Abstract
The Maf family of transcription factors is characterized by a typical bZip structure; these transcription factors act as important regulators of the development and differentiation of many organs and tissues, including the kidney. The Maf family consists of two subgroups that are characterized according to their structure: large Maf transcription factors and small Maf transcription factors. The large Maf subgroup consists of four proteins, designated as MAFA, MAFB, c-MAF and neural retina-specific leucine zipper. In particular, MAFA is a distinct molecule that has been attracting the attention of researchers because it acts as a strong transactivator of insulin, suggesting that Maf transcription factors are likely to be involved in systemic energy homeostasis. In this review, we focused on the regulation of glucose/energy balance by Maf transcription factors in various organs.
Keywords: Cell, Insulin, MAFA, Microarray, siRNA
Core tip: This manuscript demonstrates that Maf transcription factors are likely to be involved in the regulation of hormonal systems related to glucose metabolism, with regulation by Maf transcription factors likely occurring near the start of the cascade or acting directly on the expression of genes in coordination with other factors in multiple organs and tissues. The Maf family plays diverse roles as transcription factors in the establishment of energy balance in peripheral organs, such as the pancreas, liver, and adipose tissue.
INTRODUCTION
Maf is a family of oncogenes that were first discovered in the genome of the avian transforming retrovirus, AS42[1,2]. Maf-related proteins have been identified in many species and exhibit a universally recognized DNA binding site, enabling the proteins to act as transcription factors. These Maf transcription factors are well known to play active roles in many organs, tissues, and cells for the development, differentiation and establishment of specific functions, including effects in the pancreas[3], lens[4], myeloma cells[5], and cartilage[6,7].
The Maf family has two distinct subgroups that are categorized according to their molecular size: small Maf transcription factors (150-160 amino acids: MAFF, MAFG, and MAFK), and large Maf transcription factors (240-340 amino acids: MAFA, MAFB, c-MAF, and NRL). Small Maf transcription factors lack a transactivation domain, and these protein products form homodimers or heterodimers within the Maf family or with other transcription factors, inducing transactivation factors[2,8]. A complex regulatory network is known to link small Maf transcription factors with other regulatory proteins[9,10]. On the other hand, large Maf proteins consist of a family of transcription factors characterized by a typical bZip structure, which is a motif for protein dimerization and DNA binding[11,12]. Several reports have revealed that Maf proteins are involved in the essential functions of developing, differentiating and establishing the function of cells, tissues and organs. These transcription factors reportedly regulate several distinct developmental processes, cell differentiation, and the establishment of cell functions; for example, the mouse Mafb gene is responsible for the segmentation of the hindbrain[13], while c-Maf has been identified in the liver, renal tubules[14], adipocytes, and muscle. In this review, we will mainly focus on large Maf transcription factors and their roles in the regulation of various organs, as well as their effects on energy balance.
MAF TRANSCRIPTION FACTORS AND PANCREATIC β CELLS
One of the large Maf transcription factors, transcription factor MAFA, is an interesting molecule among the Maf family members since it promotes the differentiation of pancreatic β cells[15,16]. Several reports have also indicated that Mafa activates the insulin gene C1 element, contributing to β cell function and differentiation[17,18]. The formation of β cells has been described in detail in several reports and has been summarized in reviews. Two types of large Maf transcription factors, MAFA and transcription factor MAFB, are known to coordinate with each other and with other transcription factors and related genes to induce the generation and differentiation of β cells[3,19]. MAFB is known to function as a transcription factor in many tissues and organs and has been detected in the pancreas. MAFB was initially identified as a transactivator of β cells, acting on the glucagon gene G1 element. Further studies subsequently revealed that MAFB can be detected in both α and β cells during the early phase of development, followed by a reduction in expression and then a switch to mainly MAFA expression[20-22]. An additional study has demonstrated that the loss of Mafa causes a decrease in insulin gene expression in glucotoxic β cells[23], while MAFA deficient mice could not activate insulin transcription, even though the insulin content of the β cells was not significantly diminished[24]. Recently, Hang et al[25] described the collaboration of MAFA and MAFB in the development of pancreatic β cells in greater detail[25,26]. As for the transcription factor c-MAF, which is known to play a role in hematopoietic cell differentiation, its expression has been confirmed in the pancreas and is thought to be involved in α cell differentiation and function[27].
Previously, Maf transcriptional factors could be shown to stained in premature and mature pancreas tissue in our report[28]. Cells that stained positive for Maf transcription factors were diffusely localized in premature pancreas tissue, with some cells exhibiting double staining. The staining pattern for each Maf protein was different: unlike, MAFA-positive cells, which exhibited a diffuse staining pattern, MAFB and c-MAF were stained prominently in the branching ducts and acinar buds. Subsequently, MAFA and MAFB were stained more intensely in the islet areas of adult pancreas tissue, suggesting that Maf transcription factors are involved in the differentiation and acquisition of pancreatic endocrine cells, coordinating with each other in some situations (Figure 1). In contrast, non-endocrine composite cells of the pancreas, such as acinar cells and ductal cells, may also be affected by several Maf transcriptional factors during their maturation and differentiation process. More interesting observation is that cells positive stained for Maf transcription factor continued to be detected not only in the islets but around the ductal and interstitial area after maturation.
Figure 1.
