Abstract
Background
Detection of hypermethylated circulating tumor DNA has the potential to be a minimally invasive, low cost, and reproducible method for cancer detection.
Methods
We evaluated serum from 100 patients with known head and neck squamous cell carcinoma (HNSCC) and 50 healthy control patients for three previously described methylation targets, EDNRB, p16 and DCC, using quantitative methylation specific PCR (qMSP).
Results
EDNRB hypermethylation was identified in the serum of 10% HNSCC patients but in none of the control patients. DCC hypermethylation was detected in two serum samples from cancer patients that also amplified EDNRB and one of these samples also had p16 hypermethylation. ENDRB hypermethylation was statistically significant by Fisher’s exact test (p=0.03) when comparing HNSCC to controls.
Conclusions
Serum EDNRB hypermethylation is highly specific but not sensitive serum biomarker for HNSCC.
Introduction
The identification of molecular markers in body fluids for cancer detection is an area of intense research because of the promise panels of these biomarkers have for allowing the detection of a variety of solid tumors. Body fluids can carry whole cells, as well as protein, deoxyribonucleic acid (DNA), and ribonucleic acid (RNA) from tumors that can be readily identified by a variety of testing modalities. Examples of relevant body fluids used for detection include sputum for lung cancer diagnosis,1, 2 urine for renal and bladder cancers3, and saliva for head and neck squamous carcinoma (HNSCC),4–6 breast fluid collected by fine needle aspiration,7 and serum.8 Circulating biomarkers are attractive for cancer screening because they are blood-based tests that are minimally invasive, relatively low-cost and easily repeatable.
An epigenetic pathway of transcriptional inactivation for many tumor suppressor genes (TSG) includes CpG island hypermethylation within promoter regions.9, 10 This pathway has been identified in many different cancers and recent studies have focused on promoter hypermethylation in HNSCC.11, 12 Promoter hypermethylation in tissue samples can be detected by using real-time quantitative methylation-specific polymerase chain reaction (RT-Q-MS PCR). This methodology allows for an objective, robust, and rapid assessment of promoter methylation status. The ability to quantify methylation provides the potential for determination of a threshold level of methylation to improve sensitivity and specificity in detection of tumor-specific signal.13–15
The detection of DNA methylation in body fluids also has the potential to distinguish high-risk subjects that harbor occult cancers and have a higher risk for development of solid tumors in a wide variety of human cancers. Based on our group’s previous experience detecting aberrant methylation in salivary rinses16, 17 we conducted a study to investigate the ability of three of our most promising salivary detecting methylation probes to detect altered methylation in the serum of pretreatment HNSCC patients. Endothelin receptor type B (EDNRB), deleted in colorectal carcinoma (DCC) and cyclin-dependent kinase inhibitor 2A (CDKN2A or p16) were screened for hypermethylation in serum from HNSCC patients and normal control volunteers.
Methods
Serum samples
Samples were obtained from HNSCC patients presenting with a previously untreated squamous cell carcinoma from the oral cavity, larynx, or pharynx. Patients were evaluated and enrolled in a research study protocols from 1994 to 2003 in the Department of Otolaryngology–Head and Neck Surgery at Johns Hopkins Medical Institutions, Baltimore. Serum samples from these patients were collected before any cancer treatment, while the primary tumor was present. Patients were selected for candidacy for the study on basis of ability to provide adequate tumor sample, blood, salivary rinses, and availability for long-term follow-up. Serum samples from healthy patients were obtained through a head and neck cancer screening protocol. All individuals in the head and neck cancer screening protocol were called by phone once a year after enrollment and interviewed to determine interval changes in tobacco and alcohol consumption and health history including new cancer diagnosis. Patients with premalignant oral cavity lesions, a history of any type of cancer, or who had developed any type of cancer at the 1-year follow-up were excluded from this study. Both experimental protocols were approved by the Johns Hopkins Medical Institutions Institutional Review Board and written informed consent was obtained from all enrolled subjects.
