Abstract
The serine, glycine, one carbon (SGOC) metabolic network is implicated in cancer pathogenesis but its general functions are unknown. We carried out a computational reconstruction of the SGOC network and then characterized its expression across thousands of cancer tissues. Pathways including methylation and redox metabolism exhibited heterogeneous expression indicating a strong context dependency of their usage in tumors. From an analysis of coexpression, simultaneous up- or down-regulation of nucleotide synthesis, NADPH and glutathione synthesis was found to be a common occurrence in all cancers. Finally, we developed a method to trace the metabolic fate of serine using stable isotopes, high-resolution mass spectrometry and a mathematical model. Although the expression of single genes didn’t appear indicative of flux, the collective expression of several genes in a given pathway allowed for successful flux prediction. Together these findings identify expansive and heterogeneous functions for the SGOC metabolic network in human cancer.
Introduction
Serine and glycine are nutrients that fuel metabolic pathways including one carbon metabolism and sulfur metabolism. This metabolic unit referred to as the serine, glycine and one carbon (SGOC) network provides an integration point in cellular metabolism that allows for cells to achieve diverse biological functions by converting serine and glycine into several metabolic outputs. These outputs include building blocks for nucleotide, lipid, and protein synthesis. They also include polyamine synthesis and work in the maintenance of redox status as determined by glutathione biosynthesis and NADPH production (Circu and Aw, 2010; Fan et al., 2014; Lewis et al., 2014a; Murphy et al., 2011; Tedeschi et al., 2013). The network also provides the substrates for methylation reactions that may have relevance to maintaining cellular epigenetic status (Gut and Verdin, 2013; Teperino et al., 2010).
Recent work has pointed to new roles of the SGOC network in cancer pathogenesis (Chaneton et al., 2012; Jain et al., 2012; Ma et al., 2013; Maddocks et al., 2013; Scuoppo et al., 2012; Zhang et al., 2012). Whereas a subset of cancer cells increase de-novo serine biosynthesis (Locasale et al., 2011; Possemato et al., 2011), other cancers benefit from an increased serine and glycine uptake rate which allows them to metabolize these amino acids for their biosynthetic needs (Jain et al., 2012; Maddocks et al., 2013). Importantly, a recent study showed that serine but not glycine is critical in providing the one-carbon units required for biosynthesis of nucleotides in some cancer cells (Labuschagne et al., 2014). A critical role for the mitochondrial folate pathway in rapidly proliferating cancer cells has also been recently elucidated (Nilsson et al., 2014). Furthermore, the importance of one-carbon metabolism in NADPH production through the oxidation of folates was demonstrated in cancer cells in a number of recent studies (Fan et al., 2014; Lewis et al., 2014a; Tedeschi et al., 2013; Vazquez et al., 2011). These studies showed that in addition to providing cells with nucleotide units, one-carbon metabolism has an important role in redox balance. Despite these advances, the general coordinated usages and different contexts in which serine and glycine flux contributes to different metabolic functions within and across cancer types and normal tissues remain largely unknown.
Previous studies have analyzed the expression levels of metabolic pathways across a variety of cancer types using meta-analysis approaches (Hu et al., 2013; Nilsson et al., 2014; Tedeschi et al., 2013; Vazquez et al., 2013). These studies have identified tumor-associated changes in gene expression across human metabolism including one carbon metabolism. Whereas previous work has characterized expression of the SGOC network effectively as a pathway (Hu et al., 2013) and thus one or two data points or as a single series of individual genes (Nilsson et al., 2014), we attempted to carefully examine with higher resolution the coordination and context-dependence of functionally distinct pathways of serine utilization. We further investigate our computational findings experimentally by tracing the metabolic fate of serine and connecting these observations to expression patterns in the network. Together we identify several novel context dependent utilizations of serine in human cancers.
Results
Reconstruction of the human SGOC network
We constructed a network that represents the metabolism of serine and glycine through one carbon metabolism and other immediate pathways including the transsulfuration pathway that together lead to defined cellular outputs. This network, collectively referred to the SGOC network, was generated first by curating all human metabolic genes from the Kyoto Encyclopedia of Genes and Genomes (KEGG) (Kanehisa and Goto, 2000). Each gene involved in the KEGG-defined pathways, Glycine - Serine and Threonine metabolism, Cysteine and Methionine metabolism, and Folate biosynthesis was then selected. We subsequently included genes involved in adjacent chemical reactions (edges) of the selected genes (nodes), and these nodes were then connected to the selected edges to allow for a contiguous sequence of chemical reactions. Finally, we pruned this network to exclude each isolated node as well as enzymes that carry out chemical reactions involved in other pathways that we concluded to be non-specific to SGOC metabolism (Figure 1A, Methods).
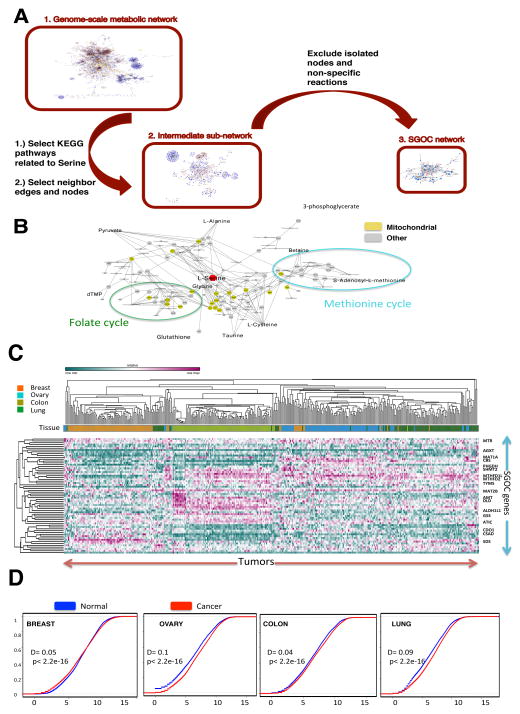
Figure 1. Reconstruction of the functional SGOC network.
A.) The entirety of the human metabolic network is first considered using a list of all metabolic genes from KEGG. A sub network involving all human serine, glycine, and one carbon metabolism related genes are considered obtained from KEGG. Reaction paths from this network were used to create the final SGOC network. B.) Schematic of resulting network and its decomposition into functional outputs (mitochondrial isoforms are shown in yellow). (See also Table S2)
C.) Expression patterns of SGOC network genes across four cancer types (ovarian, lung, colorectal, and breast) visualized with unsupervised hierarchical clustering (100 randomly chosen samples from each cancer type from TCGA were used). (See also Figure S1).
D.) Cumulative density plots showing the distribution of SGOC expression levels across four tumor types and their corresponding normal tissues (GENT).
As a result, the network comprises of a set of sixty-four genes involving core metabolic reactions and enzymes and isoenzymes (Figure 1B). Inputs of the network include de novo serine and glycine metabolism from glucose and the import of serine and glycine from the extracellular space. Outputs of the network include purine, pyrimidine, lipid, glutathione, redox, taurine, and methylation metabolism. In practice the network is compartmentalized into cytosolic/nuclear and mitochondrial components that can be to some extent captured in these reaction sequences when metabolites are shuttled in and out of the mitochondria.
