Abstract
Antibiotics target conserved bacterial cellular pathways or growth functions and therefore cannot selectively kill specific members of a complex microbial population. Here, we develop programmable, sequence-specific antimicrobials using the RNA-guided nuclease Cas91, 2 delivered by a bacteriophage. We show that Cas9 re-programmed to target virulence genes kills virulent, but not avirulent, Staphylococcus aureus. Re-programming the nuclease to target antibiotic resistance genes destroys staphylococcal plasmids that harbor antibiotic resistance genes3, 4 and immunizes avirulent staphylococci to prevent the spread of plasmid-borne resistance genes. We also demonstrate the approach in vivo, showing its efficacy against S. aureus in a mouse skin colonization model. This new technology creates opportunities to manipulate complex bacterial populations in a sequence-specific manner.
Advances in DNA sequencing technologies are revealing the diversity of complex microbial populations in different environments. They are also providing evidence for the contributions that individual species make to both populations and environments. Perhaps the most striking example of this is the human microbiome and its influence on human health5, 6. Studying the microbiome has not only shown the importance of certain species for human health, but has also revealed the undesired side-effects of traditional antimicrobials (including antibiotics) that lack killing specificity. Side-effects of antibiotic use include promoting the emergence of antibiotic resistance and important negative effects on human health7. There is a pressing need to develop species-specific, selective antibiotics that can be used to manipulate complex microbial consortia such as the microbiome
Cas9 is a dsDNA nuclease present in the type II CRISPR (Clustered Regularly Interspaced Short Palindromic Repeat) immune system of bacteria, that uses a 20-nt small RNA guide (the CRISPR RNA, crRNA) to specify the site of cleavage10. We and others recently showed that reprogramming the Cas9 nuclease against bacterial genomic sequences is lethal, most likely due to the introduction of irreparable chromosomal lesions8, 9. This observation led us to explore the possibility of using this nuclease as a sequence-specific antimicrobial, a tool that would allow selective killing of one or more bacterial species within a heterogeneous population. To achieve this, the type II CRISPR system has to be delivered to as many (if not all) target cells as possible, without the need for selection, and in a manner that can easily be used to treat bacterial populations in their natural environment. Bacteriophages naturally package their DNA into capsids which can then inject their content into host bacteria. Therefore we opted to deliver the cas9 gene and its RNA guide/s sequences using a phagemid, which is a plasmid that is designed to be packaged in phage capsids11 (Fig. 1a). This strategy was in part inspired by the recent discovery of a phage carrying its own CRISPR system12.
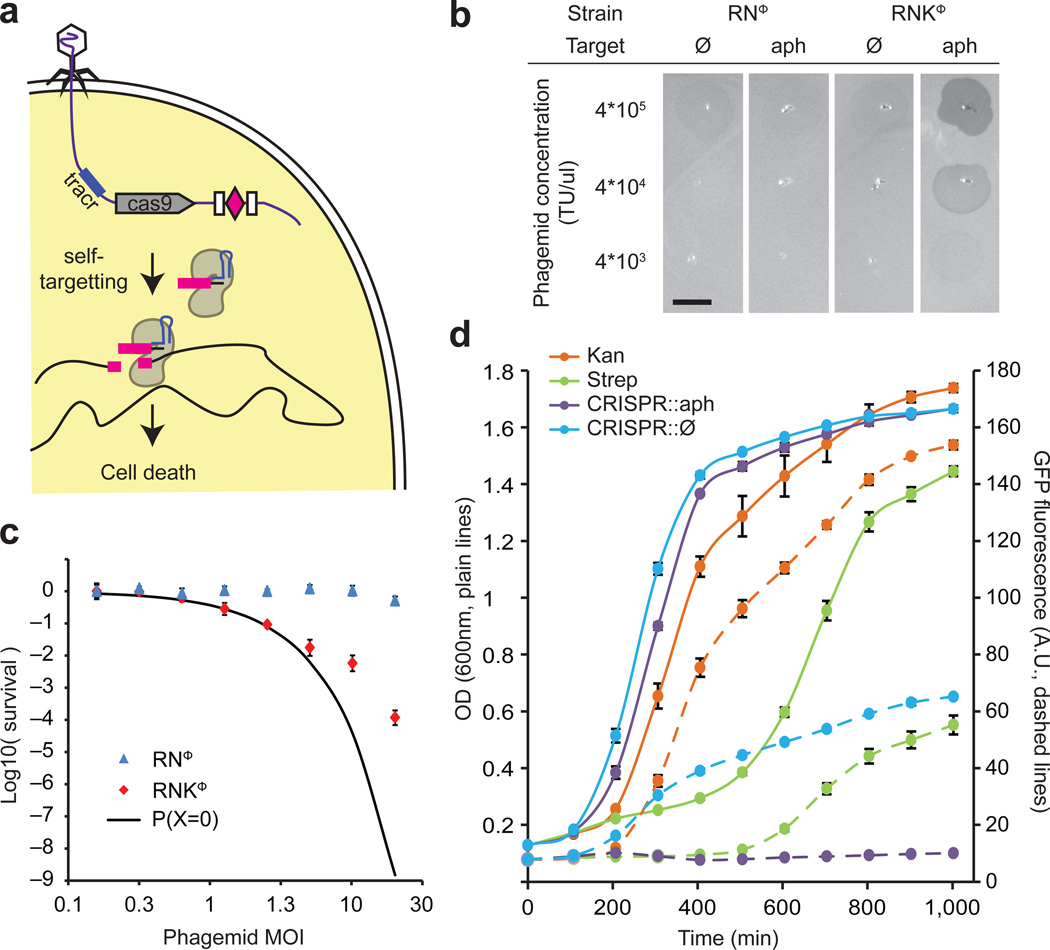
Figure 1.
