SUMMARY
Acute myeloid leukemia (AML) is an aggressive and lethal blood cancer maintained by rare populations of leukemia stem cells (LSCs). Selective targeting of LSCs is a promising approach for treating AML and preventing relapse following chemotherapy, and developing such therapeutic modalities is a key priority. Here, we show that targeting telomerase activity eradicates AML LSCs. Genetic deletion of the telomerase subunit Terc in a retroviral mouse AML model induces cell cycle arrest and apoptosis of LSCs, and depletion of telomerase-deficient LSCs is partially rescued by p53 knockdown. Murine Terc−/− LSCs express a specific gene expression signature that can be identified in human AML patient cohorts and is positively correlated with patient survival following chemotherapy. In xenografts of primary human AML, genetic or pharmacological inhibition of telomerase targets LSCs, impairs leukemia progression, and delays relapse following chemotherapy. Together, these results establish telomerase inhibition as an effective strategy for eliminating AML LSCs.
INTRODUCTION
Acute myeloid leukemia (AML) is a highly prevalent and lethal blood cancer. The 5-year overall survival is less than 45% for patients below 60 years and less than 10% for patients older than 60 (Szer, 2012). Leukemia stem cells (LSCs) are key mediators for chemotherapy resistance and relapse in AML (Gentles et al., 2010; Ishikawa et al., 2007). LSCs are defined functionally by having the ability to initiate, maintain and serially propagate AML, and to differentiate into committed progeny that lack this ability (Bonnet and Dick, 1997; Krivtsov et al., 2006; Lane and Gilliland, 2010). Therefore, depleting LSCs represents a key therapeutic strategy to prevent relapse and improve the long-term outcomes of AML therapy.
LSCs possess limitless self-renewal that is engendered by oncogenic activation of numerous pathways including the HoxA cluster (Krivtsov et al., 2006), Wnt-beta catenin (Heidel et al., 2012; Wang et al., 2010), or through telomerase activation (Gessner et al., 2010). The telomerase holoenzyme consists of a reverse transcriptase subunit (TERT), an RNA template subunit (TERC), and a protective shelterin scaffold. In the absence of telomerase activity and alternative telomere lengthening pathways, cellular division results in the loss of telomere sequences, telomere uncapping and finally in the activation of cellular checkpoints that are similar to those induced by DNA double-stranded breaks (Celli and de Lange, 2005; Okamoto et al., 2013).
Many human cancers, including AML, are characterized by robust telomerase activity and shortened telomeres relative to the normal cellular counterpart (Aalbers et al., 2013; Bernard et al., 2009; Drummond et al., 2005; Gessner et al., 2010), and a requirement of telomerase has recently been described for the development of chronic myeloid leukemia induced by BCR-ABL (Vicente-Duenas et al., 2012). These findings have identified telomerase as a potential therapeutic target in cancer, and have motivated the development of telomerase inhibitors. The 13-mer antisense oligonucleotide imetelstat (Geron Corporation, CA) is a competitive inhibitor that binds to the RNA template (TERC) of the telomerase holoenzyme and has shown in vitro efficacy in a number of tumor models (Herbert et al., 2005). Imetelstat has recently entered clinical trials for the treatment of myeloproliferative neoplasms with remarkable efficacy (Tefferi et al, American Soc. Hematol. 2013, Abstract 662). Interestingly, mutations in telomerase have been described in patients with AML (Aalbers et al., 2013; Calado et al., 2009) and constitutional marrow failure associated with genetic telomeropathies have a high risk of developing leukemias (Kirwan and Dokal, 2008), suggesting that telomere shortening may also predispose to malignancy.
Targeting AML LSCs through telomerase inhibition is an attractive proposition, however it has not yet been determined whether telomerase inhibition is effective in LSCs. Moreover, telomerase inhibition could potentially cause further genomic instability and increased mutagenesis, allowing the emergence of adaptive changes (Hu et al., 2012). To determine whether telomerase is required for AML LSC function, we performed functional LSC analysis in well-characterized murine models of AML in the presence or absence of telomerase. Furthermore, using xenograft transplantation assays, we examined the effects of genetic and pharmacological inhibition of telomerase in human AML in vivo. This study provides strong evidence that telomerase is essential for the maintenance of LSC function, and identifies the modulation of telomerase as a novel LSC-specific therapy.
RESULTS
Telomerase deficiency impairs in vitro self-renewal and delays AML onset in murine AML
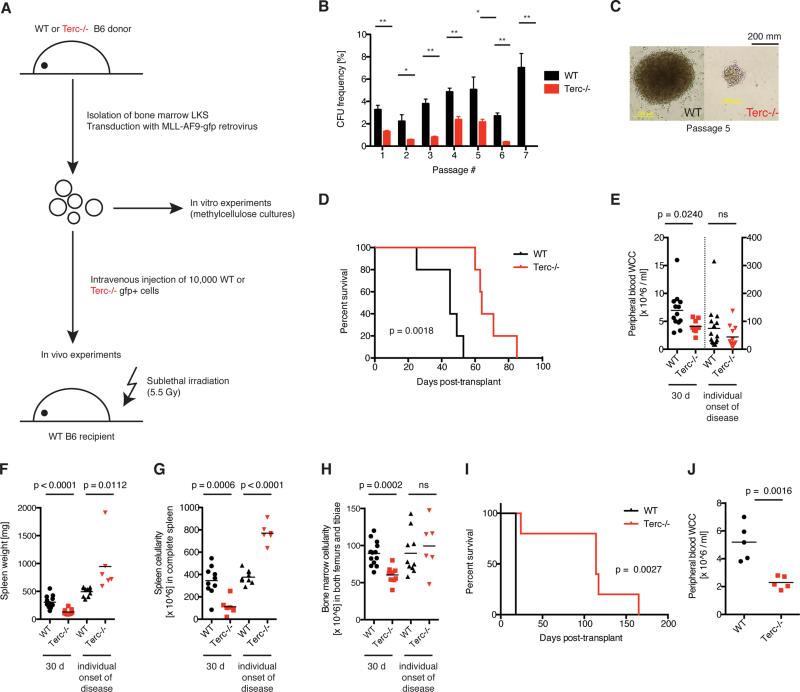
To determine the role of telomerase on AML cell function, bone marrow (BM) LKS+ cells from either G3 Terc−/− or WT donors were transformed with a retroviral MLL-AF9 construct (Figure 1A). Reduced telomere length was confirmed in Terc−/−compared to WT MLL-AF9 cells (Figure S1A-C). Moreover, MLL-AF9 transformation increased telomerase activity and reduced telomere length (Figure S1D-E). In vitro colony forming assays revealed reduced colony numbers in Terc−/−compared to WT MLL-AF9 cells and complete absence of colonies in the Terc−/−condition after 7 passages (Figure 1B). Terc−/− AML colonies were characteristically smaller when compared to WT AML (Figure 1C). These findings suggest that telomerase deficiency leads to the gradual loss of AML cells in vitro.
Figure 1. Telomerase deficiency impairs in vitro self-renewal and delays AML onset.
A) Experimental scheme. BM LKS+ cells were isolated from WT or G3 Terc−/− mice, transduced with MLL-AF9-GFP retrovirus in vitro to generate leukemia stem cells (LSC) and plated into methylcellulose or transplanted into irradiated recipient mice. B) Colony-forming assay of WT (black bars) or Terc−/− MLL-AF9 LSCs (red bars). Values represent colony-forming unit frequencies ± SE, calculated as number of colonies per input cells (%). Statistically significant differences according to Student's T test; *: p < 0.05; **: p < 0.01; n = 3. C) Week 5 WT and Terc−/− MLL-AF9 LSC colony morphology. D) Median survival was 45 vs. 64 days post-transplant for recipients of WT (black line) and Terc−/− (red line) MLL-AF9 LSCs, respectively, P = 0.0018, Mantel-Cox Test. N = 5. E) PB WCC, F) spleen weight and G) spleen WCC, and H) BM WCC from recipients of WT (black) or Terc−/− (red) MLL-AF9 LSCs 30 days post-transplant (left) and again at the onset of AML (right). Each point represents an individual animal and the black line represents the mean. P-values calculated by Student's T test are displayed, 2-3 independent experiments. I) Survival and J) PB WCC of mice injected with WT or Terc−/− Aml1Eto + KrasG12C AML. N = 5 per group.
See also Figure S1.
In order to determine the role of telomerase for AML in vivo, equal numbers of WT or Terc−/− MLL-AF9-transformed LKS+ were transplanted into syngeneic WT recipients. In both WT and Terc−/−, AML developed with full penetrance, however Terc−/− AML displayed delayed latency (Figure 1D). Thirty days after transplantation, peripheral blood (PB) white cell count (WCC), BM WCC, spleen weight and spleen cellularity were lower in Terc−/− compared to WT AML recipients (Figure 1E-H, left datasets). At individual disease onset, the AML phenotype and disease burden were similar between WT and Terc−/− conditions (Figures 1E, H, right datasets), and spleen weight and WCC were found to be higher in Terc−/− AML, likely reflecting the longer duration of disease (Figures 1F, G, right datasets). These findings demonstrate that telomerase deficiency delays the onset of leukemia, but is not essentially required for the initiation of AML in vivo.
