Abstract
Whereas P-bodies are intimately linked to the cytoplasmic RNA decay machinery, stress granules harbor stalled translation initiation complexes that accumulate upon stress-induced translation arrest. In this Chapter, we reflect on the relationship between P-bodies and stress granules. In mammalian cells, the two structures can be clearly distinguished from each other using specific protein or RNA markers, but they also share many proteins and mRNAs. While the formation of P-bodies and stress granules is coordinately triggered by stress, their assembly appears to be regulated independently by different pathways. Under certain types of stress, P-bodies frequently dock with stress granules, and overexpressing certain proteins that localize to both structures can cause P-body/stress granule fusion. Currently available data suggest that these self-assembling compartments are controlled by flux of mRNAs within the cytoplasm, and that their assembly mirrors the translation and degradation rates of their component mRNAs.
12.1 Stress Granules Assemble When Translation Initiation Is Stalled
While P-bodies (PBs) assemble around the key enzymes of cytoplasmic RNA degradation, stress granules (SGs) assemble around essential components of the translation machinery. Heat shock or heat stress granules, characterized as reversible aggregates of ribonucleoprotein complexes containing untranslated mRNA, were initially described in 1989 in tomato cell cultures (Nover et al. 1989). In the late 1990s, reversible aggregates of mRNPs were “re-discovered” in mammalian cells (Kedersha et al. 1999) and dubbed mammalian stress granules to acknowledge the presumed connection to the plant studies. Ironically, it was recently reported that the original tomato heat stress granules do not contain mRNA after all (Weber et al. 2008) although plants can also assemble both SGs and PBs. Thus, in hindsight, the first descriptions of “modern” SGs are relatively recent (Kedersha et al. 1999, 2000, 2002).
Mammalian SGs were originally defined as large cytoplasmic mRNA aggregates that become microscopically visible when global protein synthesis is inhibited in response to different types of stress. The original definition was updated upon discovering that SGs are aggregates of stalled or abortive preinitiation complexes and associated RNA-binding proteins (RNA-BPs). Heat shock, oxidative stress, viral infection, UV irradiation, or energy depletion all cause polysomes to disassemble, owing to the inhibition of translation initiation while elongation and termination rates remain normal (Fig. 12.1a, b). Blocked initiation is most commonly driven by the phosphorylation of the translation initiation factor eIF2, a trimeric GTP-binding protein that delivers initiator tRNAiMet to the small 40S ribosomal subunit (Holcik and Sonenberg 2005). eIF2 thereby allows the initiating 40S subunit within the 48S pre-initiation complex to scan the beginning of the mRNA for the AUG start codon. When phosphorylated by one of four stress-responsive kinases on its α-subunit, eIF2 no longer dissociates from its GDP exchange factor eIF2B, and thus cannot be recharged with tRNAiMet.
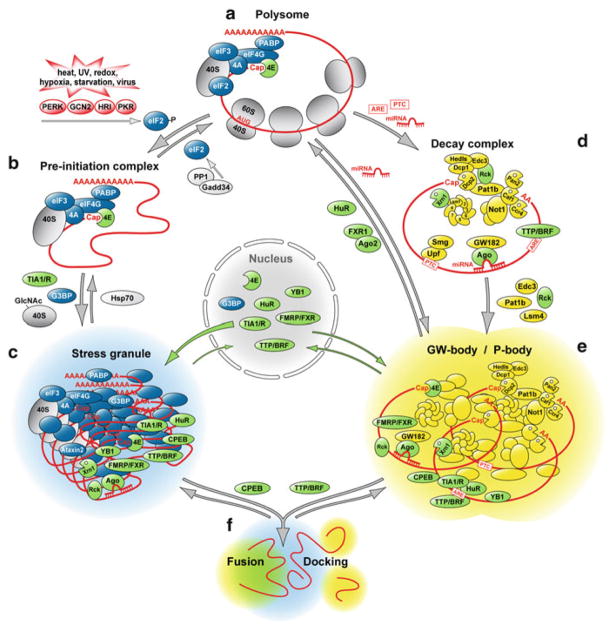
Fig. 12.1.
Scheme of mRNP complexes forming stress granules and P-bodies. (a) Actively translating mRNAs are capped, polyadenylated and form polysomes. (b) Under conditions of severe stress, global mRNA translation is inhibited by phosphorylation of eIF2 through activation of stress-responsive kinase PERK, GCN2, HRI or PKR. As a consequence, polysomes disassemble, and translation pre-initiation complexes accumulate. The protein phosphatase PP1 together with its regulatory subunit Gadd34 dephosphorylates eIF2 and thereby re-activates translation. (c) RNA-binding proteins such as TIA1 and G3BP contain aggregation-prone domains that drive the assembly of stalled pre-initiation complexes into cytoplasmic stress granules. The conjugation of N-acetylglucosamine (GlcNAc) to ribosomal proteins is also important for stress granule assembly. When stressful conditions are overcome, the chaperone Hsp70 facilitates disassembly of stress granules. (d) mRNAs are specifically degraded if they have acquired a premature termination codon (PTC), associate with miRNAs or contain AU-rich elements (AREs). Such mRNAs are either cleaved by an endonculease or subject to rapid deadenylation, which induces decapping and degradation through the 5′-3′ exoribonuclease Xrn1. (e) mRNAs targeted for decay together with key enzymes of cytoplasmic RNA degradation assemble in processing (P)-bodies. The RNA helicase Rck and aggregation-prone proteins such as Pat1, Edc3 and Lsm4 are important for P-body formation. miRNAs can also suppress the translation of target mRNAs by recruiting them to P-bodies, from where such mRNAs can also exit and re-engage in translation. (f) P-bodies are frequently observed in close physical contact to stress granules as if they were docking. The over-expression of certain RNA-binding proteins further enhances the association between P-bodies and stress granules, and causes fusion of the two structures. Such activity is observed for the cytoplasmic polyadenylation element binding protein (CPEB1) as well as for tristetraprolin (TTP) and butyrate response factor-1 (BRF1), two zinc finger proteins that accelerate the degradation of ARE-containing mRNAs. Stress granule proteins are shown in blue, P-body proteins in yellow, and proteins that localize to both structures are shown in green
The arrest of translation initiation causes ribosomes to run off their mRNAs and 48S pre-initiation complexes to accumulate (Fig. 12.1b). In a subsequent step, stalled pre-initiation complexes can then form large aggregates that become microscopically visible as SGs (Fig. 12.1c). Accordingly, SGs contain poly(A)-mRNA, 40S, but not 60S ribosomal subunits, as well as most translation initiation factors such as eIF3, eIF4A, eIF4E, eIF4G, and the cytoplasmic poly(A)-binding protein (PABP) (Kedersha et al. 2002; Kimball et al. 2003). The use of different translation inhibitors that either freeze or disassemble polysomes suggested that mRNAs in SGs are not static, but rather remain in a dynamic equilibrium with polysomal mRNA (Kedersha et al. 2000). Photobleaching studies have directly confirmed that the mRNPs within SGs are indeed in a highly dynamic flux (Kedersha et al. 2000, 2005; Mollet et al. 2008). In addition to components of the translation initiation apparatus, numerous RNA-BPs accumulate in SGs including PABP, TIA1, TIAR, FMRP, FXR1, and G3BP (Kedersha et al. 1999, 2002; Tourriere et al. 2003; Mazroui et al. 2002). The TIA proteins and G3BP contain aggregation-prone domains, which participate in the aggregation process that underlies SG assembly (Gilks et al. 2004; Tourriere et al. 2003). Ataxin-2, a protein that interacts with PABP, is also involved in SG formation (Nonhoff et al. 2007). Moreover, posttranslational modifications such as the dephosphorylation of G3BP (Tourriere et al. 2003) and the conjugation of O-linked N-acetylglucosamine to ribosomal proteins (Ohn et al. 2008) are important for SG assembly. Nevertheless, the molecular details of the actual aggregation process during SG formation are not well understood.