Expression of the SGOC network in human cancers and normal tissues
Having constructed this network, we then investigated its expression in several cancer types and corresponding normal tissues. We considered both The Cancer Genome Atlas (TCGA) data across Breast, Ovarian, Lung, and Colorectal cancers (Cancer Genome Atlas, 2012a, b; Cancer Genome Atlas Research, 2011, 2012) and the Gene Expression in Normal and Tumor (GENT) database (Shin et al., 2011) that contains larger numbers of normal tissue samples. We chose these cancer types for two reasons: 1.) The three major types Breast, Lung, and Colon represent major sources of cancer mortality with ovarian cancer exhibiting expression patterns similar to that of subtypes of breast cancer observed to have enhanced requirements for serine metabolism (Locasale et al., 2011; Possemato et al., 2011) and 2.). Each of the three major cancer types considered has been shown to exhibit clinical response to chemotherapies that target nodes within the one carbon metabolism network (Chabner and Longo, 2011). For each tumor type, expression data are available across hundreds of tumors allowing for extensive statistical characterization.
We first assessed the global expression of the genes in the network (Table 1, Table S1). A hierarchical clustering across the cancer types revealed clustering based largely on tissue type (Figure 1C) consistent with the results of principal component analysis (PCA) (Figure S1). Major exceptions were observed for lung cancer samples as well as a subset of breast cancers that clustered with ovarian cancer in which case breast cancers lacking estrogen receptor exhibited expression patterns indistinguishable from those of ovarian cancer. Next, an analysis of the global properties of the network was considered. In ovarian, colon, and lung cancers, the overall distribution of SGOC network gene expression is shifted toward higher expression levels compared to the levels in corresponding normal tissue (Figure 1D). However, in breast cancer, the broad range of the cumulative density plot shows a higher variability in the expression levels of SGOC genes compared to normal breast tissue (Figure 1D). In all cancer types, the distribution of SGOC expression differed between tumor and normal (Kolmogorov-Smirnov p-value < 2.2e-16). We next considered the variability between and within cancer types to further identify cancer contexts of the network (Table 1). These contexts include high expression in one tumor type relative to others, large variation in a single tumor type, high expression in tumor versus corresponding normal tissue, and high variability in tumor vs. normal (Table S1). The resulting calculations revealed that genes predominantly involved in de novo nucleotide synthesis pathways are over-expressed in all four cancer types compared to the corresponding normal tissues (Table 1). This result is in agreement with the previous literature emphasizing the importance of nucleotide biosynthesis to rapidly proliferating cells (Hu et al., 2012; Wilson et al., 2012). Furthermore, consistent with a recent observation (Nilsson et al., 2014) we also observe methylenetetrahydrofolate dehydrogenase 2 (NADP+ dependent) (MTHFD2) and serine hydroxymethyltransferase 2 (SHMT2), two mitochondrial enzymes that contribute to nucleotide metabolism, to be consistently overexpressed in cancers compared to corresponding normal tissues (Table 1). Furthermore, when SGOC enzymes with a mitochondrial isoform were compared to their cytosolic isoforms, the mitochondrial isoforms showed stronger up-regulation in tumors (Fisher’s exact test p-value= 0.02), demonstrating the importance of mitochondrial compartment in tumor metabolism (Supplementary Methods). However, some cancer type-specific expression patterns were also observed. For instance cystathionine beta-synthase (CBS), serine dehydratase (SDS), and glutathione synthetase (GSS) were expressed more highly in ovarian, breast, and colon tumors, respectively (Table 1). These three genes also showed very little within-cancer type variation in contrast to other genes. Cross normal tissue comparisons revealed high expression of thymidylate synthetase (TYMS) in normal colon and alanine-glyoxylate aminotransferase (AGXT) in normal breast (Table 1). Together, the variation in genes within tumor types and in comparison to corresponding normal tissues suggested many newly identified context dependent expression patterns in the network.
Table 1. SGOC expression analysis across tissues.
The SGOC genes are listed in the first column with mitochondrial enzymes denoted by “m”. The second column shows the cancer type with the highest average expression across the 4 cancers in the study. In the significance column, Mann-Whitney-Wilcoxon p-values smaller than the genome-level Bonferroni threshold (2.8 e 10-6) are denoted by an “x” and effect sizes larger than 0.3 are denoted by a “+” sign. The fourth and fifth columns summarize similar analysis across the 4 normal tissue types. The sixth and seventh columns summarize the results of tumor vs. normal comparisons. In the sixth column, tissue types in which a significant over-expression (p-value < 2.8 e 10-6) was detected in tumors are listed. The fifth column specifies corresponding effect sizes by a “+” if larger than 0.3 and a “−” otherwise. “NA” denotes missing data.(See also Table S1).
| Name | T-T highest | T-T significance | N-N highest | N-N significance | T-N overexp. (pvalue<2.8e10-6) | T-N significance |
|---|---|---|---|---|---|---|
| ADC | breast | x+ | breast | x+ | breast, ovary, lung | −, −, − |
| AGXT1 | colon | x+ | breast | x+ | Lung | − |
| AGXT2(m) | breast | x+ | breast | x+ | colon, lung | −, − |
| AHCY | colon | x | colon | x+ | breast, colon, lung | −, +, + |
| AHCYL1 | ovary | x+ | breast | x+ | ovary, colon | −, − |
| ALAS1(m) | colon | x | colon | x+ | Ovary | − |
| ALAS2(m) | lung | x | breast | x+ | Colon | − |
| AMT(m) | colon | x | ovary | x | - | |
| ANPEP | colon | + | breast | x+ | Ovary | − |
| ATIC | colon | x | colon | x+ | breast, colon, lung | +, +, + |
| BHMT | lung | x | breast | x+ | ovary, lung | −, − |
| BHMT2 | breast | x+ | breast | x+ | ovary, lung | −, − |
| CBS | ovary | x+ | ovary | breast, ovary, lung | −, −, + | |
| CDO1 | breast | x+ | breast | x+ | Colon | − |
| CHDH(m) | colon | x | breast | x+ | ovary, colon, lung | −, −, + |
| CP | ovary | x | lung | x | ovary, lung | −, + |
| CSAD | breast | x+ | breast | x+ | - | |
| CTH | colon | x+ | colon | x+ | lung | + |
| DHFR | colon | x | colon | x+ | breast, ovary, colon, lung | −, −, +, + |
| DLD(m) | colon | x+ | colon | x+ | breast, ovary | −. − |
| DMGDH(m) | ovary | x+ | breast | x+ | colon | − |
| FPGS(m) | ovary | + | breast | x+ | ovary, colon, lung | −, +, − |
| FTCD | ovary | x | breast | x+ | ovary, colon, lung | −, −, − |
| GAD1 | lung | x | breast | x+ | colon, lung | +, + |
| GAD2 | lung | breast | x+ | colon, lung | −, − | |
| GAMT | breast | x | breast | x+ | ovary, lung | −, + |
| GART | colon | x | breast | x | breast, ovary, colon, lung | −, +, +, + |
| GATM | breast | x+ | breast | x+ | lung | − |
| GCAT(m) | lung | colon | x | breast, colon, lung | −, −, + | |
| GGH | colon | x | colon | x+ | breast, ovary, colon, lung | −, −, −, + |
| GLDC(m) | ovary | x+ | breast | x+ | ovary, lung | −, + |
