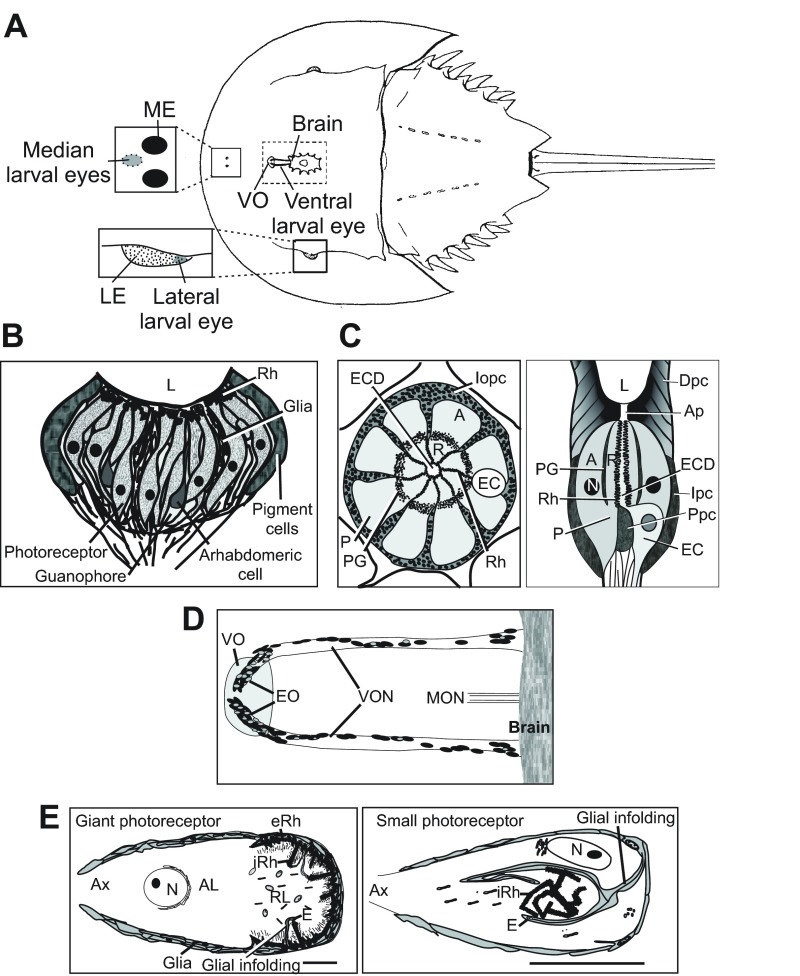
Schematic of Limulus showing the locations of its eyes and schematics of ME, LE and VE retinas and photoreceptors. (A) Dorsal view of Limulus showing the locations of its eyes. Upper box on left: enlargement to show the locations of the median ocelli and the fused median larval eyes located under the carapace between them. Lower box on left: enlargement to show the location of the lateral larval eye at the posterior edge of each lateral compound eye. Central cut-away shows the location of the brain and ventral optic nerves that extend anteriorly from the brain and terminate at the ventral organ visible on the ventral cuticle (LE, lateral eye; ME, median eye; VO, ventral organ) (modified from Calman and Battelle, 1991). (B) Schematic of a longitudinal section through a median ocellus showing clusters of photoreceptors, arhabdomeric cells and guanophores separated by partitions of glial and pigment cells. Rhabdoms of ME photoreceptors are located close to the lens. Pigment cells surround the retina (L, location of the lens; Rh, rhabdoms). Glial cells also surround the base of the photoreceptor layer (modified from Jones et al., 1971). (C) Left: schematic of a cross-section of a LE ommatidium showing photoreceptors surrounding a central eccentric cell dendrite. Intraommatidial pigment cells surround the photoreceptors and extend into partitions between them. Right: schematic of a longitudinal section through an ommatidium from a LE fixed during the day in the light (A, arhabdomeral segment; Ap, aperture; Dpc, distal pigment cells; EC, eccentric cell body; ECD, eccentric cell dendrite; Ipc, intraommatidial pigment cells; L, lens; N, nucleus; P, photoreceptor cell; PG, photoreceptor pigment granules; Ppc, proximal pigment cells; R, rhabdomeral segment; Rh, rhabdom) [based on Fahrenbach (Fahrenbach, 1969; Fahrenbach, 1975)]. (D) Schematic of ventral optic nerves projecting anteriorly from the brain and ending beneath the ventral organ on the ventral cuticle. The ovals scattered along the optic nerves and clustered at the end organ and near the brain represent cell bodies of giant and smaller ventral photoreceptors (EO, end organs; MON, median optic nerves; VO, ventral organ; VON, ventral optic nerves. (E) Diagrams of ventral photoreceptors. Left: giant ventral photoreceptor; scale bar, 50 μm. Right: small ventral photoreceptor with an internal rhabdom. A second type of small ventral photoreceptor is organized much like the giant ventral photoreceptors. Both types of small ventral photoreceptors are approximately the same size. Scale bar, 20 μm. AL, arhabdomeral lobe; Ax, axon; E, efferent terminal; eRh, external rhabdom; iRh, internal rhabdom; N, nucleus; RL, rhabdomeral lobe). Based on Clark et al. (Clark et al., 1969), Calman and Chamberlain (Calman and Chamberlain, 1982) and Herman (Herman, 1991).
Each ME has a single lens below which lie elongated photoreceptors with rhabdoms close to the base of the lens (Fig. 1B). The structure and function of MEs has been examined in detail in a number of studies (Nolte and Brown, 1969; Nolte and Brown, 1970; Nolte and Brown, 1972; Lall, 1970; Jones et al., 1971; Fahrenbach and Griffin, 1975; Behrens and Fahy, 1981). The ME retina consists of a number of loosely organized photoreceptor clusters, referred to as pseudo-ommatidia, containing UV-sensitive and visible light-sensitive photoreceptors, guanophores – cells containing reflective crystals of guanine – and usually one arhabdomeric cell. The relative number of UV compared with visible light-sensitive photoreceptors varies widely among photoreceptor clusters, but on average, about 70% of ME photoreceptors are sensitive to UV light. Arhabdomeric cells are electrically coupled to photoreceptors, generate action potentials when photoreceptors depolarize and are thought to be principally responsible for transmitting visual information from the eye to the brain. Partitions between photoreceptor clusters are formed by pigment cells, guanophores and glial cells. Glial cells also penetrate photoreceptors and line the base of the photoreceptor layer. A dense layer of pigment cells surrounds the periphery of the ME cup (Fig. 1B). The shape of the ME lens and organization of the underlying retina suggests that the eye is not image-forming (Fahrenbach, 1975), but the eye conveys directional information (Lall and Chapman, 1973) and the large aperture of the lens suggests that the eye is adapted to operating in low light conditions (Fahrenbach, 1975).
LEs in an adult animal consist of over 1000 well-organized ommatidia (Fig. 1C). Below each conical lens is a cluster of five to 12 photoreceptors, or retinular cells, that encircle the dendrites of one to three eccentric cells. LE eccentric cells are thought to be functionally equivalent to ME arhabdomeric cells (Waterman and Wiersma, 1954). Pigment cells are another major cell type in LE
List of abbreviations
- c-opsin
ciliary opsin
- LE
lateral compound eye
- Lp
Limulus polyphemus
- ME
median eye of ocellus
- Ops
opsin
- ORF
open reading frame
- PCR
polymerase chain reaction
- PerOps
peropsin
- RGR
retinal G-protein-coupled receptor
- r-opsin
rhabdomeric opsin
- UTR
untranslated region
- UV
ultraviolet
- VE
ventral eye
ommatidia. Distal pigment cells surround the base of the lens, the aperture at the base of the lens and distal portions of retinular cells. Intraommatidial pigment cells surround retinular cells at their mid-region and project into partitions between them. Proximal pigment cells are located at the base of ommatidia and extend processes into partitions between retinular cells (Fahrenbach, 1968; Fahrenbach, 1969).
Larval eyes, also called rudimentary eyes, appear in the embryo before the median ocelli and lateral compound eyes develop and persist in the adult (Harzsch et al., 2006). Of these, the ventral larval eyes (VEs) have been studied most extensively (Fig. 1D). In an adult animal, VEs consist of a pair of optic nerves that extend anteriorly from the brain and terminate at the ventral organ that can be seen on the ventral cuticle. Giant and small photoreceptors (Fig. 1E) are scattered along the length of each optic nerve and cluster near the brain and in an end organ at the distal end of each nerve. VEs contain no pigment cells, but the photoreceptor cell bodies are ensheathed by several layers of glial cells that also project into photoreceptor cell bodies (Clark et al., 1969; Calman and Chamberlain, 1982; Herman, 1991). Lateral and median larval eyes have been studied much less extensively, but the best evidence indicates that they contain the same cell types as the VEs (Millecchia et al., 1966; Fahrenbach, 1970; Battelle et al., 2014).
We previously examined the expression of three visible light-sensitive Limulus opsins (Ops), LpOps1, LpOps2 and LpOps5 (Smith et al., 1993; Dalal et al., 2003; Katti et al., 2010), and a UV-sensitive opsin (LpUVOps1) (Battelle et al., 2014). LE retinular cells and giant ventral photoreceptors each express the three visible light-sensitive opsins, LpOps1, -2 and -5. LpOps1 and LpOps2 cannot be distinguished with antibodies (Battelle et al., 2001), therefore we refer to them here as LpOps1-2. LpOps5 clusters to a different opsin clade from LpOps1 and LpOps2 (Katti et al., 2010), but its spectral sensitivity is similar to that of LpOps1-2 (Battelle et al., 2014). LpUVOps1 is expressed in each of the three types of Limulus eyes (Battelle et al., 2014): in many ME photoreceptors, as was anticipated from electrophysiological studies (Nolte and Brown, 1969; Nolte and Brown, 1970), and unexpectedly, in small VE photoreceptors where it is co-expressed with LpOps5, and in LE eccentric cells. LE eccentric cells have long been considered to be secondary visual cells (Waterman and Wiersma, 1954). However, the discovery that eccentric cells express LpUVOps1 suggests that they also may be intrinsically photosensitive to UV light.
A puzzling aspect of our previous results is that although transcripts for LpOps1, LpOps2 and LpOps5 are routinely detected by PCR in ME cDNA preparations (Katti et al., 2010) (B.-A.B., personal observation), and LpOps2 was originally cloned from a ME cDNA library (Smith et al., 1993), we have been unable to detect LpOps1-2 or LpOps5 in rhabdoms of ME photoreceptors using antibodies that detect these opsins in rhabdoms of LE and VE photoreceptors (Battelle et al., 2001; Katti et al., 2010; Battelle et al., 2013; Battelle et al., 2014). Yet we know from physiological studies that MEs are sensitive to both UV and visible light. These findings led us to propose that other, not yet identified visible light-sensitive opsins are expressed in ME photoreceptors.
In the present study we report that in a transcriptome analysis of ME (Speiser et al., 2014), we detected two rhabdomeric opsins (r-opsins) that had not been identified previously in transcriptomes of VE and LE, and in a BLAST analysis of a recent assembly of the Limulus genome (http://www.ncbi.nlm.nih.gov/bioproject/PRJNA20489), we detected an additional r-opsin sequence closely related to one found in the ME transcriptome. We call these opsins LpOps6, LpOps7 and LpOps8. We show that these are predicted long wavelength-sensitive opsins, present evidence that they are ME specific and show that all three are expressed in the same photoreceptors.
We also characterize a Limulus peropsin/RGR-like sequence we call Limulus peropsin1 (LpPerOps1). Peropsins are an enigmatic group belonging to a large clade of opsins called ‘RGR/Go’ (Plachetzki et al., 2007; Feuda et al., 2012) or Group IV opsins (Porter et al., 2012). The founding members were identified in cDNA libraries of human and mouse eyes and shown by immunocytochemistry to be expressed in retinal pigmented epithelium (Sun et al., 1997). Their functions are largely unknown. The most closely related gene to peropsin in the human genome is RGR (RPE-retinal G-protein-coupled receptor) (Jiang et al., 1993), a photoisomerase (Chen et al., 2001). More recently, peropsin/RGR-like sequences were identified in transcriptomes of two species of chelicerates (Nagata et al., 2010; Eriksson et al., 2013). In the present study, we examine the phylogenetic relationship of LpPerOps1 with other peropsins and RGRs, we demonstrate that LpPerOps1 transcripts are expressed in each of the three types of Limulus eyes and show that the encoded protein is expressed in membranes of cells closely associated with photoreceptors in each eye type.