To validate these findings in other subtypes of AML, G3 Terc−/− or WT cells were transformed with either AML1-ETO/ KRASG12C (Chou et al., 2011; Zhao et al., 2014), CDX2 (Scholl et al., 2007) or BCR-ABL/ NUP98-HOXA9 (Dash et al., 2002). AML1-ETO/ KRASG12C Terc−/− AML had prolonged survival and all Terc−/− AMLs showed reduced disease burden and impaired in vivo expansion of AML (Figure 1I-J, Figure S1F-K).
Telomerase deficiency eradicates functional LSCs in murine AML
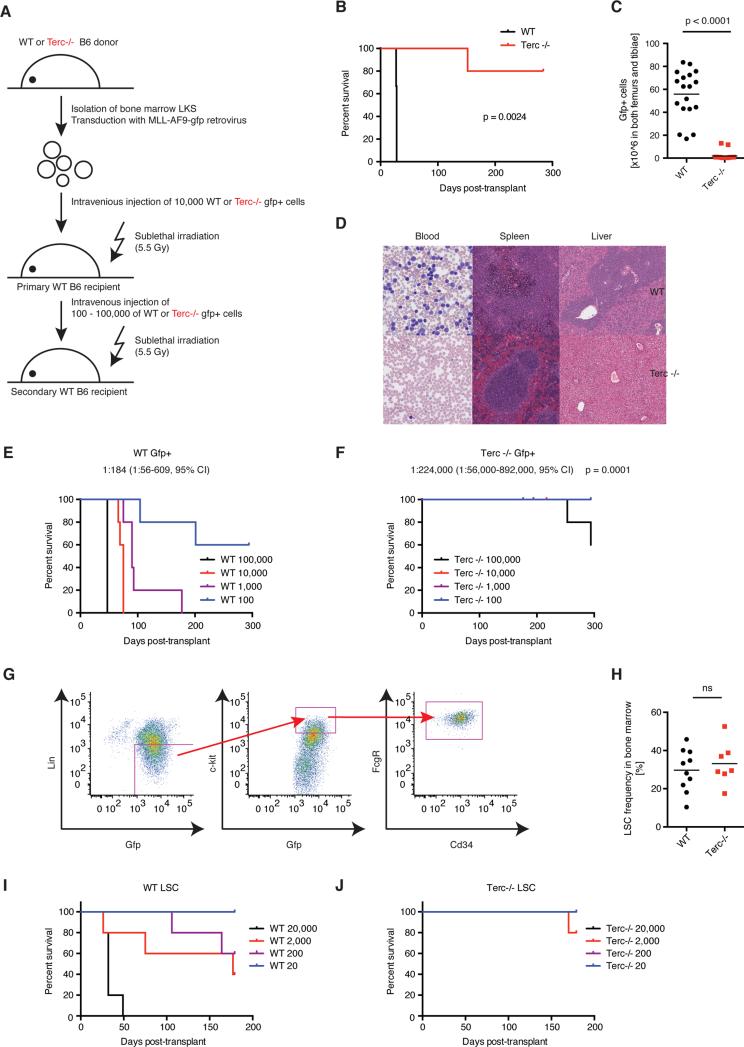
In vivo serial transplantation represents the gold standard for the analysis of long-term LSC function. We therefore isolated viable AML cells from leukemic WT or Terc−/−mice (GFP+, Sytox Blue negative) and transplanted these cells into syngeneic irradiated secondary recipients (Figure 2A). All WT AML recipients developed AML with rapid onset whereas secondary recipients of Terc−/− AML cells rarely developed AML (Figure 2B). Terc−/− AML secondary recipients showed fewer GFP+ cells (Figure 2C) and less leukemic blasts in PB, spleen and liver (Figure 2D). Consistent with these findings, other parameters of AML severity were also reduced, e.g. spleen size, weight, splenic AML cell engraftment, BM AML cell engraftment, PB WCC and circulating AML cells (Figure S2A-F). Using the MLL-AF9 model, in vivo limiting dilution analysis determined that the LSC frequency in Terc−/− AML was 1:224,000 compared to 1:184 in WT AML, representing a reduction of greater than 1000-fold (Figure 2E, F). LSC frequency (linnegGFP+KithighCD34+FcGR+; Figure 2G (Krivtsov et al., 2009)) was similar between primary WT and Terc−/− MLL-AF9 LKS+ recipients demonstrating that genetic perturbation of telomerase compromises LSC function without preventing the initial generation of LSCs (Figure 2H). Finally, limiting dilution analysis of purified LSCs confirmed the observed loss of function in Terc−/− vs. WT LSCs (Figure 2I, J). LSC frequency was markedly reduced in Terc−/−AML1-ETO/ KRASG12C AML and Terc−/− BCR-ABL/NUP98-HOXA9 AML compared to WT controls (Figure S2G-H, K). Furthermore, secondary transplantation of Terc−/− AML1-ETO/ KRASG12C AML or Terc−/− BCR-ABL/NUP98-HOXA9 AML demonstrated prolonged survival and diminished AML expansion compared to WT controls (Figure S2I-J, L-M).
Figure 2. Telomerase deficiency eradicates functional LSCs in murine AML.
A) Experimental scheme. AML cells (viable GFP+ from BM) were isolated from primary recipients at disease onset and injected into secondary recipients. B) Median survival was 28 days after transplantation of 20000 WT AML cells vs. not reached for Terc−/− AML (follow up >280 days), p = 0.0024 according to Mantel-Cox Test. N = 5, representative of 2 independent experiments. C) Engraftment of BM-derived WT (black) or Terc−/− AML cells (red) 28 days post-transplant (absolute GFP+ cells in both femurs and tibiae in each individual animal). The calculated mean is displayed. P < 0.0001, Student's T test. N = 18 from three independent experiments. D) Histologic analysis of PB, spleen and liver morphology in WT and Terc−/− AML 28 days post-transplant. E) Limiting dilution analysis of WT vs. (F) Terc−/− MLL-AF9 LSCs frequency in secondary transplants injected with 100, 1000, 10000 or 100000 viable AML cells (Sytox-, GFP+). N = 5. LSCs frequency was 1:184 for WT (95% confidence interval, 1:56-609) and 1:224000 for Terc−/− AML (95% confidence interval 1:56000-892000), p = 0.0001 using Poisson analysis. G) Gating strategy for LSCs using FACS, lineagelowGFP+Kit+FcGR+CD34+. H) LSC frequency in the BM from primary recipients injected with WT (black symbols) or Terc−/− MLL-AF9 transformed LKS+ (red symbols) at AML onset. N = 7-10 from three independent experiments. I) Limiting dilution analysis of WT or (J) Terc−/− MLL-AF9 LSCs in secondary transplants injected with 20 (blue), 200 (purple), 2000 (red) or 20000 (black) viable LSCs (lineagelowGFP+Kit+FcGR+). N = 5.
See also Figure S2.
Telomerase deficiency in LSCs is associated with cell cycle arrest, p53 activation and programmed cell death
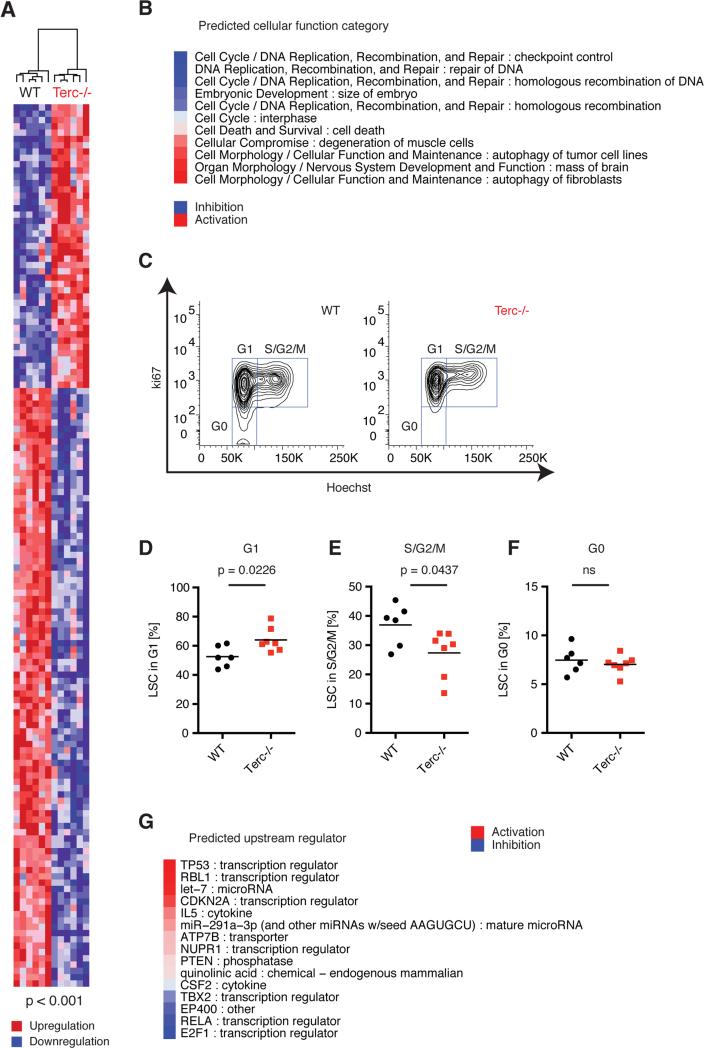
In order to gain insights into the mechanisms underlying the susceptibilities of Terc−/−LSCs in vivo, we performed transcriptional profiling of purified Terc−/− and WT LSCs from leukemic primary recipients. 140 probe sets were differentially expressed between Terc−/− and WT LSCs (using a p-value cutoff of 0.001; Figure 3A). These expression changes predicted abnormal cell cycle and aberrant DNA replication, recombination and repair including homologous recombination, altered cell morphology, cellular function and maintenance, and activated programmed cell death in Terc−/− LSC compared to WT LSC (Figure 3B). The expression of MLL-AF9 binding targets was unchanged (data not shown). Flow cytometric analysis of WT and Terc−/− LSCs confirmed cell cycle abnormalities with an increased proportion of Terc−/− LSC remaining in G1 phase followed by a concomitant reduction in S/G2/M phases when compared to WT LSC (Figure 3C-F). To identify transcription factor candidates that could mediate the observed cellular defects, we performed upstream regulator analysis and found activation of key transcription factors such as p53, p107 and p16/19, together with inhibition of E2f1, p65 and Ep400 (Figure 3G). Terc−/− LSCs exhibited marked chromosomal instability with end-to-end chromosomal fusions that were not seen in WT LSCs (Figure S3A). Western blot analyses on phosphorylated p53 (Ser15) and phosphorylated FoxO3a (Ser253) suggested a clonal dichotomy in Terc−/− LSCs regarding p53 regulation (Figure S3B) showing only minor overlay between the upstream regulators identified (Figure S3C) although activation of p53 signaling was predicted in both independent Terc−/− LSC clones (Figure S3D). Interestingly, downstream p53 target gene expression was highly similar between the different Terc−/− LSC clones although differential regulation of some additional p53 targets was identified (Figure S3E). The p53 regulatory networks were enriched in the ATR/ATM signaling pathway modeling activated ATR and inhibited ATM signaling that links activated p53 signaling with the inhibition of homologous recombination, G1 and S phase arrest, and apoptosis (Figure S3F).