12.2 Stress Granules and P-Bodies Are Distinct Structures
In mammalian cells, SGs can be clearly distinguished from PBs, although both contain non-polysomal mRNPs. PBs are formed from mRNAs targeted for degradation (Sheth and Parker 2003; Cougot et al. 2004; Franks and Lykke-Andersen 2007) (Fig. 12.1d), and PB assembly is driven by a distinct set of aggregation-prone proteins that include Lsm4, Edc3 and Pat1b (Ozgur et al. 2010; Teixeira and Parker 2007; Decker et al. 2007) (Fig. 12.1e). By light microscopy, small numbers of PBs are detected in most somatic cells under normal conditions (Bashkirov et al. 1997; van Dijk et al. 2002; Ingelfinger et al. 2002; Eystathioy et al. 2002), whereas SGs only emerge in response to severe translation arrest induced by stress or energy starvation (Kedersha et al. 1999, 2000). Electron microscopy of SGs and PBs (Souquere et al. 2009; Yang et al. 2004; Gilks et al. 2004) confirms the general observations made at the light level: PBs exhibit a compact, dense substructure that may contain a fibrillar component, while SGs are larger, more irregular, looser and rather granular structures that often contain small regions of cytoplasm. Importantly, both structures lack any limiting membrane.
PBs can be distinguished from SGs because numerous proteins are specific for either of the two structures (Kedersha and Anderson 2009). On the SG side, translation factors (such as eIF3b, eIF4A, eIF4G) and RNA-BPs such as PABP and G3BP can be used as specific markers (Kedersha et al. 1999, 2002, 2005; Tourriere et al. 2003). On the PB side, components of the cytoplasmic RNA degradation machinery such as Dcp2, Dcp1 or Hedls serve as reliable marker proteins (van Dijk et al. 2002; Kedersha et al. 2005; Ozgur et al. 2010). GW-bodies share many proteins with PBs, yet seem to preferentially form around miRNA effector proteins such as GW182 and Argonaute (Eystathioy et al. 2002). Since PBs and GW-bodies are morphologically not distinct in many cases (Liu et al. 2005a, b), we will treat them as one entity in this Chapter.
Although PBs and SGs can contain the same species of mRNA (Kedersha et al. 2005), the two compartments differ with regard to the state of the mRNA they contain: In SGs, mRNAs are polyadenylated and can be easily visualized by in situ hybridization with oligo-dT probes (Kedersha et al. 1999, 2000). Moreover, the presence of eIF4G and PABP suggests that SG-associated mRNAs might be circularized. In contrast, mRNAs in PBs lack a poly(A) tail, and deadenylation is thought to be an early step in the assembly of PBs, or in the recruitment of mRNAs to pre-existing PBs (Zheng et al. 2008). This difference indicates that mRNAs in SGs are translationally stalled, but not subject to immediate degradation. In PBs, however, most mRNAs seem to be specifically primed for decay (Sheth and Parker 2003, 2006; Franks and Lykke-Andersen 2007; Cougot et al. 2004).
12.3 Parallels Between P-Bodies and Stress Granules
Despite important differences, PBs and SGs also have several features in common: (1) They are both cytoplasmic, seemingly amorphous RNA-protein aggregates that are not surrounded by any membrane as determined by electron microscopy (Gilks et al. 2004; Eystathioy et al. 2002; Souquere et al. 2009), (2) both are induced by stress conditions (Kedersha et al. 1999; Teixeira et al. 2005; Raaben et al. 2007; Wilczynska et al. 2005), (3) growth in size of both SGs and PBs depends on retrograde transport along microtubules (Loschi et al. 2009), (4) both SGs and PBs are in exchange with polysomes and contain translationally stalled mRNAs that can re-engage in translation (Bhattacharyya et al. 2006; Kedersha et al. 2000; Cougot et al. 2004), and (5) there is a large number of proteins, listed in Table 12.1, which localize to both PBs and SGs.
Table 12.1.