Sample Selection
Samples were selected serum samples from 100 HNSCC patients and 50 healthy, cancer free controls for inclusion in this study from larger pools of previously collected samples. Every attempt was made to match both cohorts for age, gender, race, tobacco usage, and alcohol usage (Table 1). Mean age was 58.3 years in the HNSCC group and 56.2 years in the control group, and the groups were well matched for race and gender. The sera were predominantly from Caucasian patients and there was a higher incidence of both smoking and tobacco usage in the cancer group. Head and neck squamous cell carcinomas from the oral cavity, oropharynx, larynx and hypopharynx were included, with oral cavity and oropharyngeal tumors most strongly represented.
Table 1.
Patient characteristics.
| Cancer Patients (No. of patients=100) | Healthy Controls (No. of patients=50) | P-value | |
|---|---|---|---|
| Age | |||
| Mean (Std Dev) | 58.3 (11.7) | 56.2 (15.3) | 0.8 |
| Gender | |||
| Male | 77 (77%) | 29(58%) | 0.02 |
| Female | 23 (23%) | 21 (42%) | |
| Race | |||
| Caucasian | 90 (90%) | 47 (94%) | 0.59 |
| African American | 8 (8%) | 3 (6%) | |
| Asian | 2 (2%) | 0 (0%) | |
| Smoking Status | |||
| Never | 0 (0%) | 22 (44%) | <0.0001 |
| Former | 75 (75%) | 12 (24%) | |
| Current | 24 (24%) | 14 (28%) | |
| Unknown | 1 (1%) | 2 (4%) | |
| Alcohol Status | |||
| Never | 0 (0%) | 14 (28%) | <0.0001 |
| Former | 14 (14%) | 3 (6%) | |
| Current | 86 (86%) | 26 (52%) | |
| Unknown | 0 (0%) | 7 (14%) | |
| Tumor Site | |||
| Oropharynx | 46 (46%) | - | - |
| Oral cavity | 34 (34%) | - | - |
| Larynx | 14 (14%) | - | - |
| Hypopharynx | 2 (2%) | - | - |
| Unknown | 4 (4%) | - | - |
| Tumor Stage | |||
| I | 11 (11%) | - | - |
| II | 9 (9%) | - | - |
| III | 11 (11%) | - | - |
| IV | 69 (69%) | - | - |
DNA extraction
The blood was centrifuged at 2,000 × g for 10 minutes at room temperature, and 1 mL aliquots of serum/plasma samples were stored at −80°C. DNA extracted from the serum by digestion with 50 μg/mL proteinase K (Boehringer) in the presence of 1% SDS at 48°C for 2 days, followed by phenol/chloroform extraction and ethanol precipitation, and finally dissolved in 30 to 60 μL of Tris-EDTA (2.5 mmol/L ethylene diamine tetra-acetic acid [EDTA] and 10 mmol/L Tris-Hydrochloride).
Bisulfite treatment
Bisulfite conversion of 2 ug of genomic DNA from all samples was conducted using the Qiagen EpiTect bisulfite conversion kit using the manufacturer’s protocol. (Qiagen, Valencia, CA).
Target gene selection
Genes selected for this study, came from studies previously done by the authors to develop panels for early detection of HNSCC in other body fluids.16, 17 EDNRB, DCC and p16 hypermethylation probes were chosen based on previous specificity for HNSCC.
Quantitative methylation-specific polymerase chain reaction
The bisulfite-modified DNA was used as a template for fluorescence-based real-time polymerase chain reaction (PCR), as previously described.18 In brief, primers and probes were designed to specifically amplify the bisulfite-converted DNA for the β-actin gene (ACTB) and all genes of interest (primers and probes sequences are available on previous publications; DCC and p1616; EDNRB17). The ratios between the values of the gene of interest and the internal reference gene (ACTB), was obtained by Taqman analysis taking into account the PCR efficiency. Results were used as a measure of the relative quantity of methylation in a particular sample (value for the gene of interest/value for the reference gene × 100).