Sequence-specific killing of S. aureus by a phagemid-delivered CRISPR system. (a) The ΦNM1 phage delivers the pDB121 phagemid to S. aureus cells. pDB121 carries the S. pyogenes tracrRNA, cas9 and a programmable CRISPR array sequence. Expression of cas9 and a self-targeting crRNA leads to chromosome cleavage and cell death. (b) Lysates of pDB121 phagemid targeting the aph-3 kanamycin resistance gene or a non-targeting control are spotted on top-agar lawns of either RNΦ or RNKΦ cells. Scale bar, 5mm (c) Treatment of RNΦ (blue triangles) or RNKΦ (red diamonds) with pDB121::aph at various MOI. Survival is calculated as the ratio of CFU recovered after treatment to CFU from an untreated sample of the same culture (mean ± s.d.). The black curve represents the probability that a cell does not receive a phagemid making the assumption that all cells have the same chance of receiving phagemid. (d) Time course of treatment of RNKΦ/pCN57 (GFP reporter plasmid) cells in a mixed culture with non-targeted RNΦ cells. Plain lines show OD and dashed lines GFP. Kanamycin (25 ug/ml) is shown in orange, streptomycin (10 ug/ml) in green, pDB121::aph (MOI ~20) in purple, pDB121::Ø (MOI ~20) in blue.
We tested whether this technology could selectively kill antibiotic-resistant and virulent Staphylococcus aureus strains. Staphylococci are both predominant members of the human skin microbiota13 and one of the most common causes of nosocomial infections14. The recent increase in staphylococcal pathogenicity is largely due to the the transfer of antibiotic resistance and virulence genes via conjugative plasmids and other mobile genetic elements that has led to the rise of hospital- and community-acquired methicillin- and vancomycin-resistant Staphylococcus aureus (MRSA and VRSA, respectively) strains that are very difficult to treat3, 4. To investigate whether Cas9 cleavage of chromosomal sequences is sufficient to kill staphylococci, we inserted Streptococcus pyogenes cas9, tracrRNA (trans-activating crRNA, a small RNA required for crRNA biogenesis10) and a minimal CRISPR array optimized for one-step cloning of crRNA sequences, into the staphylococcal vector pC194 (ref.15), generating pDB114. This plasmid was programmed to target the aph-3 kanamycin resistance gene. The resulting construct, pDB114::aph, was transformed either into S. aureus RN4220 (ref.16) or RNK, an isogenic derivative carrying a kanamycin resistance gene in the chromosome. Transformation efficiency of RNK cells was at least 2 orders of magnitude lower than RN4220 (see Supplementary Fig. 1). We interpret this as showing sequence specific, Cas9-mediated killing of staphylococci, similarly to the results previously obtained for killing of other bacteria (Streptococcus pneumoniae9, 17, Escherichia coli8, 9 and Salmonella enterica8).
In order to develop a phagemid system suitable for Staphylococcus aureus, we cloned a ~2kb fragment containing the rinA, terS and terL genes and packaging site from the staphylococcal ΦNM1 phage18 into plasmid pC194, to yield the phagemid pDB91. To assess the efficiency of packaging of pDB91 in ΦNM1 capsids, a transduction assay was performed. RN4220 cells containing pDB91 were infected with ΦNM1, and the lysate was used to transduce RN4220 cells previously lysogenized with ΦNM1, referred to as RNΦ. The lysogenic strain is resistant to superinfection with wild-type phage thereby allowing us to detect only phagemid transduction. We determined that a lysate with a titer of 1.6×107 plaque forming units (PFU)/µl contained 3.8×106 transfer units (TU)/µl, showing that our phagemid is packaged in 24% (TU/PFU) of the particles. We cloned the CRISPR sequences of pDB114 and pDB144::aph into the pDB91 phagemid to obtain pDB121 and pDB121::aph, respectively. The proportion of phage particles that contained phagemids was substantially lower (2.9% for pDB121, see Supplementary Fig. 2) than that for pDB91, most likely due to the larger size of pDB121 (10.3 kb vs. 5.3 kb), but remains sufficiently high to facilitate delivery to a large number of cells (Supplementary Fig. 2). When spotted on a lawn of RNKΦ cells, but not on a RNΦ lawn, pDB121::aph elicited strong growth inhibition (Fig. 1b). Conversely, the non-targeting pDB121 phagemid did not produce inhibition of either strain.
In order to quantify the observed killing, infection experiments were performed at different multiplicities of infection (MOI). Here, we define the MOI as the number of TU per recipient cell. In targeting conditions cells are killed when the MOI becomes greater than one, while non-targeted cells remain unaffected (Fig. 1c). At an MOI of 20, the survival rate is 1.1×10−4. Assuming that all cells have an equal chance of taking up phagemid DNA, we can estimate the number of cells in the population that will not be injected with a CRISPR system and are thus expected to survive. The Poisson distribution with a mean of 20 gives an expected survival rate of only 1.5×10−9, which is substantially lower than the survival rate observed. Four different effects could explain this discrepancy: i) the injection of the phagemid in the recipient cells might not be completely random, i.e. some cells might be more likely to receive phagemids than others; ii) after it has been received, the phagemid might sometimes be lost through segregation before it kills the cells; iii) cells might receive phagemids carrying a defective CRISPR system; iv) cells could survive CRISPR targeting through the introduction of mutations in the target sequence.