RESULTS
Characterization of transcripts encoding three previously unidentified Limulus r-opsins
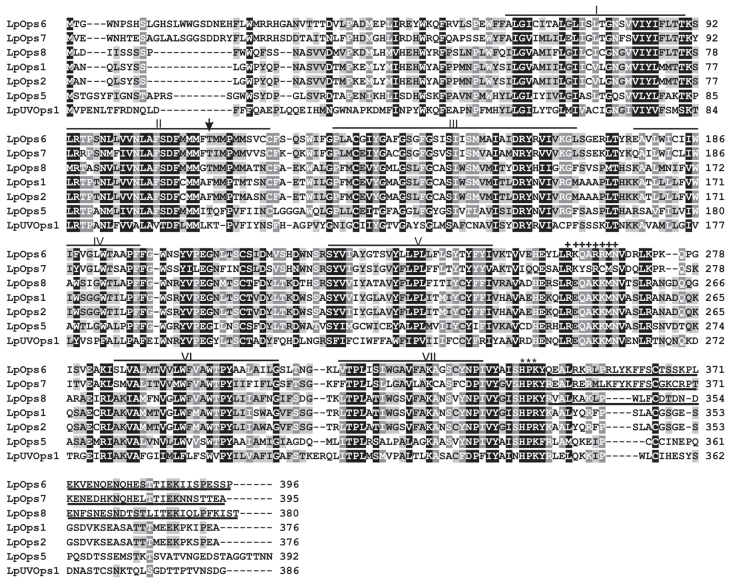
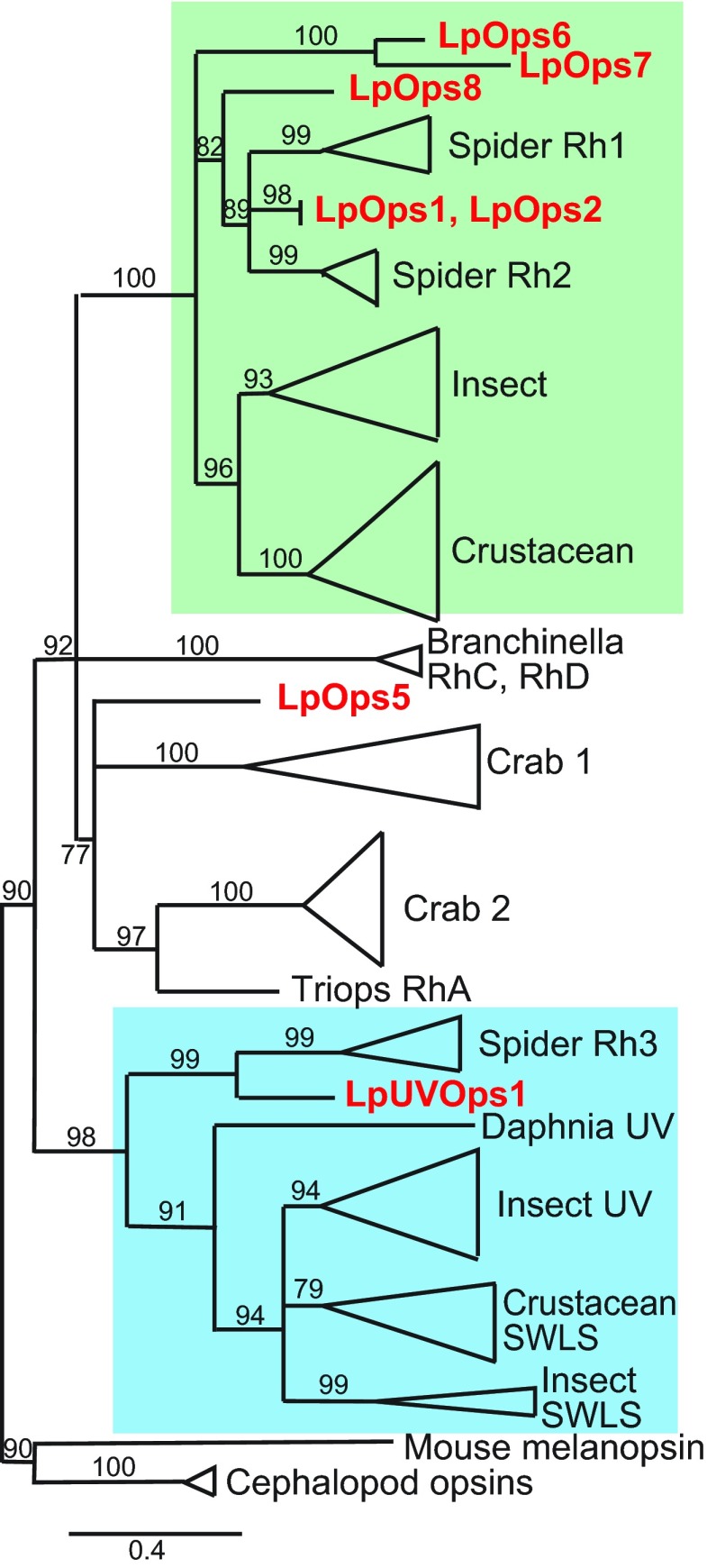
We identified full-length sequences encoding two previously unidentified opsins in a ME transcriptome (Speiser et al., 2014) (LpOps6 and -8: accession numbers KM538950 and KM538952, respectively) and an additional sequence 61% identical to LpOps6 at the amino acid level in an assembly of the Limulus genome (http://www.ncbi.nlm.nih.gov/bioproject/PRJNA20489) (LpOps7: accession number KM538951). All were cloned from ME cDNA and their sequences confirmed. An alignment of LpOps6, -7 and -8 with previously characterized Limulus opsins (LpOps1-2, LpOps5 and LpUVOps1) confirmed that they are r-opsins (Fig. 2). Their sequences contain seven predicted transmembrane domains, a conserved lysine in transmembrane VII that aligns with the predicted chromophore binding site in LpOps1 (K218), serine/threonine-rich C-termini and a triplet of amino acids (HPR/K) aligning with HPR/K334 in LpOps1 (Fig. 2, asterisks), a characteristic of opsins that activate the Gq/11 α class of GTP-binding proteins. Furthermore, a string of eight amino acids aligning with R246–N253 in LpOps1 that is conserved in many arthropod opsins [R(E/D)QAKKM(N/G)] (Porter et al., 2007) is also conserved in LpOps6 and -8 (87 and 100%, respectively), but less well in LpOps7 (50%) (Fig. 2, plus signs). All three are predicted visible light-sensitive opsins because they lack a lysine at the site equivalent to E90 in bovine rhodopsin (Fig. 2, arrow), a characteristic of UV light-sensitive r-opsins (Salcedo et al., 2003), and in a phylogenic analysis (Fig. 3) they cluster with other arthropod long wavelength-sensitive opsins.
Fig. 2.
Clustal W alignment of the predicted amino acid sequences of Limulus opsins 6, -7 and -8 (LpOps6, LpOps7 and LpOps8) with Limulus opsins 1, 2 and 5 (LpOps1, LpOps2, LpOps5) and UV opsin1 (LpUVOps1) show that LpOps6, -7 and -8 are predicted visible light-sensitive r-opsins. Positions of the transmembrane domains, indicated by the lines above the sequences, are estimated from an alignment of bovine rhodopsin (Palczewski et al., 2000). The arrow indicates the site equivalent to E90 in bovine rhodopsin. A lysine at this site in r-opsins is responsible for conferring UV sensitivity. Asterisks denote a triplet of amino acids characteristic of opsins that activate Gq/11 α GTP-binding proteins. The ‘+’ symbols denote a string of eight amino acids conserved in many arthropod r-opsins. The C-terminal sequence of LpOps6 underlined is the antigen used to produce monoclonal antibodies directed against LpOps6 (anti-LpOps6). The C-terminal sequences of LpOps7 and -8 underlined were expressed to test the specificity of anti-LpOps6. The antigen used to generate the anti-LpUVOps1 used in this study was described previously (Battelle et al., 2014).
Fig. 3.
A phylogenetic analysis of selected arthropod opsins shows that LpOps6, -7 and -8 fall in a clade along with other long wavelength-sensitive arthropod opsins. Accession numbers for the sequences used to generate the tree are given in supplementary material Table S2. The tree was constructed using a maximum likelihood analysis of amino acid sequences. Numbers on the branches indicate aLRT (approximate likelihood-ratio test) values for nodes supported by more than 70%. Major clades have been collapsed for clarity. Long wavelength-sensitive opsins are highlighted in green and UV short wavelength-sensitive opsins in blue. Limulus sequences are in red. Rh, rhodopsin; SWLS, short wavelength-sensitive.
With primers designed to amplify full-length sequences of LpOps 6, -7 and -8 (supplementary material Table S1), transcripts encoding each were amplified from cDNA prepared from ME but not from LE or VE (Fig. 4A). Because LpOps7 was not detected in the ME transcriptome, as were LpOps6 and -8, and the assembled Limulus genome indicates it is encoded on a single exon, we also probed for LpOps7 transcripts in a ME RNA preparation after it had been incubated in parallel with and without reverse transcriptase (Fig. 4B). An appropriately sized product was obtained from cDNA; no product was amplified in the no-reverse transcriptase control. This indicates that the LpOps7 PCR product from ME did not originate from genomic DNA. Thus LpOps6, -7 and -8 transcripts are present in ME and may be ME specific.
Fig. 4.
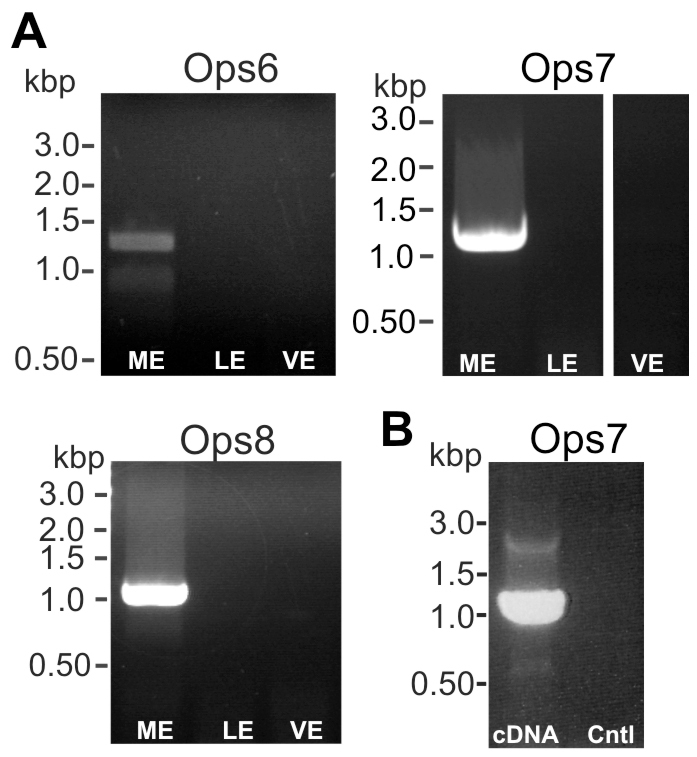
Transcripts encoding LpOps6, -7 and -8 were detected in ME cDNA but not cDNA prepared from LE or VE. (A) cDNA prepared from ME, VE and LE were probed for LpOps6, -7 and -8 using primers to amplify the full-length transcript (LpOps6-F1/R1; LpOps7-F1/R1; LpOps8-F1/R1; supplementary material Table S1). Each primer set amplified an appropriately sized product from ME cDNA and the identity of each was verified by sequencing. No products were amplified from LE or VE cDNA. (B) ME RNA (120 μg) was added to each of two reverse transcription reactions, one with reverse transcriptase and the other without. The products were then probed for LpOps7 using the primers listed in A. An appropriately sized product was amplified from cDNA but not from the no-reverse transcriptase control (Cntl).