Figure 3. Telomerase deficiency in LSCs is associated with cell cycle arrest, p53 activation and programmed cell death.
A) Unsupervised hierarchical clustering heatmap displaying differentially expressed genes (p < 0.001) comparing Terc−/− vs. WT LSCs. Red represents higher gene expression (“upregulation”) and blue zones represent lower gene expression (“downregulation”). B) Cellular functions were predicted based on the overlap of differentially expressed genes using Ingenuity Pathway Analysis (IPA), ranked based on the calculated Z-score and pre-selected on p < 0.01. Blue represents predicted inhibition and red represents predicted activation of the respective cellular function category in Terc−/− versus WT condition. C) Representative cell cycle analysis of BM WT or Terc−/− LSC using Ki-67 and Hoechst to discriminate G0, G1 and S/G2/M phases. D) Quantification of G1, E) S/G2/M and F) G0 (quiescent) cell cycle phases in LSCs at AML onset (primary recipients, n = 6-7, three independent experiments). G) Activation and inhibition of upstream regulators predicted using IPA. The upstream regulators were ranked based on Z-score and pre-selected based on the p-value for overlay: p < 0.01. Red color symbolizes predicted activation, and blue color symbolizes predicted inhibition of the upstream regulator.
See also Figure S3.
Telomerase deficiency-mediated loss of LSCs is p53 dependent
The upstream regulator analysis above revealed p53 as top candidate mediating the detrimental effects of telomerase inhibition on LSC function. To functionally validate this, we depleted p53 in WT or Terc−/− LSCs using a retroviral shRNA construct (Dickins et al., 2005). Primary WT or Terc−/− MLL-AF9 LKS+ cells were transfected with p53.shRNA or non-targeting (sh-luc) control. Knockdown of p53 in Terc−/−LSCs resulted in partial rescue of colony forming capacity in vitro (Figure 4A) although was insufficient to rescue the detrimental effects of telomerase deficiency during late in vitro passage (Figure S4A). Terc−/− LSCs exhibited increased apoptosis compared to WT cells, and this was partially prevented by p53 knockdown (Figure 4B, C). LSC function is greatly enriched in Kit+Gr1low AML stem cells compared to KitlowGr1high differentiated AML cells (Wang et al., 2010). Telomerase deficient LSCs had higher Gr1 expression (Figure 4D, E) and reduced Kit expression (Figure 4F) demonstrating enforced differentiation of Terc−/− compared to WT LSCs. Knockdown of p53 resulted in complete rescue of Kit expression, and partial rescue of Gr1 expression in Terc−/− LSC (Figure 4D-F).
Figure 4. Telomerase deficiency-mediated loss of LSCs is p53 dependent.
WT and Terc−/− MLL-AF9 LSCs were transduced with shRNA targeting p53 (shp53) or luciferase control (shluc). A) Colony forming assay of wt-shluc (black), wt-shp53 (blue), terc-shluc (red) and terc-shp53 (purple) during passage 5. Number of colonies per input cells (%). B) Representative flow cytometric analysis of LSC apoptosis (lineage-, Kithigh, annexin V+). C) Quantification of apoptotic LSCs. D) Representative flow cytometric analysis of LSCs (Kithigh) vs. differentiated cells (Gr1high). Median fluorescent intensity of Gr1 (E) and Kit (F). G) Representative flow cytometry plots and H) quantification of LSCs (LineageNegKithigh) from BM of primary recipients injected with WT or Terc−/− LSCs transduced with shp53 or shluc. Mean ± SE. N = 3, 2 independent experiments. Statistical significance by Student's T test. I) Survival analysis. Asterisks (*) denote G3 cohorts with the shortest telomeres. Median survival was 34 (WT-shluc), 34 (WT-shp53), 56 (Terc-shluc), 48 (Terc-shp53), undefined (Terc-shluc*), 187.5 (Terc-shp53*) days. Terc-shluc vs. Terc-shp53: P = 0.1396; Terc-shluc* vs. Terc-shp53*: P = 0.0404 according to Mantel-Cox Test. N = 3-8.
See also Figure S4.
To investigate the functional significance of p53 activation in Terc−/− AML, we transplanted sh-p53.MLL-AF9 LKS+ from WT or Terc−/− mice. As a control sh-luc.MLL-AF9 LKS+ from WT or G3 Terc−/− mice were also transplanted. Concordant with the in vitro results, shp53 Terc−/− AML exhibited increased Kit expression and enrichment for LSC frequency (Figure 4G, H). Functionally, G3 Terc−/− sh-p53.MLL-AF9 LKS+ showed accelerated disease onset compared to shluc.MLL-AF9 control. We have observed some variegation in telomere lengths between cohorts of G3 Terc−/− mice, so this experiment was repeated using G3 Terc−/− mice demonstrating the shortest telomeres (Terc-shluc* and Terc-shp53*; Figure S4B). AML initiation was significantly impaired in Terc-shluc* cells and conversely, Terc-shp53* had markedly accelerated disease onset (Figure 4I). These data demonstrate that the detrimental effects of telomerase loss on LSC function are telomere length and p53 dependent. Finally, in serial transplantation, sh-p53 was able to completely rescue LSC function in Terc−/− LSCs with the longer telomeres (Figure S4B, C).
These data demonstrate that p53 is required for the detrimental effects of telomerase deficiency on LSCs, however additional pathways may also act to constrain LSC function in the context of dysfunctional telomerase.
Telomerase-deficient LSCs undergo genetic crisis and apoptosis after enforced cell cycle progression
To investigate the cellular mechanisms underlying the observed impaired function of Terc−/− LSCs, we examined the initial engraftment potential and subsequent cell fate decisions during secondary transplantation (Figure 5A). Homing efficiencies were similar between WT and Terc−/− LSC (Figure 5B, C), and although there were no differences in early expansion of 100,000 WT and Terc−/− LSCs (Figure 5D, E), rapid onset of AML occurred only in the WT but not in the Terc−/− condition (Figure 2E, F). Terc−/− LSC engraftment progressively decreased demonstrating that Terc−/− LSCs are severely limited in their ability to expand after enforced replicative stress (Figure 5E and Figure S2J, M). Cell cycle analysis revealed fewer quiescent and G1 Terc−/−LSCs with concordant increase in S/G2/M phases as compared to WT LSCs (Figure 5F-I). Critically shortened telomeres have been associated with chromosomal instability and the activation of DNA damage. Consistent with this, phosphorylation of the histone variant H2AX as a sensitive marker for DNA double-stranded breaks was increased in Terc−/− compared to WT LSC (Figure 5J, K), and end-to-end chromosomal fusions were seen only in Terc−/− LSCs (Figure S3A). Terc−/− LSCs also showed extensive apoptosis (Figure 5L, M). In contrast to these detrimental effects of telomerase deficiency on LSC fate, we observed relative preservation of normal LTHSC and a quantitative but stable reduction in normal LT-HSC function (Figure S5A-B). Together, these data demonstrate depletion of functional LSCs through dysfunctional telomeres driving chromosomal instability, DNA damage and apoptosis upon enforced replicative stress, thereby preventing AML progression in vivo.
Figure 5. Telomerase-deficient LSCs undergo genetic crisis and apoptosis after enforced cell cycle progression.