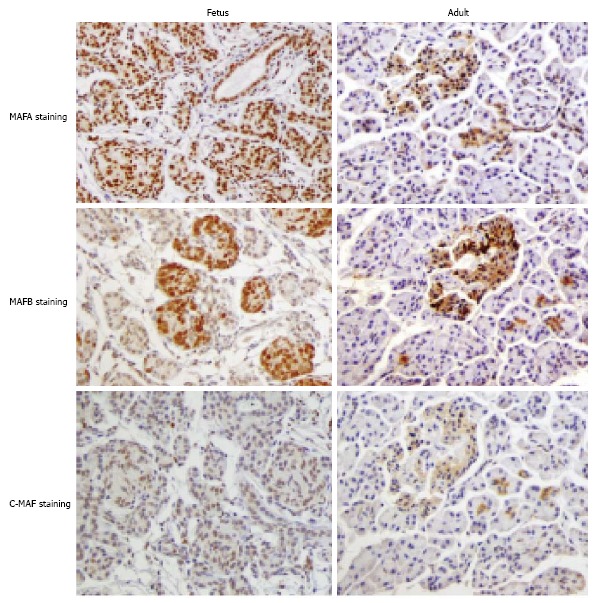
Immunostaining for MAFA, MAFB, and c-MAF in fetal and adult human pancreas tissues. An immunohistochemical analysis was performed using primary antibodies against MAFA (BL1069; Bethyl Laboratories, Inc.), MAFB (P20; Santa Cruz Biotechnology, Inc.), and c-MAF (M153; Santa Cruz Biotechnology, Inc.). The details are described in reference[28]. Samples of human normal fetal tissue (female, 20 wk, catalog No. T2244188, Lot No. A607380) and adult pancreas tissue (male, 23 years, catalog No. T2234188, Lot No. A604382) were purchased from BioChain. The fetal pancreas tissues were diffusely stained for the Maf transcription factors, and characteristic histological differences were observed between the fetal and adult tissues, with a more intense staining pattern observed in the islet areas of the adult pancreas tissue.
ACTIVITIES OF MAF TRANSCRIPTION FACTORS BEYOND β CELLS
Despite the details that have been revealed regarding the activities of Maf transcription factors in β-cell function and differentiation, the precise mechanism and coordination of Mafs and other transcriptional factors regarding the regulation of insulin production and its activity remain unknown. The precursors of pancreatic endocrine cells and the mechanism of β cell replication in the islets have been reported[29-31]. However, several types of Maf transcription factors are likely to be implicated in both the pancreatic endocrine cell lineage and interaction with other transcription factors. Each Maf transcriptional factor were often co-stained in one endocrine cell in immature pancreas.
The network of targeted genes and transcription factors, including several Maf transcription factors, needs to be clarified as part of efforts to accelerate β cell regeneration or preparation for cell therapy. Maf transcriptional factors are reportedly expressed in other tissues and cells, for example, epithelial cells and lymphocytes[32,33], where they accelerate specific cell function and differentiation. Lumelsky et al[34] reported the development of embryonic stem cells into insulin-producing cells in the pancreas[34], while Kawai et al[1] described the mechanism of β cell replication in islets. In addition, several reports have described the existence of tissue-specific stem cells in the pancreas[35,36]. Recently, several reports have discussed the more efficient production of β cells (glucose-sensitive and insulin-secreting cells) through the introduction of a combination of transcription factors, including Maf transcription factors, or the use of induced pluripotent stem cells[37,38].
Large Maf transcription factors have been identified during the development of the pancreas, and the expressions of these large Maf transcription factors exhibited different localizations in newborn and adult pancreas tissues, which differ in their endocrine characteristics. Thus, Maf transcription factors may contribute to establish all the cells in pancreatic tissue, including cells involved in endocrine cell differentiation, such as α and β cells, exocrine cells, and ductal cells.
MAF TRANSCRIPTION FACTORS AND THE KIDNEY
In the kidney, large Maf transcription factors may be implicated in both normal development and pathophy-siological processes responsible for kidney disease. We previously reported the expression profiles of large Maf transcription factors in the kidney. We have reported the expression of c-Maf mRNA levels in mouse kidney tissue from embryonic day 12 (E12) until 1 or 4 wk after birth. c-Maf mRNA was firstly expressed at E16 in the proximal tubules and continued to be expressed until 4 wk after birth. Meanwhile, MAFB expression has been identified in the glomeruli (Figure 2)[39,40].
Figure 2.
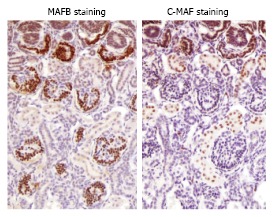
Immunostaining for MAFB and c-MAF in fetal human kidney tissue. An immunohistochemical analysis was performed using primary antibodies against MAFA (BL1069; Bethyl Laboratories, Inc.), MAFB (P20; Santa Cruz Biotechnology, Inc.), and MAF c-Maf (M153; Santa Cruz Biotechnology, Inc.). The details are described in reference[40]. A sample of human normal fetal kidney tissue (male, 25 wk) was purchased from BioChain (catalog No. T8244431, Lot No. A606275). Glomerular podocyte lesions stained positive for MAFB, and while the proximal tubules stained positive for c-MAF.