Distribution of LpOps6 in ME
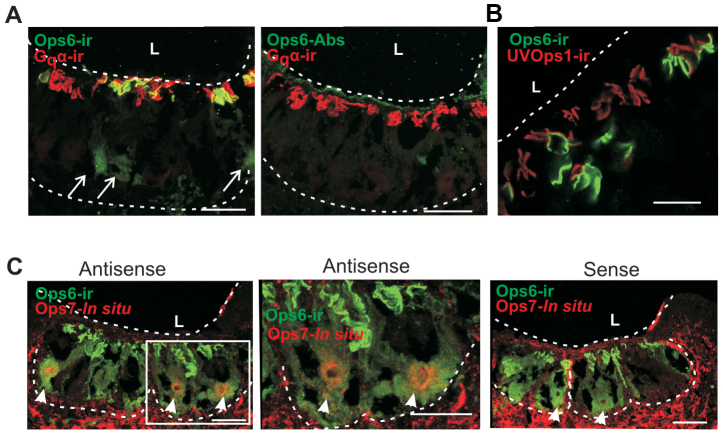
We examined the distribution of LpOps6 using a monoclonal antibody (anti-LpOps6) generated against the C-terminal sequence of LpOps6 underlined in Fig. 2 that showed no cross-reactivity with the C-terminal sequences of LpUVOps1, LpOps7 or LpOps8 (Fig. 5A). On western blots of membranes from LE, VE and ME, anti-LpOps6 immunostained a single band from ME with an apparent molecular mass of 40 kDa (Fig. 5B). This is close to the mass of LpOps6 predicted from its amino acid sequence (43 kDa). We detected no LpOps6-immunoreactivity (ir) in lanes containing LE or VE membranes, although LE and VE photoreceptor members were clearly present as indicated by the LpOps5-ir bands in these lanes. LpOps5 typically migrates as a doublet at 32 and 35 kDa (Katti et al., 2010). LpOps5-ir bands with greater mass can be attributed to opsin oligomers, which often form in sodium dodecyl sulfate (Fliesler, 1993). We detected no LpOps5-ir in ME membranes, which is consistent with previous findings (Katti et al., 2010). On ME sections, we detected LpOps6-ir in some but not all rhabdoms identified by Gqα-ir (Fig. 6A). We also observed LpOps6-ir in some photoreceptor cell bodies. All LpOps6-ir was eliminated when the antibody was pre-incubated with antigen (Fig. 6A, Ops6-Abs), indicating that immunostaining is specific. ME sections double-labelled for LpOps6-ir and LpUVOps1-ir showed that these opsins do not co-localize (Fig. 6B); therefore LpOps6 is not co-expressed with LpUVOps1 in ME photoreceptors.
Fig. 5.
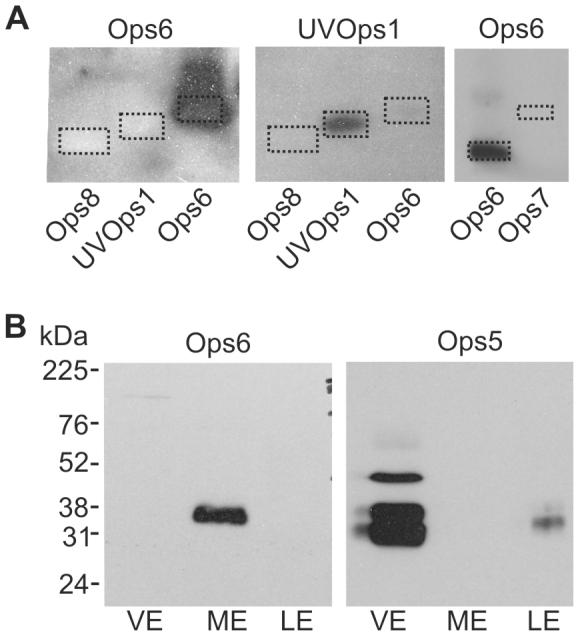
Anti-LpOps6, which does not cross-react with LpOps7, -8 or LpUVOps1, immunostained a single band in membrane preparations from ME but not LE or VE. (A) Left-hand and center panels: chemiluminescent images of western blots of C-terminal polypeptides, 1 μg each, from LpOps8, LpUVOps1 and LpOps6 that were probed with anti-LpOps6 (1:25 dilution) stripped and re-probed with anti-LpUVOps1 (1:1000). The dotted rectangles show the locations of the polypeptides as visualized with Fast Green staining. Anti-LpOps6 did not immunostain the C-terminal polypeptide from either LpOps8 or LpUVOps1, and anti-LpUVOps1 did not immunostain the C-terminal polypeptide from either LpOps8 or LpOps6. Right-hand panel: polypeptides encoding the C-terminal sequences of LpOps6 and -7 (1 μg each) were blotted together and probed with anti-LpOps6 as described above. The dotted rectangles show the locations of the polypeptides as visualized with Fast Green staining. The chemiluminescent image shows that anti-LpOps6 did not immunostain LpOps7. (B) Western blots of membranes from the equivalent of 2.5 VE, 1 ME and 0.3 LE were probed with anti-LpOps6 (1:25 dilution) stripped, then reprobed with anti-LpOps5 (1:350 dilution). A single LpOps6-ir band with an apparent molecular weight of about 40 kDa was detected in membranes from ME. No LpOps6-ir bands were detected in membranes from LE or VE, although the LpOps5-ir detected in the LE and VE lanes indicated that opsin-containing membranes were present. LpOps5 typically migrates as a doublet of about 32 and 35 kDa; higher immunostained bands are opsin oligomers. No LpOps5 was detected in ME membranes, consistent with previous findings (Katti et al., 2010).
Fig. 6.
LpOps6 is expressed in some, but not all, ME rhabdoms and is co-expressed with LpOps7. All images are of longitudinal sections of ME, ‘L’ indicates the position of the lens and dashed lines outline the photoreceptor layer. (A) Maximum projections (20–24 μm stacks) of images from sections incubated with anti-Gqα (1:1000 dilution, red) to show all rhabdoms and anti-LpOps6 (1:25 dilution, green) or anti-LpOps6 (1:25 dilution) that had been pre-incubated with antigen (Ops6-Abs). Gqα-ir rhabdoms are located close to the base of the lens. LpOps6-ir is detected in some but not all rhabdoms and in some photoreceptor cell bodies (arrows). Pre-incubating anti-LpOps6 with antigen (Ops6-Abs) eliminated all LpOps6-ir from rhabdoms and photoreceptor cell bodies. Scale bars, 100 μm. (B) A single optical section incubated with anti-LpOps6 (1:25 dilution, green) and anti-LpUVOps1 (1:50 dilution, red). LpOps6-ir is detected in fewer rhabdoms than LpUVOps1, and LpOps6-ir and LpUVOps1-ir do not co-localize. Scale bar, 50 μm. (C) Maximum projections (27–28 μm stacks) of images from sections incubated with anti-LpOps6 (1:10 dilution, green) and antisense and sense probes targeting the 3′-UTR of LpOps7 (red). The central image is a higher power view of the photoreceptor cluster within the boxed area seen on the left. A number of LpOps6-ir cells are seen in sections incubated with antisense and sense probes. Staining with the LpOps7 antisense probe was observed near the nucleus of LpOps6-ir cells (arrow heads in antisense images). No staining was seen around the nuclei of LpOps6-ir cells with the LpOps7 sense probe (arrow heads, sense image). Results indicate that LpOps7 is co-expressed with LpOps6. Staining beneath and between photoreceptor clusters was observed with LpOps7 antisense and sense probes; therefore it is considered non-specific. Scale bars, 100 μm.
Distribution of LpOps7 and LpOps8 transcripts in ME
We tested the distribution of LpOps7 on ME sections double labelled for LpOps7 transcripts and LpOps6 protein. This assay is possible because, as was shown in Fig. 6A, LpOps6-ir is detected in ME photoreceptor cell bodies as well as rhabdoms. To assure probe specificity, the LpOps7 probe targeted its 3′-untranslated region (UTR) instead of its coding region because the transcripts of LpOps6 and LpOps7 are 70% identical within their coding regions.
We consistently detected LpOps7 transcripts in LpOps6-ir photoreceptors and not in other photoreceptors (Fig. 6C), indicating that LpOps7 is co-expressed with LpOps6 in ME photoreceptors and not with LpUVOps1. Both antisense and sense probes targeting LpOps7 transcripts stained glia and other cells surrounding photoreceptor clusters; therefore we consider this staining non-specific.
ME sections double-labelled with antisense probes targeting LpOps8 and LpUVOps1 transcripts showed that LpOps8 is expressed in a population of ME photoreceptors different from those expressing LpUVOps1 (Fig. 7A). However, we consistently detected LpOps8 transcripts in LpOps6-ir photoreceptors (Fig. 7B) and in photoreceptors expressing LpOps7 transcripts (Fig. 7C). We detected no heterogeneity among photoreceptors expressing visible light-sensitive opsins, as has been found in butterfly (Kitamoto et al., 1998), i.e. subsets of photoreceptors expressing different combinations of visible light-sensitive opsins. Rather, our evidence indicates that visible light-sensitive ME photoreceptors consistently express all three visible light-sensitive opsins, and that none of these opsins is expressed together with UV opsins.
Fig. 7.
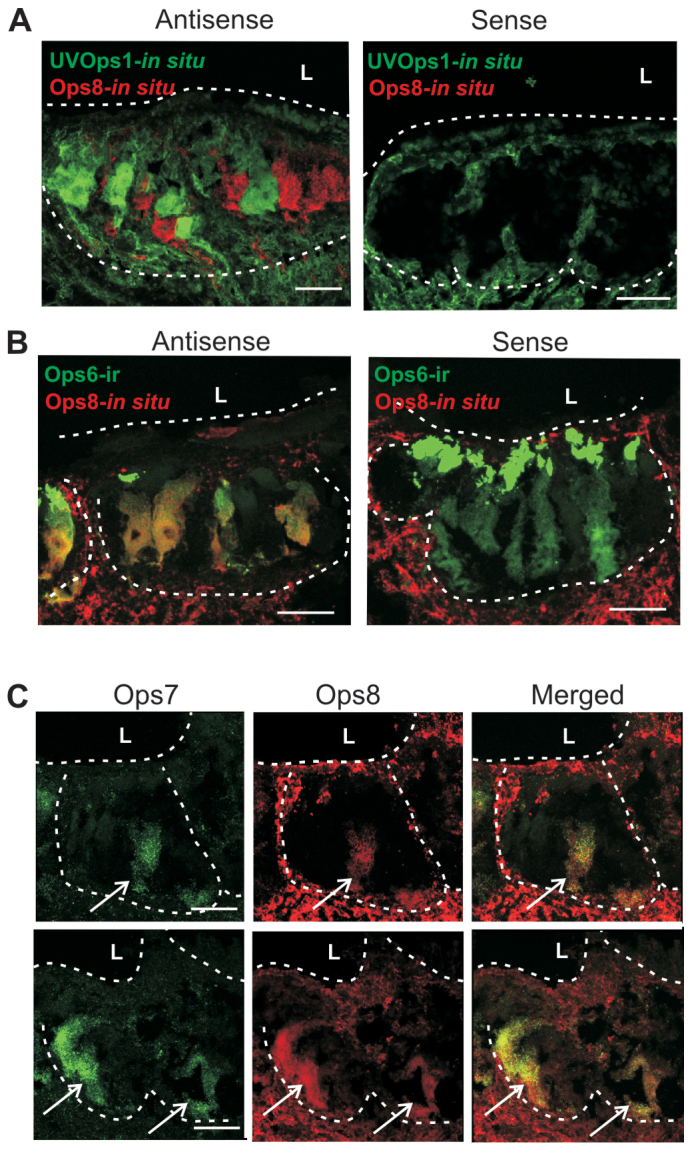
In ME retinas, LpOps8 is not co-expressed with LpUVOps1 but is co-expressed with LpOps6 and LpOps7. All images are of longitudinal sections of ME, ‘L’ indicates the position of the lens and dashed lines outline the photoreceptor layer. (A) Sections were incubated with antisense and sense probes targeting LpUVOps1 (green) and LpOps8 (red). Probes were visualized as maximum projections (10–14 μm stacks). Each antisense probe labelled photoreceptor cell bodies but the labels did not overlap, indicating that LpOps8 is not co-expressed with LpUVOps1. Neither sense probe labelled photoreceptor cell bodies; however, both labelled tissue between and below photoreceptor clusters. This staining is considered non-specific. (B) Sections were processed for in situ hybridization using antisense and sense probes targeting LpOps8 (red), then immunostained for LpOps6-ir (green, 1:10 dilution). Images are maximum projections (20–30 μm stacks). The LpOps8 antisense probe was routinely detected in photoreceptor cell bodies that also immunostained for LpOps6, indicating that LpOps8 is co-expressed with LpOps6. No photoreceptor cell body staining was observed with the LpOps8 sense probe; however, non-specific staining was detected below and between photoreceptor clusters. Scale bars, 100 μm. (C) Sections incubated with antisense probes targeting LpOps7 and LpOps8. Photoreceptor cell clusters from two different retinas are outlined with dashed lines. Antisense probes targeting LpOps7 and -8 routinely labelled the same cells in the photoreceptor layer. Arrows, double labelled cells in the photoreceptor layer: a single cell in the upper panels and two cells in the lower panels. No single labelled cells were detected. Scale bars, 100 μm.