Proteins common to P-bodies and stress granules
| Protein | Function | References |
|---|---|---|
| Ago1 | Argonaute-1, component of the RNA-induced silencing complex | Sen and Blau (2005); Leung et al. (2006) |
| Ago2 | Argonaute-2, component of the RNA-induced silencing complex, endonuclease activity associated with siRNAs | Sen and Blau (2005); Leung et al. (2006); Pare et al. (2009); Gallois-Montbrun et al. (2007) |
| APOBEC3G | Cytidine deaminase involved in RNA editing, antiviral function | Wichroski et al. (2006); Kozak et al. (2006); Gallois-Montbrun et al. (2007) |
| CPEB1 | Cytoplasmic polyadenylation element-binding protein, inhibitor of translation, overexpression causes fusion of PBs and SGs | Wilczynska et al. (2005) |
| eIF4E | Translation initiation factor, cap-binding protein | Kedersha et al. (2005); Andrei et al. (2005) |
| FAST | TIA1-interacting protein, splicing regulator, antiapoptotic and pro-inflammatory, mostly in PBs, some in SGs | Kedersha et al. (2005) |
| FMRP/FXR | RNA-BPs involved in translational control of specific mRNAs | Vasudevan and Steitz (2007); Mazroui et al. (2002); Didiot et al. (2009); Kim et al. (2006) |
| HuR | RNA-BP, enhances mRNA stability and regulates translation | Gallouzi et al. (2000) and N.K., unpublished observations |
| Importin 8 | Required for import of Ago2 into nucleus | Weinmann et al. (2009) |
| Lin28 | RNA-BP, blocks let7 miRNA processing | Balzer and Moss (2007) |
| Lsm14 | Also Rap55, SCD6 (S. cerevisiae), Tral (D. melanogaster), CAR-1 (C. elegans), involved in cytokinesis and endoplasmic reticulum organization | Yang et al. (2006) |
| MEX3B | RNA-BP, regulator of mRNA translation and germline development in C. elegans | Courchet et al. (2008) |
| Musashi1 | RNA-BP, neuronal stem cell maintenance | Kawahara et al. (2008) |
| PCBP2 | Also hnRNP-E2, αCP2, poly(rC)-binding protein, involved in control of mRNA stability and translation | Fujimura et al. (2008) |
| Rck | Also DDX6, Dhh1 (S. cerevisiae), Me31B (D. melanogaster), SGH-1 (C. elegans), RNA helicase involved in mRNA translation | Wilczynska et al. (2005) |
| Roquin | RNA-BP and suppressor of autoimmunity, enhances mRNA decay | Athanasopoulos et al. (2010); Glasmacher et al. (2010) |
| Smaug | RNA-BP, control of mRNA translation and decay | Eulalio et al. (2007); Baez and Boccaccio (2005) |
| TIA1/TIAR | RNA-BPs, alternative splicing and inhibition of mRNA translation | Kedersha et al. (1999) Fig. 12.2 and N.K., unpublished observations |
| TTP/BRF1 | RNA-BPs, enhance mRNA decay, overexpression causes fusion of PBs and SGs | Kedersha et al. (2005) |
| Xrn1 | Exoribonuclease 1, cytoplasmic 5′-3′ exoribonuclease, mostly in PBs, some in SGs | Bashkirov et al. (1997); Kedersha et al. (2005) |
| YB1 | RNA and DNA-BP, involved in transcription, translation and mRNA stability | Yang and Bloch (2007) |
Proteins commonly observed in both compartments include the cytoplasmic cap-binding protein eIF4E (Kedersha et al. 2005; Andrei et al. 2005), the RNA helicase Rck (Wilczynska et al. 2005), which is involved in suppressing translation, and the argonaute proteins Ago1 and Ago2 (Sen and Blau 2005; Leung et al. 2006; Pare et al. 2009; Gallois-Montbrun et al. 2007) that play a central role in miRNA- and siRNA-induced silencing of mRNA expression. Several other RNA-BPs that localize to both PBs and SGs are known to control either the stability or translation rate of their target mRNAs: the cold shock domain containing nucleic acid-binding protein YB1 (Yang and Bloch 2007), the poly(rC)-binding protein PCBP2 (Fujimura et al. 2008), Roquin—an RNA-BP important for suppressing autoimmunity (Athanasopoulos et al. 2010; Glasmacher et al. 2010), Smaug—an RNA-BP essential in embryonic development (Eulalio et al. 2007; Baez and Boccaccio 2005), and CPEB1—an RNA-BP that controls cytoplasmic mRNA polyadenylation during oocyte maturation (Wilczynska et al. 2005). Dual localization was also reported for several RNA-BPs with affinity to AU-rich elements (AREs), regulatory elements that typically destabilize mRNAs or repress their translation. This includes the zinc finger proteins TTP and BRF1—both enhancers of mRNA degradation (Kedersha et al. 2005), the translation repressors TIA1 and TIAR (Kedersha et al. 1999, Fig. 12.2 and N.K., unpublished observations), FMRP and FXR1 (Vasudevan and Steitz 2007; Mazroui et al. 2002; Didiot et al. 2009; Kim et al. 2006), as well as HuR, typically a stabilizer and activator of mRNA translation (Gallouzi et al. 2000 and N.K., unpublished observations). Surprisingly, the 5′–3′ exoribonuclease Xrn1 not only localizes to PBs, where most mRNA decay enzymes are concentrated, but small amounts of Xrn1 can also be detected in SGs (Bashkirov et al. 1997; Kedersha et al. 2005). Does the localization of Xrn1 suggest that mRNA decay may also occur in SGs? For the bulk of mRNAs, this is unlikely. First, many mRNAs are stabilized under stress conditions in both yeast and mammalian cells (Fan et al. 2002; Hilgers et al. 2006; Bollig et al. 2002). Second, mRNAs in SGs have retained their poly(A) tails (Kedersha et al. 1999), suggesting that they are protected from deadenylation, which is generally the first step of mRNA degradation. Third, Xrn1 cannot degrade capped mRNA, and the decapping enzyme Dcp2 does not localize to SGs. Hence, SGs appear to serve as sites where mRNAs are protected from degradation. Two RNA-BPs, HuR and ZBP1, were in fact proposed to play an active role in stabilizing mRNAs in SGs (Gallouzi et al. 2000; Stohr et al. 2006).
Fig. 12.2.
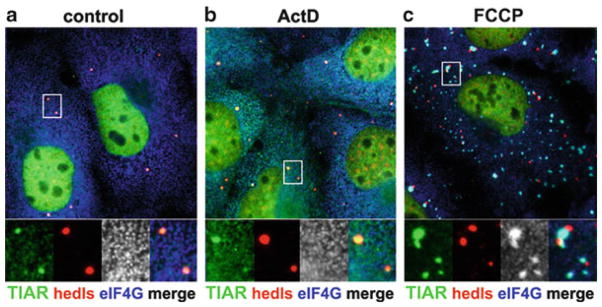
Immunofluorescence micrograph of P-bodies docking to stress granules. African green monkey COS7 kidney cells were (a) grown under normal conditions, (b) exposed to 5 μg/mL actinomycin D (ActD) for 1 h, or (c) treated with carbonyl cyanide p-trifluoromethoxyphenylhydrazone (FCCP) in glucose-free media for 1 h. Cells were then fixed and stained for TIAR (green), the stress-granule specific marker protein eIF4G (blue), and the P-body-specific protein Hedls/GE-1 (red). Actinomycin D treatment does not induce stress granules but causes TIAR accumulation at P-bodies, whereas FCCP treatment triggers stress granule formation and promotes docking of P-bodies with stress granules
So what might be the function of Xrn1 in SGs? Interestingly, the miRNA- and siRNA associated endonoclease Ago2 was found to move from PBs to SGs in response to stress (Leung et al. 2006), and it requires miRNA in order to do so. Thus, it is conceivable that the 3′ fragment generated by siRNA-induced cleavage of an mRNA is degraded by Xrn1 at SGs.