The Mafb mouse gene (also known as Kreisler and Krml) is known for its role in hindbrain patterning. Sadl et al[41] showed that mice homozygous for the kr (enu) mutation develop renal disease, in which the glomerular podocytes are affected, resulting in nephrotic syndrome. The fusion and effacement of the podocyte foot processes were observed histologically, and MAFB was shown to be essential for the cellular differentiation of the podocytes. Since the podocytes of the kr (enu) homozygotes differentiated abnormally, the homozygotes exhibited proteinuria, as is observed in nephrotic syndrome. The authors speculated that MAFB acted during the final stages of glomerular development, i.e., the transition between the capillary loop and the mature stages, and downstream of the Pod1 basic domain helix-loop-helix transcription factor[41]. In Mafb-knockout mice, renal dysgenesis with abnormal podocyte differentiation and tubular apoptosis were prominent, accompanied by the suppression of F4/80 expression in mature macrophages[42].
A prominent phenotypic feature of c-Maf - knockout mice is a small cell volume of the kidney proximal tubules and hepatocytes[40]. The precise mechanism underlying this dysregulation of cell structure formation has not been clarified, but the c-Maf transcriptional factor has been suggested to contribute to the embryonic cell development and differentiation of at least the proximal tubules and hepatocytes. The mRNA expression profile in kidney tissue from Maf-knockout mice, as evaluated using a DNA microarray, showed that the plasma level of glutathione peroxidase 3 (GPx-3) was predominantly downregulated. Since GPx-3 is an antioxidant enzyme, C-MAF may be related to the antioxidant system mediating the modulation of GPx-3 in the kidney[14]. Recently, c-Maf-inducing protein (also known as c-Mip; protein designation, CMIP), a pleckstrin homology (PH) and leucine-rich repeat (LRR)-domain-containing protein, has been identified; CMIP inactivates GSKbeta and interacts with RelA, a key member of the NF-kappaB family. Interestingly, the expression of CMIP (c-Maf-inducing protein) was increased in the podocytes of patients with idiopathic nephrotic syndromes[43]. Membrane nephropathy is characterized by nephrotic-range proteinuria in clinical and subepithelial deposits of immune complex in the basement membrane of the glomerulus. The primary cause of the disease has not been clarified, but antibodies against podocytes located on the outer layer of the basement membrane of the glomeruli form complexes that lead to deposits. A recent report has shown that CMIP was overexpressed in podocytes in an experimental glomerulonephritis rat model exhibiting heavy proteinuria and membranous nephropathy in human. This overexpression was suppressed by im-munological treatment resulting in a reduction of proteinuria; thus, while the role and significance of CMIP in podocytes and how it induces massive proteinuria have not yet been elucidated, CMIP or c-MAF-related transcriptional activities may deregulate podocyte function and cause proteinuria[44].
The expression of MAFA in the kidney is uncertain; however, based on the UniGene databank, human MAFA is also expressed in the kidney, lung, and blood. One interesting report described transgenic mice with a disease in which the hybridized gene complex resulted in MAFA deficiency and MAFK overproduction in pancreatic β cells. The phenotype of these transgenic mice was severe diabetes with large amount of proteinuria. A histological examination showed a reduction in β cells in the pancreas and typical histological diabetic nephropathy accompanied by a characteristic nodular lesion in the glomeruli.
The combination of several Maf dysfunctions generate diabetes with diabetic nephropathy, suggesting one possible mechanism for the onset of diabetic nephropathy. Consequently, these mouse models mice may suggest a mechanism for disease onset and could be useful in investigations of treatments for diabetic nephropathy[45].
MAF TRANSCRIPTION FACTORS AND THE CENTRAL NERVOUS SYSTEM
Energy balance in humans is well regulated by multiple organized systems, and the central nervous system (CNS) is likely to be involved and to play important roles in these systems[46]. The CNS contributes to the maintenance of energy balance in part by controlling feeding behavior and also by changing biological conditions and the homeostasis of intra-body conditions. Systemic adjustments of the metabolic state are achieved by the coordination of the CNS and peripheral effector organs. In the CNS, transcription factors are involved in the regulation of behavior and intra-body biological conditions[47]. Calorie intake is sensed in the CNS, altering the expression of signal transduction-mediating transcription factors. These responses are then translated into intra-CNS hormones (resulting in changes in eating behavior) and peripheral hormones take out including insulin and leptin, which function to regulate energy balance by direct effects on peripheral organs in coordination with calorie intake and consumption balance.
In our previous mouse study, the Mafa mRNA level was significantly downregulated in brain tissue in vivo, as observed using a siRNA technique, and changes in the gene profile in the CNS were screened[48] (Figure 3). The results showed distinct effects on gene expressions in brain tissue, and some of the affected genes were related to eating behavior and energy consumption, such as growth hormone and arginine vasopressin. In addition, several interesting genes and related gene products were identified in this profiling. Pro-melanin-concentrating hormone (Pro-MCH) regulates body weight[49], and changes in its expression can alter the susceptibility to fat metabolism[50,51]. Orexin is a topical neuronal peptide that regulates arousal and sleep[52], and defective orexin producing neuronal cells cause narcolepsy. Orexin may work in the brain-gut network, which regulates appetite during wakefulness[53]. In addition, pro-opiomelanocortin-alpha (an alpha-melanocyte stimulating hormone) and gastrin-releasing peptide, which are important neuropeptides regulating eating behavior and modifying the excretion of several hormones required for food digestion, have also been implicated in the above-mentioned network.