Fluorogenic PCR reactions were carried out in a reaction volume of 20 μL consisting of 600 nmol/L of each primer; 200 μmol/L of probe; 0.75 U of platinum Taq polymerase (Invitrogen); 200 μmol/L of each dATP, dCTP, dGTP, and dTTP; 200 nmol/L of ROX Dye reference (Invitrogen); 16.6 mmol/L of ammonium sulfate; 67 mmol/L of Trizma (Sigma); 6.7 mmol/L of magnesium chloride; 10 mmol/L of mercaptoethanol; and 0.1% dimethyl sulfoxide. Three microliters of treated DNA solution were used in each real-time MSP reaction. Amplifications were carried out in 384-well plates in a 7900 Sequence Detector System (Perkin–Elmer Applied Biosystems). Thermal cycling was initiated with a first denaturation step at 95°C for 2 minutes, followed by 45 cycles at 95°C for 15 seconds and 60°C or 62°C for 1 minute. Leukocytes from a healthy individual were methylated in vitro with excess SssI methyltransferase (New England Biolabs) to generate completely methylated DNA, and serial dilutions of this DNA were used for constructing the calibration curves on each plate.
Each reaction was done in triplicate, the average of the triplicate was considered for analysis. Results for Q-MSP was analyzed considering the quantity of methylation (normalized to ACTB expression) and considering methylation as a binary event, in which any quantity of methylation in a sample would be considered as positive. Samples for which the ACTB control did not amplify were removed from the study and replaced with new samples.
Statistical analysis
Statistical analyses for Q-MSP results were performed using Stata® 11 data analysis and statistical software (StataCorp, College Station, Texas). Methylation levels of EDNRB, DCC, and p16 between healthy subjects and HNSCC patients were compared using the Wilcoxon rank sum test and the frequency or marker presence was compared using Fisher’s exact test.
Binary indicators of EDNRB, DCC, and p16 methylation in HNSCC salivary rinses were used to study their association with two time-to-event clinical outcomes: overall survival and local-recurrence-free survival. Local recurrence free survival time was defined as the time elapsed from the date of completion of therapy to the date of local recurrence. Death without local recurrence is censored at the time of death. Other factors such as gender, tobacco exposure, HPV positivity, primary site location, overall stage, as well as T and N classification were also examined. For each prognostic factor, proportional hazards model was used to estimate the hazard ratio and the corresponding 95% confidence interval (CI). P-values were obtained from the log-rank test. All tests were two-sided with significance level set at 0.05.
Results
Head and neck cancer and control patient characteristics
HNSCC patients were predominantly male (77%) and Caucasian (90%) with a median age of 58.3 years. Current or past tobacco and alcohol consumption was found in 100% of patients in the HNSCC cohort, compared to control patients where 44% and 33% were non-smokers and non-drinkers, respectively (Table 1). The healthy control patients were more equally distributed among the sexes with only a small male predominance (58%), when compared to HNSCC samples where 77% were male. Age and race were able to be closely matched in this study (Table 1).
Evaluation of methylation levels in serum samples
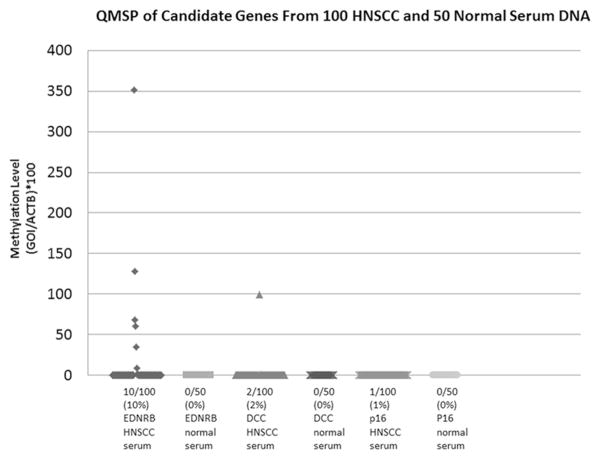
Q-MSP was conducted on all tumor and control samples using primer-probe pairs designed to detect hypermethylated EDNRB, DCC and p16 genes. Detection of hypermethylation was interpreted in a binary fashion with any amplification being considered positive. Ten serum samples from cancer patients had amplification of EDNRB. Two of these ten samples also had amplified DCC. One of these two samples also had amplification of p16. There was no amplification of any of the genes in serum from healthy control patients (Figure 1). The expression levels of EDNRB was significantly higher among cancer patients than controls (p=0.02), but the difference was not significant with DCC or p16 (p=0.32, and 0.48, respectively) (Table 2). The ability of the EDNRB probe to differentiate between HNSCC and normal control serum samples was found to be statistically significant by Fisher’s exact test (p=0.03), however neither DCC (p=0.55) nor p16 (p=0.99) were found to have statistical significance.