LpOps6 protein and LpOps7 and -8 transcripts are not detected in LE or VE
We did not detect LpOps6-ir in LE retinular cell rhabdoms or eccentric cell dendrites or in the rhabdoms of small or giant ventral photoreceptors (supplementary material Fig. S1). We also did not detect LpOps7 or LpOps8 transcripts in LE photoreceptors, eccentric cells or VE photoreceptors (supplementary material Fig. S2). These findings are consistent with the PCR (Fig. 4) and western blot results (Fig. 5), and provide further evidence that the expression of LpOps6, -7 and -8 is ME specific.
Characterization of a Limulus peropsin/RGR (LpPerOps1, accession number KM538953)
Starting with a peropsin-like fragment identified in a transcriptome of VE, we amplified and cloned full-length sequences 59% identical and 75% similar to spider peropsins from VE and CNS cDNA (Fig. 8A). We call this sequence LpPerOps1. When we screened the Limulus genome with this sequence we found another sequence that is 58% identical and 67% similar at the amino acid level. We call this sequence LpPerOps2. An alignment of the predicted amino acid sequences of LpPerOps1 and -2 is shown in supplementary material Fig. S3. When we probed for LpPerOps1 and LpPerOps2 with PCR in the same VE, LE and ME cDNA preparations using specific primers predicted to amplify across an intron, we obtained an anticipated 1.1 kbp LpPerOps1 product from each cDNA and confirmed the identity of each by sequencing, but no product from any of the cDNAs using LpPerOps2 primers (Fig. 9A). This suggests that LpPerOps1, but not LpPerOps2, is expressed in each eye type.
Fig. 8.
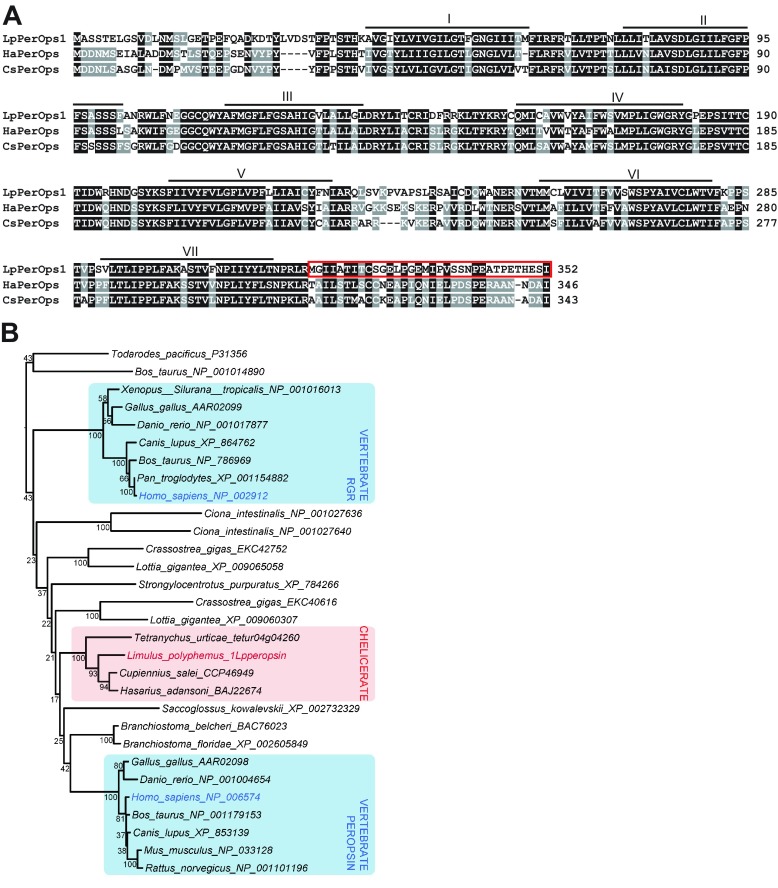
Limulus peropsin 1 (LpPerOps1) is similar to spider peropsins but the position of the chelicerate peropsin clade relative to vertebrate peropsins and RGRs cannot be resolved. (A) Clustal W alignment of LpPerOps1 with peropsins from spiders (Ha, Hasaruis adansoni; Cs Cupiennius salei). Transmembrane domains estimated from hydrophobicity plots are indicated. The region boxed in red is the polypeptide used to generate anti-LpPerOps1. (B) Maximum likelihood phylogenetic analysis of LpPerOps1 with 27 members of the RGR/peropsin clade (Hering and Mayer, 2014) plus two out groups – Bos Taurus c-opsin (Palczewski et al., 2000) and Todarodes r-opsin (Murakami and Kouyama, 2008). Bootstrap values from 1000 pseudoreplications are shown at nodes.
Fig. 9.
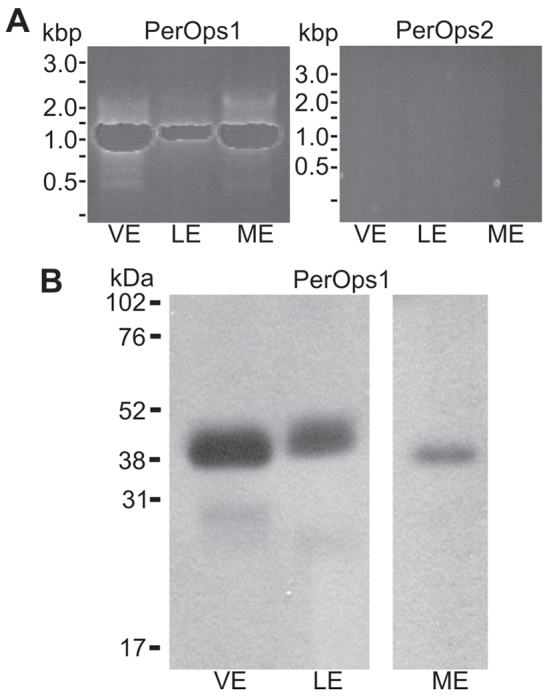
Transcripts encoding LpPerOps1, but not LpPerOps2, are detected in VE, LE and ME cDNA, and LpPerOps1 is present in membrane preparations from each eye type. (A) cDNA prepared from VE, LE and ME was probed for LpPerOps1 using primers to amplify the full-length transcript (LpPerOps1F6/R5, supplementary material Table S1) and an appropriately sized product (1.1 kbp) was amplified from each. The identity of the product was confirmed by sequencing. No product was amplified from the same cDNA preparations when probed for LpPerOps2 using primers (LpPerOps2 F4/R4, supplementary material Table S1) based on the genomic sequence. Additional PCR reactions with different primer pairs based on the genomic sequence also produced no product. (B) Western blots of membrane preparations from the equivalent of 2.5 VE, 0.2 LE and 2.5 ME were immunostained with anti-LpPerOps1 (1:250 dilution). A single major immunostained band with an apparent molecular mass of about 40 kDa was observed in preparations from each eye type.
Our phylogenetic analysis was conducted to test the relationship of LpPerOps1 with other presumptive peropsins from chelicerates, the only arthropods in which peropsin-like sequences have been detected, and the peropsins and RGR (RPE-retinal G-protein-coupled receptor) proteins from deuterostomes and molluscs (Fig. 8B). The Limulus peropsin/RGR-like sequence forms a clade with the three peropsins from chelicerates (100%), and is sister to the two spider peropsins (93%). Our results are similar to previous analyses that cannot confidently place the chelicerate peropsin-like sequences in a specific position between peropsin and RGR.
LpPerOps1 is present in membranes of cells surrounding photoreceptors in each eye type
On western blots of membrane preparations from each Limulus eye type, a monoclonal antibody directed against the C-terminus of LpPerOps1 (anti-LpPerOps1) immunostained a single band with an apparent molecular mass of about 40 kDa, which is close to the predicted molecular mass of 39.3 kDa for LpPerOps1 (Fig. 9B). These bands were eliminated when we pre-incubated the antibody with antigen (not shown), indicating that the immunostaining is specific. These results show that LpPerOps1 is expressed in VE, LE and ME membranes.
Low power images of VE end organs revealed LpPerOps1-ir in cells surrounding photoreceptor cell bodies identified by Gqα-ir in rhabdoms (Fig. 10, VE). In LE cross-sections, we detected LpPerOps1-ir in the location of pigment cells that surround photoreceptors (Fig. 10, LE). In longitudinal sections of ME (Fig. 10, ME), we observed LpPerOps1-ir in cells that surround the photoreceptor layer and in partitions between photoreceptor clusters. Higher power views of the rhabdomeral layer, which is located just below the lens, also show extensive LpPerOps1-ir between rhabdoms. No LpPerOps1-ir was detected with antibody we had pre-incubated with antigen (PerOps1-Abs), indicating that immunostaining is specific.
Fig. 10.
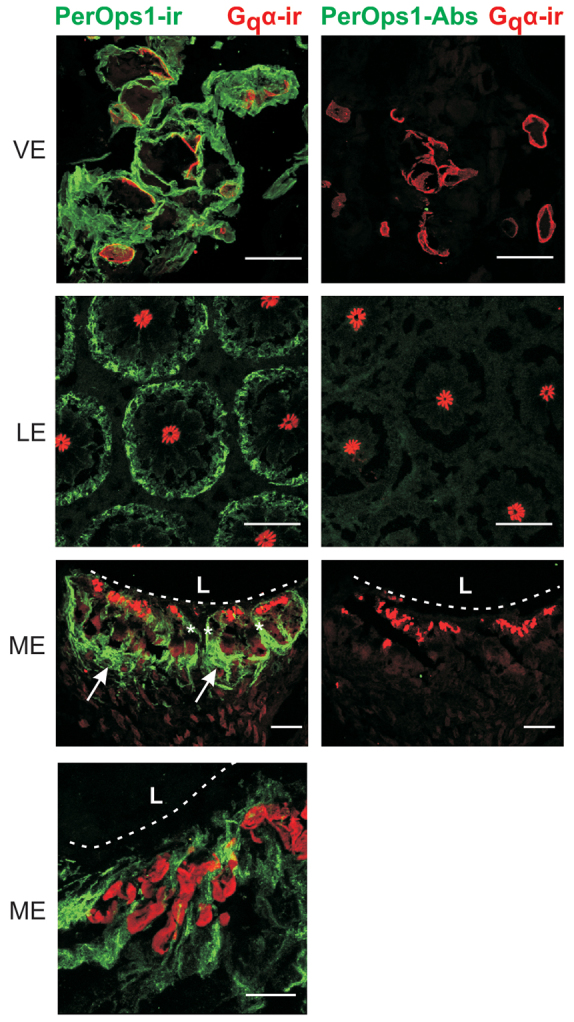
LpPerOps is expressed in cells closely associated with photoreceptors in each eye type. Sections of VE near the EO, cross-sections of LE and longitudinal sections of ME were immunostained with anti-Gqα (red, 1:1000 dilution) to visualize rhabdoms and anti-LpPerOps1 (green, 1:100 dilution) or anti-LpPerOps1 that had been incubated with antigen (PerOps1-Abs). Each image is a maximum projection (10–15 μm stacks). LpPerOps1-ir was detected in each eye type. In VE, it is in cells surrounding photoreceptor cell bodies, and in LE it is at the periphery of ommatidia where pigment cells are located. In ME, LpPerOps1-ir is in glia that surround the photoreceptor layer (arrows) and between clusters of photoreceptors (white asterisk). LpPerOps1-ir glia are also located in the rhabdomeral layer of the ME close to the lens (see high power image). Pre-incubating anti-LpPerOps1 with antigen (LpPerOp1-Abs) eliminated LpPerOps1-ir. L, lens; dashed line, base of lens. Scale bar for VE, LE and low power ME, 100 μm. Scale bar for higher power ME, 25 μm.