In order to investigate the nature of the survivor cells, colonies recovered after treatment were re-streaked on chloramphenicol plates (pDB121 carries a chloramphenicol resistance marker). We found that 6/8 colonies were still sensitive to chloramphenicol, supporting scenario i) or ii) (indistinguishable in this chloramphenicol sensitivity assay). The remaining 2/8 chloramphenicol-resistant colonies were further analyzed to determine whether the aph-3 target and/or the CRISPR system were still intact. We found that they contained plasmids incapable of CRISPR targeting in which the cas9 gene was deleted (see Supplementary Fig. 3a and b). None of the cells (8/8) able to escape phagemid treatment contained target mutations (see Supplementary Fig. 3c), indicating that such mutations happen at a frequency lower than 1 in 1.3×105 cells. This preliminary experiment shows that survivor colonies either did not receive the phagemid, lost the phagemid, or received a phagemid containing a defective CRISPR system. In all cases survivors are still sensitive to a second round of treatment, and their numbers were reduced using a higher MOI (data not shown).
Following treatment with conventional antibiotics that eradicate most members of a bacterial community, the incidence and spread of resistance is fueled by the lack of competitor bacteria in the treated niche. As demonstrated above, sequence-specific killing is not exempt from the generation of cells that escape treatment, however our strategy has the benefit of killing only a fraction of the whole population, which could potentially leave other members of the community (including those of the same species) to colonize the niche following treatment and thereby limit the growth of resistant organisms. To investigate the effects of sequence-specific killing on populations, we transformed the pCN57 GFP reporter plasmid19 into RNKΦ in order to monitor the growth of RNKΦ when grown in co-culture with RNΦ during different treatments (Fig 1d). The mixed RNΦ / RNKΦ culture was grown in a plate reader that measured OD and GFP, and flow cytometry was performed at the end of the experiment to confirm the results (Supplementary Fig. 4a). Treatment with pDB121::aph killed RNKΦ cells only, stopping the increase of the GFP signal. In contrast, RNKΦ kept growing when treated with spacerless control (Fig. 2d, compare the dashed purple line with the dashed blue line) We also compared our sequence-specific antimicrobial with traditional antibiotics. Kanamycin kills the RNΦ cells leaving only RNKΦ cells in the culture, resulting in a strong fluorescence signal (dashed orange line). A near-MIC concentration of streptomycin temporarily stops the growth of both RNΦ and RNKΦ cells but after 6 hours resistant RNΦ and RNKΦ cells resume growth (green lines). Therefore in this specific experiment the sequence-specific treatment was better than a non-specific, traditional antibiotic in limiting the incidence of resistant bacteria. We also carried out the same experiment in a monoculture of RNKΦ cells (Supplementary Fig. 4a). In these conditions, the RNKΦ cells that survive the treatment resume growth after 7 hours, and the sequence-specific treatment fares no better than 10 µg/ml of streptomycin. Altogether these experiments highlight the potential benefits of sequence-specific killing with respect to the emergence and establishment of resistant strains. When applied to a mixed bacterial population, a sequence-specific antimicrobial allows non-targeted cells to keep growing, which may in turn prevent or reduce the capacity for growth of the small proportion of targeted cells that survive the treatment.
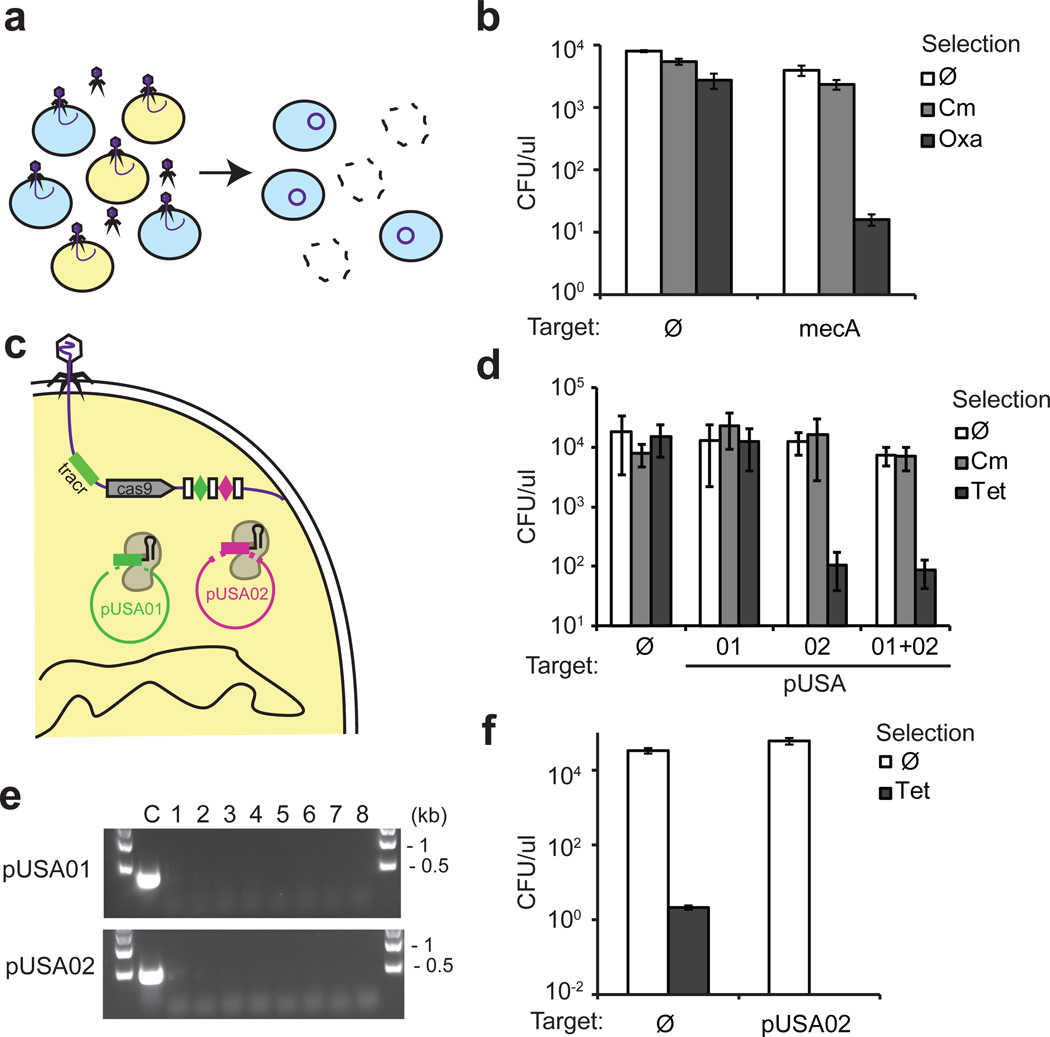
Figure 2.