Figure 1.
EDNRB, DCC and p16 QMSP results in HNSCC and normal serum DNA.
Table 2.
EDNRB, DCC and p16 methylation signal for tumor and normal serum DNA samples.
| Mean | Median | P-value* | |
|---|---|---|---|
| EDNRB HNSCC serum | 6.55 | 0 | p=0.02 |
| EDNRB Normal serum | 0 | 0 | |
| DCC HNSCC serum | 1.01 | 0 | p=0.32 |
| DCC Normal serum | 0 | 0 | |
| p16 HNSCC serum | 0.000009 | 0 | p=0.48 |
| p16 Normal serum | 0 | 0 |
Wilcoxon rank-sum test
Interestingly, previous hypermethylation studies performed on primary tumors and oral salivary rinses from HNSCC patients showed concordant results. Only twenty-two of the serum samples evaluated in this study were previously used to evaluate promoter hypermethylation in salivary rinses and tumors from the same HNSCC patients. Nine out of the 10 HNSCC serum samples that amplified EDNRB in this study, also showed similar promoter hypermethylation in both primary tumor and salivary rinse samples from the same HNSCC patients. Of the 12 remaining HNSCC serum samples from patients tested in previous studies, 6 failed to amplify EDNRB in neither serum, saliva or tumor, 4 showed hypermethylation in both tumor and saliva but not serum, and 2 were positive in either the tumor or the saliva and not the serum samples.16, 17 These preliminary results suggest that promoter hypermethylation found in body fluids may closely mirror the methylation changes in primary tumors.
Methylation and clinical outcomes
We examined the association of the serum methylation markers and various clinical outcomes with overall survival and local recurrence free survival. As previously outlined, univariate analysis was conducted comparing the demographic characteristics of cancer patient serum samples revealed a statistically significant association between decreased local recurrence free survival and oral cavity primary tumor site (Table 3), and improved overall survival and HPV positivity, oropharyngeal primary site and low T classification, but no association with overall tumor stage or N classification (Table 4). No significant association was found between local recurrence-free survival or overall survival and serum positivity for any of the EDNRB, DCC and p16 hypermethylation probes (Tables 3 and 4).
Table 3.
Univariate analysis for local recurrence-free survival.
| P-value | Hazard Ratio | 95% CI | |
|---|---|---|---|
| Methylation Markers | |||
| Any positive (EDNRB/DCC/p16) | 0.77 | 1.35 | [0.17, 10.5] |
| Other risk factors | |||
| Gender (Male vs. Female) | 0.88 | 0.9 | [0.24, 3.33] |
| Smoking status – continuous for at least 1 year (Yes vs. No) | 0.19 | 2.79 | [0.61, 12.75] |
| Overall Stage | 0.38 | ||
| 1 | 0 | - | |
| 2 | reference | [0.32, 6.86] | |
| 3 | 0.53 | [0.05, 5.85] | |
| 4 | 0.67 | [0.15, 3.13] | |
| HPV (positive vs. negative) | 0.34 | 0.56 | [0.17, 1.86] |
| Primary site | 0.005 | ||
| Oral cavity | reference | ||
| Oropharynx | 0.11 | [0.02, 0.50] | |
| Larynx | 0.36 | [0.08, 1.71] | |
| Unknown primary | 0 | - | |
| T-classification | 0.12 | ||
| 1 | reference | ||
| 2 | 2.9 | [0.53, 15.86] | |
| 3 | 1.99 | [0.28, 14.12] | |
| 4 | 6.63 | [1.21, 36.37] | |
| Tx | 0 | - | |
| N-classification | 0.31 | ||
| 0 | reference | ||
| 1 | 4.07 | [0.57, 28.95] | |
| 2A | 0 | - | |
| 2B | 2.67 | [0.55, 12.87] | |
| 2C | 1.51 | [0.14, 16.68] | |
Table 4.