Figs 11 and 12 provide more detailed views of LpPerOps1 expression in VEs and LEs. In VEs, LpPerOps1 transcripts and the expressed protein are detected most intensely in glial cells surrounding photoreceptor cell bodies. The bulk of the axon is only weakly labelled (Fig. 11). Fig. 11 also shows that each photoreceptor type in the VE is surrounded by LpPerOps1-ir glia: giant photoreceptors (Fig. 11B), small photoreceptors with external rhabdoms (Fig. 11C, left panel) and small photoreceptors with internal rhabdoms (Fig. 11C, right panel).
Fig. 11.
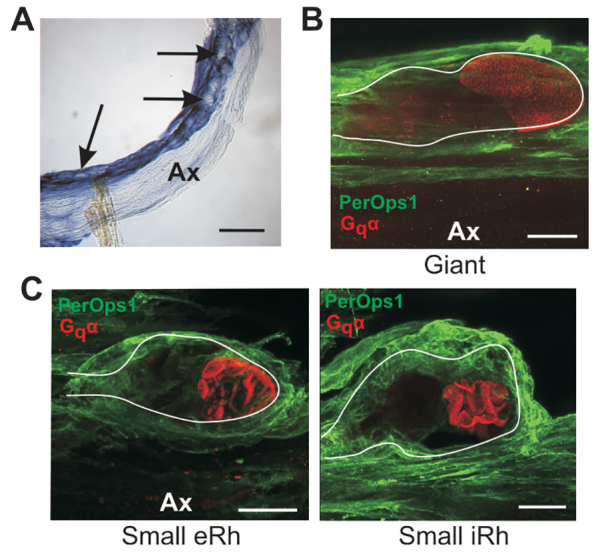
LpPerOps1 is expressed in glia surrounding giant VE photoreceptors and both types of small VE photoreceptors. (A) In situ hybridization showing the location of LpPerOps1 transcripts in a whole mount of VE nerve. Probes were visualized with an anti-digoxigenin-alkaline phosphatase antibody, followed by the NBT/BCIP reaction. The region shown is near the VE end organ. The antisense probe labelled glia surrounding clusters of photoreceptor cell bodies (arrows). Labelling was much less intense elsewhere in the axon (Ax). VE nerves incubated with sense probe were not stained. Scale bar, 500 μm. (B) Maximum projection (38 μm stack) from a giant photoreceptor cell body immunostained as described in the legend to Fig. 10 with anti-Gqα (red) to show rhabdoms and anti-LpPerOps1 (green). This cell body was located along the edge of the axon (Ax) and its approximate periphery is outlined. Glia surrounding the cell body show bright LpPerOps1-ir, whereas LpPerOps1-ir elsewhere in the axon is much less intense. Scale bar, 50 μm. (C) Maximum projections of cell bodies of two small VE photoreceptor cell bodies, one with an external rhabdom (eRh, 17 μm stack) and another with an internal rhabdom (iRh, 27 μm stack), immunostained as described in B. The approximate periphery of each cell body is outlined. Both cells are tightly surrounded by LpPerOps1-ir glia. The cell with the external rhabdom (eRh) was located along the edge of an axon (Ax) that shows little LpPerOps1-ir; the cell with the internal rhabdom (iRh) was located within a cluster of somata near the end organ. Scale bars, 25 μm.
Fig. 12.
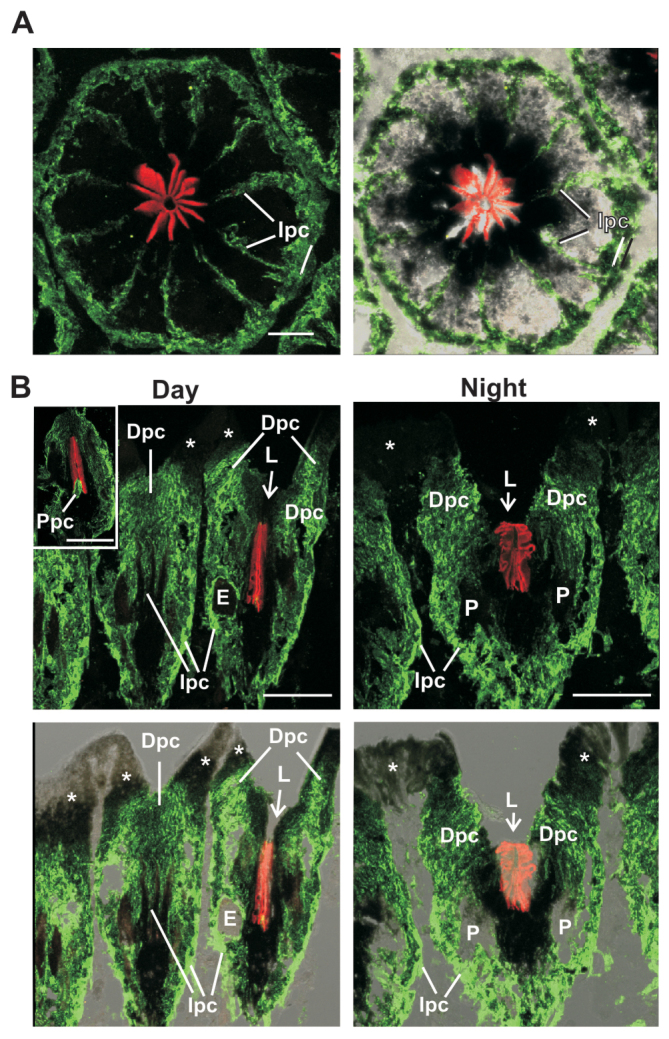
In LE, LpPerOps1 is expressed in distal, intraommatidial and proximal pigment cells. (A) Maximum projection (11 μm stack) through a cross-section of a LE ommatidium immunostained with anti-LpPerOps1 (green) and anti-Gqα (red) as described in the legend to Fig. 10. Left: fluorescent image. Right: fluorescent image superimposed on a brightfield image to show the location of pigment cells. LpPerOps1-ir is detected in pigment cells surrounding ommatidia and in intraommatidial pigment cell processes within partitions between photoreceptors. Photoreceptors are also heavily pigmented but show no LpPerOps1-ir. Scale bar, 25 μm. (B) Maximum projections of longitudinal sections of LE fixed during the day in the light (18 μm stack) or during the night in the dark (22 μm stack). Upper panels: fluorescent images. Lower panels: fluorescent images superimposed on a brightfield image of the section to show the locations of pigment cells and the aperture at the base of the lens. LpPerOps-ir is detected in each of the three types of pigment cells identified by Fahrenbach (Fahrenbach, 1968; Fahrenbach, 1969). Dpc, distal pigment cells; E, eccentric cell body; Ipc, intraommatidial pigment cells; L, position of the lens; P, photoreceptor cell body; Ppc, proximal pigment cells; arrow, aperture at the base of the lens. White asterisks indicate LpPerOps1 negative cells that line the distal lens and cornea between ommatidia.
In LEs, LpPerOps1 is expressed in pigment cells. Three types of pigment cells have been identified in LEs (Fahrenbach, 1968; Fahrenbach, 1969), and the distribution of LpPerOps1-ir (Fig. 12) suggests that it is expressed in each. A cross-section of a LE ommatidium at about mid-level shows LpPerOps1-ir in intraommatidial pigment cells that surround retinular cells at the periphery of the ommatidium and extend processes into the partitions between retinular cells (Fig. 12A). Longitudinal sections of eyes fixed during the day in the light (daytime eyes) and during the night in the dark (night-time eyes) revealed LpPerOps1-ir in distal pigment cells adjacent to where the base of the lens was located and adjacent to the aperture at the base of the lens (Fig. 12B). In daytime eyes, LpPerOps1-ir immediately adjacent to the aperture is often obscured by the high concentration of pigment at this location. However, LpPerOps1-ir is evident in this region in night-time eyes when pigment in distal pigment cells is less concentrated near the aperture (Fahrenbach, 1968). In fortuitous sections through ommatidia from daytime eyes (Fig. 12B, day: inset), we also observed bright LpPerOps1-ir at the base of the rhabdom in the location of proximal pigment cells. We did not detect LpPerOps1-ir in cells that line distal regions of the lens or the cornea between ommatidia.
DISCUSSION
In the present study we characterized three predicted visible light sensitive r-opsins (LpOps6, -7 and -8) that are expressed in MEs but not LEs or VEs, and showed that all three are co-expressed in a population of ME photoreceptors that are distinct from those expressing LpUVOps1. We also characterized a peropsin/RGR (LpPerOps1) and showed it is expressed in membranes of cells closely associated with photoreceptors in each Limulus eye type.
Ocellar-specific expression of visible light-sensitive opsins
We classify LpOps6, -7 and -8 as visible light-sensitive opsins based on their sequences and their phylogenic relationship to other arthropod opsins (Figs 2 and 3). We consider them ME-specific because they are expressed in MEs (Figs 4, 5, 6 and 7) but not in LEs or VEs (Fig. 4; supplementary material Figs S1 and S2). A separate question is whether the visible light-sensitive opsins expressed in the LEs and VEs (LpOps1-2 and -5) are expressed in MEs. A puzzling finding mentioned in the Introduction is that although we routinely detect LpOps1-2 and -5 transcripts in ME cDNA with PCR using primers that amplify across an intron (Smith et al., 1993; Katti et al., 2010), we do not detect LpOps1-2 or -5-ir in ME rhabdoms using antibodies that clearly detect these proteins in LE and VE (Katti et al. 2010; Battelle et al., 2013) (present study, Fig. 5B). There are several possible explanations for these conflicting results. Our ME RNA preparations may be contaminated by RNA from median larval eyes that are located close to the median ocelli (Fig. 1). Photoreceptors in median larval eyes, like those in ventral larval eyes, express LpOps1-2 and -5 (B.-A.B., not shown). LpOps1-2 and -5 transcripts may be expressed in ME photoreceptors but not translated, or they may be expressed at very low levels. LpOps1-2 and -5 transcripts were not detected in ME photoreceptors by in situ hybridization (B.-A.B., not shown). Despite the puzzling PCR results, our present study provides strong evidence that the visible light-sensitive opsins expressed in Limulus MEs are different from those expressed in LE and larval eyes.
Ocellar-specific expression of visible light-sensitive opsins has been described in insects (Pollock and Benzer, 1988; Spaethe and Briscoe, 2005; Velarde et al., 2005; Henze et al., 2012) and crustaceans (Oakley and Huber, 2004). The current study suggests that this opsin expression pattern is consistent among the visual systems of three of the four major groups of arthropods: insects, crustaceans and chelicerates. By contrast, in Limulus and some insects, e.g. honeybees (Velarde et al., 2005), bumblebees (Spaethe and Briscoe, 2005) and fruit flies (Pollock and Benzer, 1988), the same UV opsin is expressed in ocelli and compound eyes, whereas crickets express an ocellar-specific UV opsin (Henze et al., 2012).
The three visible light-sensitive opsins in Limulus MEs are co-expressed
Our combined immunocytochemical and in situ data (Figs 6 and 7) show that all three ME-specific, visible light-sensitive opsins identified in this study are co-expressed. These findings, taken together with previous results, show that all visible light-sensitive photoreceptors in Limulus eyes express more than one opsin. LE retinular cells and giant VE photoreceptors each express three: LpOps1, LpOps2 and LpOps5 (Katti et al., 2010); small VE photoreceptors express two: LpUVOps1 and LpOp5 (Battelle et al., 2014); and visible light-sensitive ME photoreceptors express three: LpOps6, LpOps7 and LpOps8 (the present study). Opsin co-expression seems the rule in Limulus photoreceptors rather than the exception. The sequences of some co-expressed Limulus opsins are very similar to one another (e.g. LpOps1 and LpOps2; LpOps6 and LpsOps7) and may be products of recent gene duplications, but others are from different clades (e.g. LpOps1-2 and LpOps5; LpOps5 and LpUVOps1). The evolutionary processes that have produced this complex pattern of opsin co-expession in Limulus, an animal in which there is no evidence for color vision, are not clear.