A) Experimental scheme. Animals were analyzed 16h post-transplant to study homing efficiency, or at three weeks post-transplant for cell fate analysis. B) Representative flow cytometry and C) quantification of 16h GFP+ percentage engraftment in total BM from both femurs and tibiae of each individual sample from WT (black) or Terc−/− LSC recipients (red). P = 0.1757, Student's T test. N = 11-17 from two independent experiments. D-E) Time-course analysis of GFP+ engraftment of WT and Terc−/−secondary recipients injected with 100, 1000, 10000 or 100000 BM-derived AML (mean ± SE, N = 5). F) Representative flow cytometry and quantification of cell cycle of LSC in (G) G0, (H) G1 and (I) S/G2/M. N=16-17. J) Representative flow cytometric analysis and (K) quantification of gamma-H2AX in diploid WT (black) or Terc−/− (red) LSCs. N = 16-17. L) Representative apoptosis flow cytometry (GFP+lineage-KitHighannexin V+) and M) Quantification of apoptotic LSCs in the BM. N = 16-17. All cell fate experiments from three independent experiments. Each point represents an individual biological replicate, significance by Student's T test.
See also Figure S5.
A telomerase-deficient gene signature predicts outcome in human AML
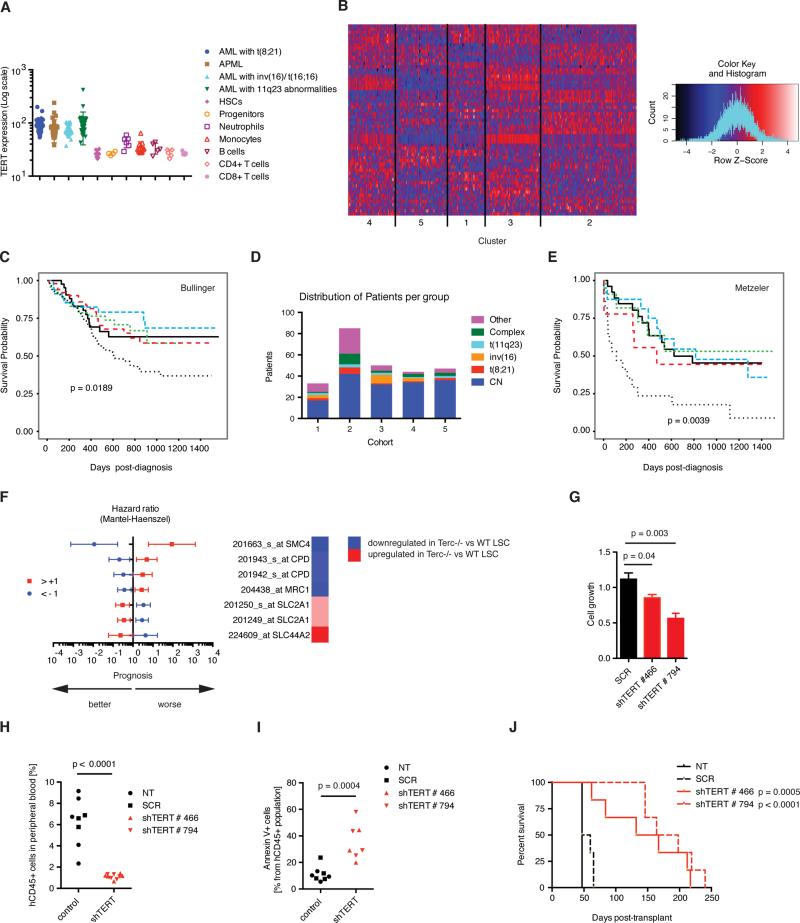
To determine the role of telomerase in human AML, we examined TERT expression in human AML subtypes vs. normal controls. TERT was highly overexpressed in human AML subtypes including AML with t(8;21), AML with inv(16), acute promyelocytic leukemia and AML with MLL-rearrangements (Figure 6A)(Bagger et al., 2012). We next sought to determine whether the gene expression changes found in telomerase-deficient LSCs could be identified in distinct subgroups of human AML. We used the top 140 differentially expressed genes between Terc−/− and WT LSCs and identified the human homologues of these genes to perform unsupervised hierarchical clustering analysis of a large, defined cohort of human AML (Figure 6B; Bullinger et al. Manuscript in preparation). Five distinct clusters could be identified, and one of these clusters showed a particularly adverse patient outcome after standard chemotherapy (Figure 6C). The adverse prognosis cluster exhibited fewer cases of cytogenetically normal (CN, intermediate prognosis) AML with a trend to more complex karyotype (adverse prognosis) samples (p=0.07, Figure 6D, Figure S6A-D). Therefore, these results were tested and validated in a secondary independent dataset comprising exclusively CN AML (Figure 6E; (Metzeler et al., 2011)). To find the key genes that were responsible for driving these prognostic differences, we used random forest modeling to identify a short list of 40 genes with a significant impact on survival and then tested the impact of these genes individually on prognosis. These genes were preferentially found in pathways regulating DNA damage, replication and repair (Supplementary Figure 6E). We identified five genes (SMC4, CPD, MRC1, SLC2A1 and SLC44A2) that individually predicted survival in human AML. Interestingly, the expression profiles resembling telomerase deficiency were correlated with favorable prognosis (Figure 6F). In summary, the correlation analyses performed have identified a telomerase-deficient gene signature consisting of five novel AML survival candidates predicting favorable outcome in AML.
Figure 6. A telomerase-regulated gene signature identifies a cohort of human AML with adverse clinical prognosis.
A) In silico analysis of TERT expression levels in human AML and purified hematopoietic cells (servers.binf.ku.dk/hemaexplorer/). B) Hierarchical clustering analysis of AML patients by the Terc signature (top 140 genes differentially regulated between Terc−/− and WT LSCs according to p-value). Heatmap showing expression intensities of the probesets in the Bullinger dataset. Blue denotes low and red shows high expression intensities. C) Survival analysis of the Bullinger dataset of the 5 classes determined by hierarchical clustering analysis by the Terc signature. D) Cytogenetic analysis within the gene set clusters from the Bullinger dataset. E) Survival analysis of these clusters within the Metzeler dataset (cytogenetically normal samples). F) Hazard ratios in the Metzeler dataset of the five genes that had been identified by random forest modeling in the Bullinger dataset. The population was divided into AML patient groups with either high expression intensities of the probeset of interest (deviating at least by log2>1 from the average expression intensity of the probeset of interest within the total population (red symbols), or low expression intensities (log2<-1 from average expression intensity blue symbols). The heatmap on the right displays the corresponding directionality of regulation of the respective gene in the murine model (Terc−/− LSC vs. WT LSC). G) Analysis of MM6 transduced and stably selected for the expression of two different shRNA constructs targeting TERT (shTERT#466 or shTERT#794), or scrambled (SCR) controls. Mean ± SE, three biological replicates. H) Engraftment of MM6 in NSGS peripheral blood (human CD45% 47 days post-transplant). Each point represents an individual biological replicate. NT = non-transduced control. N = 5 (NT, shTERT) or 3 (SCR). I) Quantification of apoptotic MM6 (mean ± SE, N≥3). J) Survival analysis of NSGS injected with 10,000 viable MM6 from each NT, SCR, shTERT#466, or shTERT#795 condition. N = 6. All cell line data collected from two independent experiments.
See also Figure S6.
Genetic telomerase deficiency impairs human AML LSC function
The data from the murine AML models described above identified telomerase as a bona fide target to deplete LSCs in vivo. To confirm these findings in human AML, we examined the effects of genetic and pharmacological telomerase depletion in AML cell lines and primary human AML samples. Initially, we investigated the genetic perturbation of telomerase using short hairpins targeting the enzymatic subunit TERT in the MLL-AF9 containing human AML cell line Monomac6 (MM6). Reduced levels of TERT were confirmed at the transcript and protein level with two independent shRNA constructs (Figure S6F,G). TERT knockdown correlated with reduced AML cell growth in vitro (Figure 6G). We then performed xenograft transplantation experiments to determine the effect of telomerase insufficiency on human AML cell growth in vivo. Equivalent cell numbers from either non-transduced, non-targeting shRNA control or TERT-shRNA transduced MM6 cells were transplanted into irradiated NOD.Cg-Prkdcscid Il2rgtm1Wjl Tg(CMV-IL3,CSF2,KITLG)1Eav/MloySzJ (NSGS) recipients (Wunderlich et al., 2010). The expansion of MM6 cells was significantly reduced in both TERT-shRNA conditions compared to non-targeting and non-transduced control conditions as determined by the analysis of hCD45+ cells in the peripheral blood (Figure 6H), and H&E staining of spleen sections (Figure S6H). Concordant with the primary murine data, we observed a significant increase of apoptotic MM6 cells and loss of G0/G1 cell cycle phases in both TERT-shRNA conditions compared to controls (Figure 6I, Figure S6 I-K). Finally, TERT knockdown was correlated with increased survival of MM6:NSGS xenografts compared to controls (Figure 6J). These data demonstrate that genetic perturbation of telomerase is detrimental for human AML.