| GNMT | breast | x | breast | - | ||
| GOT1 | colon | x+ | colon | x+ | breast | − |
| GOT2(m) | colon | x+ | ovary | x | breast, colon, lung | −, −, − |
| GPT | colon | x+ | colon | x+ | - | |
| GPT2 | ovary | x | breast | x+ | colon, lung | +, + |
| GSS | colon | x+ | colon | x+ | breast, colon, lung | −, −, + |
| IL4I1 | ovary | x | ovary | breast, ovary, colon, lung | −, −, −, + | |
| MAT1A | ovary | x+ | ovary | x | colon | − |
| MAT2A | ovary | x+ | lung | ovary, colon | +, − | |
| MAT2B | colon | x | breast | x | - | |
| MPST(m) | colon | x | colon | x+ | ovary | − |
| MTFMT (m) | colon | x | ovary | - | ||
| MTHFD1 | colon | x+ | breast | x+ | colon, lung | −, + |
| MTHFD1L (m) | lung | x | ovary | x | colon, lung | +, + |
| MTHFD2 (m) | lung | x | colon | x | breast, ovary, colon, lung | +, +, +, + |
| MTHFR | ovary | x+ | breast | x | - | |
| MTHFS | lung | x | ovary | breast, colon, lung | −, −, − | |
| MTR | ovary | x+ | ovary | x+ | colon | + |
| NAGS(m) | colon | x | colon | lung | − − | |
| PDPR(m) | ovary | x+ | breast | x+ | ovary, colon | −, − |
| PHGDH | ovary | x | breast | x+ | colon, lung | +, − |
| PIPOX | ovary | x | ovary | x+ | colon, lung | +, + |
| PPCS | lung | ovary | x | breast, lung | −, + | |
| PPIG | ovary | x+ | ovary | x | - | |
| PSAT1 | ovary | x | breast | x+ | ovary, colon, lung | +, +, + |
| PSPH | lung | ovary | x | breast, colon | −, + | |
| SARDH(m) | breast | x+ | breast | x+ | lung | − |
| SDS | breast | x+ | breast | x+ | breast, ovary, colon, lung | −, −, −, + |
| SHMT1 | ovary | x | breast | x+ | lung | − |
| SHMT2(m) | ovary | x+ | ovary | x | breast, ovary, colon, lung | +, −, +, + |
| TYMS | colon | colon | x+ | breast, colon, lung | +, −, + | |
| ALDH1L1 | colon | x | breast | x+ | ovary, colon, lung | −, +, + |
| ALDH1L2 (m) | NA | NA | breast | x+ | ovary, colon, lung | +, +, + |
Serine utilization in the SGOC network
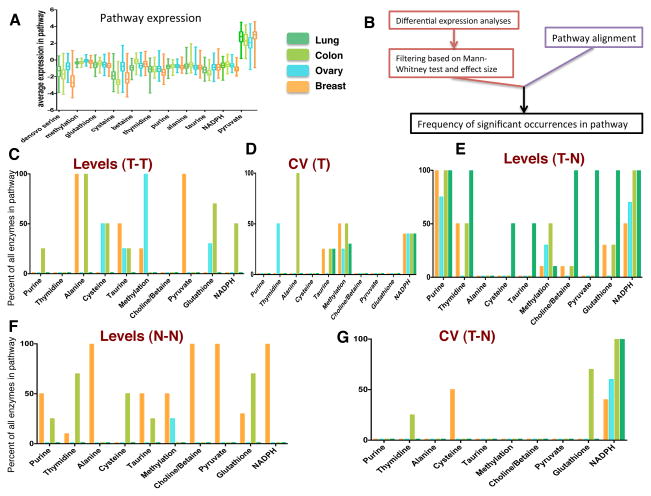
After analyzing the expression of individual genes, we considered a functional analysis of the metabolic outputs of the network. We first introduced a framework for understanding the different metabolic fates of serine. We decomposed the SGOC network into pathways that utilize serine as an input and achieve a distinct biological function as an output (Methods). We also added de novo serine biosynthesis as an additional pathway due to its implications in cancer (Locasale, 2013). For each pathway we analyzed the overall range of expression (Figure 2A). It was observed that there is large within-cancer variability in expression of pathways especially across different breast tumors (Figure 2A). Furthermore, mean pathway expression was similar across cancer types for some pathways such as methylation, whereas there were significant differences in other pathways including de novo serine biosynthesis (Figure 2A). Next we developed an algorithm to evaluate the expression along each functional pathway (Figure 2B). We first decomposed each biologically distinct unit of the network into a set of genes. Then, the mean, median, and variation of expression for each gene comprising each pathway was computed and statistics were evaluated across the population of tumors (Methods). Since the pathways varied in length, we normalized the obtained values to the number of genes contained in the pathway.
Figure 2. Functional SGOC pathway utilization across four cancer types.
A.) Boxplots showing average pathway expression across four cancer types (TCGA).
B) Summary for quantification of pathway utilization analyses.
C–G.) Barplots denoting the collective expression of a given pathway route in the SGOC network. C.) Pathways hyper-utilized in a single cancer type relative to others. D.) Pathways exhibiting high variation within cancer types. E.) Pathways overexpressed in tumor versus corresponding normal tissue. F.) Pathways highly expressed in one normal tissue relative to others. G.) Pathways with high variation in tumor versus normal tissue.
We considered this pathway analysis with several contrasts: expression levels in one cancer relative to other tumor types (T-T), variability in individual tumor (T) types, over-expression of the pathway in tumor relative to normal tissue (T-N), expression of the pathway in one normal tissue relative to other tissue types (N-N), variability in tumor vs. corresponding normal tissue (T-N CV). Each of these analyses provides unique information relevant to cancer. For example, high expression in the normal tissue may indicate a predisposition for the usage of the pathway in a particular cancer type. Furthermore, a high variability indicates possible selective usage or overexpression in some context of tumorigenesis such as a particular mutational event. Due to the tremendous heterogeneity within cancer types, a high within-cancer-type variability is important to know since it implies that different sub-populations of samples behave differentially with respect to a certain pathway, which suggests their potential use as biomarkers, provides further context for the use of the pathway, and finally possibly yields some predictive capacity for evaluating the response to agents that target the pathway. A comparison across tumor tissues shows that several pathways are overexpressed relative to other tumors with breast and ovarian cancer exhibiting higher expression of specific components of sulfur related metabolism (Figure 2C). High variability across each cancer type was observed throughout the network but the taurine, methylation, and NADPH pathways showed highest variability compared to other pathways (Figure 2D). However, when comparing expression in normal and tumor tissue, only routes related to nucleotide and redox metabolism were commonly upregulated in tumors consistent with previous analysis of individual genes (Figure 2E). Interestingly, in comparing individual genes and pathways, we found that high relative levels of expression in one normal tissue did not necessarily overlap with high relative levels of expression in tumor tissue, indicating shifts in metabolism that are not directly due to differences in tissues of origin (Figure 2F). This observation is also apparent in comparing the variability in expression of tumor and normal cancer types (Figure 2G). Together these findings identify novel relationships of context dependent utilizations of the SGOC network.
Co-occurrence of pathway utilization within the network
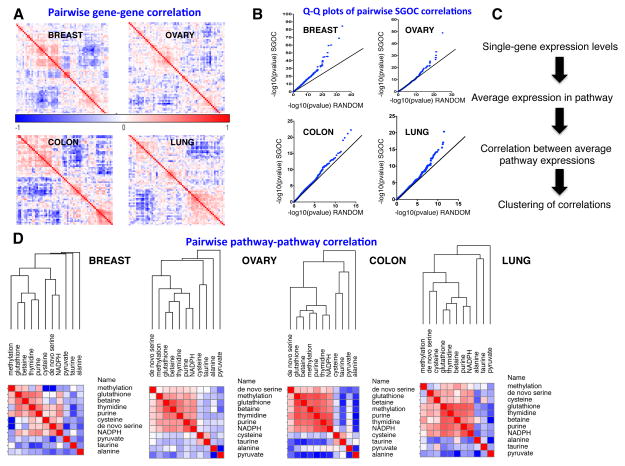
Since in metabolism, the output of one branch of the network is coupled to the output of all other branches, we hypothesized that there could exist correlations in the expression of sets of genes leading to metabolic outputs of the network. To investigate this possibility we computed a similarity matrix for each of the cancer types. Clustering of each similarity matrix revealed co-occurring expression patterns in each cancer type suggesting coordinated utilization of certain enzymes (Figure 3A). A global analysis of these pairwise correlations revealed that the genes in the SGOC network were significantly more correlated compared to randomly chosen genes as revealed by quantile-quantile (QQ) plots (Figure 3B). To our knowledge this is the first systematic demonstration of coordinated expression of metabolic genes in a defined metabolic pathway in humans.