Figure 3.
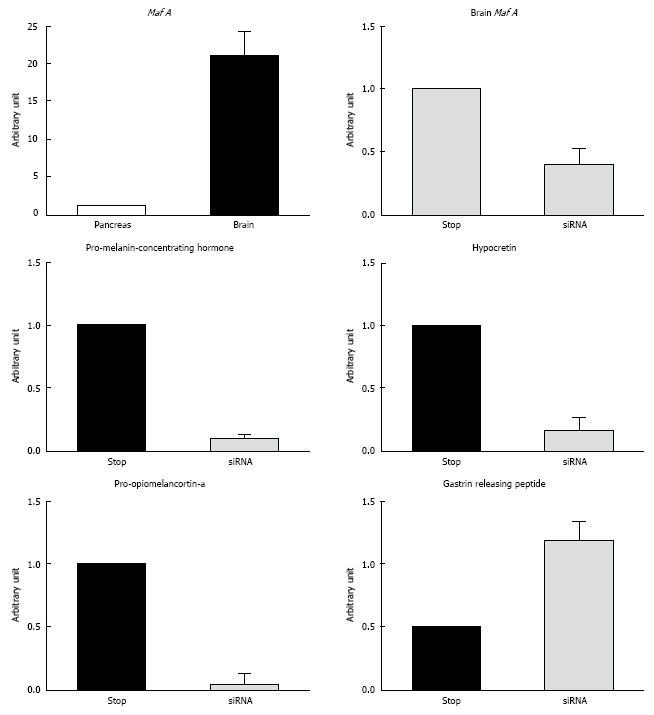
Suppression of MafA mRNA by siRNA in the brain and resulting alternation of related genes. A designed small interfering RNA (siRNA) oligomer for mouse Mafa was intravenously injected using the hydrodynamic method according to a procedure described by Hamar et al[58]. A DNA microarray analysis was then performed using Affymetrix GeneChip technology. The mRNA levels were quantified using real-time PCR. The details of the experiment have been described previously. Expression level of Mafa mRNA in the brain. The expression level of Mafa mRNA in the brain was 20 times higher than that of Mafa mRNA in the pancreas, as assessed using real-time PCR. Suppression of Mafa in mice using siRNA in the brain. The mRNA expression level take out in the brain tissue are shown. The Mafa mRNA expression level was significantly downregulated by the siRNA. Pro-melanin-concentrating hormone, Hypocretin, and Pro-opiomelanocortin-a were downregulated, and Gastrin-releasing peptide was upregulated, as assessed using real-time PCR with specific primers.
Thus, MAFA is a strong transactivator of insulin in peripheral organs and pancreas, while the modulation of MAFA mRNA expression in the CNS induces change in related genes resulting in upregulation and downregulation of neuropeptides that influence appetite, behavior, arousal, and sleep.
MAF TRANSCRIPTION FACTORS AND ADIPOCYTES
Adipocytes develop from mesenchymal stem cells in adipose tissue and various other tissues. Mesenchymal stem cells destined to become adipocytes develop and differentiate into mature adipocytes as a result of transcriptional regulation. PPARγ is known to coordinate with members of the C/EBP family to exert well-documented and important functions at different time points during adipocyte differentiation[54]. Siersbæk et al[55] also reported transcriptional networks for adipogenesis take out in which two waves of transcriptional cascades composed the adipogenetic pathway. Maf transcription factors are likely to be involved in this process, and Serria et al[33] reported that the expression of c-MAF is downregulated during 3T3-L1 cell differentiation and proliferation. Furthermore, an age-related decrease in the expression of c-MAF in mesenchymal cells has been reported, and present evidence indicates that c-MAF regulates mesenchymal cell bifurcation into osteoblasts and adipocytes. A role of c-MAF in osteogenesis and adipogenesis was also observed in c-Maf-knockout mice[56].
As discussed previously, MAFA may be involved in the differentiation of both adipocytes and pancreatic β cells. To explore the role of MAFA in adipose tissue, alterations in the expressions of MAFA-related genes in a cultured adipocyte cell line, 3T3-L1, were observed after Mafa mRNA interference had been induced[57]. Mafa mRNA suppression induced morphological changes in 3T3-L1 cells during differentiation. As shown in Figure 4, the cytoplasm of spindle-shape cells expanded after differentiation and lipid droplets formed in mature adipocytes, as revealed by the presence of red droplets of Oil Red O stain in the cytoplasm. This morphological change was not observed during Mafa siRNA suppression, and no expansion of the cytoplasm was observed. Since lipid droplet formation is essential for adipocyte differentiation, MAFA may play a critical role in the process of adipocyte differentiation. The expression levels of peroxisome proliferator-activated receptor (PPARγ2) and CCAAT/enhancer-binding protein (C/EBPα) were recognized as being essential for the differentiation and function of 3T3-L1 cells. PPARγ 2 plays a leading role in the synthesis and accumulation of lipid droplets in adipocytes, and C/EBPα is critical for the establishment of insulin sensitivity[54]. At the molecular level, the mRNA expression levels of the PPARγ gene or the C/EBP gene, which encode master adipogenic transcription factors, were markedly suppressed by Mafa-siRNA treatment, i.e., by the suppression of MAFA expression. In conclusion, adiopocyte differentiation and formation is regulated by a network of multiple transcription factors, and Maf transcription factors are likely to be involved, in coordination with other transcription factors.