Targeting antibiotic resistance genes and plasmids in an MRSA strain. (a) Treatment of a mixed population of RNΦ and USA300Φ results in killing of the targeted USA300 MRSA strain and delivery of an immunizing phagemid to the rest of the population. (b) pDB121::mecA specifically kills USA300Φ in a mixed population. Exponentially growing USA300Φ and RNΦ cells were mixed 1:1 and treated with pDB121 at an MOI of ~5. Cells were plated either on a non-selective medium, on chloramphenicol-containing medium to measure the proportion of cells receiving the phagemid treatment, or on oxacillin-containing medium to measure the proportion of USA300Φ cells in the population (mean ± s.d.). (c) The CRISPR array sequence is programmed to target the pUSA01 and pUSA02 plasmids simultaneously. (d) USA300Φ was treated with pDB121 lysates targeting each plasmid individually or in combination. Cells were plated either on a non-selective medium, on chloramphenicol-containing medium to measure the proportion of cells receiving the phagemid treatment, or on tetracycline-containing medium to measure the proportion of cells cured of pUSA02 (mean ± s.d.). (e) Plasmid curing was confirmed by the lack of PCR amplification with plasmid specific oligonucleotides in 8 independent CFUs after treatment with the double targeting construct. (f) A population of RNΦ cells was immunized against plasmid horizontal transfer by treatment with the pUSA02-targeting pDB121 phagemid. 30 min after treatment, the population is transduced with a ΦNM1 stock grown on USA300. Cells are plated either without selection or on tetracycline to measure transduction efficiency of the pUSA02 plasmid (mean ± s.d.).
We produced a phagemid targeting the methicillin resistance gene mecA20, pDB121::mecA to eradicate MRSA strains from a mixed population of bacteria (Fig. 2a). This phagemid was used to treat the clinical isolate S. aureus USA300Φ (ref.3) in a mixed culture with RNΦ cells (both ΦNM1 lysogens). Exponentially growing USA300Φ and RNΦ cells were mixed 1:1 and treated with pDB121::mecA at an MOI of ~5. Cells were plated either on a non-selective medium, oxacillin-containing medium to measure the proportion of USA300Φ cells in the population, or on chloramphenicol-containing medium to measure the proportion of cells receiving the phagemid treatment (Fig. 2b). The proportion of USA300Φ dropped from 50% before treatment to 0.4% after treatment, while no significant decrease could be observed in the control experiment using the non-targeting pDB121 phagemid. Plasmids are the main source of antibiotic resistance and virulence genes in pathogenic bacteria. The USA300 strain carries three plasmids, pUSA01-3 (ref.3), with the pUSA02 plasmid conferring tetracycline resistance. We designed phagemids that target pUSA01, pUSA02 or both (pUSA03 is unstable, data not shown) and tested them for their ability to cure these plasmids from the population (Fig. 2c). In all cases, treating USA300Φ with the phagemid preparation did not result in cell death (Fig. 2d, CFU counts without selection similar to the non-targeting control). However, more than 99.99 % of the cells become sensitive to tetracycline (Fig. 2d), the majority of them due to the loss of pUSA02 (Fig. 2e). In order to investigate the dynamics of plasmid curing, we performed a time course experiment showing a dramatic drop in tetracycline resistant CFU immediately after the cells and the phagemid are mixed (Supplementary Fig. 5). This indicates that pUSA02 is rapidly cleaved and degraded, most likely as soon as a phagemid enters the cell. Many bacterial virulence plasmids can transfer horizontally and spread antibiotic resistance4 so we tested whether phagemid treatment could be used to immunize naïve staphylococci against pUSA02 transfer. To achieve this, an exponentially growing culture of RNΦ cells was treated with a phagemid targeting the pUSA02 plasmid or a non-targeting control. After 30 minutes, treated cells were infected with a ΦNM1 phage lysate grown on USA300 cells that can transduce pUSA02. Transduction efficiency was measured by selecting for tetracycline resistance. While pUSA02 could readily be transferred to cells treated with the control phagemid, no tetracycline resistant colonies were recovered after cells were treated with the targeting phagemid (Fig. 2f), demonstrating efficient immunization against plasmid transfer.
One advantage of using Cas9-mediated killing is the possibility of programming the nuclease with two or more crRNA guides in order to target different chromosomal and/or plasmid sequences. This strategy could limit the rise of resistant clones that escape phagemid treatment through the generation of target mutations, and also expand the range of targeted cells. To illustrate the multiplex capabilities of the CRISPR-Cas9 antimicrobial, we expanded the CRISPR array carried by the phagemid to produce a second crRNA targeting either the superantigen enterotoxin sek gene21 or another region of the mecA gene. These arrays were all able to kill USA300 cells with comparable efficiencies. (Supplementary Fig. 6).. Altogether, these results demonstrate that delivery of the sequence-specific Cas9 nuclease dramatically reduces the plasmid content in a bacterial population without killing the host, can be used to immunize non-pathogenic strains against the transfer of virulence and/or antibiotic-resistant plasmids and can be easily reprogrammed to target multiple sequences.