Figure 3. Correlations across reaction paths in the SGOC network.
A.) Hierarchically-clustered similarity matrices for four cancer types based on pairwise Spearman correlations. Modularity in the co-expression network is apparent. B.) Overview of algorithm that collapses co-regulated genes into reaction paths. C.) Quantile-Quantile (QQ) plots showing p-values from gene-gene spearman correlations in SGOC compared to randomly chosen pair of genes. D.) Correlations in expression of transcripts in one pathway in reference to another. Results are organized by hierarchical clustering using Spearman correlations in linkage similarities.
Next, we considered the normalized mean expression of the pathway routes that illustrated the extent of co-occurrence in the network (Figure 3C). When assessed across the four cancer types, it is found that correlations emerge along specific pathways. Surprisingly many of these correlations were largely found to be independent of tissue type demonstrating the existence unique metabolic programs within each tissue. A core group of pathways involving glutathione, NADPH, and nucleotide metabolism show correlations in their expression suggesting that a coupling between de novo nucleotide and redox metabolism occurs. De novo serine synthesis correlated only with cysteine metabolism in some cancer types indicating that the condensation of serine and entry into the transsulfuration pathway is a major usage of serine in cells with enhanced de novo serine biosynthesis. Also in all cancer types, taurine and nucleotide metabolism anti-correlated with one another indicating that their usages are orthogonal in cancers.
Serine-derived metabolic fluxes in the network
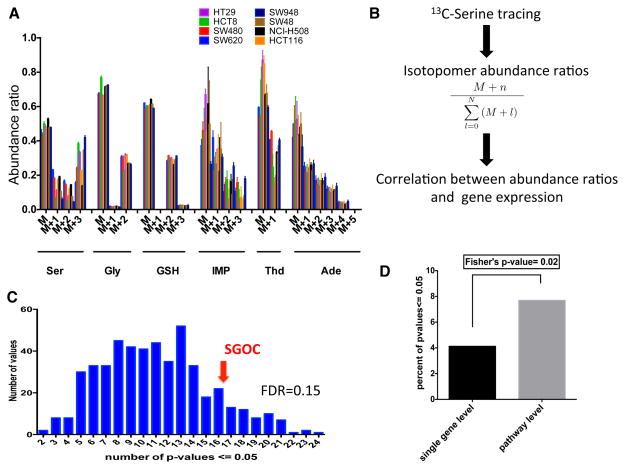
Gene expression in human clinical samples offers an unbiased assessment of the expression of the network in cancer. However gene expression does not necessarily determine metabolic phenotype that ultimately involves metabolic flux or the rate of flow of a metabolite from one point in the network to another. To investigate the relationship of metabolic flux with gene expression, we first developed a 13C serine-based isotopomer mass spectrometry method. A panel of cancer cells was incubated with 13C labeled serine and relative abundances of all mass isotopomers (molecules differing only in the extent of their isotopic composition) of each metabolite were measured from the integrated ion current. Uniformly labeled serine (U-13C Serine) was present in the media in a 1:1 proportion with respect to unlabeled serine in the culture medium. Therefore, at steady state, as expected a substantial portion of the serine is unlabeled (Figure S2). Furthermore, M+1 glycine was detected near the natural abundance level in our experiment (Figure S2), suggesting that glycine cleavage does not happen in reverse at a substantial rate. The eight human colon cancer cell lines used in this study consistently showed labeling of glutathione and de novo purine and thymidine synthesis intermediates, while little or no label was detected on methionine, pyruvate, alanine, betaine and taurine (Figure 4A, Figure S3). Since gene expression is available for each of these cell lines, we could then ask to what extent patterns in gene expression could be related to these isotope patterns. Using abundance ratios of mass isotopomers as surrogates for the corresponding fluxes, we studied correlations between labeling patterns and gene expression first at the single gene level and then at the level of pathways (Figure 4B). We asked whether gene expression within the network could predict isotope labeling patterns. We found that the expression of individual genes in the SGOC network could to some extent predict fluxes (Figure 4C, FDR q-value 0.15). Notably, results from a Fisher’s exact test indicate that gene expression at the pathway level is more strongly correlated with fluxes compared to single gene level (Figure 4D). In summary, results from these data indicate that fluxes can be determined directly to an extent from gene expression in cancer cells but this requires knowledge of the overall expression of the pathway. To our knowledge this is the first study in a mammalian system that provides an example of the relationship between gene expression and flux.
Figure 4. Flux analysis of serine metabolism.
A.) Abundance ratios for labeled isotopomers detected across the 8 colon cancer cell lines. B.) Overview of mass isotopomer analyses. C.) Histogram of the number of significant correlations between labeled SGOC metabolite isotopomers and expression of randomly selected networks of genes. Expression of SGOC genes shows a higher than random correlation (FDR=0.15). D.) Quantification of the frequency of significant correlations between SGOC expression and isotopomer labeling at single genes level vs. pathway level using a Fisher’s exact test. (See also Figure S3).
Interaction between de novo nucleotide and glutathione biosynthesis
We then further investigated the relationship between de novo nucleotide metabolism and glutathione synthesis (Figure 5A) in colorectal cancer cells. We considered the relationship between thymidine and glutathione synthesis, purine and glutathione synthesis, and purine and thymidine synthesis (Figure 5A). In each case, the expression levels of key genes in the pathway provided little information about the pathway relationships. However, when comparing pathway expression, strong correlations in nucleotide synthesis and glutathione biosynthesis exist. This phenomenon was observed both in TCGA colon cancer data as well as in gene expression data from the 8 colon cancer cell lines in the study. Experimentally, this result is also apparent in that labeling of de novo nucleotide synthesis intermediates was highly correlated with glutathione labeling (Figure 5A). Together these findings point to a model where increases in nucleotide biosynthesis are coupled to flux to glutathione synthesis whereas other pathway routes from serine are not correlated with nucleotide synthesis (Figure 5B). Since NADPH synthesis is coupled to the redox balance of oxidized and reduced glutathione, this finding provides a possible connection to a recent finding indicating that one carbon metabolism is a major source of NADPH in cells (Fan et al., 2014). In fact, we also observe a positive association between NADPH synthesis with nucleotide and glutathione synthesis at the pathway level (Figure S4).
Figure 5. Interaction between de novo nucleotide and glutathione synthesis.
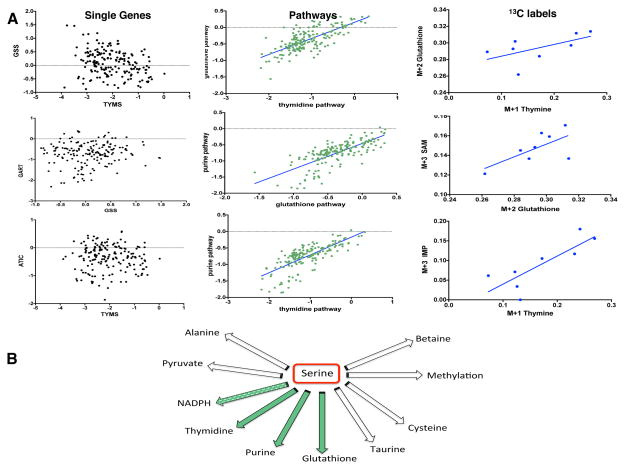
A.) Thymidine and Glutathione (top panel): The left plot shows weak association between TYMS and GSS expression (R-squared=0.094). The middle plot shows significantly higher correlation between thymidine and glutathione at average pathway expression level (R-squared= 0.53). The right plot shows association between glutathione and thymine label ratios (R-squared=0.35). Purine and Glutathione (middle panel): The left plot shows weak association between phosphoribosylglycinamide formyltransferase (GART) and GSS expression at single gene level (R-squared=0.01). The middle plot shows significantly higher correlation between purine and glutathione at average pathway expression level (R-squared=0.54). The right plot shows association between glutathione and S-adenosyl-methionine (SAM) - a purine intermediate- label ratios (R-squared=0.44). Thymidine and Purine (bottom panel): The left plot shows weak association between TYMS and IMP cyclohydrolase (ATIC) expression at single gene level (R-squared=0.001). The middle plot shows significantly higher correlation between thymidine and purine at average pathway expression level (R-squared=0.55). The right plot shows association between inosine-monophosphate (IMP) -a purine intermediate- and thymine label ratios (R-squared=0.67).