The only classical photoreceptor type in Limulus eyes in which opsin co-expression has not yet been detected is the UV photoreceptor in MEs. Our finding that none of the visible light-sensitive opsins in ME is co-expressed with LpUVOps1 (Figs 6 and 7) is consistent with electrophysiological studies that detected no ME photoreceptors with dual sensitivity to UV and visible light (Chapman and Lall, 1967; Nolte and Brown, 1969; Lall, 1970).
The functional consequences of opsin co-expression are largely unknown except in photoreceptors where the co-expressed opsins have distinctly different spectral sensitivities. In these instances opsin co-expression is thought to broaden the spectral sensitivity of the photoreceptor. A number of examples come from insect photoreceptors (Kitamoto et al., 1998; Arikawa et al., 2003; Sison-Mangus et al., 2006; Mazzoni et al., 2008; Hu et al., 2014). The small VE photoreceptors in Limulus provide another example. Small VE photoreceptors exhibit two peaks of spectral sensitivity, one at 350 nm and the other at 520 nm, and their relative sensitivity to UV and visible light probably changes with a diurnal rhythm (Battelle et al., 2014).
For photoreceptors in which spectrally similar opsins are co-expressed (Sakamoto et al., 1996; Rajkumar et al., 2010; Ogawa et al., 2012), other functional consequences must be hypothesized. In Limulus, LpOps1-2 and -5, which have similar spectral sensitivities (Battelle et al., 2014), are co-expressed in LE retinular cells and giant ventral photoreceptors (Katti et al., 2010). However, interestingly, the relative levels of LpOps1-2 and LpOps5 proteins in rhabdoms change dramatically from day to night under the influence of light and signals from an internal circadian clock (Katti et al., 2010; Battelle et al., 2013; Battelle, 2013). Changes in the relative levels of these co-expressed opsins in rhabdoms will not alter the spectral tuning of the cell, but might influence the dynamics of the photoresponse.
The individual spectral sensitivities of LpOps6, -7 and -8 are not yet known, but the spectral sensitivity of visible light-sensitive photoreceptors in MEs has been compared with those of LE retinular cells and giant VE photoreceptors (Nolte and Brown, 1969). The peak sensitivity of all three photoreceptor types is similar (about 520 nm); however, the visible light-sensitive photoreceptors in MEs are more sensitive at longer wavelengths than LE retinular cells or giant VE photoreceptors. This indicates that the combined spectral sensitivity of LpOps6, -7 and -8 peaks at about 520 nm, which is similar to LpOps1-2 and -5, but that their combined sensitivity extends further into the red. We do not yet know the relative expression levels of LpOps6, -7 and -8 in ME rhabdoms or whether their relative levels change with a diurnal rhythm.
What are the functions of UV- and visible light-sensitive photoreceptors in Limulus MEs?
Limulus clearly detect UV light with their MEs. Adult Limulus are positively phototaxic to UV light detected with their MEs (Lall and Chapman, 1973), and when MEs are stimulated with UV light, UV-sensitive photoreceptors depolarize and arhabdomeric cells, to which UV-sensitive photoreceptors are electrically coupled, produce trains of action potentials (spikes) that propagate to the brain (Nolte and Brown, 1972). Because UV light attenuates sharply with water depth, it has been speculated that the animal uses its MEs as depth detectors to help guide them to shallow water and beaches for reproduction (Lall and Chapman, 1973).
The effects of visible light on ME output are less well understood. Limulus exhibit no detectable behavioral response when MEs are stimulated with visible light (Lall and Chapman, 1973) and visible light photoreceptors are not electrically coupled to arhabdomeric cells. However, when long wavelength light is applied during a prolonged UV stimulus, arhabdomeric cells hyperpolarize and spike activity is suppressed (Nolte and Brown, 1972). Therefore, the visible component of natural light may modulate ME output that is primarily driven by UV light.
Peropsins among arthropods
Limulus has two peropsin genes, but only LpPerOps1 is expressed in the eyes. In a phylogenetic analysis, it clusters with high confidence with three other chelicerate peropsins (Fig. 8). Limulus is considered an early branching sister group to arachnids (Edgecombe and Legg, 2014); thus peropsin expression may have been a characteristic of euchelicerates. Peropsins have so far not been detected in the transcriptomes or genomes of other members of the Panarthropod group (Onychophora, Tardigrada, Pancrustaceans) (Eriksson et al., 2013; Hering and Mayer, 2014); therefore if peropsins were among the opsins in the Panarthropoda repertoire, this opsin clade has been lost in three major arthropod lineages.
Does LpPerOps1 play a role in vision?
Little is known about the functions of persopsins in vision, and they may differ depending on the species. The distributions of peropsins in eyes may provide clues, but this information is limited (Sun et al., 1997; Bailey and Cassone, 2004; Nagata et al., 2010; Eriksson et al., 2013). In the present study we demonstrate a close association between cells expressing LpPerOps1 and the cell bodies and rhabdoms of photoreceptors in each of the three, very different types of Limulus eyes (Figs 10 and 11). This leads us to propose that LpPerOps1 plays a role in vision, and it may be a retinal photoisomerase. This hypothesis is based on the similarity between LpPerOps1 and spider peropsin, a bistable photopigment that in the dark binds all-trans retinal which is converted to 11-cis retinal by light (Nagata et al., 2010). Furthermore, LpPerOps1 and other chelicerate peropsins are related to RGR proteins (Fig. 8), which are photoisomerases (Chen et al., 2001).
The proposed retinal-photoisomerase activity of LpPerOps1 may not be important for the photoresponse itself because r-opsins and peropsins are both bistable photopigments from which the chromophore is not readily released. However, a retinal-photoisomerase may be important for recycling chromophores released from rhodopsin proteins that are internalized and degraded during light-dependent rhabdom shedding (Wang et al., 2010; Wang et al., 2012). In Limulus, rhabdom shedding and renewal is dramatic. In LE retinular cells, for example, roughly 50% of rhabdomeric LpOps1-2 is shed during the day in response to light and signals from an internal circadian clock, and much appears to be degraded through a lysosomal pathway (Chamberlain and Barlow, 1979; Sacunas et al., 2002; Katti et al., 2010). However, by 4 h after sunset, the same amount of LpOps1-2 that was removed during the day is restored to rhabdoms (Battelle, 2013; Battelle et al., 2013). A similar daytime loss of r-opsins has been documented for VE photoreceptors (Katti et al., 2010). If LpPerOps1 is a photoisomerase, its location in membranes of cells immediately surrounding Limulus photoreceptors positions it appropriately to play a role in chromophore recycling, although other photoisomerases in Limulus eyes could also serve this function (Smith et al., 1992).
Other evidence suggests that peropsins are not required for chromophore recycling in all chelicerates. Spider photoreceptors, especially those in the eyes of nocturnal spiders, undergo dramatic light-dependent rhabdom shedding and night-time rhabdom renewal, similar to that described in Limulus (Blest, 1978; Uehara et al., 1993; Grusch et al., 1997). In the spider Cupiennius salei, dramatic rhabdom shedding is observed in its principal and secondary eyes (Grusch et al., 1997), yet peropsin transcripts were only detected in the secondary eyes (Eriksson et al., 2013). In the jumping spider Hasarius adansoi, peropsin is expressed in the principal eye but it is located in cells quite distant from photoreceptor cell bodies and rhabdoms, making it unlikely that it plays a significant role in chromophore recycling (Nagata et al., 2010).
LpPerOps1 could also function in signalling. In LEs, distal pigment cells surrounding the aperture at the base of the lens undergo dramatic day–night structural changes, producing an aperture that is long and narrow during the day and short and wide during the night (Chamberlain and Barlow, 1987). These changes are driven by signals from an internal circadian clock and amplified by light. The effect of the clock can be mimicked by octopamine, the neurotransmitter released from the clock-driven efferent neurons that innervate the pigment cells, and activation of a cAMP cascade (Battelle et al., 1982; Battelle and Evans, 1984; Battelle, 2002; Dalal and Battelle, 2010). The receptor responsible for the effect of light is unknown. In addition, pigment granules within distal and intraommatidial pigment cells change their distribution in response to light (Fahrenbach, 1975; Fahrenbach, 1968). LpPerOps1 could be the light receptor that drives pigment cell structural changes and pigment migration within pigment cells. Clearly much more work is required to establish a role for peropsins in vision.
In summary, this study significantly extends our understanding of the diversity of opsins expressed in Limulus eyes, preparations that have long been used for studies of basic mechanisms of vision, and contributes to our broader understanding of opsin expression in arthropods. The number of different visible light-sensitive opsins expressed in Limulus eyes is a surprise because, in the visible range, the sensitivity of each eye type peaks at about 520 nm, and unlike many other chelicerates, in Limulus, there is no evidence for color vision. These findings raise questions about evolutionary mechanisms that may have led to the diversification of Limulus opsins. The discovery of a peropsin in the eyes of Limulus, an early branching sister group to arachnids, adds to evidence that this clade of opsins has been retained in chelicerate linages while lost in other arthropods, and raises new questions regarding the functions of these opsins in vision.
MATERIALS AND METHODS
Reagents
Unless otherwise specified, reagents were purchased from Fisher Scientific (Pittsburgh, PA, USA) or Sigma-Aldrich (St Louis, MO, USA).
Opsin cloning
The full-length open reading frames (ORFs) of LpOps6 and -8 were identified in a ME transcriptome. A BLAST search of an assembly of the Limulus genome (ftp://ftp.ncbi.nlm.nih.gov/genbank/genomes/Eukaryotes/invertebrates/) with these sequences revealed an additional sequence 61% identical to LpOps6 at the amino acid level. We call this sequence LpOps7. The ORFs of all three were amplified from ME cDNA prepared from RNA isolated with RNeasy (Qiagen, Valencia, CA, USA) and reverse transcribed with SuperScript III-First Strand Synthesis System for RT-PCR (Life Technologies, Grand Island, NY, USA). Primers were specific for the 5′-end beginning with the predicted initiation methionine and the 3′-end, ending with the stop codon (supplementary material Table S1). The sequences of cloned LpOps6 and -8 clones matched exactly those identified in the transcriptome; the sequence of cloned LpOps7 matched the genomic sequence.
A 647-nucleotide fragment with high homology to the 3′-end of peropsin from jumping spider (Hasarius adansoni, accession number AB525082) was identified in a 454 transcriptome analysis of cDNAs from VE and central nervous system (CNS). The full-length ORF extending into the 3′- and 5′-UTRs was obtained using a RACE (Rapid Amplification of cDNA Ends) strategy with the VE library as template (5′ RACE primers: PerOps1-R1 and CAP followed by PerOps1-R3 and CAP; 5′ RACE primers: PerOps-F1 and TRSALu4 followed by PerOps-F2 and Lu4NS) (supplementary material Table S1). The full-length ORF with portions of the 5′- and 3′-UTRs was amplified from VE and adult CNS cDNA with primers PerOpsF6 and PerOpsR5. Three separate clones were sequenced in forward and reverse directions to obtain a consensus sequence. A BLAST search of the Limulus genome with this sequence revealed the LpPerOps1 sequence plus an additional, similar sequence we call LpPerOps2. Primers based on the predicted coding region of LpPerOps2 were used in PCR assays for this transcript in ME, LE and VE cDNA. PCR reactions were performed using LATaq polymerase (Takara, Madison, WI, USA) and an Eppendorf Mastercycler (Hauppague, NY, USA).
Phylogenetic analyses
The predicted amino acid sequences of LpOps6, -7 and -8 were aligned with previously characterized Limulus opsins and other arthropod opsins downloaded from GenBank (http://ncbi.nlm.nih.gov/Genbank/). Sequences were analysed and the graphical representation of the phylogenic tree produced as described previously (Battelle et al., 2014). Accession numbers for sequences included in this tree are in supplementary material Table S2.
The phylogenetic analysis of LpPerOps1 was conducted using the Osiris phylogenetics package (Oaklely et al., 2014) within Galaxy (Giardine et al., 2005). The predicted amino acid sequences of LpPerOps1, 27 members of the RGR/peropsin clade (Hering and Mayer, 2014) plus two out groups – Bos Taurus c-opsin (Palczewski et al., 2000) and Todarodes r-opsin (Murakami and Kouyama, 2008) both with solved crystal structures – were aligned with MUSCLE (Edgar, 2004). To search for the Maximum Likelihood phylogeny, RAxML version 7.4 (Stamatakis, 2006) was used, assuming a GTR+G model, and 100 bootstrap pseudoreplications were conducted to gauge node stability.