12.4 Docking and Fusion of P-Bodies with Stress Granules
With certain types of stress, one can frequently observe PBs grouped around SGs. For instance, treatment of various human cell lines with sodium arsenite, an inhibitor of the citric acid cycle and inducer of oxidative stress, causes many PBs to cluster around SGs (Wilczynska et al. 2005; Kedersha et al. 2005; Souquere et al. 2009). Similarly, the mitochondrial poison FCCP causes PBs to group around SGs in certain cell types (Fig. 12.2). In contrast, heat shock or clotrimazol, which induces energy starvation by displacing hexokinase from the mitochondrial outer membrane, induce SGs but do not cause SG-PB association. SGs induced by overexpression of SG components display transient contacts with PBs, as shown by live imaging. PBs appear to dock with SGs as they touch them for short periods of time (min) before they separate again (Kedersha et al. 2005). When examined by electron microscopy, PBs in close proximity to arsenite-induced SGs remained distinct: PBs retain their dense structure as opposed to the more granular appearance of SGs (Souquere et al. 2009).
Contacts between PBs and SGs can be dramatically stabilized by increasing the expression levels of specific proteins (Fig. 12.1f). Overexpression of TTP or BRF1, two RNA-BPs that can both target ARE-containing mRNAs to PBs for degradation (Franks and Lykke-Andersen 2007), causes tight clustering of PBs around and within SGs (Kedersha et al. 2005). Interestingly, this phenomenon is linked to the activity of these proteins: When TTP is phosphorylated at two specific serine residues that cause binding of 14-3-3 adaptor proteins, its mRNA destabilizing activity is reduced, and TTP no longer localizes to SGs (Stoecklin et al. 2004). Likewise, clustering of PBs around SGs is disrupted when TTP is phosphorylated (N.K., unpublished data). One way to interpret these data is that active TTP/BRF1 recruits stalled mRNPs in SGs for translocation into PBs. If this happens at a high rate upon overexpression of TTP/BRF1, the two compartments may literally fuse. Indeed, live microscopy reveals that PBs no longer dissociate from SGs in cells overexpressing TTP or BRF1 (Kedersha et al. 2005).
Another dual SG/PB protein, CPEB1, can also increase contacts between PBs and SGs. When CPEB1 is overexpressed, PBs are found to tightly cluster around SGs (Wilczynska et al. 2005). In some cells, PB components were observed to completely redistribute into SGs, effectively abolishing the distinction between the two compartments (Wilczynska et al. 2005). A similar observation was made after inhibiting the expression of the RNA helicase Rck. Knock down of Rck leads to the loss of PBs and, in stressed cells, causes the PB protein Dcp1 to re-localize in SGs (Serman et al. 2007). These examples of tight PB/SG clustering induced by manipulating the expression of a defined group of proteins argue that contacts between PBs and SGs do not simply result from random collisions of moving entities. Rather, it is tempting to speculate that specific proteins and/or RNAs are exchanged between PBs and SGs during such contacts.
What could be the molecular basis of PB-SG docking and fusion? One possibility is that an increased flux of mRNPs from PBs to SGs, or vice versa, may cause docking or fusion of the two compartments. The TTP/BRF1 proteins might act in this way by promoting the transfer of mRNPs from SGs to PBs for rapid deadenylation and degradation. Since TTP associates with polysomes (Brooks et al. 2002), but also binds to proteins of the decapping complex (Fenger-Gron et al. 2005), it could form transient bridges between stalled 48S-complexed mRNAs in SGs and core proteins in PBs. This model is consistent with photo-bleaching studies indicating that the interactions of TTP with SGs are very fleeting (Kedersha et al. 2005).
CPEB1 might also cause SG-PB fusion by increasing the flux of mRNPs between the two compartments. CPEB proteins recognize cytoplasmic polyadenylation elements within the 3′UTR of mRNAs, and by interacting with a poly(A) polymerase, mediate cytoplasmic polyadenylation of mRNAs both in germline and somatic cells (Richter 2007). Given that CPEB1 localizes to PBs in unstressed somatic cells, one may speculate that CPEB1 re-adenylates mRNAs in PBs. CPEB1 might thereby rescue mRNAs from degradation in PBs and promote their transfer to SGs, which could cause PB-SG fusion.
A second mechanism to explain PB-SG docking or fusion is that scaffold proteins within PBs and SGs may co-aggregate. Such co-aggregation could occur when chaperones that normally assist in dissolving aggregates, e.g., chaperones of the Hsp70 family, become limiting. Elevated expression of chaperones during heat shock could explain why SG-PB docking or fusion is typically not seen under these conditions (Kedersha et al. 2005).
A third possibility is that the cytoskeleton might connect PBs with SGs. Indeed, both SGs and PBs can be associated with the microtubule network, and motor proteins were found to influence both the assembly and movement of SGs and PBs (Loschi et al. 2009; Aizer et al. 2008). Extensive SG-PB docking induced by some mitochondrial poisons such as FCCP (Fig. 12.2) could indicate that an energy-dependent, motor-driven step is needed for their separation.
12.5 Mammalian Stress Granules and P-Bodies Form Independently
The ability of PBs and SGs to dock and fuse under certain conditions raises the question whether the two compartments are related. Since PBs exist in unstressed cells lacking SGs, it is clear that “basal” PBs do not depend on SGs for their assembly. Additional genetic data provided clear evidence that stress-induced PBs form independently of SGs. Mouse embryonic fibroblast (MEFs) expressing a non-phosphorylatable mutant of eIF2α do not form SGs under conditions of arsenite-induced oxidative stress, whereas PBs are induced several fold (Kedersha et al. 2005).
The reverse question is whether SGs are formed out of PBs under conditions of severe stress. Using video microscopy of mammalian cells, we do not observe SGs emerging at or growing out of PBs (N.K., unpublished observations), whereas other labs report that some but not all SGs appear to grow out of preexisting PBs (Mollet et al. 2008). This apparent contradiction may be due to the different marker proteins used: the Mollet study used stably expressed GFP-CPEB in HeLa cells as their SG marker, whereas our studies used stably expressed GFP-G3BP in U2OS cells. Since CPEB is present in both SGs and PBs (Wilczynska et al. 2005), its overexpression possibly enhances the interaction between nascent SGs and PBs, which G3BP does not do. Regardless, both studies agree that some SGs can and do arise independently of PBs.
Another way to address this question is to test whether SGs would still form in the absence of PBs. This experiment, however, turns out to be challenging. Knock down of Lsm4 abolishes PBs under normal growth conditions, but does not prevent the formation of either PBs or SGs under stress conditions (Kedersha et al. 2005). A surprising effect is observed with Rck, an RNA helicase generally viewed as an inhibitor of translation. Reducing expression levels of Rck very efficiently prevents the assembly of PBs under normal conditions, whereas in stressed cells, Rck knock down causes the PB-specific protein Dcp1 to re-localize in SGs (Serman et al. 2007). This would suggest that SGs do form in the absence of PBs, but that PB proteins have a tendency to co-aggregate with SG proteins when they lack factors required for canonical PB assembly. Again, the distinction between PBs and SGs becomes blurred once the experimenter starts to manipulate important regulators of translation and mRNA decay.
