Abstract
Background and objective
Bacterial invasion into pulps of primary teeth can lead to infection and premature tooth loss in children. This pilot study aimed to explore whether the microbiota of carious exposures of dental pulps resembles that of carious dentin or that of infected root canals.
Design
Children with severe early childhood caries were studied. Children were consented and extent of caries, plaque, and gingivitis measured. Bacteria were sampled from carious lesion biofilms and vital carious exposures of pulps, and processed by anaerobic culture. Isolates were characterized from partial sequences of the 16S rRNA gene and identified by comparison with taxa in the Human Oral Microbiome Database (http://www.HOMD.org). The microbiotas of carious lesions and dental pulps were compared using univariate and multivariate approaches.
Results
The microbiota of cariously exposed pulps was similar in composition to that of carious lesion biofilms except that fewer species/taxa were identified from pulps. The major taxa identified belonged to the phyla Firmicutes (mainly streptococci) and Actinobacteria (mainly Actinomyces species). Actinomyces and Selenomonas species were associated with carious lesions whereas Veillonella species, particularly Veillonella dispar was associated with pulps. Other bacteria detected in pulps included Streptococcus mutans, Parascardovia denticolens, Bifidobacterium longum, and several Lactobacillus and Actinomyces species. By principal, component analysis pulp microbiotas grouped together, whereas those in caries biofilms were widely dispersed.
Conclusions
We conclude that the microbiota of cariously exposed vital primary pulps is composed of a subset of species associated with carious lesions. Vital primary pulps had a dominant Firmicutes and Actinobacteria microbiota which contrasts with reports of endodontic infections which can harbor a gram-negative microbiota. The microbiota of exposed primary pulps may provide insight into bacterial species at the forefront of caries invasion in dentinal lesions that can invade into the pulp and the nature of species that need suppressing for successful pulp therapy.
Keywords: 16S rRNA, culture, Streptococcus mutans, Parascardovia
Severe early childhood caries (ECC) is a devastating dental infection that disproportionately affects children with oral health disparities (1, 2). If left untreated, carious lesions can advance into the pulp and lead to root canal infections, abscess formation, tooth fracture, and tooth loss (3). Other complications of ECC, particularly severe ECC, include an elevated risk for new carious lesions (4, 5), high treatment costs, and increased time needed for therapy (6, 7).
The microbiota of ECC is complex and includes acidogenic and acid-tolerant species (8, 9) that are candidate pathogens for caries progression. Gram-negative species are also detected in deep dentinal caries (10, 11) and in pulp and root canal infections of primary teeth (12–14). In our studies using combined cultural and molecular methods, we detected a new Bifidobacteriaceae species, Scardovia wiggsiae, as significantly associated with severe ECC (9). The microbiota of pulpal and root canal infection in children has included molecular assay of selected, mainly gram negative species (12, 13, 15–17). A study using anaerobic culture detected mainly gram-positive species (18), whereas culture of root canals of extracted teeth reported a dominant gram-negative infection (14). Thus, while the microbiota associated with infected deciduous pulps and root canals has been studied, few studies have focused on the vital coronal pulp microbiota using open-ended approaches incorporating current molecular microbial taxonomies.
In the current pilot study, we compared the cultivable microbiota of exposed vital pulps with that of carious lesions in deciduous teeth. We hypothesized that taxa and species found in the exposed pulps would reflect the microbiota at the advancing front of carious lesions. Alternatively the microbiota of pulps might comprise reservoirs of mainly gram-negative species that may contribute to endodontic infections. Optimal therapy to prevent spread of infection from the pulp would depend on the nature of the microbiota detected.