B.) Schematic showing serine metabolic outputs. Results from tracing serine in vitro suggest a simultaneous flux going through de novo nucleotide and glutathione synthesis pathways (green arrows) in the colon cancer cell lines studied when little or no flux goes from serine to the other pathways. NADPH labeling could not be assayed in our experimental set-up using 13C-serine. (See also Figure S4).
Mathematical modeling of pathway fluxes
Although labeling patterns can sometimes be used to infer the extent of flux through a pathway, in general this is not always the case. Fluxes ultimately are required to be estimated from network models that account for the isotope patterns. We therefore considered a mathematical model of the Mass Isotopomer Distributions (MIDs) to estimate quantitative values for the fluxes in the network (Figure 6A, Methods). A two-compartment Serine-Glycine-Methylenetetrahydrofolate (Ser-Gly-meTHF) metabolic model was used to fit experimental 13C MIDs of the SGOC network to determine metabolic fluxes involving transport and exchange relative to extracellular serine transport (Ftr-ser=1). The metabolic network includes cellular serine production via de novo synthesis from 3-phosphoglycerate, extracellular serine uptake, reversible cytosolic SHMT1 and mitochondrial SHMT2 fluxes, mitochondrial glycine cleavage system activity (GCS), serine, glycine and meTHF exchange fluxes between cytosolic and mitochondrial compartments, dilution fluxes for cytosolic and mitochondrial one-carbon metabolite pool represented in the model by meTHF, de novo and salvage formation pathways for adenine (Ade representing purines) and thymidine (Thd representing pyrimidines), and glutathione production flux. An analysis of the fluxes across each cell line provided direct quantitative information about the distribution of fluxes across the network (Figure 6B). For example flux from glycine to serine varied over an order of magnitude suggesting that some cells may be able to compensate for differences in serine availability through the uptake of glycine. Across the cell lines, results from the analyses of estimated fluxes show a positive relationship between the de novo purine and thymidine pathway fluxes consistent with the pathway expression levels across the 8 cell lines (Figure 6C). Furthermore, using estimated flux through SHMT1 as a surrogate for glutathione production flux, a positive relationship is detected between glutathione and thymidine synthesis both at the flux level as well as the pathway expression level (Figure 6D). Similarly, purine biosynthesis shows a positive association with glutathione production (Figure 6E). Together these findings both identify the heterogeneity of pathway fluxes across the network in a set of cell lines and provide confirmation of correlations that exist in the utilization of serine for specific metabolic functions.
Figure 6. Mathematical modeling estimates fluxes in purine and pyrimidine synthesis pathways.
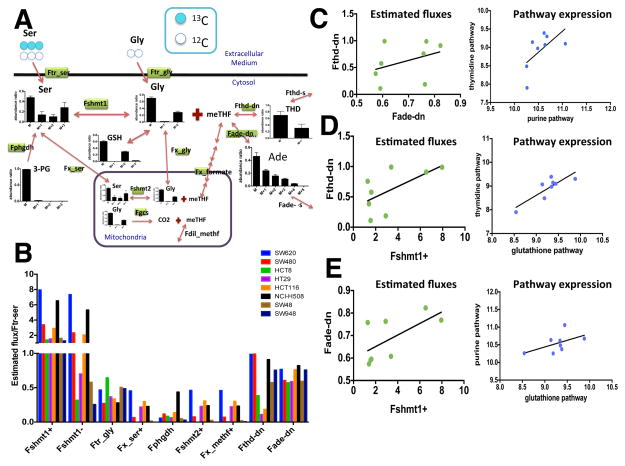
A.) Schematic of the model that was used for flux estimation. Serine, glycine, and formate are shuttled in and out of the mitochondria whereas folate is not. Plots show mass isotopomer distributions (MID) for metabolites that were detected experimentally. Fluxes that were estimated are labeled by green rectangles.
B.) Barplots of estimated fluxes with respect to the serine transport flux (Ftr-ser=1) across the 8 cell lines. (Fshmt1+: forward flux through SHMT1; Fshmt1−: reverse flux through SHMT1; Ftr-gly: Glycine transport flux; Fx-ser+: serine exchange flux; Fphgdh: de novo serine synthesis flux; Fshmt2+: forward flux through SHMT2; Fx-methf+: meTHF exchange flux through formate; Fthd-dn: de novo thymidine synthesis flux; Fade-dn: de novo adenine synthesis flux)
C.) Purine and thymidine pathways are correlated. Estimated fluxes for de novo thymidine and adenine synthesis are positively associated (R-squared=0.14) (left). Expression of the purine pathway is positively correlated with that of the de novo thymidine pathway (R-squared=0.4) across the 8 colon cancer cell lines (right).
D.) Glutathione and thymidine pathways are correlated. Estimated fluxes for de novo thymidine and glutathione synthesis (SHMT1+) are positively associated (R-squared=0.39) (left). Expression of the glutathione pathway is positively correlated with that of the de novo thymidine pathway (R-squared=0.7) across the 8 colon cancer cell lines (right).
E.) Glutathione and purine pathway correlations. Estimated fluxes for de novo adenine and glutathione synthesis (SHMT1+) are positively associated (R-squared=0.42) (left). Expression of the glutathione pathway is positively correlated with that of the de novo purine pathway (R-squared=0.28) across the 8 colon cancer cell lines (right). (See also Figure S2 and Table S3).
Discussion
Our findings present the first comprehensive systems-level analysis of the expressions patterns, metabolic fluxes, and interrelationships of serine metabolizing pathways in numerous cancer contexts and together delineate the likely roles of the SGOC network in several cancer types. While there is no general global overexpression of the entire network in cancer as found in other pathways such as glycolysis, the expression of the network differs in more complex ways. Heterogeneity is strikingly apparent with for example breast cancers showing both overall under- and over- expression of multiple pathway components. Components contributing to nucleotide synthesis are commonly upregulated as previously reported (Hu et al., 2013; Jain et al., 2012), but other pathways exhibit no general features of over- and under- expression across cancer types. In normal tissues, many of these components are constituently expressed with variability suggesting that the usage of a particular component in the normal tissue may confer predisposition of its usage in cancer.
When considering co-expression of the network, strong correlations in individual genes and expression of the pathways were observed. The most common co-existence was the expression of genes contributing to nucleotide metabolism, glutathione and NADPH synthesis. This was also observed in direct measurements of pathway fluxes for glutathione and nucleotides. Since glutathione is the essential component of cellular redox maintenance, concomitant synthesis of glutathione and nucleotides likely provides a redox environment necessary for nucleotide synthesis and repair. In fact, several studies in plants have demonstrated that redox regulation by glutathione has an important role during nucleotide synthesis and cell division (Belmonte et al., 2003; Belmonte et al., 2005; Stasolla, 2010). By performing co-expression analyses on the serine pathways, we provided new insights into the biology of the SGOC network in cancer. In fact, our finding that glutathione synthesis is associated with nucleotide synthesis appears novel in the context of human cancer, and could suggest that cancer cells utilize a redox balance mechanism in parallel to the up-regulation of biosynthetic pathways.