Tissue distribution of the opsin transcripts
PCR
Aliquots of cDNA prepared from eight MEs, one LE and eight VEs were probed for LpOps6, -7 and -8 with primers designed to amplify the full-length ORF of each. No-reverse transcriptase controls were also prepared. These cDNAs were also probed with primers specific for LpPerOps1 and -2 using primers that are predicted to amplify across exon/intron boundaries.
In situ hybridization
Antisense and sense digoxigenin or fluoroscein-labelled RNA probes were generated from the full-length coding regions of LpOps7, -8 and LpPerOps1 and also the 3′-UTR of LpOps7 with T3 or T7 RNA polymerases and labelling protocols from Roche Applied Science (Penzberg, Germany). Sense and antisense probes targeting the 3′-UTR of LpOps7, 695 nucleotides long (amplified with primers LpOps7-F6 and LpOps7-R7), were produced because LpOps7 is 70% identical to LpOps6 in the coding region. This probe was used on ME tissue where LpOps6 is also expressed to avoid possible hybridization to LpOps6 transcripts.
VEs were dissected in Limulus saline (Warren and Pierce, 1982), fixed for 2 h on ice in 4% paraformaldehyde in phosphate-buffered saline (PBS: 11.9 mmol l−1, pH 7.2, 0.5 mol l−1 NaCl) and processed for in situ hybridization as whole mounts (Jezzini et al., 2005). The probe was visualized with Fab fragments of an anti-digoxigenin-alkaline phosphatase antibody, followed by the NBT/BCIP reaction (Roche Applied Science). LEs and MEs were dissected, fixed, sectioned and processed for in situ hybridization as described previously (Battelle et al., 2014). In some experiments, antibody directed against LpOps6 was applied after the in situ protocol was complete. Sections were incubated in primary antibody overnight at 4°C and in secondary antibody for either 2 h at room temperature or overnight at 4°C.
Antibody production
LpOps6 and LpPerOps1
cDNA encoding the C-terminal sequences of LpOps6 (Q349–G396), LpOps7 (R349–A395), LpOps8 (R337–T380) (Fig. 2) and LpPerOps1 (M318–I352) (Fig. 8) were subcloned into pET28a (Novagen, EMD Chemicals, Gibbston, NJ, USA) at the HindIII and NdeI sites. Each was expressed and purified by affinity chromatography (PrepEase Histidine Tagged Protein Purification Kit, Affymetrix, Santa Clara, CA, USA). The LpOps6 and LpPerOps1 sequences were used to produce batteries of monoclonal antibodies as described previously (Katti et al., 2010; Battelle et al., 2014). The C-terminal polypeptides of LpOps7, -8 and LpUVOps1 (Battelle et al., 2014) were used to test the specificity of the antibody generated against LpOps6. Monoclonal antibody 5001-2-8 (isotype IgG1), called here anti-LpOps6, and LpPerOps1-3-50 (isotype IgG2B), called here anti-LpPerOps1, were used in all experiments.
SDS-PAGE
Proteins were separated on NuPAGE 4-12% Bis-Tris Mini Gels (Novex, Life Technologies, Grand Island, NY, USA) using the manufacturer's protocol.
Preparing membranes, quantifying antigens, western blotting and immunostaining western blots
These protocols have been described previously (Battelle et al., 2001; Katti et al., 2010). Before immunostaining, protein bands on the western blots were located by staining the blots with 0.001% Fast Green (Sigma-Aldrich) in 30% methanol and 7% acetic acid, then detained with 30% methanol and 7% acetic acid. Limulus eye membranes were prepared from daytime, light-adapted eyes.
Tissue fixation for immunostaining, immunostaining and pre-absorbing antibodies
These protocols have been detailed elsewhere (Katti et al., 2010). In addition to anti-LpOps6 and anti-LpPerOps1 described above, tissues were immunostained with a rabbit polyclonal antibody directed against Gq/11 α (C-19, Santa Cruz Biotechnology, Santa Cruz, CA, USA) (Munger et al. 1996) and a monoclonal antibody directed against LpUVOps1 (57-62, isotype IgG2A, Kappa) (Battelle et al., 2014). AlexaFluor-labelled secondary antibodies were purchased from Life Technologies.
Confocal microscopy
Fluorescent images of in situ and immunocytochemical assays were collected using a Leica confocal microscope (Leica SP5, Leica Microsystems, Mannheim, Germany). Double-labelled sections were analysed with sequential scans. Images to be compared directly were collected during a single confocal session using identical settings.
Image preparation
Images were intensified in CorelDrawX3 or Adobe Photoshop CS2 (Adobe Systems Inc., San Jose, CA, USA). Images to be compared were adjusted as a unit. Figures were assembled in CorelDrawX3.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Jennifer Blythe and Leanne Adams for technical assistance, and W. C. Smith and K. S. Alligood for helpful comments on an earlier draft of the manuscript. S. R. Saraf was a participant in the Whitney Laboratory's National Science Foundation-sponsored Research Experience for Undergraduates program.
FOOTNOTES
Competing Interests
The authors declare no competing or financial interests.
Funding
This work was supported by the National Science Foundation (IOS 1146175 and DIB 0648969 to B.-A.B., DMR-1121053 and CNS-0960316 to the University of California at Santa Barbara's Center for Scientific Computing at the California NanoSystems Institute and Materials Research Laboratory, EAGER-10457 to T.H.O. and DEB 1355230 to Douglas Eernisse, T.H.O. and D.I.S.), the National Institutes of Health (EY021721) and Research to Prevent Blindness grants to the University of Florida Ophthalmology Core Facility and the National Human Genome Research Institute (HG003079 to Richard K. Wilson, The Genome Institute at Washington University, St Louis, for providing the Limulus polyphemus 2.1.2 assembly).
Supplementary material
Supplementary material available online at http://jeb.biologists.org/lookup/suppl/doi:10.1242/jeb.116087/-/DC1
References
- Arikawa K., Mizuno S., Kinoshita M., Stavenga D. G. (2003). Coexpression of two visual pigments in a photoreceptor causes an abnormally broad spectral sensitivity in the eye of the butterfly Papilio xuthus. J. Neurosci. 23, 4527-4532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey M. J., Cassone V. M. (2004). Opsin photoisomerases in the chick retina and pineal gland: characterization, localization, and circadian regulation. Invest. Ophthalmol. Vis. Sci. 45, 769-775. [DOI] [PubMed] [Google Scholar]
- Battelle B. A., Evans J. A., Chamberlain S. C. (1982). Efferent fibers to Limulus eyes synthesize and release octopamine. Science 216, 1250-1252. [DOI] [PubMed] [Google Scholar]
- Battelle B. A. (2002). Circadian efferent input to Limulus eyes: anatomy, circuitry, and impact. Microsc. Res. Tech. 58, 345-355. [DOI] [PubMed] [Google Scholar]
- Battelle B. A. (2013). What the clock tells the eye: lessons from an ancient arthropod. Integr. Comp. Biol. 53, 144-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battelle B. A., Evans J. A. (1984). Octopamine release from centrifugal fibers of the Limulus peripheral visual system. J. Neurochem. 42, 71-79. [DOI] [PubMed] [Google Scholar]
- Battelle B. A., Dabdoub A., Malone M. A., Andrews A. W., Cacciatore C., Calman B. G., Smith W. C., Payne R. (2001). Immunocytochemical localization of opsin, visual arrestin, myosin III, and calmodulin in Limulus lateral eye retinular cells and ventral photoreceptors. J. Comp. Neurol. 435, 211-225. [DOI] [PubMed] [Google Scholar]
- Battelle B. A., Kempler K. E., Parker A. K., Gaddie C. D. (2013). Opsin1-2,G(q)α and arrestin levels at Limulus rhabdoms are controlled by diurnal light and a circadian clock. J. Exp. Biol. 216, 1837-1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battelle B.-A., Kempler K. E., Harrison A., Dugger D. R., Payne R. (2014). Opsin expression in Limulus eyes: a UV opsin is expressed in each eye type and co-expressed with a visible light-sensitive opsin in ventral larval eyes. J. Exp. Biol. 217, 3133-3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrens M. E., Fahy J. L. (1981). Slow potentials in non-spiking optic-nerve fibers in the peripheral visual-system of Limulus. J. Comp. Physiol. 141, 239-247. [Google Scholar]
- Blest A. D. (1978). Rapid synthesis and destruction of photoreceptor membrane by a dinopid spider – daily cycle. Proc. R. Soc. B 200, 463-483. [Google Scholar]
- Calman B. G., Battelle B.-A. (1991). Central origin of the efferent neurons projecting to the eyes of Limulus polyphemus. Vis. Neurosci. 6, 481-495. [DOI] [PubMed] [Google Scholar]
- Calman B. G., Chamberlain S. C. (1982). Distinct lobes of Limulus ventral photoreceptors. II. Structure and ultrastructure. J. Gen. Physiol. 80, 839-862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamberlain S. C., Barlow R. B., Jr (1979). Light and efferent activity control rhabdom turnover in Limulus photoreceptors. Science 206, 361-363. [DOI] [PubMed] [Google Scholar]
- Chamberlain S. C., Barlow R. B., Jr (1987). Control of structural rhythms in the lateral eye of Limulus: interactions of natural lighting and circadian efferent activity. J. Neurosci. 7, 2135-2144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman R. M., Lall A. B. (1967). Electroretinogram characteristics and the spectral mechanisms of the median ocellus and the lateral eye in Limulus polyphemus. J. Gen. Physiol. 50, 2267-2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P., Hao W., Rife L., Wang X. P., Shen D., Chen J., Ogden T., Van Boemel G. B., Wu L., Yang M., et al. (2001). A photic visual cycle of rhodopsin regeneration is dependent on Rgr. Nat. Genet. 28, 256-260. [DOI] [PubMed] [Google Scholar]
- Clark A. W., Millecchia R., Mauro A. (1969). The ventral photoreceptor cells of Limulus. I. The microanatomy. J. Gen. Physiol. 54, 289-309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalal J. S., Battelle B. A. (2010). Circadian regulation of Limulus visual functions: a role for octopamine and cAMP. Curr. Zool. 56, 518-536. [Google Scholar]
- Dalal J. S., Jinks R. N., Cacciatore C., Greenberg R. M., Battelle B. A. (2003). Limulus opsins: diurnal regulation of expression. Vis. Neurosci. 20, 523-534. [DOI] [PubMed] [Google Scholar]
- Edgar R. C. (2004). MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5, 113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgecombe G. D., Legg D. A. (2014). Origins and early evolution of arthropods. Palaeontology 57, 457-468. [Google Scholar]
- Eriksson B. J., Fredman D., Steiner G., Schmid A. (2013). Characterisation and localisation of the opsin protein repertoire in the brain and retinas of a spider and an onychophoran. BMC Evol. Biol. 13, 186-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahrenbach W. H. (1968). The morphology of the eyes of Limulus. I. Cornea and epidermis the compound eye. Z. Zellforsch. Mikrosk. Anat. 87, 278-291. [DOI] [PubMed] [Google Scholar]
- Fahrenbach W. H. (1969). Morphology of eyes of Limulus. 2. Ommatidia of compound eye. 93, 451-487. [DOI] [PubMed] [Google Scholar]
- Fahrenbach W. H. (1970). The morphology of the Limulus visual system. 3. The lateral rudimentary eye. Z. Zellforsch. Mikrosk. Anat. 105, 303-316. [DOI] [PubMed] [Google Scholar]
- Fahrenbach W. H. (1975). The visual system of the horseshoe crab Limulus polyphemus. Int. Rev. Cytol. 41, 285-349. [DOI] [PubMed] [Google Scholar]
- Fahrenbach W. H., Griffin A. J. (1975). The morphology of the Limulus visual system. VI. Connectivity in the ocellus. Cell Tissue Res. 159, 39-47. [DOI] [PubMed] [Google Scholar]