Figure 4.
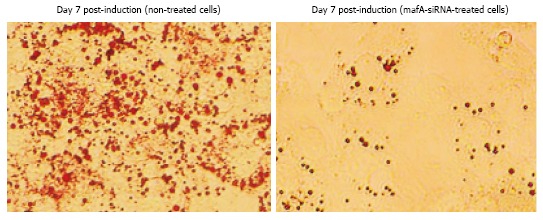
Comparison between histological changes and the Oil-Red-O staining of stop-mafA-siRNA- and mafA-siRNA-treated cells. Mouse 3T3-L1 pre-adipocytes were induced to differentiate, and Mafa SiRNA was transfected using a transfection reagent. The morphological appearances of the pre-adipocyte culture before induction and 7 d after induction were then compared. The morphology of the 3T3-L1 cells was directly observed, and lipid droplets were stained using Oil Red O. Oil Red O staining was compared between untreated and Mafa-siRNA-treated cells. Intracellular lipid staining was not observed in the Mafa-siRNA-treated cells.
ACKNOWLEDGMENTS
The authors are grateful to Atsuko Teraoka, Mayuko Tsuboi and Ai Munekawa for their technical assistance.
Footnotes
P- Reviewer: Yamaguchi M S- Editor: Ji FF L- Editor: A E- Editor: Lu YJ
Supported by A Grant-in-Aid for Scientific Research of Japan (C; 17591443: Tsuchiya M, C; 26461243: Tsuchiya K).
Conflict-of-interest: The authors have no conflict of interest directly relevant to the content of this article.
Open-Access: This article is an open-access article which was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution Non Commercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: http://creativecommons.org/licenses/by-nc/4.0/
Peer-review started: July 19, 2014
First decision: September 4, 2014
Article in press: December 10, 2014
References
- 1.Kawai S, Goto N, Kataoka K, Saegusa T, Shinno-Kohno H, Nishizawa M. Isolation of the avian transforming retrovirus, AS42, carrying the v-maf oncogene and initial characterization of its gene product. Virology. 1992;188:778–784. doi: 10.1016/0042-6822(92)90532-t. [DOI] [PubMed] [Google Scholar]
- 2.Nishizawa M, Kataoka K, Goto N, Fujiwara KT, Kawai S. v-maf, a viral oncogene that encodes a “leucine zipper” motif. Proc Natl Acad Sci USA. 1989;86:7711–7715. doi: 10.1073/pnas.86.20.7711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Matsuoka TA, Zhao L, Artner I, Jarrett HW, Friedman D, Means A, Stein R. Members of the large Maf transcription family regulate insulin gene transcription in islet beta cells. Mol Cell Biol. 2003;23:6049–6062. doi: 10.1128/MCB.23.17.6049-6062.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sakai M, Imaki J, Yoshida K, Ogata A, Matsushima-Hibaya Y, Kuboki Y, Nishizawa M, Nishi S. Rat maf related genes: specific expression in chondrocytes, lens and spinal cord. Oncogene. 1997;14:745–750. doi: 10.1038/sj.onc.1200869. [DOI] [PubMed] [Google Scholar]
- 5.Chesi M, Bergsagel PL, Shonukan OO, Martelli ML, Brents LA, Chen T, Schröck E, Ried T, Kuehl WM. Frequent dysregulation of the c-maf proto-oncogene at 16q23 by translocation to an Ig locus in multiple myeloma. Blood. 1998;91:4457–4463. [PubMed] [Google Scholar]
- 6.MacLean HE, Kim JI, Glimcher MJ, Wang J, Kronenberg HM, Glimcher LH. Absence of transcription factor c-maf causes abnormal terminal differentiation of hypertrophic chondrocytes during endochondral bone development. Dev Biol. 2003;262:51–63. doi: 10.1016/s0012-1606(03)00324-5. [DOI] [PubMed] [Google Scholar]
- 7.Omoteyama K, Ikeda H, Imaki J, Sakai M. Activation of connective tissue growth factor gene by the c-Maf and Lc-Maf transcription factors. Biochem Biophys Res Commun. 2006;339:1089–1097. doi: 10.1016/j.bbrc.2005.11.119. [DOI] [PubMed] [Google Scholar]
- 8.Fujiwara KT, Kataoka K, Nishizawa M. Two new members of the maf oncogene family, mafK and mafF, encode nuclear b-Zip proteins lacking putative trans-activator domain. Oncogene. 1993;8:2371–2380. [PubMed] [Google Scholar]
- 9.Blank V. Small Maf proteins in mammalian gene control: mere dimerization partners or dynamic transcriptional regulators? J Mol Biol. 2008;376:913–925. doi: 10.1016/j.jmb.2007.11.074. [DOI] [PubMed] [Google Scholar]
- 10.Kannan MB, Solovieva V, Blank V. The small MAF transcription factors MAFF, MAFG and MAFK: current knowledge and perspectives. Biochim Biophys Acta. 2012;1823:1841–1846. doi: 10.1016/j.bbamcr.2012.06.012. [DOI] [PubMed] [Google Scholar]