Gene expression measurements are typically thought to not be useful surrogates in cells for flux measurements that are the ultimate phenotypes of interest in studying metabolism. However, we show that pathway-level gene expression to a large extent in the SGOC network can be used as a predictor of experimentally measured fluxes. This is a very important finding in general given the wealth of publically available gene expression data, and the technical limitations associated with performing large-scale metabolomics analyses on human tumor samples. Therefore this work hopefully provides motivation for further comparative analysis of gene expression and flux distributions in biological samples.
Methods
Cell culture and metabolite extraction
Colon cancer cells (SW620, SW480, HCT8, HT29, HCT116, NCI-H508, SW48, and SW948) were cultured as previously described (Liu et al., 2014). For 13C-serine tracing experiments, cells were seeded in 6-well plate at a density of 2×105 to 5×105 cells per well. After overnight incubation in 37 °C with 5% CO2, full growth media were removed, and cells were washed with 2 ml PBS before the addition of RPMI media supplemented with 10% dialyzed and heat inactivated FBS, 100 U/ml penicillin, 100 mg/ml streptomycin and 13C-U-serine (3 mg/100 ml medium, Cambridge Isotope Laboratory) such that in the final medium, 50% of serine is 13C labeled. After a 24 hour incubation, the cells were harvested as previously described (Liu et al., 2014).
Mass spectrometry and Liquid chromatography
The qExactive Mass Spectrometer (QE-MS) coupled to liquid chromatography (Ultimate 3000 UHPLC) was used for metabolite separation and detection as previously described (Liu et al., 2014). Raw data collected from LC-Q Exactive MS was processed with Sieve 2.0 (Thermo Scientific). Relevant instrument parameters are contained in a previous work (Liu et al., 2014).
Network construction and gene expression analyses
The network reconstruction was carried out using the Cytoscape plugin – Metscape (Gao et al., 2010). The SGOC network was generated first by curating all human metabolic genes from the Kyoto Encyclopedia of Genes and Genomes (KEGG) (Kanehisa and Goto, 2000). All genes involved in the KEGG-defined pathways Glycine, Serine and Threonine metabolism, Cysteine and Methionine metabolism, and Folate biosynthesis were selected. We subsequently included genes involved in adjacent chemical reactions (edges) of the selected genes (nodes), and these nodes were then connected to the selected edges to allow for a contiguous sequence of chemical reactions. Finally, we manually excluded all genes that involve non pathway-specific chemical reactions that thus take part in many metabolic pathways implying that their function need not be confined to 1-C metabolism (reactions including: ATP-dependent; tRNA synthesis; Methylation; Co-A; CO2 and NH3; aldehyde dehydrogenases; pyruvate metabolism) (see Table S2 for the complete list of genes excluded).
Gene expression across human tumors was analyzed using level 3 TCGA (The Cancer Genome Atlas) mRNA data (Cancer Genome Atlas, 2012a, b; Cancer Genome Atlas Research, 2011, 2012). Data from AgilentG4502A microarray chips were collected for all TCGA breast (BRCA), ovarian (OV), colon (COAD), and lung (LUSC, LUAD) samples corresponding to four cancer types. Level 3 data contain combined probe signals for each gene and samples were LOWESS normalized using a reference RNA sample (cy5/cy3)(Yang et al., 2002), therefore, poor probe binding was accounted by reporting the ratio mRNA from sample to that of the standard. Mean and median expression levels were calculated for each of the 64 genes in the SGOC network across each cancer type.
For evaluating high expression of one cancer type relative to other cancer types, we considered the following. For each gene, the cancer type that had the highest median expression was grouped (“high expression”) and all samples from the other three cancer types were pooled into one group (“low expression”). The Mann-Whitney Wilcoxon (Wilcoxon) test was then performed to compare gene expression between the “high expression” vs. “low expression “ groups. The Bonferroni method for multiple hypothesis correction was then applied to determine the significant p-values. Effect sizes for the Wilcoxon test are calculated as r=Z/sqrt(N) where N is the total number of samples and Z is the Z-score for the Wilcoxon test. Genes with a significant p-value but a small effect size (r < 0.3) were also considered insignificant. The result of this analysis is a set of genes highly expressed in one cancer type.
For tumor vs. normal comparisons, Affymetrix U133Plus2 expression data from the GENT (Gene Expression across Normal and Tumor tissues) dataset were used for breast, ovarian, colon, and lung cancer as well as normal tissue samples. The data were preprocessed using the MAS5 algorithm and then normalized to a target density of 500 to correct for batch effects according to GENT specifications (Shin et al., 2011). Each cancer type was compared to its corresponding normal tissue using the same statistical approach (Wilcoxon test with Bonferroni and effect size corrections). Comparisons of expression in one normal tissue type relative to other normal tissue types were conducted on these data using the same statistical criterion.
For an analysis of variability in expression, the coefficient of variation (CV) was calculated for each gene across each tissue type. CV comparisons for each case were carried out using the same criteria starting with the Wilcoxon test.
Pathway definitions and analysis
We considered the amino acid serine as the input source of one-carbon units to the SGOC network and decomposed the network into biochemical pathways that consume serine in different reactions and result in a distinct biological output.
The enzymes in each of the pathways from serine metabolism are as follows (mitochondrial isoforms are denoted by “m”):
Methylation: SHMT1/SHMT2 (m) – MTHFR- MTR/BHMT/BHMT2-MAT1A/MAT2A/MAT2B
Thymidine: SHMT1/SHMT2(m) – AMT(m) -TYMS- DHFR
Purine: SHMT1/SHMT2(m) - MTHFD1/MTHFD2(m) /MTHFD1L(m) - ATIC/GART
NADPH: SHMT1/SHMT2(m) - MTHFD1/MTHFD2(m) /MTHFD1L(m) - ALDH1L1/ALDH1L2(m)
Alanine: AGXT1/AGXT2(m)
Glutathione: SHMT1/SHMT2(m) - GSS
Cysteine: CBS- CTH
Taurine: CBS- CTH- CDO1- CSAD
Betaine/Choline: SHMT1/SHMT2(m) - CHDH(m)
Pyruvate: SDS
To compare these pathways in reference to the statistical analysis that we conducted for each gene, we considered an overall pathway score (frequency of occurrences). For each pathway, the number of significant genes, normalized to the number of genes in that pathway, was computed for each of the statistical analyses that we performed. Results are reported in Figure 2.
For the analysis of network covariance, similarity matrices were constructed based on pairwise Spearman correlations between expression levels of the 64 SGOC metabolic genes across each one of the four cancer types (Lung, Breast, Colon, and Ovarian) in the study. In order to visualize these correlations in the context of serine-fate pathways, average pathway expression levels were measured for all tumor samples in study, and similarity matrices were made based on Spearman correlations between average pathway expressions separately in each cancer type. All clustering calculations were carried out using the Gene-E package (Broad Institute www.broadinstitute.org/cancer/software/GENE-E/).
For comparison of pairwise correlations between expression levels of SGOC genes to that expected by chance, we used a randomization method. We randomly picked 64 genes from the genome and calculated pairwise correlations of those genes and repeated this for 100 iterations. Finally, we used the average of sorted p-values form the 100 iterations and plotted the results against the sorted p-values from SGOC pairwise correlations separately for each cancer type (quantile-quantile plots).