Pharmacological targeting of telomerase impairs human AML LSC function and prolongs survival in primary AML xenografts
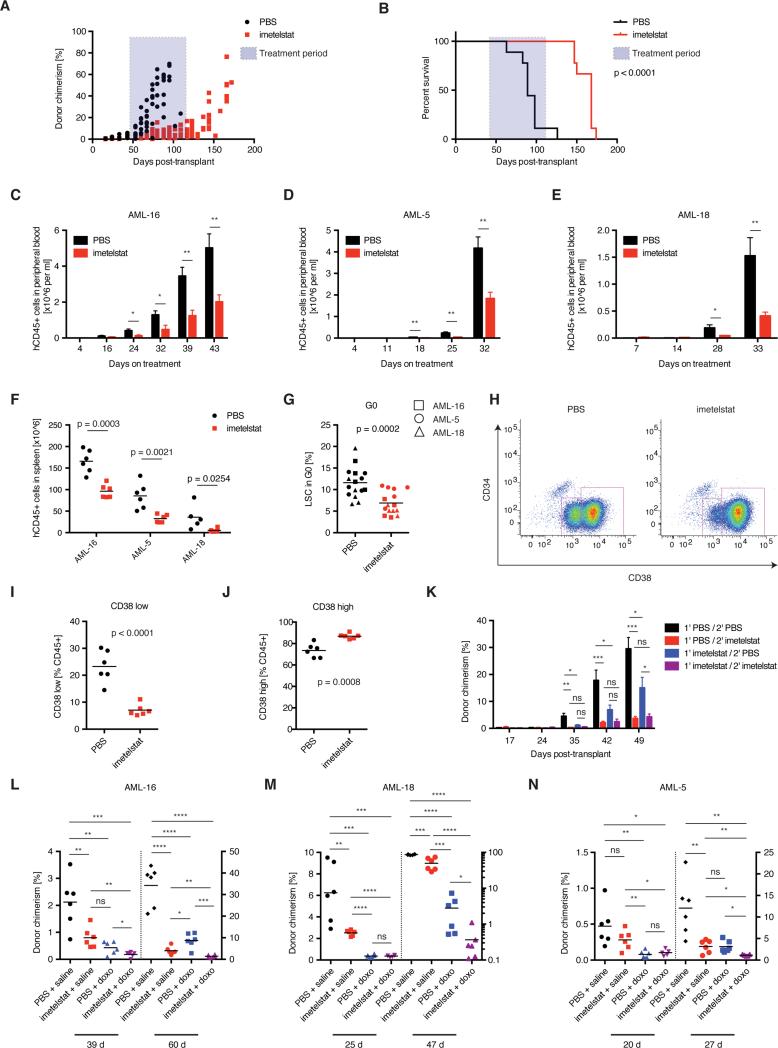
In order to investigate whether pharmacologic inhibition of telomerase could serve as therapeutic strategy in AML, we obtained imetelstat (Geron Corporation), a novel antisense oligonucleotide that specifically targets the RNA component of the telomerase complex. For optimal translational relevance, we examined the efficacy of imetelstat on a primary AML patient sample containing the MLL-AF9 oncogene, using the NSGS xenograft model. AML engraftment was confirmed by flow cytometry prior to the commencement of imetelstat therapy and found to be similar between groups. Subsequent administration of imetelstat prevented the in vivo expansion of AML cells, and ongoing therapy prevented the development of AML when compared to vehicle-treated xenografts (Figure 7A). All vehicle-treated xenografts succumbed to disease at 12 weeks post-transplant when AML engraftment in peripheral blood had reached 60%. Reduced engraftment of AML remained significant, even 5 weeks after imetelstat treatment had been suspended. All imetelstat treated mice eventually progressed after the cessation of treatment with a rising human AML cell frequency in peripheral blood and visible signs of leukemia (Figure 7B). Imetelstat treatment was associated with prolonged overall survival compared to vehicle control (Figure 7B). Notably, imetelstat was also able to prevent the expansion of primary patient derived AML xenografts in an additional three samples tested, demonstrating the broad applicability of this therapeutic strategy (Figure 7C-F, Figure S7A, Table S1).
Figure 7. Pharmacological targeting of telomerase impairs human AML LSC function and prolongs survival in primary AML xenografts.
A) Donor chimerism (hCD45%) and B) survival analysis of tertiary human MLL-AF9 AML patient (AML-X):NSGS transplants treated with telomerase inhibitor imetelstat or vehicle control. N = 9; two independent experiments. Statistical differences in survival were calculated by Mantel-Cox test. Expansion of AML patient samples measured in the PB of C) normal cytogenetics AML with FLT3-ITD (AML-16), D) complex cytogenetics subtype (AML-5), and E) MLL-AF9 AML (AML-18) xenografts. Mean ± SE, N = 6. F) Splenic infiltration of AML for AML-16 , AML-5, and AML-18. N = 6. G) G0 cell cycle flow cytometry of CD45+ cells from AML-16, AML-18, and CD45+CD34+ cells from AML-5:NSGS xenografts. N = 6. P = 0.0002. H) Example CD34/CD38 flow plot on splenic cells from imetelstat or vehicle-treated AML-16:NSGS xenografts. I) CD38low and J) CD38high AML cells. N = 6. K) Secondary transplant of hCD45+ cells from imetelstat (blue) or vehicle-treated (black) primary AML-16:NSGS xenografts. The secondary recipients were treated with imetelstat (red or purple, respectively). *: p < 0.05; **: p < 0.01; ***: p < 0.001. Donor chimerism analysis of AML-16 (L), AML-18 (M) and AML-5:NSGS xenografts (N) treated with vehicle controls (black), imetelstat and vehicle control (red), doxorubicin and vehicle control (blue), or doxorubicin in combination with imetelstat (purple) during the early and late progression phases of AML. N = 5-6, 3 biologically independent samples. *: p < 0.05; **: p < 0.01; ***: p < 0.001; ****:p < 0.0001. All statistical significances were determined using Student's T test unless stated otherwise.
See also Figure S7 and Table S1.
We next examined the effect of imetelstat on LSCs in vivo. All samples exhibited significant reduction in G0 cell cycle, consistent with enforced cell cycle entry or failure to re-enter quiescence (Figure 7G, Figure S7D-F) and increased DNA damage (Figure S7B, C). In AML-16 (normal cytogenetics sample) we observed a significant reduction in the phenotypic LSC population (CD38low) and increase in differentiated blasts (CD38high) (Figure 7H-J). Concordant with these findings, we observed a significant delay in the engraftment and proliferation of AML cells in secondary recipients, and ongoing imetelstat treatment effectively prevented in vivo expansion of this sample (Figure 7K). We next examined a patient derived xenograft sample (AML-5) with complex cytogenetics (predicting a dismal clinical outcome). Even in this sample imetelstat prevented AML expansion, although in this case, there was no observed depletion in immunophenotypic LSCs or delay in disease relapse after transplantation (Figure S7G-J). Interestingly, the AML patient sample with the highest sensitivity to imetelstat in vivo (i.e. AML-X) showed enrichment for the Terc−/− LSC gene expression signature (Figure S7K). Principal component analysis based on the Terc−/−LSC gene expression signature clustered individual AML patients together and separated AML-X furthest away from the least sensitive sample, AML-5 (Figure S7L).
Finally, we tested whether imetelstat could impair disease relapse after chemotherapy, a highly clinically relevant model of LSC function. Transplanted recipient mice were treated with doxorubicin chemotherapy, imetelstat, a combination of imetelstat and chemotherapy, or vehicle control. Chemotherapy alone caused a striking depletion in AML cells initially, however there was rapid relapse and expansion of the residual hCD45+ AML cells (Figure 7L-N). In contrast, chemotherapy combined with imetelstat impaired hCD45+ cell expansion in recipient mice, an effect that was more pronounced than treatment with imetelstat alone (Figure 7L-N). No significant hematological toxicity was seen in blood counts from NSGS mice during imetelstat treatment (Figure S7M-O).
These data demonstrate the efficacy of pharmacologic inhibition of telomerase using imetelstat in human AML to delay or prevent relapse by targeting AML stem cells in vivo. Together, these data reveal a requirement for telomerase in murine and human LSC function and identify telomerase as a tractable therapeutic target in human AML.
DISCUSSION
LSCs represent an elusive, yet critical therapeutic target for the improved survival of patients and long-term eradication of AML. LSCs possess virtually unlimited self-renewal capacity, evidenced by the ability to be propagated inexhaustibly in mice during serial transplantation (Hope et al., 2004). By using syngeneic murine AML models, human cell lines and primary human AML samples, we showed that AML LSCs require telomerase for self-renewal and AML maintenance in vivo.
In murine models of AML, genetic deletion of the RNA component of the telomerase complex (Terc) resulted in the loss of in vitro and in vivo self-renewal, best exemplified by a markedly reduced LSC frequency and the failure of in vivo AML expansion. This LSC depletion was mediated by telomere shortening and enforced replicative stress through serial transplantation leading to genetic instability, cell cycle arrest and massive apoptosis. LSCs proliferate slowly (Ishikawa et al., 2007), and it is believed that restricted cell cycle activity allowing DNA damage response is essential for AML maintenance and the prevention of LSC self-renewal exhaustion (Viale et al., 2009). Interestingly, the detrimental effects of telomerase loss were most evident in LSCs, as demonstrated by a progressive increase in the effect on self-renewal during serial passaging in vitro and serial transplantation in vivo.
The maintenance of an immature LSC population correlates strongly with LSC function (Sykes et al., 2011; Wang et al., 2010). Differentiation and apoptosis are induced in telomerase-deficient Kit+ LSC populations suggesting that these pathways are common mediators of LSC depletion. Moreover, the enforced differentiation of Terc−/− LSCs is consistent with a previous study in which DNA damage mediated by gamma irradiation or telomerase deficiency induced adult stem cell differentiation, thereby limiting self-renewal (Wang et al., 2012). Together, these data provide strong mechanistic evidence that telomerase deficiency depletes LSCs via the DNA damage and differentiation / apoptosis cascade.