Univariate analysis for overall survival.
| P-value | Hazard Ratio | 95% CI | |
|---|---|---|---|
| Methylation Markers | |||
| Any positive (EDNRB/DCC/P16) | 0.63 | 1.35 | [0.41, 4.43] |
| Other risk factors | |||
| Gender (Male vs. Female) | 0.74 | 1.15 | [0.52, 2.54] |
| Smoking status – continuous for at least 1 year (Yes vs. No) | 0.35 | 1.75 | [0.79, 3.88] |
| Overall Stage | 0.77 | ||
| 1 | ref | ||
| 2 | 1.61 | [0.36, 7.32] | |
| 3 | 1.15 | [0.23, 5.70] | |
| 4 | 1.75 | [0.52, 5.91] | |
| HPV (positive vs. negative) | 0.01 | 0.38 | [0.17, 0.84] |
| Primary site | 0.0003 | ||
| Oral cavity | reference | ||
| Oropharynx | 0.11 | [0.02, 0.50] | |
| Larynx | 0.36 | [0.08, 1.71] | |
| Unknown primary | 0 | - | |
| T-classification | <0.0001 | ||
| 1 | reference | ||
| 2 | 3.58 | [1.07, 11.97] | |
| 3 | 3.71 | [1.11, 12.37] | |
| 4 | 10.97 | [3.56, 33.82] | |
| Tx | 0.73 | [0.08, 6.67] | |
| N-classification | 0.7 | ||
| 0 | reference | ||
| 1 | 1.44 | [0.40, 5.17] | |
| 2A | 0.75 | [0.17, 3.39] | |
| 2B | 0.84 | [0.38, 1.85] | |
| 2C | 1.65 | [0.58, 4.73] | |
Discussion
Aberrant promoter hypermethylation has been proposed as a means for detection of tumor-specific cells in body fluids and exfoliated cells in solid tumors, including HNSCC. In previous studies, we have evaluate a large sample size of both controls and HNSCC patients by using an expanded panel of methylated promoter regions to determine the ability of Q-MSP to detect tumor-specific promoter methylation in salivary rinses. In this study we evaluated three of the top genes identified from these studies to evaluate their ability to differentiate serum from cancer and healthy control patients. Given the sensitivity of the Q-MSP technique used to detect the presence of methylated alleles in a background of normal at a threshold of 1/1,000 to 1/10,000, this strategy allowed us to differentiate serum from a subset of cancer patients with 100% specificity. It has previously been shown that a combination of 3 or 4 genes is able to provide a sensitivity of cancer detection ranging from 24.0% to 35.1% with a specificity ranging from 90.0% to 97.1%.16, 19 EDNRB proved to be the most specific probe with specificity of 100%, but only recognized 10% or HNSCC samples. Interestingly, the addition of DCC and p16 probes did not improve the sensitivity of our study since they only amplified in samples that also had EDNRB hypermethylation. DCC and p16 were previously shown to have high specificity for HNSCC samples in oral rinses16 but this did not hold true for serum DNA as shown in this study.
Interestingly, EDNRB promoter hypermethylation found in HNSCC serum samples mirrored those of both oral salivary rinses and primary tumor tissue from the same HNSCC patients.16, 17 Only a limited number of HNSCC serum samples used in this screening study were also used in previous studies looking at hypermethylation in HNSCC oral salivary rinses and primary tumor tissue, but it does suggest that once methylation changes occur in the tumor they may also be present in secreted body fluids of HNSCC patients. These results are encouraging to continue the effort to actively identify and test potential HNSCC biomarkers in body fluids, which potentially can then be used for diagnostic and prognostic purposes. Further studies with larger sample sizes are needed to evaluate promoter hypermethylation changes in matched body fluid samples and primary tumors to help with these efforts.
The results of this screening study highlight the challenges of finding highly specific and sensitive markers for HNSCC as well as other solid tumors. As promoter hypermethylation patterns in individual tumors and even normal tissues show variation depending on specific altered molecular pathways, the use of multiple genes will provide greater applicability and coverage for diverse tumors when compared with a single gene for general detection use. This study was unable to increase the sensitivity by combining results from multiple probes but this is likely due to the overall low levels and frequency of hypermethylation in the HNSCC samples for the DCC and p16 probes.