Finally, mRNA expression data for the 8 colon cancer cell lines in study were obtained from Broad Institute Cancer Cell Line Encyclopedia (Barretina et al., 2012) and similar SGOC pathway expression analyses were performed on the data for comparisons with isotope labeling and flux data. Correlation between expression levels of SGOC genes with label ratios in Serine, Glycine, Thymidine, Glutathione, and IMP (representing purines) were calculated across the eight cell lines. As a control, label ratios were also correlated with expression of same-size sets of genes randomly picked from the genome. A histogram of all significant p-values (p<0.05) was generated from the results of the 500 simulations. Furthermore, gene expression at the pathway level (average across pathways shown) was also correlated with detected isotope enrichments. Single-genes and pathways were associated with fluxes by plotting the percent of significant correlations (p-value< 0.05) between gene expression and label ratios for each case. All calculations were carried out using R. (URL http://www.R-project.org/)
13C Mass-isotopomer distribution model
A two-compartment (mitochondria and cytosol) Serine-Glycine-Methylenetetrahydrofolate (Ser-Gly-meTHF) flux model was used to fit experimental 13C mass isotopomer distributions (MIDs) of the SGOC associated metabolites to determine intercellular metabolic fluxes, transport and exchange fluxes relative to extracellular serine transport flux. The model was formalized using two types of mass balance equations: 1) mass balance for total metabolite concentration; and 2) 13C mass-isotopomer mass balance for labeled metabolites based on the network depicted in Figure 6A and corresponding atom distribution matrices. MID equations were derived in the similar manner as equations for bonded cumulative isotopomers as described previously(Shestov et al., 2012). In terms of the ordinary differential equations, the model describes the rates of loss and creation of particular labeled and unlabeled metabolite that forms after incubation of labeled serine in extracellular media. Those equations are based on the flux balance of metabolites and take the general form (e.g. for parallel unimolecular reactions):
| (1) |
where metabolite M is downstream of another metabolites Sj. The total outflux ΣFk balances total influx ΣFj. [M] represents the total pool size of metabolite M, while μ(i) and σj(i) represent the I mass-isotopomer fraction of metabolite M (M+I mass-isotopomer) and metabolite Sj (S+I mass-isotopomer), respectively. The number of labeled C atoms in M molecule, I, changes between 0 and N, where N is the total number of C atoms in metabolites. At steady state the left term of equation (1) is equal to zero resulting in a set of algebraic equations. For labeled [U-13C] serine experiments in RPMI medium, the fitted experimental steady state mass-isotopomers (combined cytosolic and mitochondrial) were four isotopomer forms of serine, three forms of glycine, four forms of glutathione which serves as a readout for cytosolic glycine patterns, three forms of thymidine, and six isotopomer forms of adenine together making a total of 20 steady state mass-isotopomers. Thymidine and adenine isotopomers reflect pyrimidine and purine metabolism, respectively. Eleven fluxes were determined relative to serine transport flux, which are represented in Table S3: de novo serine synthesis, two unidirectional rates for reversible cytosolic and mitochondrial SHMT fluxes, unidirectional glycine uptake flux, inter-compartmental serine, glycine, and one-carbon pool (meTHF) exchange fluxes, glycine cleavage system (GCS) activity, mitochondrial dilution flux for meTHF due to dimethylglycine and sarcosine contribution. Also the fraction of the de novo thymidine and adenine production along with salvage contribution was calculated. For each metabolite MIDs, 13C natural abundance of 13C isotope (1.08%) was taken into account. There was no need to correct for 15N natural abundance (0.38%) due to the mass resolution used (70,000) that is able to separate 13C and 15N for the molecules considered. Solving a system of non-linear differential equations in terms of whole/fragmented mass-isotopomers, with the Runge-Kutta 4th order procedure (Matlab, Natick, MA), yields time courses for all possible 13C mass-isotopomers (e.g. serine and glycine). A quadratic cost function was used to quantify differences between measurements and estimated results for labeled steady state data and to select the corresponding vector of fluxes that minimizes the cost function in 13C serine experiments using a simplex algorithm. Mean-square convergence was confirmed by verifying that goodness-of-fit values were close to expected theoretical values. To overcome potential local minima we used several sets of initial random fluxes.
We estimated errors for the obtained values using a Monte Carlo simulation method as previously described (Shestov et al., 2007). Initial values for all parameters were chosen close to the HCT116 cell line fluxes as a reference cell line. 13C mass-isotopomer values for Ser, Gly, GSH, Thd and Ade were then generated by solving differential equations describing the model with initial value of fluxes. For each Monte-Carlo run, random Gaussian noise with mean zero and standard deviation σ = 0.01 was added to the steady-state of these 13C mass-isotopomers. Finally, the MIDs for each metabolite were computed and used for fitting with the SGOC metabolic model to obtain the values of relative metabolic fluxes. This procedure was repeated 500 times to obtain histograms for each parameter. Standard deviations are reported in Table S3.
Supplementary Material
Acknowledgments
We gratefully acknowledge support from the National Institutes of Health (R00CA168997 to JWL, T32GM007617 to MM) and the International Life Sciences Institute. We thank Andrew Clark, Mona Khalaj, and members of the Locasale group for helpful comments and Jaaved Mohammed, Zheng Ser, and Seon-Young Kim for help with data processing.
Footnotes
Conflict of interest
The authors declare that they have no conflicting interests.
References
- Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, Wilson CJ, Lehar J, Kryukov GV, Sonkin D, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–607. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belmonte M, Stasolla C, Loukanina N, Yeung EC, Thorpe TA. Glutathione modulation of purine metabolism in cultured white spruce embryogenic tissue. Plant Sci. 2003;165:1377–1385. [Google Scholar]
- Belmonte MF, Stasolla C, Katahira R, Loukanina N, Yeung EC, Thorpe TA. Glutathione-induced growth of embryogenic tissue of white spruce correlates with changes in pyrimidine nucleotide metabolism. Plant Sci. 2005;168:803–812. [Google Scholar]
- Cancer Genome Atlas N. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012a;487:330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas N. Comprehensive molecular portraits of human breast tumours. Nature. 2012b;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research N. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research N. Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012;489:519–525. doi: 10.1038/nature11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chabner B, Longo DL. Cancer chemotherapy and biotherapy: principles and practice. Philadelphia: Wolters Kluwer Health/Lippincott Williams & Wilkins; 2011. [Google Scholar]