Importantly, these results were validated in human AML using both AML cell lines and primary human AML samples. Genetic knockdown of the enzymatic subunit of telomerase (hTERT) delayed leukemia onset in the MM6 xenograft model resulting in improved survival. Consistent with the primary murine data, the in vivo expansion of hTERT-knockdown MM6 cells was limited by the induction of apoptosis. We therefore tested the response of mice engrafted with primary human AML cells to treatment with the novel antisense oligonucleotide telomerase inhibitor imetelstat. Strikingly, treatment with imetelstat prevented the in vivo expansion of AML cells and this was sustained during treatment. Although the AML was not fully eradicated by this therapy, the in vivo proliferation was controlled, raising the possibility that telomerase inhibition may be used to prolong remission by preventing LSC expansion and hence clinical relapse. These findings are of direct and immediate clinical relevance, as imetelstat has entered early phase clinical trials for a number of malignancies.
To further investigate the mechanism of LSC loss after telomerase deletion, gene expression analysis revealed widespread transcriptional changes involving DNA damage repair pathways, homologous recombination and cell death. Unlike others (Bernt et al., 2011; Dawson et al., 2011; Zuber et al., 2011), we did not observe any loss of the MLL-fusion gene expression signature, suggesting that MLL-AF9 binding to target genes is not affected by telomerase loss. In contrast, activation of p53 was predicted in functionally impaired Terc−/− LSCs. Genetic depletion of p53 rescued some of the effects of telomere loss, most notably LSC apoptosis and enforced differentiation. However, p53 depletion was insufficient to completely rescue LSC function, suggesting that other pathways co-operate to constrain AML LSCs in telomerase-deficient cells. The p53 pathway is required for chemotherapy sensitivity (Zuber et al., 2009), and TP53 is mutated in many complex karyotype AML patients leading to chemotherapy resistance and poor prognosis (2013; Rucker et al., 2012). Telomerase inhibition may retain some function even in p53 mutated AML due to the additional p53-independent effects of telomerase deficiency.
Telomerase is also active and required for long-term HSC function. Early generation telomerase deficient mice can engraft and repopulate lethally irradiated recipients, however this function is lost after the 3rd round of transplantation (Allsopp et al., 2003). Moreover, late generation (G4/G5) telomerase deficient mice have critically shortened telomeres, loss of HSCs and premature ageing (Blasco et al., 1997). We used G3 telomerase deficient mice in which telomeres are shortened, however have minimal phenotypic aberrations. These mice showed relative preservation of LT-HSC frequency and quantitative but stable reduction in LT-HSC function. These data are consistent with other reports demonstrating that HSCs from G3 telomerase deficient mice have a quantitative reduction in function (Choudhury et al., 2007). These studies raise legitimate concerns about the effective therapeutic window of telomerase inhibitors, however it should be noted that most existing AML chemotherapy regimens are limited by severe myelosuppression particularly in elderly patients. The effects of telomerase inhibitors on DNA damage repair pathways may also predispose to leukemogenesis or second malignancies, however given the dismal long-term survival in elderly AML patients, there is an urgent need to test novel, highly effective therapies in these patients. Notably, in the xenograft transplantation experiments we did not observe hematological toxicity of imetelstat-treated mice compared to controls. Only properly conceived and designed clinical trials, with long-term follow up, can accurately determine the efficacy and tolerability of these compounds in AML patients.
Interestingly, we identified a telomerase-related gene expression signature that reproducibly clustered primary human AML samples into discrete groups. Most importantly, this signature identified a poor prognostic subgroup of patients with AML. Correlation analysis revealed 5 genes (SMC4, CPD, MRC1, SLC2A1 and SLC44A2) with the most powerful impact on survival and provocatively, these genes were all located within a pathway controlling DNA damage and repair. Three out of the five genes identified represent annotated c-MYC target genes (i.e MRC1, CPD and SLC2A1). C-MYC is a proto-oncogene and its role in tumor cell biology has been extensively investigated. Two recent papers used novel bromodomain inhibitors as potent therapies against MLL-rearranged AML and the inhibition of Myc targets is proposed as a key component of this therapeutic success (Dawson et al., 2011; Zuber et al., 2011). Furthermore, SMC4 is a member of the cohesin family of genes that are recurrently mutated in human AML, although the functional role of abnormal cohesin expression or mutations in AML remains unknown (2013). Together, these data suggest that the regulation of DNA repair pathways within LSCs may also govern response to AML treatment.
In conclusion, modulation of the telomerase complex is a novel and highly effective strategy for targeting LSCs in AML. Clinical trials should address the efficacy of telomerase inhibitors such as imetelstat as a novel consolidation strategy to prevent relapse or potentially together with chemotherapy to improve outcomes in AML patients.
EXPERIMENTAL PROCEDURES
Plasmids
The MSCV-MLL-AF9-GFP and MSCV-MLL-AF9-neo constructs were previously described (Lane et al., 2011). The v2luc construct was a gift from Dr. M. Milsom (DKFZ Heidelberg). MSCV-NUP98-HOXA9, MSCV-BCR-ABL and MSCV-AML1ETO were obtained from Dr. D.G. Gilliland (Boston, MA). MSCV-KRasG12C and MSCV-CDX2 were obtained from Dr S. Froehling and Dr C. Scholl (NCT, Heidelberg, Germany). The lmp.shP53 and lmp.shLuc constructs were a gift from Dr. Ross Dickins (WEHI, Melbourne).
Mouse models
Terc−/− mice were obtained from Jackson Laboratories (Bar Harbor, ME) and have been maintained on C57Bl/6J background (http://jaxmice.jax.org/strain/004132.html). Third generation of homozygous (G3 Terc−/−) were used. C57Bl/6J recipient mice were purchased from Animal Resources Centre, Australia. NSGS mice, i.e NOD.Cg-Prkdcscid Il2rgtm1Wjl Tg(CMV-IL3,CSF2,KITLG)1Eav/MloySzJ were imported from Jackson Laboratories. All mice were kept pathogen-free in the animal facility of QIMR Berghofer Medical Research Institute. NSGS mice received autoclaved and Baytril-treated (Provet Queensland PTY LTD) water. All mouse experiments had been approved by the institutional ethics committee protocol A11605M.
Fluorescence-activated cell sorting and analysis
LKS+ were purified from BM after red blood cell lysis (BD Pharmlyse, BD Biosciences) and were stained with a lineage cocktail comprising of Cd3e (145-2C11), Cd5 (53-7.3), Ter119 (TER-119), Gr-1 (RB6-8C5), B220 (RA3-6B2) and Mac-1 (M1/70), lineage-depleted using biotin-binder Dynabeads (Invitrogen) and stained with streptavidin-ApcCy7, Kit (2B8) and Sca-1 (D7). LSCs were isolated from BM after RBC lysis and stained with a biotinylated lineage cocktail containing Gr-1 (RB6-8C5), Ter119 (TER-119), B220 (RA3-6B2), Cd3e (145-2C11), Cd5 (53-7.3) and Sca-1 (D7), and subsequently with streptavidin, Kit (2B8), Cd16/32 (93) and Cd34 (RAM34). Cells were sorted with a FACSAria instrument (Becton Dickinson). Flow cytometry analysis was performed on a FACS LSR Fortessa (BD Biosciences). Post-acquisition analyses were performed using FlowJo software V9.2.3 (Treestar, CA). For live-dead cell discrimination, Sytox blue (Invitrogen) was used. For cell cycle and DNA damage analysis, cells were then fixed and permeabilized (Fix & Perm, GAS-004; Invitrogen) according to the manufacturer's instructions, and incubated with Ki-67 (B56) and gH2AX (20E3; Cell Signaling). Hoechst 33342 (Invitrogen) was used at 20 mg/ml in PBS containing 2% fetal bovine serum. For apoptosis analysis, cells were stained with Annexin V (BD Biosciences) in Annexin V binding buffer according to the manufacturer's recommendations.
MLL-AF9 retroviral BM transplantation assay
Purified LKS+ populations were transduced with MLL-AF9-GFP or MLL-AF9-neo containing retrovirus and transplanted into 5.5Gy irradiated WT recipients via lateral tail vein. For serial transplantation assay, BM or splenic GFP+ cells were injected into irradiated WT recipients. Mice were monitored for external (hunched posture, reduced movement, ruffled fur) and internal (WCC) signs of leukemia. For homing assay, 1 x 10^6 cells were injected per recipient, and BM was analyzed 16 h post-transplant.
Blood analysis
Blood was collected into EDTA-coated tubes and analyzed on a Hemavet 950 analyzer (Drew Scientific). PB smears were stained with Wright-Giemsa staining according to the manufacturer's protocol (Bio-scientific PTY LTD).
Colony forming assay
Transformed LKS+ were plated in M3434 cytokine-enriched methylcellulose (Stem Cell Technologies), counted and passaged after 5-7 days.
Histology
Tissues were fixed in 10% neutral buffered formalin, embedded in paraffin, and stained with hematoxylin and eosin. Images of histological slides were obtained on a ScanScope FL (Aperio).
Cell culture
MM6 cell line was obtained from Dr S. Froehling, NCT, Heidelberg, Germany. Cells were cultured in RPMI supplemented with 10% fetal calf serum, 2 mM glutamine, 200 U/ml penicillin and 200 ug/ml streptomycin, 2 x non-essential amino acids and 5 ml OPI supplement (Sigma) per 500 ml medium.