12.6 Con-Fusion with P-Bodies, EGP-Bodies and Stress Granules in Yeast
S. cerevisiae also forms SG-like aggregates in response to glucose deprivation (Buchan et al. 2008; Hoyle et al. 2007) or heat shock (Grousl et al. 2009). When induced by glucose deprivation, these aggregates are more accurately termed EGP-bodies because they contain eIF4E, eIF4G and Pab1, the PABP ortholog in yeast (Hoyle et al. 2007), but lack eIF3 and small ribosomal subunits, both of which are present in stalled 48S pre-initation complexes, which define metazoan SGs. Yeast EGP-bodies share similarities with mammalian SGs in that they contain polyadenylated mRNA, eIF4E, eIF4G1, eIF4G2 and Pab1, as well as Pub1, Ngr1 and Pbp1, the yeast orthologs of mammalian TIA1, TIAR and ataxin-2, respectively. The assembly of yeast EGP-bodies upon glucose removal depends on Pub1 and Pbp1 (Buchan et al. 2008), similar to the importance of the two respective orthologs, TIA1 and ataxin-2, for mammalian SG formation (Gilks et al. 2004; Nonhoff et al. 2007). There are, however, important differences between yeast EGP-bodies and mammalian SGs. eIF3 subunits are defining components of mammalian SGs (Kedersha et al. 2005), whereas eIF3 is absent from yeast EGP-bodies induced by glucose deprivation (Buchan et al. 2008; Hoyle et al. 2007). Moreover, formation of yeast EGP-bodies does not require eIF2α phosphorylation. Interestingly, severe heat shock was reported to cause eIF3 to localize to “yeast SGs” (Grousl et al. 2009), but the relationship between glucose starvation-induced EGP-bodies and heat shock-induced “yeast SGs” remains to be elucidated. Based on the localization of ribosomal protein Rps30A (Grousl et al. 2009), 40S ribosomal subunits were also proposed to localize to heat shock-induced “yeast SGs.” However, this remains to be verified, and in stark contrast to mammalian SGs, there is no clear evidence that “yeast SGs” contain either ribosomal subunits or stalled pre-initiation complexes.
What adds to the confusion is that yeast EGP-bodies show a much closer spatial connection to PBs. After glucose deprivation, about half of all EGP-bodies in yeast overlap with PBs, whereas the remaining ones do not contain PB markers (Buchan et al. 2008; Hoyle et al. 2007). Moreover, the formation of EGP-bodies is clearly stimulated by factors that are important for PB formation such as Edc3, Lsm4, Pat1 and Dhh1, the Rck ortholog in yeast (Buchan et al. 2008). The authors of this study also observed that both PBs and EGP-bodies become larger and more numerous in yeast strains lacking Dcp1 or Xrn1. These data indicate that EGP-bodies are closely related to PBs in budding yeast. Based on live imaging, one study came to the conclusion that EGP-bodies form independently of PBs (Hoyle et al. 2007), whereas another study observed that EGP bodies form next to and require pre-existing PBs (Buchan et al. 2008). The difficulties of distinguishing between “yeast SGs,” EGP-bodies and PBs in S. cerevisiae suggests that mRNP aggregates may in fact have a variable composition. According to this model, each granule is positioned within a continuum that ranges from a typical PB containing mRNA decay factors to a canonical SG comprised of stalled translation pre-initiation complexes.
12.7 Conclusions
Stress-induced assembly of SGs and PBs represents a profound reorganization of the cytoplasm. Both structures illustrate the cell’s ability to form compartments by means of transient protein aggregation. While SGs form as a result of stress-induced translation arrest and polysome disassembly, SG formation itself is not required for the inhibition of translation. Likewise, PBs harbor almost all enzymes of the basic RNA degradation machinery in the cytoplasm, yet the assembly of microscopically visible PBs is not required for mRNA degradation. Rather, SGs and PBs create cytoplasmic domains at which mRNPs remain partially assembled, possibly allowing their mRNAs to re-engage in translation more efficiently once stressful conditions are overcome. Contacts between PBs and SGs are not random collisions, but result from the specific activity of RNA-BPs exemplified by TTP, CPEB and Rck. Docking and fusion of PBs with SGs could be the result of enhanced trafficking of mRNPs between the two compartments. An important task for future research will be to visualize trafficking at the level of single mRNPs.
In addition to being hubs of mRNP trafficking, PBs and SGs may serve as platforms that allow integration of signaling events within the cytoplasm. The sequestration of signaling molecules such as RACK1 and TRAF2 in SGs indicates a role of SGs in signal transduction (Arimoto et al. 2008; Kim et al. 2005). Given the tight link between SGs, PBs and stress, we envision that protein aggregation at PBs and SGs helps in coordinating the various processes that together shape an integrated stress response: the adjustment of mRNA and protein expression to a survival mode, the re-direction of energy resources and metabolites to damage control, the activation of catabolic programs to cope with periods of starvation, and the timing as to when cell death should be induced in the event that all other strategies fail. Exploring the broader role of PBs and SGs as part of integrated stress responses will require careful dissection of the localization, activity, interactoins and functions of individual proteins that are associated with PBs and SGs.
Acknowledgments
This work was supported by young investigator grant HZ-NG-210 from the Helmholtz Gemeinschaft, and RO1 AI 033600 from the National Institutes of Health.
Contributor Information
Georg Stoecklin, Email: g.stoecklin@dkfz.de, Helmholtz Junior Research Group Posttranscriptional Control of Gene Expression, German Cancer Research Center (DKFZ), DKFZ-ZMBH Alliance, Im Neuenheimer Feld 280, Heidelberg 69120, Germany.
Nancy Kedersha, Division of Rheumatology, Immunology and Allergy, Brigham and Women’s Hospital and Harvard Medical School, One Jimmy Fund Way, Boston, MA 02115, USA.