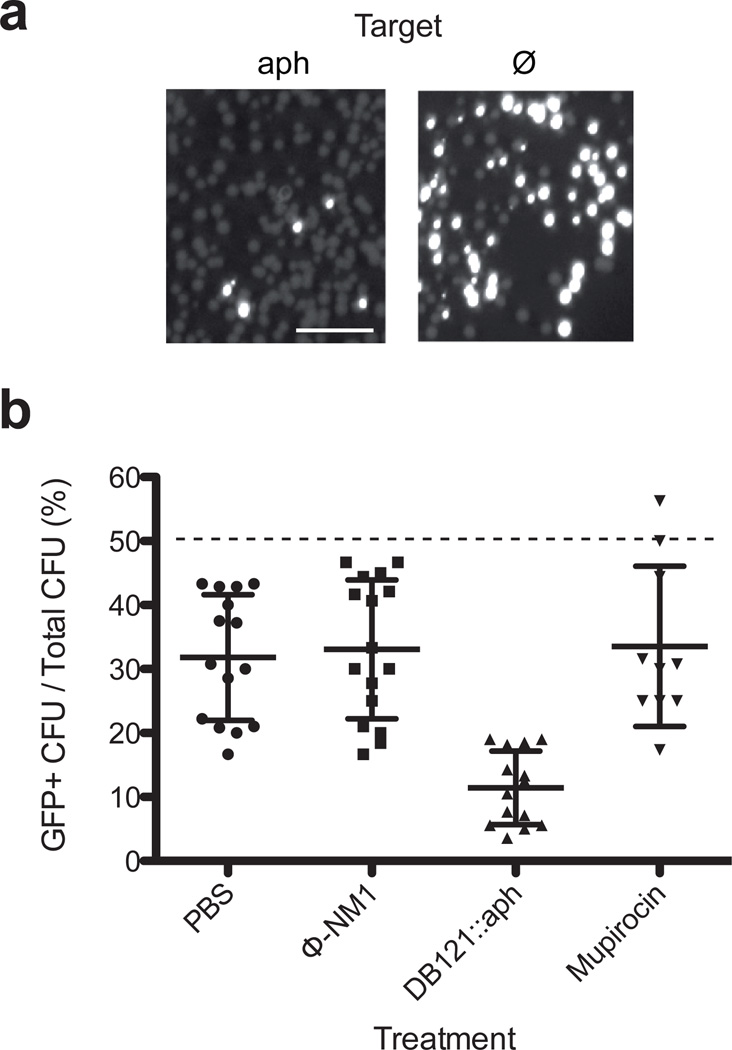
In order to demonstrate that CRISPR-Cas9 treatment can be used to selectively kill staphylococci in vivo, we tested it in a mouse skin colonization model22, 23. The backs of CD1 mice were shaved and treated with depilatory cream to expose the skin. An area on the back was colonized with 105 cells of a 1:1 mixture of RNΦ and RNKΦ bacteria, the latter harboring the pCN57 plasmid to facilitate detection of targeted cells by measuring green fluorescence. Following colonization, infected areas were treated with pDB121::aph, a control containing phiNM1 phage but no phagemid, a topical 2 % mupirocin ointment (commonly used in clinical settings to decolonize patients from staphylococci) or streptomycin (200 mg/mouse). After 24 hr the treated skin was dissected and homogenized to enumerate staphylococci (Supplementary Table 1). The proportion of RNKΦ cells in the population was measured as the ratio of GFP CFU to total CFU (Fig. 3a). Treatment with pDB121::aph resulted in a decrease in the proportion of RNKΦ cells from 50 % to 11.2 % (+/− 1.2 %) that was significantly different from all the other treatment conditions (t-test, p<0.001) (Fig. 3b). In comparison, the systemic streptomycin treatment decolonized the mice of all staphylococci (not shown), but the mupirocin left 6/10 mice colonized with RNΦ and RNKΦ cells in similar proportions. Unexpectedly, a small decrease in the proportion of RNKΦ cells (from the expected theoretical 50 %) was also observed for mice treated with the control phage or saline, possibly due to a fitness disadvantage of RNKΦ/pCN57 cells when growing on the mouse skin.
Figure 3.
Sequence-specific killing of kanamycin resistant S. aureus in a mouse skin colonization model. Mice skin was colonized with a 1:1 mixture of 105 RNΦ and RNKΦ cells carrying the pCN57 GFP reporter plasmid, followed by treatment at 1 hour with phosphate buffer saline (n=15), ΦNM1 (n=16), pDB121::aph (n=14) or mupirocin (n=10). After 24 hours the skin from the treated area was excised, homogenized and serial dilutions of the homogenate was plated on mannitol salt agar (a) Pictures of two representative plates exposed to a wavelength enabling visualization of GFP. Scale bar, 1 cm. (b) The proportion of RNKΦ cells in the population was measured as the proportion of green fluorescent cfu on the plates. Data points indicate individual mice; black lines represent the mean ± SD.