In vitro cell growth analysis
For in vitro cell growth analysis, MM6 cells were seeded into a 96-well plate at a density of 2500 cells per 100 ul final volume. Cells were analyzed using Celltiter 96 aqueous non-radioactive cell proliferation assay (MTS) according to the manufacturer's instructions (Promega).
Gene expression profiling and bioinformatics analysis
WT and G3 Terc−/− MLL-AF9 LSC were purified from primary recipients at AML onset. RNA was extracted using Qiagen RNeasy Micro kit, pre-amplified using the Illumina Total Prep RNA amplification kit (Ambion) and hybridized on Illumina MouseWG-6 v2 beadchip array. The data were imported with IDATreader (version 0.2.0, http://www.compbio.group.cam.ac.uk/software/idatreader) to R from Illumina GenomeStudio, after default trimming and collapsing of beads. The data were normalized with lumi (Du et al., 2008). Probes were reannotated as described previously (Barbosa-Morais et al., 2010) (using version: mm9_V1.0.0_Aug09) and filtered for poor or unspecific probes as described (Ritchie et al., 2011). Human homologues were acquired using HomoloGene (http://www.ncbi.nlm.nih.gov/homologene, build 67). The microarray dataset has been deposited in NCBI's Gene Expression Omnibus GEO Series accession number GSE63241 (http://www.ncbi.nlm.nih.gov/geo).
Unsupervised Hierarchical clustering was performed in R by Hartigan-Wong algorithm (Hartigan and Wong, 1979), with 10 random starts, for robust clustering. Random survival forest models were build with censored and uncensored survival observations (Ishwaran and Kogalur, 2010). Importance was assessed by permutation and the forest contained 20,000 trees.
Xenograft transplantation experiments
Primary AML samples were obtained from patients with AML, after informed consent in accordance with the Declaration of Helsinki. Ficoll density gradient was then used to recover viable mononuclear cells. MM6 cells or primary human AML cells were injected via the lateral tail vein into 2.8 Gy irradiated NSGS recipients.
Drug treatment studies
NSGS mice were treated with 15 mg/kg imetelstat or vehicle control via the intraperitoneal route three times per week for the period of time specified in the respective experiment. For chemotherapy studies, doxorubicin (1.5mg/kg) was administered intravenously for three consecutive days starting from 24h post-transplant in strict 24 hours intervals, and non-irradiated recipients were used. For chemotherapy plus imetelstat combination studies, the first imetelstat injection was administrated on day 3 post-transplant directly after doxorubicin injection. Human AML cell engraftment was monitored by staining of peripheral blood with human-specific CD45 staining (H130) against mouse Cd45.1 and subsequent flow cytometric analysis on LSR Fortessa (BD Biosciences).
Statistical analysis
Log-rank (Mantel-Cox) test was used to determine p values for all Kaplan-Meier survival analyses. Unpaired, two-tailed Student's T test was used for all analyses comparing 2 experimental groups. Poisson statistics was used to determine LSC frequencies in the limiting dilution experiments.
Supplementary Material
HIGHLIGHTS.
Telomerase deficiency eradicates LSC function upon enforced replication
Terc−/− LSCs are eradicated via cell cycle arrest and apoptosis
A Terc−/− LSC gene expression signature predicts an improved outcome in human AML
Imetelstat prevents the expansion of human AML LSCs in patient derived xenografts
ACKNOWLEDGEMENTS
We acknowledge the generous contribution of imetelstat, provided by Geron Corporation, Menlo Park, CA. We gratefully acknowledge the technical assistance of Amity Roberts, Glen Boyle, Grace Chojnowski and Paula Hall (flow cytometry), the members of the QIMR-Berghofer Animal House. We thank Stephen Sykes, Mark Smyth, Kelli MacDonald and members of the Hill and MacDonald laboratories for their helpful discussion.
S.W.L. and G.R.H. have received funding from the Leukaemia Foundation, Cure Cancer Australia Foundation and the National Health and Medical Research Council. S.W.L. is the recipient of the Rhys Pengelly Fellowship in Leukaemia Research from In Vitro Technologies. C.B., G.R.H. and S.W.L. have received funding from the Rio Tinto Ride to Conquer Cancer. S.A. acknowledges funding from the National Institutes of Health (NIH; CA66996 and CA140575).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 2013;368:2059–2074. doi: 10.1056/NEJMoa1301689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aalbers AM, Calado RT, Young NS, Zwaan CM, Wu C, Kajigaya S, Coenen EA, Baruchel A, Geleijns K, de Haas V, et al. Telomere length and telomerase complex mutations in pediatric acute myeloid leukemia. Leukemia. 2013;27:1786–1789. doi: 10.1038/leu.2013.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allsopp RC, Morin GB, DePinho R, Harley CB, Weissman IL. Telomerase is required to slow telomere shortening and extend replicative lifespan of HSCs during serial transplantation. Blood. 2003;102:517–520. doi: 10.1182/blood-2002-07-2334. [DOI] [PubMed] [Google Scholar]
- Bagger FO, Rapin N, Theilgaard-Monch K, Kaczkowski B, Jendholm J, Winther O, Porse B. HemaExplorer: a Web server for easy and fast visualization of gene expression in normal and malignant hematopoiesis. Blood. 2012;119:6394–6395. doi: 10.1182/blood-2012-05-427310. [DOI] [PubMed] [Google Scholar]
- Barbosa-Morais NL, Dunning MJ, Samarajiwa SA, Darot JF, Ritchie ME, Lynch AG, Tavare S. A re-annotation pipeline for Illumina BeadArrays: improving the interpretation of gene expression data. Nucleic Acids Res. 2010;38:e17. doi: 10.1093/nar/gkp942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard L, Belisle C, Mollica L, Provost S, Roy DC, Gilliland DG, Levine RL, Busque L. Telomere length is severely and similarly reduced in JAK2V617F-positive and -negative myeloproliferative neoplasms. Leukemia. 2009;23:287–291. doi: 10.1038/leu.2008.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernt KM, Zhu N, Sinha AU, Vempati S, Faber J, Krivtsov AV, Feng Z, Punt N, Daigle A, Bullinger L, et al. MLL-rearranged leukemia is dependent on aberrant H3K79 methylation by DOT1L. Cancer Cell. 2011;20:66–78. doi: 10.1016/j.ccr.2011.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blasco MA, Lee HW, Hande MP, Samper E, Lansdorp PM, DePinho RA, Greider CW. Telomere shortening and tumor formation by mouse cells lacking telomerase RNA. Cell. 1997;91:25–34. doi: 10.1016/s0092-8674(01)80006-4. [DOI] [PubMed] [Google Scholar]
- Bonnet D, Dick JE. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat Med. 1997;3:730–737. doi: 10.1038/nm0797-730. [DOI] [PubMed] [Google Scholar]
- Calado RT, Regal JA, Hills M, Yewdell WT, Dalmazzo LF, Zago MA, Lansdorp PM, Hogge D, Chanock SJ, Estey EH, et al. Constitutional hypomorphic telomerase mutations in patients with acute myeloid leukemia. Proc Natl Acad Sci U S A. 2009;106:1187–1192. doi: 10.1073/pnas.0807057106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celli GB, de Lange T. DNA processing is not required for ATM-mediated telomere damage response after TRF2 deletion. Nat Cell Biol. 2005;7:712–718. doi: 10.1038/ncb1275. [DOI] [PubMed] [Google Scholar]
- Chou FS, Wunderlich M, Griesinger A, Mulloy JC. N-Ras(G12D) induces features of stepwise transformation in preleukemic human umbilical cord blood cultures expressing the AML1-ETO fusion gene. Blood. 2011;117:2237–2240. doi: 10.1182/blood-2010-01-264119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhury AR, Ju Z, Djojosubroto MW, Schienke A, Lechel A, Schaetzlein S, Jiang H, Stepczynska A, Wang C, Buer J, et al. Cdkn1a deletion improves stem cell function and lifespan of mice with dysfunctional telomeres without accelerating cancer formation. Nat Genet. 2007;39:99–105. doi: 10.1038/ng1937. [DOI] [PubMed] [Google Scholar]
- Dash AB, Williams IR, Kutok JL, Tomasson MH, Anastasiadou E, Lindahl K, Li S, Van Etten RA, Borrow J, Housman D, et al. A murine model of CML blast crisis induced by cooperation between BCR/ABL and NUP98/HOXA9. Proc Natl Acad Sci U S A. 2002;99:7622–7627. doi: 10.1073/pnas.102583199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson MA, Prinjha RK, Dittmann A, Giotopoulos G, Bantscheff M, Chan WI, Robson SC, Chung CW, Hopf C, Savitski MM, et al. Inhibition of BET recruitment to chromatin as an effective treatment for MLL-fusion leukaemia. Nature. 2011;478:529–533. doi: 10.1038/nature10509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickins RA, Hemann MT, Zilfou JT, Simpson DR, Ibarra I, Hannon GJ, Lowe SW. Probing tumor phenotypes using stable and regulated synthetic microRNA precursors. Nat Genet. 2005;37:1289–1295. doi: 10.1038/ng1651. [DOI] [PubMed] [Google Scholar]