References
- Aizer A, Brody Y, Ler LW, Sonenberg N, Singer RH, Shav-Tal Y. The dynamics of mammalian P body transport, assembly, and disassembly in vivo. Mol Biol Cell. 2008;19:4154–4166. doi: 10.1091/mbc.E08-05-0513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrei MA, Ingelfinger D, Heintzmann R, Achsel T, Rivera-Pomar R, Luhrmann R. A role for eIF4E and eIF4E-transporter in targeting mRNPs to mammalian processing bodies. RNA. 2005;11:717–727. doi: 10.1261/rna.2340405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arimoto K, Fukuda H, Imajoh-Ohmi S, Saito H, Takekawa M. Formation of stress granules inhibits apoptosis by suppressing stress-responsive MAPK pathways. Nat Cell Biol. 2008;10:1324–1332. doi: 10.1038/ncb1791. [DOI] [PubMed] [Google Scholar]
- Athanasopoulos V, Barker A, Yu D, Tan AH, Srivastava M, Contreras N, Wang J, Lam KP, Brown SH, Goodnow CC, Dixon NE, Leedman PJ, Saint R, Vinuesa CG. The ROQUIN family of proteins localizes to stress granules via the ROQ domain and binds target mRNAs. FEBS J. 2010;277:2109–2127. doi: 10.1111/j.1742-4658.2010.07628.x. [DOI] [PubMed] [Google Scholar]
- Baez MV, Boccaccio GL. Mammalian Smaug is a translational repressor that forms cytoplasmic foci similar to stress granules. J Biol Chem. 2005;280:43131–43140. doi: 10.1074/jbc.M508374200. [DOI] [PubMed] [Google Scholar]
- Balzer E, Moss EG. Localization of the developmental timing regulator Lin28 to mRNP complexes, P-bodies and stress granules. RNA Biol. 2007;4:16–25. doi: 10.4161/rna.4.1.4364. [DOI] [PubMed] [Google Scholar]
- Bashkirov VI, Scherthan H, Solinger JA, Buerstedde JM, Heyer WD. A mouse cytoplasmic exoribonuclease (mXRN1p) with preference for G4 tetraplex substrates. J Cell Biol. 1997;136:761–773. doi: 10.1083/jcb.136.4.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya SN, Habermacher R, Martine U, Closs EI, Filipowicz W. Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell. 2006;125:1111–1124. doi: 10.1016/j.cell.2006.04.031. [DOI] [PubMed] [Google Scholar]
- Bollig F, Winzen R, Kracht M, Ghebremedhin B, Ritter B, Wilhelm A, Resch K, Holtmann H. Evidence for general stabilization of mRNAs in response to UV light. Eur J Biochem. 2002;269:5830–5839. doi: 10.1046/j.1432-1033.2002.03300.x. [DOI] [PubMed] [Google Scholar]
- Brooks SA, Connolly JE, Diegel RJ, Fava RA, Rigby WF. Analysis of the function, expression, and subcellular distribution of human tristetraprolin. Arthritis Rheum. 2002;46:1362–1370. doi: 10.1002/art.10235. [DOI] [PubMed] [Google Scholar]
- Buchan JR, Muhlrad D, Parker R. P bodies promote stress granule assembly in Saccharomyces cerevisiae. J Cell Biol. 2008;183:441–455. doi: 10.1083/jcb.200807043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cougot N, Babajko S, Seraphin B. Cytoplasmic foci are sites of mRNA decay in human cells. J Cell Biol. 2004;165:31–40. doi: 10.1083/jcb.200309008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courchet J, Buchet-Poyau K, Potemski A, Bres A, Jariel-Encontre I, Billaud M. Interaction with 14-3-3 adaptors regulates the sorting of hMex-3B RNA-binding protein to distinct classes of RNA granules. J Biol Chem. 2008;283:32131–32142. doi: 10.1074/jbc.M802927200. [DOI] [PubMed] [Google Scholar]
- Decker CJ, Teixeira D, Parker R. Edc3p and a glutamine/asparagine-rich domain of Lsm4p function in processing body assembly in Saccharomyces cerevisiae. J Cell Biol. 2007;179:437–449. doi: 10.1083/jcb.200704147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Didiot MC, Subramanian M, Flatter E, Mandel JL, Moine H. Cells lacking the fragile X mental retardation protein (FMRP) have normal RISC activity but exhibit altered stress granule assembly. Mol Biol Cell. 2009;20:428–437. doi: 10.1091/mbc.E08-07-0737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulalio A, Behm-Ansmant I, Schweizer D, Izaurralde E. P-body formation is a consequence, not the cause, of RNA-mediated gene silencing. Mol Cell Biol. 2007;27:3970–3981. doi: 10.1128/MCB.00128-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eystathioy T, Chan EK, Tenenbaum SA, Keene JD, Griffith K, Fritzler MJ. A phosphorylated cytoplasmic autoantigen, GW182, associates with a unique population of human mRNAs within novel cytoplasmic speckles. Mol Biol Cell. 2002;13:1338–1351. doi: 10.1091/mbc.01-11-0544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J, Yang X, Wang W, Wood WH, 3rd, Becker KG, Gorospe M. Global analysis of stress-regulated mRNA turnover by using cDNA arrays. Proc Natl Acad Sci USA. 2002;99:10611–10616. doi: 10.1073/pnas.162212399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenger-Gron M, Fillman C, Norrild B, Lykke-Andersen J. Multiple processing body factors and the ARE binding protein TTP activate mRNA decapping. Mol Cell. 2005;20:905–915. doi: 10.1016/j.molcel.2005.10.031. [DOI] [PubMed] [Google Scholar]
- Franks TM, Lykke-Andersen J. TTP and BRF proteins nucleate processing body formation to silence mRNAs with AU-rich elements. Genes Dev. 2007;21:719–735. doi: 10.1101/gad.1494707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimura K, Kano F, Murata M. Identification of PCBP2, a facilitator of IRES-mediated translation, as a novel constituent of stress granules and processing bodies. RNA. 2008;14:425–431. doi: 10.1261/rna.780708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallois-Montbrun S, Kramer B, Swanson CM, Byers H, Lynham S, Ward M, Malim MH. Antiviral protein APOBEC3G localizes to ribonucleoprotein complexes found in P bodies and stress granules. J Virol. 2007;81:2165–2178. doi: 10.1128/JVI.02287-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallouzi IE, Brennan CM, Stenberg MG, Swanson MS, Eversole A, Maizels N, Steitz JA. HuR binding to cytoplasmic mRNA is perturbed by heat shock. Proc Natl Acad Sci USA. 2000;97:3073–3078. doi: 10.1073/pnas.97.7.3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilks N, Kedersha N, Ayodele M, Shen L, Stoecklin G, Dember LM, Anderson P. Stress granule assembly is mediated by Prion-like aggregation of TIA-1. Mol Biol Cell. 2004;15:5383–5398. doi: 10.1091/mbc.E04-08-0715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glasmacher E, Hoefig KP, Vogel KU, Rath N, Du L, Wolf C, Kremmer E, Wang X, Heissmeyer V. Roquin binds inducible costimulator mRNA and effectors of mRNA decay to induce microRNA-independent post-transcriptional repression. Nat Immunol. 2010;11:725–733. doi: 10.1038/ni.1902. [DOI] [PubMed] [Google Scholar]