In this report we present the use of the programmable Cas9 nuclease as a sequence-specific antimicrobial to manipulate heterogeneous bacterial populations. Such an antimicrobial could be used to decolonize patients of antibiotic resistant bacteria such as beta-lactam or vancomycin resistant staphylococci, enterococci, enterobacteria and toxigenic clostridia. Our strategy has several advantages over both small molecule antibiotics and phage therapy. We’ve shown that it can be more efficient than an antibiotic treatment when non-targeted bacteria are free to occupy the niche. Similarly to traditional antibiotics, phages therapy will kill dangerous and innocuous bacteria indiscriminately when targeting a commensal species, potentially disturbing the microbiota and selecting for resistant organisms. Another drawback of phage therapy is that phages used are often poorly characterized. In contrast, the composition of our CRISPR antimicrobial and the function of all the genes are well understood. Undoubtedly the main obstacle to translation of this technology into a viable therapeutic is the efficient delivery of the Cas9 and its RNA guide/s into bacterial cells. As opposed to bacteriophages, which can produce hundreds of copies of themselves when they kill a cell, our phagemid system does not produce more particles after infection. This means that the amount of phagemid used in the treatment needs to be much larger than the size of the target population. Furthermore, delivery of the phagemid in an environment more complex than the mouse skin remains to be investigated. While phagemids provide a suitable delivery for some applications, difficulties associated with their purity, large-scale production and narrow host range could preclude their extensive use. Other potential delivery methods for programmable Cas9 nucleases, such as polymeric nanoparticles24 will need to be explored in future. Also, while necessary for the characterization of the technology in this study, antibiotic resistance genes need to be removed from phagemids to avoid their spread. AU please insert a line or two about the advantages over phage therapy in the concluding remarks. In spite of these caveats, this technology has many advantages over traditional antimicrobials. Besides selective killing, provided with a suitable delivery system, the built-in multiplex feature of CRISPR-Cas systems could be exploited to target several different species at the same time and/or several sequences of the same bacterium to prevent the rise of resistant mutants. Our approach can also be used to cure plasmids and other mobile genetic elements from a population without killing the host. Moreover, the technology could be easily adapted to reduce or abolish the expression of antibiotic resistance, virulence and other genes of interest without causing the death of the host using dCas9, the nuclease-defective version of Cas925, 26. These unique features create opportunities for the application of this technology in many medical, environmental and industrial settings, offering the possibility to shape complex bacterial populations.
Methods
Strains and culture conditions
S. aureus strain RN4220 (ref.16) was grown at 37C in TSB, when appropriate, with the following antibiotics: kanamycin (Kan, 25 µg/ml), chloramphenicol (Cm, 10 µg/ml) and tetracycline (Tet, 5 µg/ml). S. aureus USA300 (ref.27) was provided by the Fischetti lab. Phage ΦNM1 was isolated from the S. aureus Newman (ref.28) strain. The supernatant of an overnight culture was used to infect RN4220 in a top-agar layer. Single plaques were isolated and passaged 3 times to ensure purity. ΦNM1 lysogens of RN4220 (RNΦ) and USA300 (USAΦ) were isolated by re-streaking cells from the middle of a turbid plaque twice. Chromosomal integration of ΦNM1 was checked by both PCR and by ensuring resistance to ΦNM1 superinfection. A kanamycin resistance gene was introduced in RN4220 by using a derivative of the pCL55-itet integrative vector where the chloramphenicol resistance gene was replaced with the aphA-3 kanamycin resistance gene to produce pKL55-itet. Briefly, aphA-3 was amplified from strain crR6 (ref.17) using primers L484/L485 and pCL55-itet was amplified with primers L482/L483, followed by digestion with XhoI and ligation. Integration in the RN4220 chromosome was achieved by transformation in electrocompetent cells and selection on TSA+Kan.
Plasmid construction
To assemble the pDB91 phagemid, the rinA-terS-terL region of ΦNM1 was amplified with oligos B234/B235, and pC194 with oligos B233/ B127. PCR products were digested with KpnI and SphI followed by ligation and transformation in RN4220 competent cells. The pDB114 plasmid was constructed in two steps. First the full M1GAS S. pyogenes CRISPR02 system was cloned on pC194 by amplifying S. pyogenes genomic DNA with oligos L362/W278 and pC194 with oligos W270/W282, followed by digestion with BglII and BssSI and ligation, giving pWJ40 (ref29). The pWJ40 plasmid was then amplified with oligos B334/L410 and the BsaI CRISPR array from pCas9 with oligos L409/B333, followed by Gibson assembly30. To construct pDB121, pDB114 was amplified with oligos B351/W278 and pDB91 with oligos L316/L318, followed by Gibson assembly of the two fragments. Spacers were cloned by digestion with BsaI, and ligation of annealed oligonucleotides designed as follow: 5‘-aaac+(target sequence)+g-3’ and 5’-aaaac+(reverse complement of the target sequence)-3’, where the target sequence is 30 nt and is followed by a functional PAM (NGG). Alternatively, two spacers were cloned in a single reaction in the BsaI digested pDB121 vector. Two pairs of oligonucleotides were annealed and ligated with the vector. The pair carrying the first spacer was designed as follow: 5’-aaac+(target sequence)+GTTTTAGAGCTATG-3’ and 5’-AACAGCATAGCTCTAAAAC+(reverse complement of the target sequence)-3’, and the pair carrying the second spacer as follow: 5’-CTGTTTTGAATGGTCCCAAAAC+(target sequence)+g-3’ and 5’-aaaac+(reverse complement of the target sequence)+ GTTTTGGGACCATTCAA-3’. A list of all spacers tested in this study is provided in Supplementary Table 2, and a list of oligonucleotides in Supplementary Table 3.