Methods
Study population
Medically healthy 2- to 6-year-old children were recruited from the dental clinics at the Departments of Pediatric Dentistry at Boston and Tufts Universities. Children studied had severe ECC for whom the diet and the microbiota by culture and clonal analysis have been previously described (19–21). Children with severe ECC (22) had extensive caries in the primary dentition and were scheduled for full mouth dental rehabilitation treatment under general anesthesia. Inclusion criteria were that the child was medically healthy, had not used antibiotics within the last 3 months, and the parent or guardian was willing to consent to the child's clinical examination and microbial sampling. The study was performed in full accordance with ethical principles including the World Medical Association Declaration of Helsinki. The study design was explained to the child's parent or guardian, from whom informed consent was obtained if they were willing to participate with their child. The study design, protocol, and informed consent were approved by the Institutional Review Boards of Boston University, Tufts University, and the Forsyth Institute.
Clinical characteristics
The caries and gingival status of children were charted and biofilm from carious lesions taken using toothpicks as previously described (9) when children were under general anesthesia. Rubber dams were then placed before comprehensive restorative treatment. Vital pulp chambers that were exposed during removal of deep carious dentine were sampled using sterile paper points. Bacterial samples from carious biofilm and pulp chambers were placed in gas tight vials containing 2 ml of pre-reduced anaerobically sterilized Ringer's solution with 3 mm glass beads as previously described (9). Samples were transported to the laboratory in insulated bags with freezer blocks to keep cold.
Microbiological methods
Samples were processed within 4 h of sampling. Samples were dispersed by vortexing and underwent 10-fold serial dilutions under anaerobic gas and were plated onto enriched blood agar (Trypticase soy agar 20 g/l, Brain Heart Infusion agar 26 g/l, yeast extract 10 g/l, hemin 5 mg/l, menadione 0.5 mg/l, N-acetyl-muramic acid 10 mg/l, defibrinated sheep blood 50 ml/l, pH = 7) and on acid agar (Trypticase soy agar 20 g/l, Brain Heart Infusion agar 26 g/l, yeast extract 10 g/l, hemin 5 mg/l, with the pH adjusted to 5.0 with HCl before autoclaving). Plates were incubated anaerobically for 10–12 days in an atmosphere of 80% N2, 10% CO2, and 10% H2. Isolates were purified and characterized using partial 16S rRNA sequences as previously described (9). Briefly, partial sequences approximately 400–800 bases long from pure isolates were edited, aligned, and checked for chimeric inserts. Isolates (n=711) were identified by comparing partial 16S rRNA sequences with taxa (species) in the Human Oral Microbiome Database (HOMD) (www.homd.org) (23). Isolates were considered identified if they showed at least 98.5% similarity to taxa in HOMD.
Data analysis
Taxa from HOMD were grouped by caries or pulp origin at genus and species levels, and detection frequencies compared using Fisher's exact tests. Percentages of total isolates at species and genus levels were compared from carious biofilm and exposed pulps using the Kruskal–Wallis one-way analysis of variance test. Principal component analysis (PCA) and two-way hierarchical cluster analyses were performed on detection frequencies of species/taxa that were found in at least two children from either carious lesion or pulp categories. Principal component 1 (PC1) was plotted against principal component 2 (PC2). Hierarchical clustering was performed using the Ward method using binary data (JMP statistical discovery software, Cary, NC).
Results
Bacteria from caries and vital coronal pulps were obtained from nine children. One culture plate from caries had confluent growth so that colonies could not be isolated and processed leaving nine pulp and eight plaque samples for analysis. Child age ranged from 2.6 to 5.0 years, six of the children were boys and two were Hispanic (Table 1). Four children were Caucasian, three were Asian, and two were Black (African American). In seven of the nine children, over half of the teeth had carious lesions.
Table 1.