- Feuda R., Hamilton S. C., McInerney J. O., Pisani D. (2012). Metazoan opsin evolution reveals a simple route to animal vision. Proc. Natl. Acad. Sci. USA 109, 18868-18872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fliesler S. J. (1993). In vitro biosynthetic studies with isolated vertebrate retinas. Methods Neurosci. 15, 86-107. [Google Scholar]
- Giardine B., Riemer C., Hardison R. C., Burhans R., Elnitski L., Shah P., Zhang Y., Blankenberg D., Albert I., Taylor J., et al. (2005). Galaxy: a platform for interactive large-scale genome analysis. Genome Res. 15, 1451-1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grusch M., Barth F. G., Eguchi E. (1997). Fine structural correlates of sensitivity in the eyes of the ctenid spider, Cupiennius salei Keys. Tissue Cell 29, 421-430. [DOI] [PubMed] [Google Scholar]
- Harzsch S., Vilpoux K., Blackburn D. C., Platchetzki D., Brown N. L., Melzer R., Kempler K. E., Battelle B. A. (2006). Evolution of arthropod visual systems: development of the eyes and central visual pathways in the horseshoe crab Limulus polyphemus Linnaeus, 1758 (Chelicerata, Xiphosura). Dev. Dyn. 235, 2641-2655. [DOI] [PubMed] [Google Scholar]
- Henze M. J., Dannenhauer K., Kohler M., Labhart T., Gesemann M. (2012). Opsin evolution and expression in arthropod compound eyes and ocelli: insights from the cricket Gryllus bimaculatus. BMC Evol. Biol. 12, 163-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hering L., Mayer G. (2014). Analysis of the opsin repertoire in the tardigrade Hypsibius dujardini provides insights into the evolution of opsin genes in panarthropoda. GBE 6, 2380-2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman K. G. (1991). Two classes of Limulus ventral photoreceptors. J. Comp. Neurol. 303, 1-10. [DOI] [PubMed] [Google Scholar]
- Hu X., Leming M. T., Whaley M. A., O'Tousa J. E. (2014). Rhodopsin coexpression in UV photoreceptors of Aedes aegypti and Anopheles gambiae mosquitoes. J. Exp. Biol. 217, 1003-1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jezzini S. H., Bodnarova M., Moroz L. L. (2005). Two-color in situ hybridization in the CNS of Aplysia californica. J. Neurosci. Methods 149, 15-25. [DOI] [PubMed] [Google Scholar]
- Jiang M., Pandey S., Fong H. K. W. (1993). An opsin homologue in the retina and pigment epithelium. Invest. Ophthalmol. Vis. Sci. 34, 3669-3678. [PubMed] [Google Scholar]
- Jones C., Nolte J., Brown J. E. (1971). The anatomy of the median ocellus of Limulus. Z. Zellforsch. Mikrosk. Anat. 118, 297-309. [DOI] [PubMed] [Google Scholar]
- Katti C., Kempler K., Porter M. L., Legg A., Gonzalez R., Garcia-Rivera E., Dugger D., Battelle B. A. (2010). Opsin co-expression in Limulus photoreceptors: differential regulation by light and a circadian clock. J. Exp. Biol. 213, 2589-2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamoto J., Sakamoto K., Ozaki K., Mishina Y., Arikawa K. (1998). Two visual pigments in a single photoreceptor cell: identification and histological localization of three mRNAs encoding visual pigment opsins in the retina of the butterfly Papilio xuthus. J. Exp. Biol. 201, 1255-1261. [DOI] [PubMed] [Google Scholar]
- Lall A. B. (1970). Spectral sensitivity of intracellular responses from visual cells in median ocellus of Limulus polyphemus. Vision Res. 10, 905-909. [DOI] [PubMed] [Google Scholar]
- Lall A. B., Chapman R. M. (1973). Phototaxis in Limulus under natural conditions – evidence for reception of near-ultraviolet light in median dorsal ocellus. J. Exp. Biol. 58, 213-224. [Google Scholar]
- Mazzoni E. O., Celik A., Wernet M. F., Vasiliauskas D., Johnston R. J., Cook T. A., Pichaud F., Desplan C. (2008). Iroquois complex genes induce co-expression of rhodopsins in Drosophila. PLoS Biol. 6, e97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millecchia R., Bradbury J., Mauro A. (1966). Simple photoreceptors in Limulus polyphemus. Science 154, 1199-1201. [DOI] [PubMed] [Google Scholar]
- Munger S. D., Schremser-Berlin J. L., Brink C. M., Battelle B. A. (1996). Molecular and immunological characterization of a Gq protein from ventral and lateral eye of the horseshoe crab Limulus polyphemus. Invert. Neurosci. 2, 175-182. [DOI] [PubMed] [Google Scholar]
- Murakami M., Kouyama T. (2008). Crystal structure of squid rhodopsin. Nature 453, 363-367. [DOI] [PubMed] [Google Scholar]
- Nagata T., Koyanagi M., Terakita A. (2010). Identification and characterization of a protostome homologue of peropsin from a jumping spider. J. Comp. Physiol. A 196, 51-59. [DOI] [PubMed] [Google Scholar]
- Nilsson D. E., Kelber A. (2007). A functional analysis of compound eye evolution. Arthropod Struct. Dev. 36, 373-385. [DOI] [PubMed] [Google Scholar]
- Nolte J., Brown J. E. (1969). The spectral sensitivities of single cells in the median ocellus of Limulus. J. Gen. Physiol. 54, 636-649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolte J., Brown J. E. (1970). The spectral sensitivities of single receptor cells in the lateral, median, and ventral eyes of normal and white-eyed Limulus. J. Gen. Physiol. 55, 787-801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolte J., Brown J. E. (1972). Electrophysiological properties of cells in the median ocellus of Limulus. J. Gen. Physiol. 59, 167-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakley T. H., Huber D. R. (2004). Differential expression of duplicated opsin genes in two eyetypes of ostracod crustaceans. J. Mol. Evol. 59, 239-249. [DOI] [PubMed] [Google Scholar]
- Oakley T. H., Alexandrou M. A., Ngo R., Pankey M. S., Churchill C. K. C., Chen W., Lopker K. B. (2014). Osiris: accessible and reproducible phylogenetic and phylogenomic analyses within the Galaxy workflow management system. BMC Bioinformatics 15, 230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa Y., Awata H., Wakakuwa M., Kinoshita M., Stavenga D. G., Arikawa K. (2012). Coexpression of three middle wavelength-absorbing visual pigments in sexually dimorphic photoreceptors of the butterfly Colias erate. J. Comp. Physiol. A 198, 857-867. [DOI] [PubMed] [Google Scholar]
- Palczewski K., Kumasaka T., Hori T., Behnke C. A., Motoshima H., Fox B. A., Le Trong I., Teller D. C., Okada T., Stenkamp R. E., et al. (2000). Crystal structure of rhodopsin: a G protein-coupled receptor. Science 289, 739-745. [DOI] [PubMed] [Google Scholar]
- Plachetzki D. C., Degnan B. M., Oakley T. H. (2007). The origins of novel protein interactions during animal opsin evolution. PLoS ONE 2, e1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollock J. A., Benzer S. (1988). Transcript localization of four opsin genes in the three visual organs of Drosophila; RH2 is ocellus specific. Nature 333, 779-782. [DOI] [PubMed] [Google Scholar]
- Porter M., Cronin T., McClellan D., Crandall K. (2007). Molecular characterization of crustacean visual pigments and the evolution of pancrustacean opsins. Mol. Biol. Evol. 24, 253-68. [DOI] [PubMed] [Google Scholar]
- Porter M. L., Blasic J. R., Bok M. J., Cameron E. G., Pringle T., Cronin T. W., Robinson P. R. (2012). Shedding new light on opsin evolution. Proc. Biol. Sci. 279, 3-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajkumar P., Rollmann S. M., Cook T. A., Layne J. E. (2010). Molecular evidence for color discrimination in the Atlantic sand fiddler crab, Uca pugilator. J. Exp. Biol. 213, 4240-4248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regier J. C., Shultz J. W., Zwick A., Hussey A., Ball B., Wetzer R., Martin J. W., Cunningham C. W. (2010). Arthropod relationships revealed by phylogenomic analysis of nuclear protein-coding sequences. Nature 463, 1079-1083. [DOI] [PubMed] [Google Scholar]
- Sacunas R. B., Papuga M. O., Malone M. A., Pearson A. C., Jr, Marjanovic M., Stroope D. G., Weiner W. W., Chamberlain S. C., Battelle B. A. (2002). Multiple mechanisms of rhabdom shedding in the lateral eye of Limulus polyphemus. J. Comp. Neurol. 449, 26-42. [DOI] [PubMed] [Google Scholar]
- Sakamoto K., Hisatomi O., Tokunaga F., Eguchi E. (1996). Two opsins from the compound eye of the crab Hemigrapsus sanguineus. J. Exp. Biol. 199, 441-450. [DOI] [PubMed] [Google Scholar]
- Salcedo E., Zheng L., Phistry M., Bagg E. E., Britt S. G. (2003). Molecular basis for ultraviolet vision in invertebrates. J. Neurosci. 23, 10873-10878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sison-Mangus M. P., Bernard G. D., Lampel J., Briscoe A. D. (2006). Beauty in the eye of the beholder: the two blue opsins of lycaenid butterflies and the opsin gene-driven evolution of sexually dimorphic eyes. J. Exp. Biol. 209, 3079-3090. [DOI] [PubMed] [Google Scholar]
- Smith W. C., Friedman M. A., Goldsmith T. H. (1992). Retinoids in the lateral eye of Limulus: evidence for a retinal photoisomerase. Vis. Neurosci. 8, 329-336. [DOI] [PubMed] [Google Scholar]
- Smith W. C., Price D. A., Greenberg R. M., Battelle B. A. (1993). Opsins from the lateral eyes and ocelli of the horseshoe crab, Limulus polyphemus. Proc. Natl. Acad. Sci. USA 90, 6150-6154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spaethe J., Briscoe A. D. (2005). Molecular characterization and expression of the UV opsin in bumblebees: three ommatidial subtypes in the retina and a new photoreceptor organ in the lamina. J. Exp. Biol. 208, 2347-2361. [DOI] [PubMed] [Google Scholar]
- Speiser D. I., Pankey M. S., Zaharoff A. K., Battelle B. A., Bracken-Grissom H. D., Breinholt J. W., Bybee S. M., Cronin T. W., Garm A., Patel N. H., et al. (2014). Using Phylogenetically-Informed Annotation (PIA) to search for light-interacting genes in transcriptomes from non-model organisms. BMC Bioinformatics 15, 350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. (2006). RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22, 2688-2690. [DOI] [PubMed] [Google Scholar]
- Sun H., Gilbert D. J., Copeland N. G., Jenkins N. A., Nathans J. (1997). Peropsin, a novel visual pigment-like protein located in the apical microvilli of the retinal pigment epithelium. Proc. Natl. Acad. Sci. USA 94, 9893-9898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uehara A., Uehara K., Ogawa K. (1993). Efferent fibers and daily rhabdomal changes in the anteromedial eye of the liphistiid spider, Heptathela kimurai. Cell Tissue Res. 272, 517-522. [Google Scholar]
- Velarde R. A., Sauer C. D., Walden K. K. O., Fahrbach S. E., Robertson H. M. (2005). Pteropsin: a vertebrate-like non-visual opsin expressed in the honey bee brain. Insect Biochem. Mol. Biol. 35, 1367-1377. [DOI] [PubMed] [Google Scholar]
- Wang X., Wang T., Jiao Y., von Lintig J., Montell C. (2010). Requirement for an enzymatic visual cycle in Drosophila. Curr. Biol. 20, 93-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Wang T., Ni J. D., von Lintig J., Montell C. (2012). The Drosophila visual cycle and de novo chromophore synthesis depends on rdhB. J. Neurosci. 32, 3485-3491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren M. K., Pierce S. K. (1982). Two cell-volume regulatory systems in the Limulus myocardium – an interaction of ions and quaternary ammonium-compounds. Biol. Bull. 163, 504-516. [Google Scholar]
- Waterman T. H., Wiersma C. A. G. (1954). The functional relation between retinal cells and optic nerve in Limulus. J. Exp. Zool. 126, 59-85. [Google Scholar]
- Yokoyama S. (2000). Molecular evolution of vertebrate visual pigments. Prog. Retin. Eye Res. 19, 385-419. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.