Phage and phagemid production
Phage and phagemid stocks were produced by growing cells from an overnight culture diluted 1:50 in TSB+Cm+CaCl2 5mM until an OD600 of 0.6. The cultures were then inoculated with 10 µl of a concentrated ΦNM1 phage stock and incubated for 3 hours. Cell debris was eliminated by centrifugation and filtering of the supernatant through 0.45um filters. Phage titers were determined by serial dilution and spotting on a top-agar layer of RN4220 cells on HIA plates supplemented with 5mM CaCl2. To determine the transducing titer, serial dilutions of the phage stock were produced and used to infect a culture of RNΦ cells grown to OD~1. After 1 hour of incubation at 37C, cells were plated on TSA+Cm, and transducing units (TU) were measured from the number of CFU obtained.
Killing and plasmid curing assays
Phage stocks were produced on RN4220 cells carrying a phagemid with the desired CRISPR spacer. Recipient cells were grown in TSB to an OD600 of 0.6, diluted 10× in TSB+5mM CaCl2 and 100 µl of the culture was mixed with 100 µl of the appropriate phage stock dilution. After 1H of incubation, cells were plated on TSA. Survival rates were measured as the ratio of CFUs obtained with treatment over CFUs obtained without treatment. When appropriate, cells were also plated on TSA+Cm to measure phagemid transduction efficiency, TSA+Tet to measure pUSA02 curing, and TSA+Oxa to measure the proportion of MRSA cells in the population.
Immunization assay
RNΦ cells were diluted 1:100 in TSB and grown to OD600 of 0.2. Phagemid was added to an MOI of 10, and cells were incubated 30min to allow for establishment of the CRISPR system. The pUSA02 plasmid was transduced by infecting with a phiNM1 stock grown on USA300 cells. Cells were plated on TSB, TSB+Cm or TSB+Tet to measure transduction efficiency.
Growth curves and fluorescence measurements
Growth curves and GFP fluorescence were measured in a Tecan microplate reader. Cultures were started by diluting a ON culture 1:100 in 200 µl of TSB. Phagemid was added to an MOI of ~10 after 80min of growth.
Flow cytometry
A 1 µl aliquot of cells growing in the Tecan microplate reader were diluted into 3 ml of PBS and were used to acquire flow cytometry data using a LSR II flow cytometer (Becton Dickinson). Data were analyzed with FlowJo software (TreeStar).
Mouse skin colonization
The Rockefeller University’s Institutional Animal Care and Use Committee approved all in vivo protocols. All experiments were conducted at The Rockefeller University’s Animal housing facility, an AAALAC accredited research facility with all efforts to minimalize suffering. An adapted approach from Kugelberg et al.22 and Pastagia et al.23 was used to induce topical skin colonization with S. aureus on 6- to 8-week-old female CD1 mice (Charles River Laboratories, Wilmington, MA). Briefly, mice were anesthetized by intraperitoneal injection of ketamine (1.5 mg/animal; Fort Dodge Animal Health, Fort Dodge, IA) and xylazine (0.3 mg/animal; Miles Inc., Shawnee Mission, KS). A 2 cm2 area of the dorsum of each mouse was shaved with an electric razor; Nair depilatory cream was then applied to the shaved area for one minute and wiped away with 70% ethanol pads. The area was then tape stripped, with autoclave tape, approximately 10 times in succession, using a fresh piece of tape each time to irritate and remove the upper layers of the epidermis. The mice were topically colonized with a 2 µl mixture of cultures of S. aureus RNΦ and RNKΦ/pCN57 containing 1×105 cells in logarithmic growth phases in PBS. Animals were then immobilized under isoflurane anesthesia. After 1 hour, 10 µl of concentrated phagemid lysate containing 2×107 TU/µl was applied on the infected skin area. To obtain this concentration, crude lysates were concentrated using 100kD Amicon Ultra centrifugal filters and washed once with PBS. Additional mice were treated with either streptomycin 200 mg/mouse or 2 % mupirocin. After 24 hours, tissue from the infected skin area was excised and homogenized in 0.5 ml of PBS using the Stomacher 80. Bacterial dilutions were plated on mannitol salt agar (an S. aureus-selective medium) and TSA+Cm.
Supplementary Material
Acknowledgments
We would like to thank Assaf Raz for providing plasmid pCN57 and Daniel Mucida for assistance with flow cytometry experiments. D.B. is supported by the Bettencourt Schuller Foundation. L.A.M is supported by the Searle Scholars Program, the Rita Allen Scholars Program, an Irma T. Hirschl Award, a Sinsheimer Foundation Award and a NIH Director’s New Innovator Award (1DP2AI104556-01). V.A.F. is supported by NIH Grant AI057472.
Footnotes
Competing financial interests
A patent application (US 61/761,971, PCT/US2014/015252) has been filed related to this work. D.B., L.A.M. and X.D. are holding shares in PhageX, a company pursuing applications of this technology.
Individual contributions
D.B. and L.A.M. designed the experiments. D.B. and W.J. performed the in vitro experiments. D.B., P.M.N. and C.E. performed the animal experiments. G.W.G. isolated phage phiNM1 and constructed strain RNK. V.F. and X.D. participated in the conception of the project.