- Chaneton B, Hillmann P, Zheng L, Martin AC, Maddocks OD, Chokkathukalam A, Coyle JE, Jankevics A, Holding FP, Vousden KH, et al. Serine is a natural ligand and allosteric activator of pyruvate kinase M2. Nature. 2012;491:458–462. doi: 10.1038/nature11540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Circu ML, Aw TY. Reactive oxygen species, cellular redox systems, and apoptosis. Free radical biology & medicine. 2010;48:749–762. doi: 10.1016/j.freeradbiomed.2009.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J, Ye J, Kamphorst JJ, Shlomi T, Thompson CB, Rabinowitz JD. Quantitative flux analysis reveals folate-dependent NADPH production. Nature. 2014;510:298–302. doi: 10.1038/nature13236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J, Tarcea VG, Karnovsky A, Mirel BR, Weymouth TE, Beecher CW, Cavalcoli JD, Athey BD, Omenn GS, Burant CF, et al. Metscape: a Cytoscape plug-in for visualizing and interpreting metabolomic data in the context of human metabolic networks. Bioinformatics. 2010;26:971–973. doi: 10.1093/bioinformatics/btq048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gut P, Verdin E. The nexus of chromatin regulation and intermediary metabolism. Nature. 2013;502:489–498. doi: 10.1038/nature12752. [DOI] [PubMed] [Google Scholar]
- Hu CM, Yeh MT, Tsao N, Chen CW, Gao QZ, Chang CY, Lee MH, Fang JM, Sheu SY, Lin CJ, et al. Tumor cells require thymidylate kinase to prevent dUTP incorporation during DNA repair. Cancer cell. 2012;22:36–50. doi: 10.1016/j.ccr.2012.04.038. [DOI] [PubMed] [Google Scholar]
- Hu J, Locasale JW, Bielas JH, O’Sullivan J, Sheahan K, Cantley LC, Vander Heiden MG, Vitkup D. Heterogeneity of tumor-induced gene expression changes in the human metabolic network. Nature biotechnology. 2013;31:522–529. doi: 10.1038/nbt.2530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain M, Nilsson R, Sharma S, Madhusudhan N, Kitami T, Souza AL, Kafri R, Kirschner MW, Clish CB, Mootha VK. Metabolite profiling identifies a key role for glycine in rapid cancer cell proliferation. Science. 2012;336:1040–1044. doi: 10.1126/science.1218595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalhan SC, Hanson RW. Resurgence of serine: an often neglected but indispensable amino Acid. The Journal of biological chemistry. 2012;287:19786–19791. doi: 10.1074/jbc.R112.357194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic acids research. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labuschagne CF, van den Broek NJ, Mackay GM, Vousden KH, Maddocks OD. Serine, but Not Glycine, Supports One-Carbon Metabolism and Proliferation of Cancer Cells. Cell reports. 2014;7:1248–1258. doi: 10.1016/j.celrep.2014.04.045. [DOI] [PubMed] [Google Scholar]
- Lewis CA, Parker SJ, Fiske BP, McCloskey D, Gui DY, Green CR, Vokes NI, Feist AM, Vander Heiden MG, Metallo CM. Tracing compartmentalized NADPH metabolism in the cytosol and mitochondria of Mammalian cells. Molecular cell. 2014a;55:253–263. doi: 10.1016/j.molcel.2014.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Ser Z, Locasale JW. Development and quantitative evaluation of a high-resolution metabolomics technology. Analytical chemistry. 2014;86:2175–2184. doi: 10.1021/ac403845u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locasale JW. Serine, glycine and one-carbon units: cancer metabolism in full circle. Nature reviews Cancer. 2013;13:572–583. doi: 10.1038/nrc3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locasale JW, Grassian AR, Melman T, Lyssiotis CA, Mattaini KR, Bass AJ, Heffron G, Metallo CM, Muranen T, Sharfi H, et al. Phosphoglycerate dehydrogenase diverts glycolytic flux and contributes to oncogenesis. Nature genetics. 2011;43:869–874. doi: 10.1038/ng.890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Tao Y, Duran A, Llado V, Galvez A, Barger JF, Castilla EA, Chen J, Yajima T, Porollo A, et al. Control of Nutrient Stress-Induced Metabolic Reprogramming by PKCzeta in Tumorigenesis. Cell. 2013;152:599–611. doi: 10.1016/j.cell.2012.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddocks OD, Berkers CR, Mason SM, Zheng L, Blyth K, Gottlieb E, Vousden KH. Serine starvation induces stress and p53-dependent metabolic remodelling in cancer cells. Nature. 2013;493:542–546. doi: 10.1038/nature11743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy MP, Holmgren A, Larsson NG, Halliwell B, Chang CJ, Kalyanaraman B, Rhee SG, Thornalley PJ, Partridge L, Gems D, et al. Unraveling the biological roles of reactive oxygen species. Cell metabolism. 2011;13:361–366. doi: 10.1016/j.cmet.2011.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson R, Jain M, Madhusudhan N, Sheppard NG, Strittmatter L, Kampf C, Huang J, Asplund A, Mootha VK. Metabolic enzyme expression highlights a key role for MTHFD2 and the mitochondrial folate pathway in cancer. Nature communications. 2014;5:3128. doi: 10.1038/ncomms4128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Possemato R, Marks KM, Shaul YD, Pacold ME, Kim D, Birsoy K, Sethumadhavan S, Woo HK, Jang HG, Jha AK, et al. Functional genomics reveal that the serine synthesis pathway is essential in breast cancer. Nature. 2011;476:346–350. doi: 10.1038/nature10350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scuoppo C, Miething C, Lindqvist L, Reyes J, Ruse C, Appelmann I, Yoon S, Krasnitz A, Teruya-Feldstein J, Pappin D, et al. A tumour suppressor network relying on the polyamine-hypusine axis. Nature. 2012;487:244–248. doi: 10.1038/nature11126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shestov AA, Valette J, Deelchand DK, Ugurbil K, Henry PG. Metabolic modeling of dynamic brain (1)(3)C NMR multiplet data: concepts and simulations with a two-compartment neuronal-glial model. Neurochemical research. 2012;37:2388–2401. doi: 10.1007/s11064-012-0782-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shestov AA, Valette J, Ugurbil K, Henry PG. On the reliability of (13)C metabolic modeling with two-compartment neuronal-glial models. Journal of neuroscience research. 2007;85:3294–3303. doi: 10.1002/jnr.21269. [DOI] [PubMed] [Google Scholar]
- Shin G, Kang TW, Yang S, Baek SJ, Jeong YS, Kim SY. GENT: gene expression database of normal and tumor tissues. Cancer informatics. 2011;10:149–157. doi: 10.4137/CIN.S7226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stasolla C. Glutathione redox regulation of in vitro embryogenesis. Plant Physiol Bioch. 2010;48:319–327. doi: 10.1016/j.plaphy.2009.10.007. [DOI] [PubMed] [Google Scholar]
- Tedeschi PM, Markert EK, Gounder M, Lin H, Dvorzhinski D, Dolfi SC, Chan LL, Qiu J, Dipaola RS, Hirshfield KM, et al. Contribution of serine, folate and glycine metabolism to the ATP, NADPH and purine requirements of cancer cells. Cell death & disease. 2013;4:e877. doi: 10.1038/cddis.2013.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teperino R, Schoonjans K, Auwerx J. Histone methyl transferases and demethylases; can they link metabolism and transcription? Cell metabolism. 2010;12:321–327. doi: 10.1016/j.cmet.2010.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez A, Markert EK, Oltvai ZN. Serine biosynthesis with one carbon catabolism and the glycine cleavage system represents a novel pathway for ATP generation. PloS one. 2011;6:e25881. doi: 10.1371/journal.pone.0025881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez A, Tedeschi PM, Bertino JR. Overexpression of the mitochondrial folate and glycine-serine pathway: a new determinant of methotrexate selectivity in tumors. Cancer research. 2013;73:478–482. doi: 10.1158/0008-5472.CAN-12-3709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson PM, LaBonte MJ, Lenz HJ, Mack PC, Ladner RD. Inhibition of dUTPase induces synthetic lethality with thymidylate synthase-targeted therapies in non-small cell lung cancer. Molecular cancer therapeutics. 2012;11:616–628. doi: 10.1158/1535-7163.MCT-11-0781. [DOI] [PubMed] [Google Scholar]
- Yang YH, Dudoit S, Luu P, Lin DM, Peng V, Ngai J, Speed TP. Normalization for cDNA microarray data: a robust composite method addressing single and multiple slide systematic variation. Nucleic acids research. 2002;30:e15. doi: 10.1093/nar/30.4.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang WC, Shyh-Chang N, Yang H, Rai A, Umashankar S, Ma S, Soh BS, Sun LL, Tai BC, Nga ME, et al. Glycine decarboxylase activity drives non-small cell lung cancer tumor-initiating cells and tumorigenesis. Cell. 2012;148:259–272. doi: 10.1016/j.cell.2011.11.050. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.