- 11.Kataoka K, Nishizawa M, Kawai S. Structure-function analysis of the maf oncogene product, a member of the b-Zip protein family. J Virol. 1993;67:2133–2141. doi: 10.1128/jvi.67.4.2133-2141.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kataoka K. Multiple mechanisms and functions of maf transcription factors in the regulation of tissue-specific genes. J Biochem. 2007;141:775–781. doi: 10.1093/jb/mvm105. [DOI] [PubMed] [Google Scholar]
- 13.Giudicelli F, Gilardi-Hebenstreit P, Mechta-Grigoriou F, Poquet C, Charnay P. Novel activities of Mafb underlie its dual role in hindbrain segmentation and regional specification. Dev Biol. 2003;253:150–162. doi: 10.1006/dbio.2002.0864. [DOI] [PubMed] [Google Scholar]
- 14.Shirota S, Yoshida T, Sakai M, Kim JI, Sugiura H, Oishi T, Nitta K, Tsuchiya K. Correlation between the expression level of c-maf and glutathione peroxidase-3 in c-maf -/- mice kidney and c-maf overexpressed renal tubular cells. Biochem Biophys Res Commun. 2006;348:501–506. doi: 10.1016/j.bbrc.2006.07.111. [DOI] [PubMed] [Google Scholar]
- 15.Olbrot M, Rud J, Moss LG, Sharma A. Identification of beta-cell-specific insulin gene transcription factor RIPE3b1 as mammalian MafA. Proc Natl Acad Sci USA. 2002;99:6737–6742. doi: 10.1073/pnas.102168499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kataoka K, Han SI, Shioda S, Hirai M, Nishizawa M, Handa H. MafA is a glucose-regulated and pancreatic beta-cell-specific transcriptional activator for the insulin gene. J Biol Chem. 2002;277:49903–49910. doi: 10.1074/jbc.M206796200. [DOI] [PubMed] [Google Scholar]
- 17.Matsuoka TA, Artner I, Henderson E, Means A, Sander M, Stein R. The MafA transcription factor appears to be responsible for tissue-specific expression of insulin. Proc Natl Acad Sci USA. 2004;101:2930–2933. doi: 10.1073/pnas.0306233101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhao L, Guo M, Matsuoka TA, Hagman DK, Parazzoli SD, Poitout V, Stein R. The islet beta cell-enriched MafA activator is a key regulator of insulin gene transcription. J Biol Chem. 2005;280:11887–11894. doi: 10.1074/jbc.M409475200. [DOI] [PubMed] [Google Scholar]
- 19.Kataoka K, Shioda S, Ando K, Sakagami K, Handa H, Yasuda K. Differentially expressed Maf family transcription factors, c-Maf and MafA, activate glucagon and insulin gene expression in pancreatic islet alpha- and beta-cells. J Mol Endocrinol. 2004;32:9–20. doi: 10.1677/jme.0.0320009. [DOI] [PubMed] [Google Scholar]
- 20.Artner I, Le Lay J, Hang Y, Elghazi L, Schisler JC, Henderson E, Sosa-Pineda B, Stein R. MafB: an activator of the glucagon gene expressed in developing islet alpha- and beta-cells. Diabetes. 2006;55:297–304. doi: 10.2337/diabetes.55.02.06.db05-0946. [DOI] [PubMed] [Google Scholar]
- 21.Artner I, Blanchi B, Raum JC, Guo M, Kaneko T, Cordes S, Sieweke M, Stein R. MafB is required for islet beta cell maturation. Proc Natl Acad Sci USA. 2007;104:3853–3858. doi: 10.1073/pnas.0700013104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hang Y, Stein R. MafA and MafB activity in pancreatic β cells. Trends Endocrinol Metab. 2011;22:364–373. doi: 10.1016/j.tem.2011.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harmon JS, Stein R, Robertson RP. Oxidative stress-mediated, post-translational loss of MafA protein as a contributing mechanism to loss of insulin gene expression in glucotoxic beta cells. J Biol Chem. 2005;280:11107–11113. doi: 10.1074/jbc.M410345200. [DOI] [PubMed] [Google Scholar]
- 24.Zhang C, Moriguchi T, Kajihara M, Esaki R, Harada A, Shimohata H, Oishi H, Hamada M, Morito N, Hasegawa K, et al. MafA is a key regulator of glucose-stimulated insulin secretion. Mol Cell Biol. 2005;25:4969–4976. doi: 10.1128/MCB.25.12.4969-4976.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hang Y, Yamamoto T, Benninger RK, Brissova M, Guo M, Bush W, Piston DW, Powers AC, Magnuson M, Thurmond DC, et al. The MafA transcription factor becomes essential to islet β-cells soon after birth. Diabetes. 2014;63:1994–2005. doi: 10.2337/db13-1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hu He K, Juhl K, Karadimos M, El Khattabi I, Fitzpatrick C, Bonner-Weir S, Sharma A. Differentiation of pancreatic endocrine progenitors reversibly blocked by premature induction of MafA. Dev Biol. 2014;385:2–12. doi: 10.1016/j.ydbio.2013.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gosmain Y, Marthinet E, Cheyssac C, Guérardel A, Mamin A, Katz LS, Bouzakri K, Philippe J. Pax6 controls the expression of critical genes involved in pancreatic {alpha} cell differentiation and function. J Biol Chem. 2010;285:33381–33393. doi: 10.1074/jbc.M110.147215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tsuchiya M, Taniguchi S, Yasuda K, Nitta K, Maeda A, Shigemoto M, Tsuchiya K. Potential roles of large mafs in cell lineages and developing pancreas. Pancreas. 2006;32:408–416. doi: 10.1097/01.mpa.0000220867.64787.99. [DOI] [PubMed] [Google Scholar]