This study has obvious limitation including that the probes utilized did not investigate the complete promoter region, so the potential functional silencing of the gene is not necessarily associated with our detection of methylation. However, the aim of this study was to evaluate performance of these probes as a screening tool rather that an indicator of functional alteration in primary tumor. These results also show that overall EDNRB, DCC and p16 have high specificity but also low sensitivity, which would not be advantageous for population-based screening. However, these results may have potential use for surveillance of a high-risk population or post-treatment HNSCC patients. Finally, this is an exploratory study in a single cohort of patients. As with all screening and detection modalities, an optimum combination of genes for hypermethylation-based HNSCC detection needs to continue to be developed and then further validated in an independent cohort.
In general, we were able to define EDNRB as a target for Q-MSP detection of HNSCC with a high degree of specificity but with an accompanied low sensitivity. All of our positive samples were Stage IV cancers. This may be due to the fact that larger or more aggressive tumors have a higher degree of intratumor vascular irregularities that allow increased leakage of tumor DNA into serum. Additionally, more aggressive tumors are likely to have a higher burden of epigenetic alteration increasing the likelihood that hypermethylated DNA can be detected in these patients serum.
Conclusion
We were able to confirm an elevated rate of promoter hypermethylation detected in HNSCC patient serum by probing three gene promoters previously found to be hypermethylated in HNSCC but not in control subjects. We found that we were able to detect serum from cancer patients with 100% specificity but low sensitivity using our probe for EDNRB. Further investigation is warranted to find additional serum methylation targets that can be used as a panel to increase the sensitivity of serum-based HNSCC screening. Serum-base screening has the potential to allow for earlier detection and treatment of HNSCC. New screening modalities for HNSCC are desperately needed, since, despite advances in surgical techniques and our understanding of the pathophysiology of HNSCC, overall survival has not increased in the past 30-years.20
Acknowledgments
Financial Support: Drs. Califano is supported by a NCI/NIDCR SPORE (5P50CA096784-05) and NIDCR Challenge Grant (RC1DE020324). Drs. Mydlarz and Hennessey are funded in part by a NIH T32 Research Training Grant (T32DC000027).
Footnotes
Conflict of interest statement: None of the authors of this manuscript have any relationships, financial or otherwise, or actual, potential, or perceived conflicts of interest to disclose.
References
- 1.Belinsky SA, Liechty KC, Gentry FD, et al. Promoter hypermethylation of multiple genes in sputum precedes lung cancer incidence in a high-risk cohort. Cancer Res. 2006;66(6):3338–44. doi: 10.1158/0008-5472.CAN-05-3408. [DOI] [PubMed] [Google Scholar]
- 2.Palmisano WA, Divine KK, Saccomanno G, et al. Predicting lung cancer by detecting aberrant promoter methylation in sputum. Cancer Res. 2000;60(21):5954–8. [PubMed] [Google Scholar]
- 3.Hoque MO, Begum S, Topaloglu O, et al. Quantitative detection of promoter hypermethylation of multiple genes in the tumor, urine, and serum DNA of patients with renal cancer. Cancer Res. 2004;64(15):5511–7. doi: 10.1158/0008-5472.CAN-04-0799. [DOI] [PubMed] [Google Scholar]