- Grousl T, Ivanov P, Frydlova I, Vasicova P, Janda F, Vojtova J, Malinska K, Malcova I, Novakova L, Janoskova D, Valasek L, Hasek J. Robust heat shock induces eIF2alpha-phosphorylation-independent assembly of stress granules containing eIF3 and 40S ribosomal subunits in budding yeast, Saccharomyces cerevisiae. J Cell Sci. 2009;122:2078–2088. doi: 10.1242/jcs.045104. [DOI] [PubMed] [Google Scholar]
- Hilgers V, Teixeira D, Parker R. Translation-independent inhibition of mRNA deadenylation during stress in Saccharomyces cerevisiae. RNA. 2006;12:1835–1845. doi: 10.1261/rna.241006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holcik M, Sonenberg N. Translational control in stress and apoptosis. Nat Rev Mol Cell Biol. 2005;6:318–327. doi: 10.1038/nrm1618. [DOI] [PubMed] [Google Scholar]
- Hoyle NP, Castelli LM, Campbell SG, Holmes LE, Ashe MP. Stress-dependent relocalization of translationally primed mRNPs to cytoplasmic granules that are kinetically and spatially distinct from P-bodies. J Cell Biol. 2007;179:65–74. doi: 10.1083/jcb.200707010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingelfinger D, Arndt-Jovin DJ, Luhrmann R, Achsel T. The human LSm1-7 proteins colocalize with the mRNA-degrading enzymes Dcp1/2 and Xrnl in distinct cytoplasmic foci. RNA. 2002;8:1489–1501. [PMC free article] [PubMed] [Google Scholar]
- Kawahara H, Imai T, Imataka H, Tsujimoto M, Matsumoto K, Okano H. Neural RNA-binding protein Musashi1 inhibits translation initiation by competing with eIF4G for PABP. J Cell Biol. 2008;181:639–653. doi: 10.1083/jcb.200708004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N, Anderson P. Chapter 4 regulation of translation by stress granules and processing bodies. Prog Mol Biol Transl Sci. 2009;90C:155–185. doi: 10.1016/S1877-1173(09)90004-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha NL, Gupta M, Li W, Miller I, Anderson P. RNA-binding proteins TIA-1 and TIAR link the phosphorylation of eIF-2 alpha to the assembly of mammalian stress granules. J Cell Biol. 1999;147:1431–1442. doi: 10.1083/jcb.147.7.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N, Cho MR, Li W, Yacono PW, Chen S, Gilks N, Golan DE, Anderson P. Dynamic shuttling of TIA-1 accompanies the recruitment of mRNA to mammalian stress granules. J Cell Biol. 2000;151:1257–1268. doi: 10.1083/jcb.151.6.1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N, Chen S, Gilks N, Li W, Miller IJ, Stahl J, Anderson P. Evidence that ternary complex (eIF2-GTP-tRNA(i)(Met))-deficient preinitiation complexes are core constituents of mammalian stress granules. Mol Biol Cell. 2002;13:195–210. doi: 10.1091/mbc.01-05-0221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N, Stoecklin G, Ayodele M, Yacono P, Lykke-Andersen J, Fitzler MJ, Scheuner D, Kaufman RJ, Golan DE, Anderson P. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J Cell Biol. 2005;169:871–884. doi: 10.1083/jcb.200502088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim WJ, Back SH, Kim V, Ryu I, Jang SK. Sequestration of TRAF2 into stress granules interrupts tumor necrosis factor signaling under stress conditions. Mol Cell Biol. 2005;25:2450–2462. doi: 10.1128/MCB.25.6.2450-2462.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SH, Dong WK, Weiler IJ, Greenough WT. Fragile X mental retardation protein shifts between polyribosomes and stress granules after neuronal injury by arsenite stress or in vivo hippocampal electrode insertion. J Neurosci. 2006;26:2413–2418. doi: 10.1523/JNEUROSCI.3680-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimball SR, Horetsky RL, Ron D, Jefferson LS, Harding HP. Mammalian stress granules represent sites of accumulation of stalled translation initiation complexes. Am J Physiol Cell Physiol. 2003;284:C273–C284. doi: 10.1152/ajpcell.00314.2002. [DOI] [PubMed] [Google Scholar]
- Kozak SL, Marin M, Rose KM, Bystrom C, Kabat D. The anti-HIV-1 editing enzyme APOBEC3G binds HIV-1 RNA and messenger RNAs that shuttle between polysomes and stress granules. J Biol Chem. 2006;281:29105–29119. doi: 10.1074/jbc.M601901200. [DOI] [PubMed] [Google Scholar]
- Leung AK, Calabrese JM, Sharp PA. Quantitative analysis of Argonaute protein reveals microRNA-dependent localization to stress granules. Proc Natl Acad Sci USA. 2006;103:18125–18130. doi: 10.1073/pnas.0608845103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Rivas FV, Wohlschlegel J, Yates JR, 3rd, Parker R, Hannon GJ. A role for the P-body component GW182 in microRNA function. Nat Cell Biol. 2005a;7:1261–1266. doi: 10.1038/ncb1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Valencia-Sanchez MA, Hannon GJ, Parker R. MicroRNA-dependent localization of targeted mRNAs to mammalian P-bodies. Nat Cell Biol. 2005b;7:719–723. doi: 10.1038/ncb1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loschi M, Leishman CC, Berardone N, Boccaccio GL. Dynein and kinesin regulate stress-granule and P-body dynamics. J Cell Sci. 2009;122:3973–3982. doi: 10.1242/jcs.051383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazroui R, Huot ME, Tremblay S, Filion C, Labelle Y, Khandjian EW. Trapping of messenger RNA by Fragile X Mental Retardation protein into cytoplasmic granules induces translation repression. Hum Mol Genet. 2002;11:3007–3017. doi: 10.1093/hmg/11.24.3007. [DOI] [PubMed] [Google Scholar]