References
- 1.Jinek M, et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gasiunas G, Barrangou R, Horvath P, Siksnys V. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc. Natl. Acad. Sci. U.S.A. 2012 doi: 10.1073/pnas.1208507109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Diep BA, et al. Complete genome sequence of USA300, an epidemic clone of community-acquired meticillin-resistant Staphylococcus aureus. Lancet. 2006;367:731–739. doi: 10.1016/S0140-6736(06)68231-7. [DOI] [PubMed] [Google Scholar]
- 4.Weigel LM, et al. Genetic analysis of a high-level vancomycin-resistant isolate of Staphylococcus aureus. Science. 2003;302:1569–1571. doi: 10.1126/science.1090956. [DOI] [PubMed] [Google Scholar]
- 5.Round JL, et al. The Toll-like receptor 2 pathway establishes colonization by a commensal of the human microbiota. Science. 2011;332:974–977. doi: 10.1126/science.1206095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mazmanian SK, Round JL, Kasper DL. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature. 2008;453:620–625. doi: 10.1038/nature07008. [DOI] [PubMed] [Google Scholar]
- 7.Dethlefsen L, Huse S, Sogin ML, Relman DA. The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol. 2008;6:e280. doi: 10.1371/journal.pbio.0060280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gomaa AA, et al. Programmable removal of bacterial strains by use of genome-targeting CRISPR-Cas systems. MBio. 2013;5:e00928–e00913. doi: 10.1128/mBio.00928-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jiang W, Bikard D, Cox D, Zhang F, Marraffini LA. RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat Biotechnol. 2013;31:233–239. doi: 10.1038/nbt.2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deltcheva E, et al. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature. 2011;471:602–607. doi: 10.1038/nature09886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Melnikov AA, Tchernov AP, Fodor I, Bayev AA. Lambda phagemids and their transducing properties. Gene. 1984;28:29–35. doi: 10.1016/0378-1119(84)90084-2. [DOI] [PubMed] [Google Scholar]
- 12.Seed KD, Lazinski DW, Calderwood SB, Camilli A. A bacteriophage encodes its own CRISPR/Cas adaptive response to evade host innate immunity. Nature. 2013;494:489–491. doi: 10.1038/nature11927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grice EA, et al. Topographical and temporal diversity of the human skin microbiome. Science. 2009;324:1190–1192. doi: 10.1126/science.1171700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lowy FD. Staphylococcus aureus infections. N. Engl. J. Med. 1998;339:520–532. doi: 10.1056/NEJM199808203390806. [DOI] [PubMed] [Google Scholar]
- 15.Horinouchi S, Weisblum B. Nucleotide sequence and functional map of pC194, a plasmid that specifies inducible chloramphenicol resistance. J. Bacteriol. 1982;150:815–825. doi: 10.1128/jb.150.2.815-825.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kreiswirth BN, et al. The toxic shock syndrome exotoxin structural gene is not detectably transmitted by a prophage. Nature. 1983;305:709–712. doi: 10.1038/305709a0. [DOI] [PubMed] [Google Scholar]
- 17.Bikard D, Hatoum-Aslan A, Mucida D, Marraffini LA. CRISPR interference can prevent natural transformation and virulence acquisition during in vivo bacterial infection. Cell Host Microbe. 2012;12:177–186. doi: 10.1016/j.chom.2012.06.003. [DOI] [PubMed] [Google Scholar]
- 18.Baba T, Bae T, Schneewind O, Takeuchi F, Hiramatsu K. Genome sequence of Staphylococcus aureus strain Newman and comparative analysis of staphylococcal genomes: polymorphism and evolution of two major pathogenicity islands. J. Bacteriol. 2008;190:300–310. doi: 10.1128/JB.01000-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Charpentier E, et al. Novel cassette-based shuttle vector system for gram-positive bacteria. Appl Environ Microbiol. 2004;70:6076–6085. doi: 10.1128/AEM.70.10.6076-6085.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ubukata K, Nonoguchi R, Matsuhashi M, Konno M. Expression and inducibility in Staphylococcus aureus of the mecA gene, which encodes a methicillin-resistant S. aureus-specific penicillin-binding protein. J Bacteriol. 1989;171:2882–2885. doi: 10.1128/jb.171.5.2882-2885.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Isnard C, Malbruny B, Leclercq R, Cattoir V. Genetic Basis for In Vitro and In Vivo Resistance to Lincosamides, Streptogramins A, and Pleuromutilins (LSAP Phenotype) in Enterococcus faecium. Antimicrob Agents Chemother. 2013;57:4463–4469. doi: 10.1128/AAC.01030-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kugelberg E, et al. Establishment of a superficial skin infection model in mice by using Staphylococcus aureus and Streptococcus pyogenes. Antimicrob Agents Chemother. 2005;49:3435–3441. doi: 10.1128/AAC.49.8.3435-3441.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pastagia M, et al. A novel chimeric lysin shows superiority to mupirocin for skin decolonization of methicillin-resistant and -sensitive Staphylococcus aureus strains. Antimicrob Agents Chemother. 2011;55:738–744. doi: 10.1128/AAC.00890-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Agarwal R, Roy K. Intracellular delivery of polymeric nanocarriers: a matter of size, shape, charge, elasticity and surface composition. Ther Deliv. 2013;4:705–723. doi: 10.4155/tde.13.37. [DOI] [PubMed] [Google Scholar]
- 25.Bikard D, et al. Programmable repression and activation of bacterial gene expression using an engineered CRISPR-Cas system. Nucleic Acids Res. 2013 doi: 10.1093/nar/gkt520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qi LS, et al. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 2013;152:1173–1183. doi: 10.1016/j.cell.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McDougal LK, et al. Pulsed-field gel electrophoresis typing of oxacillin-resistant Staphylococcus aureus isolates from the United States: establishing a national database. J Clin Microbiol. 2003;41:5113–5120. doi: 10.1128/JCM.41.11.5113-5120.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bae T, Baba T, Hiramatsu K, Schneewind O. Prophages of Staphylococcus aureus Newman and their contribution to virulence. Mol Microbiol. 2006;62:1035–1047. doi: 10.1111/j.1365-2958.2006.05441.x. [DOI] [PubMed] [Google Scholar]
- 29.Goldberg GW, Jiang W, Bikard D, Marraffini LA. Conditional tolerance of temperate phages via transcription-dependent CRISPR-Cas targeting. Nature. 2014 doi: 10.1038/nature13637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gibson DG, et al. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods. 2009;6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.