- 4.El-Naggar AK, Mao L, Staerkel G, et al. Genetic heterogeneity in saliva from patients with oral squamous carcinomas: implications in molecular diagnosis and screening. J Mol Diagn. 2001;3(4):164–70. doi: 10.1016/S1525-1578(10)60668-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nunes DN, Kowalski LP, Simpson AJ. Detection of oral and oropharyngeal cancer by microsatellite analysis in mouth washes and lesion brushings. Oral Oncol. 2000;36(6):525–8. doi: 10.1016/s1368-8375(00)00045-2. [DOI] [PubMed] [Google Scholar]
- 6.Rosas SL, Koch W, da Costa Carvalho MG, et al. Promoter hypermethylation patterns of p16, O6-methylguanine-DNA-methyltransferase, and death-associated protein kinase in tumors and saliva of head and neck cancer patients. Cancer Res. 2001;61(3):939–42. [PubMed] [Google Scholar]
- 7.Lee A, Kim Y, Han K, Kang CS, Jeon HM, Shim SI. Detection of tumor markers including carcinoembryonic antigen, APC, and cyclin D2 in fine-needle aspiration fluid of breast. Arch Pathol Lab Med. 2004;128(11):1251–6. doi: 10.5858/2004-128-1251-DOTMIC. [DOI] [PubMed] [Google Scholar]
- 8.Aggarwal C, Somaiah N, Simon GR. Biomarkers with predictive and prognostic function in non-small cell lung cancer: ready for prime time? J Natl Compr Canc Netw. 2010;8(7):822–32. doi: 10.6004/jnccn.2010.0059. [DOI] [PubMed] [Google Scholar]
- 9.Clark SJ, Melki J. DNA methylation and gene silencing in cancer: which is the guilty party? Oncogene. 2002;21(35):5380–7. doi: 10.1038/sj.onc.1205598. [DOI] [PubMed] [Google Scholar]
- 10.Herman JG, Baylin SB. Gene silencing in cancer in association with promoter hypermethylation. N Engl J Med. 2003;349(21):2042–54. doi: 10.1056/NEJMra023075. [DOI] [PubMed] [Google Scholar]
- 11.Esteller M, Corn PG, Baylin SB, Herman JG. A gene hypermethylation profile of human cancer. Cancer Res. 2001;61(8):3225–9. [PubMed] [Google Scholar]
- 12.Ha PK, Califano JA. Promoter methylation and inactivation of tumour-suppressor genes in oral squamous-cell carcinoma. Lancet Oncol. 2006;7(1):77–82. doi: 10.1016/S1470-2045(05)70540-4. [DOI] [PubMed] [Google Scholar]
- 13.Bernard PS, Wittwer CT. Real-time PCR technology for cancer diagnostics. Clin Chem. 2002;48(8):1178–85. [PubMed] [Google Scholar]
- 14.Eads CA, Danenberg KD, Kawakami K, et al. MethyLight: a high-throughput assay to measure DNA methylation. Nucleic Acids Res. 2000;28(8):E32. doi: 10.1093/nar/28.8.e32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jeronimo C, Usadel H, Henrique R, et al. Quantitation of GSTP1 methylation in non-neoplastic prostatic tissue and organ-confined prostate adenocarcinoma. J Natl Cancer Inst. 2001;93(22):1747–52. doi: 10.1093/jnci/93.22.1747. [DOI] [PubMed] [Google Scholar]
- 16.Carvalho AL, Jeronimo C, Kim MM, et al. Evaluation of promoter hypermethylation detection in body fluids as a screening/diagnosis tool for head and neck squamous cell carcinoma. Clin Cancer Res. 2008;14(1):97–107. doi: 10.1158/1078-0432.CCR-07-0722. [DOI] [PubMed] [Google Scholar]
- 17.Demokan S, Chang X, Chuang A, et al. KIF1A and EDNRB are differentially methylated in primary HNSCC and salivary rinses. Int J Cancer. 2010;127(10):2351–9. doi: 10.1002/ijc.25248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harden SV, Tokumaru Y, Westra WH, et al. Gene promoter hypermethylation in tumors and lymph nodes of stage I lung cancer patients. Clin Cancer Res. 2003;9(4):1370–5. [PubMed] [Google Scholar]
- 19.Carvalho AL, Magrin J, Kowalski LP. Sites of recurrence in oral and oropharyngeal cancers according to the treatment approach. Oral Dis. 2003;9(3):112–8. doi: 10.1034/j.1601-0825.2003.01750.x. [DOI] [PubMed] [Google Scholar]
- 20.Mydlarz WK, Hennessey PT, Califano JA. Advances and Perspectives in the Molecular Diagnosis of Head and Neck Cancer. Expert Opin Med Diagn. 2010;4(1):53–65. doi: 10.1517/17530050903338068. [DOI] [PMC free article] [PubMed] [Google Scholar]