Characteristics of children sampled with severe ECC
| Child | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 |
|---|---|---|---|---|---|---|---|---|---|
| Age (years) | 4.8 | 4.8 | 3.8 | 3.5 | 5 | 2.6 | 3.3 | 2.9 | 4.3 |
| Gendera | M | M | M | F | F | M | F | M | M |
| Hispanic ethnicityb | N | Y | N | Y | Y | N | N | N | N |
| Race/ethnicity | Asian | White | White | White | White | Asian | Black | Asian | Black |
| Plaque index (mean) | 2.36 | 0 | 0 | 0 | 1.99 | 2.42 | 2.01 | 1.21 | 3 |
| Gingival index (mean) | 2.9 | 0 | 0 | 0 | 2.49 | 1.68 | 1.69 | 0 | 0.93 |
| Bleeding (% teeth) | 87.5 | 0 | 0 | 0 | 0 | 63.16 | 72.5 | 0 | 47.5 |
| Decayed surfaces | 64 | 16 | 21 | 21 | 60 | 37 | 51 | 47 | 41 |
| Decayed teeth | 19 | 12 | 9 | 9 | 17 | 11 | 12 | 12 | 12 |
M=male, F=female;
Y=Hispanic, N=not Hispanic.
A mean of 11 (range 3–17) taxa were detected in pulp samples compared to 22 (range 16–26) taxa detected in caries biofilms. One hundred and five taxa/species were identified from samples. Thirty-seven percent of the species were detected in pulp and caries, 43% of taxa were detected only in caries, and 18% only in pulp. The dominant phyla detected in both caries and pulps were Firmicutes (mainly streptococci); followed by Actinobacteria (mainly Actinomyces species) (Table 2). More taxa in the genera Actinomyces, Selenomonas, Fusobacterium, and Campylobacter species were detected in the carious lesions than from exposed pulps. Conversely, more taxa in the genus Veillonella were detected in pulps (Table 2). For the closely related taxa, Actinomyces naeslundii and Actinomyces oris, most of the isolates were identified at 99.5% or above to the consensus sequences in HOMD.
Table 2.
Microbiota by phylum and genus cultured from caries and exposed pulps
| Phylum | Genus | Caries (n=8) mean % (±SD) | Pulp (n=9) mean % (±SD) | KW test | Caries detection frequency % | Pulp detection frequency % | Fisher's exact test |
|---|---|---|---|---|---|---|---|
| Actinobacteria | 23.75 (±7.01) | 29.11 (±18.64) | 100.00 | 100.00 | |||
| Actinomyces | 19.00 (±7.67) | 8.89 (±8.19) | 0.016 | 100.00 | 77.78 | ||