- 29.Dor Y, Brown J, Martinez OI, Melton DA. Adult pancreatic beta-cells are formed by self-duplication rather than stem-cell differentiation. Nature. 2004;429:41–46. doi: 10.1038/nature02520. [DOI] [PubMed] [Google Scholar]
- 30.Ramiya VK, Maraist M, Arfors KE, Schatz DA, Peck AB, Cornelius JG. Reversal of insulin-dependent diabetes using islets generated in vitro from pancreatic stem cells. Nat Med. 2000;6:278–282. doi: 10.1038/73128. [DOI] [PubMed] [Google Scholar]
- 31.Soria B, Roche E, Berná G, León-Quinto T, Reig JA, Martín F. Insulin-secreting cells derived from embryonic stem cells normalize glycemia in streptozotocin-induced diabetic mice. Diabetes. 2000;49:157–162. doi: 10.2337/diabetes.49.2.157. [DOI] [PubMed] [Google Scholar]
- 32.Agnello D, Lankford CS, Bream J, Morinobu A, Gadina M, O’Shea JJ, Frucht DM. Cytokines and transcription factors that regulate T helper cell differentiation: new players and new insights. J Clin Immunol. 2003;23:147–161. doi: 10.1023/a:1023381027062. [DOI] [PubMed] [Google Scholar]
- 33.Serria MS, Ikeda H, Omoteyama K, Hirokawa J, Nishi S, Sakai M. Regulation and differential expression of the c-maf gene in differentiating cultured cells. Biochem Biophys Res Commun. 2003;310:318–326. doi: 10.1016/j.bbrc.2003.08.144. [DOI] [PubMed] [Google Scholar]
- 34.Lumelsky N, Blondel O, Laeng P, Velasco I, Ravin R, McKay R. Differentiation of embryonic stem cells to insulin-secreting structures similar to pancreatic islets. Science. 2001;292:1389–1394. doi: 10.1126/science.1058866. [DOI] [PubMed] [Google Scholar]
- 35.Yalniz M, Pour PM. Are there any stem cells in the pancreas? Pancreas. 2005;31:108–118. doi: 10.1097/01.mpa.0000174939.97438.9f. [DOI] [PubMed] [Google Scholar]
- 36.Soria B, Bedoya FJ, Martin F. Gastrointestinal stem cells. I. Pancreatic stem cells. Am J Physiol Gastrointest Liver Physiol. 2005;289:G177–G180. doi: 10.1152/ajpgi.00116.2005. [DOI] [PubMed] [Google Scholar]
- 37.Ham DS, Shin J, Kim JW, Park HS, Cho JH, Yoon KH. Generation of functional insulin-producing cells from neonatal porcine liver-derived cells by PDX1/VP16, BETA2/NeuroD and MafA. PLoS One. 2013;8:e79076. doi: 10.1371/journal.pone.0079076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang L, Huang Y, Guo Q, Fan X, Lu Y, Zhu S, Wang Y, Bo X, Chang X, Zhu M, et al. Differentiation of iPSCs into insulin-producing cells via adenoviral transfection of PDX-1, NeuroD1 and MafA. Diabetes Res Clin Pract. 2014;104:383–392. doi: 10.1016/j.diabres.2014.03.017. [DOI] [PubMed] [Google Scholar]
- 39.Imaki J, Onodera H, Tsuchiya K, Imaki T, Mochizuki T, Mishima T, Yamashita K, Yoshida K, Sakai M. Developmental expression of maf-1 messenger ribonucleic acids in rat kidney by in situ hybridization histochemistry. Biochem Biophys Res Commun. 2000;272:777–782. doi: 10.1006/bbrc.2000.2865. [DOI] [PubMed] [Google Scholar]
- 40.Imaki J, Tsuchiya K, Mishima T, Onodera H, Kim JI, Yoshida K, Ikeda H, Sakai M. Developmental contribution of c-maf in the kidney: distribution and developmental study of c-maf mRNA in normal mice kidney and histological study of c-maf knockout mice kidney and liver. Biochem Biophys Res Commun. 2004;320:1323–1327. doi: 10.1016/j.bbrc.2004.05.222. [DOI] [PubMed] [Google Scholar]
- 41.Sadl V, Jin F, Yu J, Cui S, Holmyard D, Quaggin S, Barsh G, Cordes S. The mouse Kreisler (Krml1/MafB) segmentation gene is required for differentiation of glomerular visceral epithelial cells. Dev Biol. 2002;249:16–29. doi: 10.1006/dbio.2002.0751. [DOI] [PubMed] [Google Scholar]