- Mollet S, Cougot N, Wilczynska A, Dautry F, Kress M, Bertrand E, Weil D. Translationally repressed mRNA transiently cycles through stress granules during stress. Mol Biol Cell. 2008;19:4469–4479. doi: 10.1091/mbc.E08-05-0499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonhoff U, Ralser M, Welzel F, Piccini I, Balzereit D, Yaspo ML, Lehrach H, Krobitsch S. Ataxin-2 interacts with the DEAD/H-box RNA helicase DDX6 and interferes with P-bodies and stress granules. Mol Biol Cell. 2007;18:1385–1396. doi: 10.1091/mbc.E06-12-1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nover L, Scharf KD, Neumann D. Cytoplasmic heat shock granules are formed from precursor particles and are associated with a specific set of mRNAs. Mol Cell Biol. 1989;9:1298–1308. doi: 10.1128/mcb.9.3.1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohn T, Kedersha N, Hickman T, Tisdale S, Anderson P. A functional RNAi screen links O-GlcNAc modification of ribosomal proteins to stress granule and processing body assembly. Nat Cell Biol. 2008;10:1224–1231. doi: 10.1038/ncb1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozgur S, Chekulaeva M, Stoecklin G. Human Pat1b connects deadenylation with mRNA decapping and controls the assembly of processing-bodies. Mol Cell Biol. 2010;30:4308–4323. doi: 10.1128/MCB.00429-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pare JM, Tahbaz N, Lopez-Orozco J, LaPointe P, Lasko P, Hobman TC. Hsp90 regulates the function of argonaute 2 and its recruitment to stress granules and P-bodies. Mol Biol Cell. 2009;20:3273–3284. doi: 10.1091/mbc.E09-01-0082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raaben M, Groot Koerkamp MJ, Rottier PJ, de Haan CA. Mouse hepatitis coronavirus replication induces host translational shutoff and mRNA decay, with concomitant formation of stress granules and processing bodies. Cell Microbiol. 2007;9:2218–2229. doi: 10.1111/j.1462-5822.2007.00951.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter JD. CPEB: a life in translation. Trends Biochem Sci. 2007;32:279–285. doi: 10.1016/j.tibs.2007.04.004. [DOI] [PubMed] [Google Scholar]
- Sen GL, Blau HM. Argonaute 2/RISC resides in sites of mammalian mRNA decay known as cytoplasmic bodies. Nat Cell Biol. 2005;7:633–636. doi: 10.1038/ncb1265. [DOI] [PubMed] [Google Scholar]
- Serman A, Le Roy F, Aigueperse C, Kress M, Dautry F, Weil D. GW body disassembly triggered by siRNAs independently of their silencing activity. Nucleic Acids Res. 2007;35:4715–4727. doi: 10.1093/nar/gkm491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheth U, Parker R. Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science. 2003;300:805–808. doi: 10.1126/science.1082320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheth U, Parker R. Targeting of aberrant mRNAs to cytoplasmic processing bodies. Cell. 2006;125:1095–1109. doi: 10.1016/j.cell.2006.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souquere S, Mollet S, Kress M, Dautry F, Pierron G, Weil D. Unravelling the ultrastructure of stress granules and associated P-bodies in human cells. J Cell Sci. 2009;122:3619–3626. doi: 10.1242/jcs.054437. [DOI] [PubMed] [Google Scholar]
- Stoecklin G, Stubbs T, Kedersha N, Wax S, Rigby WF, Blackwell TK, Anderson P. MK2-induced tristetraprolin:14-3-3 complexes prevent stress granule association and ARE-mRNA decay. EMBO J. 2004;23:1313–1324. doi: 10.1038/sj.emboj.7600163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stohr N, Lederer M, Reinke C, Meyer S, Hatzfeld M, Singer RH, Huttelmaier S. ZBP1 regulates mRNA stability during cellular stress. J Cell Biol. 2006;175:527–534. doi: 10.1083/jcb.200608071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira D, Parker R. Analysis of P-body assembly in Saccharomyces cerevisiae. Mol Biol Cell. 2007;18:2274–2287. doi: 10.1091/mbc.E07-03-0199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira D, Sheth U, Valencia-Sanchez MA, Brengues M, Parker R. Processing bodies require RNA for assembly and contain nontranslating mRNAs. RNA. 2005;11:371–382. doi: 10.1261/rna.7258505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tourriere H, Chebli K, Zekri L, Courselaud B, Blanchard JM, Bertrand E, Tazi J. The RasGAP-associated endoribonuclease G3BP assembles stress granules. J Cell Biol. 2003;160:823–831. doi: 10.1083/jcb.200212128. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- van Dijk E, Cougot N, Meyer S, Babajko S, Wahle E, Seraphin B. Human Dcp2: a catalytically active mRNA decapping enzyme located in specific cytoplasmic structures. EMBO J. 2002;21:6915–6924. doi: 10.1093/emboj/cdf678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasudevan S, Steitz JA. AU-rich-element-mediated upregulation of translation by FXR1 and Argonaute 2. Cell. 2007;128:1105–1118. doi: 10.1016/j.cell.2007.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber C, Nover L, Fauth M. Plant stress granules and mRNA processing bodies are distinct from heat stress granules. Plant J. 2008;56:517–530. doi: 10.1111/j.1365-313X.2008.03623.x. [DOI] [PubMed] [Google Scholar]
- Weinmann L, Hock J, Ivacevic T, Ohrt T, Mutze J, Schwille P, Kremmer E, Benes V, Urlaub H, Meister G. Importin 8 is a gene silencing factor that targets argonaute proteins to distinct mRNAs. Cell. 2009;136:496–507. doi: 10.1016/j.cell.2008.12.023. [DOI] [PubMed] [Google Scholar]
- Wichroski MJ, Robb GB, Rana TM. Human retroviral host restriction factors APOBEC3G and APOBEC3F localize to mRNA processing bodies. PLoS Pathog. 2006;2:e41. doi: 10.1371/journal.ppat.0020041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilczynska A, Aigueperse C, Kress M, Dautry F, Weil D. The translational regulator CPEB1 provides a link between dcp1 bodies and stress granules. J Cell Sci. 2005;118:981–992. doi: 10.1242/jcs.01692. [DOI] [PubMed] [Google Scholar]
- Yang WH, Bloch DB. Probing the mRNA processing body using protein macroarrays and “autoantigenomics”. RNA. 2007;13:704–712. doi: 10.1261/rna.411907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Jakymiw A, Wood MR, Eystathioy T, Rubin RL, Fritzler MJ, Chan EK. GW182 is critical for the stability of GW bodies expressed during the cell cycle and cell proliferation. J Cell Sci. 2004;117:5567–5578. doi: 10.1242/jcs.01477. [DOI] [PubMed] [Google Scholar]
- Yang WH, Yu JH, Gulick T, Bloch KD, Bloch DB. RNA-associated protein 55 (RAP55) localizes to mRNA processing bodies and stress granules. RNA. 2006;12:547–554. doi: 10.1261/rna.2302706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng D, Ezzeddine N, Chen CY, Zhu W, He X, Shyu AB. Deadenylation is prerequisite for P-body formation and mRNA decay in mammalian cells. J Cell Biol. 2008;182:89–101. doi: 10.1083/jcb.200801196. [DOI] [PMC free article] [PubMed] [Google Scholar]