- Drummond MW, Hoare SF, Monaghan A, Graham SM, Alcorn MJ, Keith WN, Holyoake TL. Dysregulated expression of the major telomerase components in leukaemic stem cells. Leukemia. 2005;19:381–389. doi: 10.1038/sj.leu.2403616. [DOI] [PubMed] [Google Scholar]
- Du P, Kibbe WA, Lin SM. lumi: a pipeline for processing Illumina microarray. Bioinformatics. 2008;24:1547–1548. doi: 10.1093/bioinformatics/btn224. [DOI] [PubMed] [Google Scholar]
- Gentles AJ, Plevritis SK, Majeti R, Alizadeh AA. Association of a leukemic stem cell gene expression signature with clinical outcomes in acute myeloid leukemia. Jama. 2010;304:2706–2715. doi: 10.1001/jama.2010.1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gessner A, Thomas M, Castro PG, Buchler L, Scholz A, Brummendorf TH, Soria NM, Vormoor J, Greil J, Heidenreich O. Leukemic fusion genes MLL/AF4 and AML1/MTG8 support leukemic self-renewal by controlling expression of the telomerase subunit TERT. Leukemia. 2010;24:1751–1759. doi: 10.1038/leu.2010.155. [DOI] [PubMed] [Google Scholar]
- Hartigan JA, Wong MA. Algorithm AS 136: A K-Means Clustering Algorithm. Journal of the Royal Statistical Society Series C (Applied Statistics) 1979;28:100–108. [Google Scholar]
- Heidel FH, Bullinger L, Feng Z, Wang Z, Neff TA, Stein L, Kalaitzidis D, Lane SW, Armstrong SA. Genetic and pharmacologic inhibition of beta-catenin targets imatinib-resistant leukemia stem cells in CML. Cell Stem Cell. 2012;10:412–424. doi: 10.1016/j.stem.2012.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbert BS, Gellert GC, Hochreiter A, Pongracz K, Wright WE, Zielinska D, Chin AC, Harley CB, Shay JW, Gryaznov SM. Lipid modification of GRN163, an N3'-->P5' thio-phosphoramidate oligonucleotide, enhances the potency of telomerase inhibition. Oncogene. 2005;24:5262–5268. doi: 10.1038/sj.onc.1208760. [DOI] [PubMed] [Google Scholar]
- Hope KJ, Jin L, Dick JE. Acute myeloid leukemia originates from a hierarchy of leukemic stem cell classes that differ in self-renewal capacity. Nat Immunol. 2004;5:738–743. doi: 10.1038/ni1080. [DOI] [PubMed] [Google Scholar]
- Hu J, Hwang SS, Liesa M, Gan B, Sahin E, Jaskelioff M, Ding Z, Ying H, Boutin AT, Zhang H, et al. Antitelomerase therapy provokes ALT and mitochondrial adaptive mechanisms in cancer. Cell. 2012;148:651–663. doi: 10.1016/j.cell.2011.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa F, Yoshida S, Saito Y, Hijikata A, Kitamura H, Tanaka S, Nakamura R, Tanaka T, Tomiyama H, Saito N, et al. Chemotherapy-resistant human AML stem cells home to and engraft within the bone-marrow endosteal region. Nat Biotechnol. 2007;25:1315–1321. doi: 10.1038/nbt1350. [DOI] [PubMed] [Google Scholar]
- Ishwaran H, Kogalur UB. Consistency of Random Survival Forests. Stat Probab Lett. 2010;80:1056–1064. doi: 10.1016/j.spl.2010.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirwan M, Dokal I. Dyskeratosis congenita: a genetic disorder of many faces. Clin Genet. 2008;73:103–112. doi: 10.1111/j.1399-0004.2007.00923.x. [DOI] [PubMed] [Google Scholar]
- Krivtsov AV, Feng Z, Armstrong SA. Transformation from committed progenitor to leukemia stem cells. Ann N Y Acad Sci. 2009;1176:144–149. doi: 10.1111/j.1749-6632.2009.04966.x. [DOI] [PubMed] [Google Scholar]
- Krivtsov AV, Twomey D, Feng Z, Stubbs MC, Wang Y, Faber J, Levine JE, Wang J, Hahn WC, Gilliland DG, et al. Transformation from committed progenitor to leukaemia stem cell initiated by MLL-AF9. Nature. 2006;442:818–822. doi: 10.1038/nature04980. [DOI] [PubMed] [Google Scholar]
- Lane SW, Gilliland DG. Leukemia stem cells. Semin Cancer Biol. 2010;20:71–76. doi: 10.1016/j.semcancer.2009.12.001. [DOI] [PubMed] [Google Scholar]
- Lane SW, Wang YJ, Lo Celso C, Ragu C, Bullinger L, Sykes SM, Ferraro F, Shterental S, Lin CP, Gilliland DG, et al. Differential niche and Wnt requirements during acute myeloid leukemia progression. Blood. 2011;118:2849–2856. doi: 10.1182/blood-2011-03-345165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzeler KH, Maharry K, Radmacher MD, Mrozek K, Margeson D, Becker H, Curfman J, Holland KB, Schwind S, Whitman SP, et al. TET2 mutations improve the new European LeukemiaNet risk classification of acute myeloid leukemia: a Cancer and Leukemia Group B study. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2011;29:1373–1381. doi: 10.1200/JCO.2010.32.7742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto K, Bartocci C, Ouzounov I, Diedrich JK, Yates JR, 3rd, Denchi EL. A two-step mechanism for TRF2-mediated chromosome-end protection. Nature. 2013;494:502–505. doi: 10.1038/nature11873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie ME, Dunning MJ, Smith ML, Shi W, Lynch AG. BeadArray expression analysis using bioconductor. PLoS Comput Biol. 2011;7:e1002276. doi: 10.1371/journal.pcbi.1002276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rucker FG, Schlenk RF, Bullinger L, Kayser S, Teleanu V, Kett H, Habdank M, Kugler CM, Holzmann K, Gaidzik VI, et al. TP53 alterations in acute myeloid leukemia with complex karyotype correlate with specific copy number alterations, monosomal karyotype, and dismal outcome. Blood. 2012;119:2114–2121. doi: 10.1182/blood-2011-08-375758. [DOI] [PubMed] [Google Scholar]
- Scholl C, Bansal D, Dohner K, Eiwen K, Huntly BJ, Lee BH, Rucker FG, Schlenk RF, Bullinger L, Dohner H, et al. The homeobox gene CDX2 is aberrantly expressed in most cases of acute myeloid leukemia and promotes leukemogenesis. J Clin Invest. 2007;117:1037–1048. doi: 10.1172/JCI30182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sykes SM, Lane SW, Bullinger L, Kalaitzidis D, Yusuf R, Saez B, Ferraro F, Mercier F, Singh H, Brumme KM, et al. AKT/FOXO signaling enforces reversible differentiation blockade in myeloid leukemias. Cell. 2011;146:697–708. doi: 10.1016/j.cell.2011.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szer J. The prevalent predicament of relapsed acute myeloid leukemia. Hematology Am Soc Hematol Educ Program. 2012;2012:43–48. doi: 10.1182/asheducation-2012.1.43. [DOI] [PubMed] [Google Scholar]
- Viale A, De Franco F, Orleth A, Cambiaghi V, Giuliani V, Bossi D, Ronchini C, Ronzoni S, Muradore I, Monestiroli S, et al. Cell-cycle restriction limits DNA damage and maintains self-renewal of leukaemia stem cells. Nature. 2009;457:51–56. doi: 10.1038/nature07618. [DOI] [PubMed] [Google Scholar]
- Vicente-Duenas C, Barajas-Diego M, Romero-Camarero I, Gonzalez-Herrero I, Flores T, Sanchez-Garcia I. Essential role for telomerase in chronic myeloid leukemia induced by BCR-ABL in mice. Oncotarget. 2012;3:261–266. doi: 10.18632/oncotarget.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Sun Q, Morita Y, Jiang H, Gross A, Lechel A, Hildner K, Guachalla LM, Gompf A, Hartmann D, et al. A differentiation checkpoint limits hematopoietic stem cell self-renewal in response to DNA damage. Cell. 2012;148:1001–1014. doi: 10.1016/j.cell.2012.01.040. [DOI] [PubMed] [Google Scholar]
- Wang Y, Krivtsov AV, Sinha AU, North TE, Goessling W, Feng Z, Zon LI, Armstrong SA. The Wnt/beta-catenin pathway is required for the development of leukemia stem cells in AML. Science. 2010;327:1650–1653. doi: 10.1126/science.1186624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wunderlich M, Chou FS, Link KA, Mizukawa B, Perry RL, Carroll M, Mulloy JC. AML xenograft efficiency is significantly improved in NOD/SCID-IL2RG mice constitutively expressing human SCF, GM-CSF and IL-3. Leukemia. 2010;24:1785–1788. doi: 10.1038/leu.2010.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S, Zhang Y, Sha K, Tang Q, Yang X, Yu C, Liu Z, Sun W, Cai L, Xu C, Cui S. KRAS (G12D) Cooperates with AML1/ETO to Initiate a Mouse Model Mimicking Human Acute Myeloid Leukemia. Cell Physiol Biochem. 2014;33:78–87. doi: 10.1159/000356651. [DOI] [PubMed] [Google Scholar]
- Zuber J, Radtke I, Pardee TS, Zhao Z, Rappaport AR, Luo W, McCurrach ME, Yang MM, Dolan ME, Kogan SC, et al. Mouse models of human AML accurately predict chemotherapy response. Genes Dev. 2009;23:877–889. doi: 10.1101/gad.1771409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuber J, Shi J, Wang E, Rappaport AR, Herrmann H, Sison EA, Magoon D, Qi J, Blatt K, Wunderlich M, et al. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature. 2011;478:524–528. doi: 10.1038/nature10334. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.