- 42.Moriguchi T, Hamada M, Morito N, Terunuma T, Hasegawa K, Zhang C, Yokomizo T, Esaki R, Kuroda E, Yoh K, et al. MafB is essential for renal development and F4/80 expression in macrophages. Mol Cell Biol. 2006;26:5715–5727. doi: 10.1128/MCB.00001-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang SY, Kamal M, Dahan K, Pawlak A, Ory V, Desvaux D, Audard V, Candelier M, BenMohamed F, Matignon M, et al. c-mip impairs podocyte proximal signaling and induces heavy proteinuria. Sci Signal. 2010;3:ra39. doi: 10.1126/scisignal.2000678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sendeyo K, Audard V, Zhang SY, Fan Q, Bouachi K, Ollero M, Rucker-Martin C, Gouadon E, Desvaux D, Bridoux F, et al. Upregulation of c-mip is closely related to podocyte dysfunction in membranous nephropathy. Kidney Int. 2013;83:414–425. doi: 10.1038/ki.2012.426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shimohata H, Yoh K, Fujita A, Morito N, Ojima M, Tanaka H, Hirayama K, Kobayashi M, Kudo T, Yamagata K, et al. MafA-deficient and beta cell-specific MafK-overexpressing hybrid transgenic mice develop human-like severe diabetic nephropathy. Biochem Biophys Res Commun. 2009;389:235–240. doi: 10.1016/j.bbrc.2009.08.124. [DOI] [PubMed] [Google Scholar]
- 46.Arch JR. Central regulation of energy balance: inputs, outputs and leptin resistance. Proc Nutr Soc. 2005;64:39–46. doi: 10.1079/pns2004407. [DOI] [PubMed] [Google Scholar]
- 47.Coppari R, Ramadori G, Elmquist JK. The role of trans-criptional regulators in central control of appetite and body weight. Nat Clin Pract Endocrinol Metab. 2009;5:160–166. doi: 10.1038/ncpendmet1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tsuchiya M, Tsuchiya K, Yasuda K, Fujita M, Takinishi A, Furukawa M, Nitta K, Maeda A. MafA is a Key Molecule in Glucose and Energy Balance in the Central Nervous System and Peripheral Organs. Int J Biomed Sci. 2011;7:19–26. [PMC free article] [PubMed] [Google Scholar]
- 49.Ludwig DS, Tritos NA, Mastaitis JW, Kulkarni R, Kokkotou E, Elmquist J, Lowell B, Flier JS, Maratos-Flier E. Melanin-concentrating hormone overexpression in transgenic mice leads to obesity and insulin resistance. J Clin Invest. 2001;107:379–386. doi: 10.1172/JCI10660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Qu D, Ludwig DS, Gammeltoft S, Piper M, Pelleymounter MA, Cullen MJ, Mathes WF, Przypek R, Kanarek R, Maratos-Flier E. A role for melanin-concentrating hormone in the central regulation of feeding behaviour. Nature. 1996;380:243–247. doi: 10.1038/380243a0. [DOI] [PubMed] [Google Scholar]
- 51.Shimada M, Tritos NA, Lowell BB, Flier JS, Maratos-Flier E. Mice lacking melanin-concentrating hormone are hypophagic and lean. Nature. 1998;396:670–674. doi: 10.1038/25341. [DOI] [PubMed] [Google Scholar]
- 52.Sakurai T, Amemiya A, Ishii M, Matsuzaki I, Chemelli RM, Tanaka H, Williams SC, Richardson JA, Kozlowski GP, Wilson S, et al. Orexins and orexin receptors: a family of hypothalamic neuropeptides and G protein-coupled receptors that regulate feeding behavior. Cell. 1998;92:573–585. doi: 10.1016/s0092-8674(00)80949-6. [DOI] [PubMed] [Google Scholar]
- 53.Kirchgessner AL. Orexins in the brain-gut axis. Endocr Rev. 2002;23:1–15. doi: 10.1210/edrv.23.1.0454. [DOI] [PubMed] [Google Scholar]
- 54.Farmer SR. Transcriptional control of adipocyte formation. Cell Metab. 2006;4:263–273. doi: 10.1016/j.cmet.2006.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Siersbæk R, Mandrup S. Transcriptional networks controlling adipocyte differentiation. Cold Spring Harb Symp Quant Biol. 2011;76:247–255. doi: 10.1101/sqb.2011.76.010512. [DOI] [PubMed] [Google Scholar]
- 56.Nishikawa K, Nakashima T, Takeda S, Isogai M, Hamada M, Kimura A, Kodama T, Yamaguchi A, Owen MJ, Takahashi S, et al. Maf promotes osteoblast differentiation in mice by mediating the age-related switch in mesenchymal cell differentiation. J Clin Invest. 2010;120:3455–3465. doi: 10.1172/JCI42528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tsuchiya M, Maeda A, Suzuki A, Yasuda K, Yoshida T, Nitta K, Tsuchiya K. Suppression of MafA mRNA with siRNA prevents adipose cell differentiation in 3T3-L1 cells. Int J Mol Med. 2009;23:725–732. doi: 10.3892/ijmm_00000186. [DOI] [PubMed] [Google Scholar]
- 58.Hamar P, Song E, Kökény G, Chen A, Ouyang N, Lieberman J. Small interfering RNA targeting Fas protects mice against renal ischemia-reperfusion injury. Proc Natl Acad Sci USA. 2004;101:14883–14888. doi: 10.1073/pnas.0406421101. [DOI] [PMC free article] [PubMed] [Google Scholar]