| Atopobium | 0.25 (±0.71) | 0.78 (±2.33) | 12.50 | 11.11 | |||
| Bifidobacterium | 1.25 (±1.83) | 6.67 (±20.00) | 37.50 | 11.11 | |||
| Cryptobacterium | 0.00 (±0.00) | 0.33 (±1.00) | 0.00 | 11.11 | |||
| Parascardovia | 1.00 (±2.83) | 9.56 (±17.88) | 12.50 | 33.33 | |||
| Propionibacterium | 1.00 (±2.14) | 2.89 (±4.81) | 25.00 | 33.33 | |||
| Rothia | 0.25 (±0.71) | 0.00 (±0.00) | 12.50 | 0.00 | |||
| Scardovia | 1.50 (±1.77) | 0.00 (±0.00) | 0.020 | 50.00 | 0.00 | 0.029 | |
| Bacteroidetes | 6.38 (±5.76) | 2.56 (±2.51) | 62.50 | 55.56 | |||
| Capnocytophaga | 3.00 (±3.70) | 0.00 (±1.56) | 50.00 | 22.22 | |||
| Prevotella | 3.38 (±4.17) | 0.00 (±2.17) | 50.00 | 44.44 | |||
| Firmicutes | 61.88 (±11.14) | 67.44 (±18.08) | 100.00 | 100.00 | |||
| Abiotrophia | 0.50 (±0.93) | 1.00 (±3.00) | 25.00 | 11.11 | |||
| Eubacterium [11][G-7] | 0.00 (±0.00) | 0.44 (±1.33) | 0.00 | 11.11 | |||
| Gemella | 0.75 (±1.04) | 0.33 (±1.00) | 37.50 | 11.11 | |||
| Granulicatella | 1.25 (±2.12) | 1.44 (±2.19) | 37.50 | 33.33 | |||
| Lachnoanaerobaculum | 10.25 (±7.42) | 4.22 (±8.39) | 87.50 | 44.44 | |||
| Lactobacillus | 1.25 (±2.12) | 8.89 (±15.84) | 37.50 | 44.44 | |||
| Lactococcus | 0.00 (±0.00) | 0.56 (±1.67) | 0.000 | 11.11 | |||
| Parvimonas | 0.00 (±0.00) | 0.89 (±2.67) | 0.00 | 11.11 | |||
| Selenomonas | 6.63 (±5.29) | 1.44 (±3.64) | 0.033 | 75.00 | 22.22 | 0.050 | |
| Solobacterium | 0.00 (±0.00) | 0.44 (±1.33) | 0.00 | 11.11 | |||
| Staphylococcus | 0.50 (±0.93) | 0.22 (±0.67) | 25.00 | 11.11 | |||
| Streptococcus | 35.63 (±17.04) | 32.11 (±18.91) | 100.00 | 88.89 | |||
| Veillonella | 5.25 (±6.65) | 15.22 (±9.48) | 0.033 | 75.00 | 88.89 | ||
| Fusobacteria | 2.75 (±2.38) | 0.78 (±2.33) | 0.031 | 75.00 | 11.11 | 0.015 | |
| Fusobacterium | 1.50 (±1.14) | 0.78 (±2.33) | 62.50 | 11.11 | 0.050 | ||
| Leptotrichia | 1.25 (±1.49) | 0.00 (±0.00) | 0.019 | 50.00 | 0.00 | 0.029 | |
| Proteobacteria | 5.88 (±4.55) | 0.22 (±0.67) | 0.001 | 87.50 | 11.11 | 0.003 | |
| Campylobacter | 5.88 (±4.55) | 0.22 (±0.67) | 0.001 | 87.50 | 11.11 | 0.003 | |
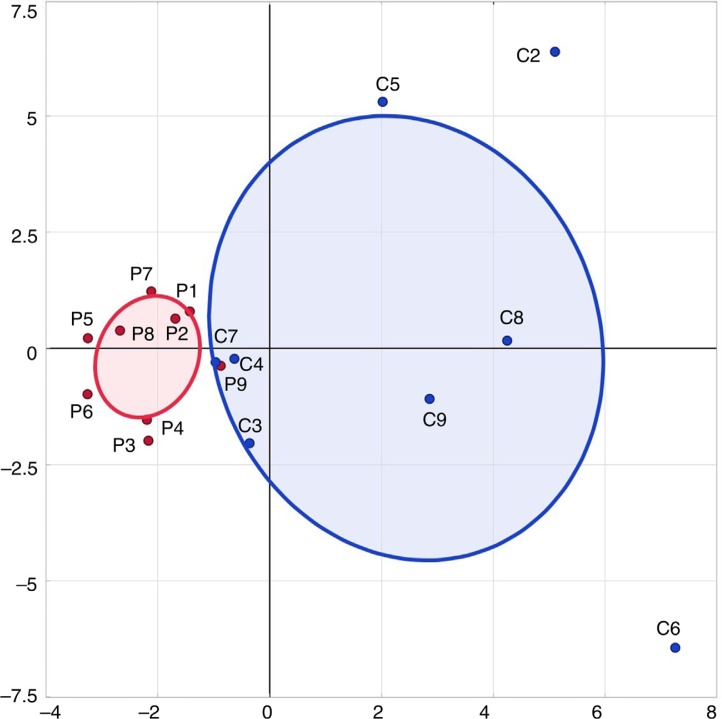
Sixty-three taxa were identified in two or more children and this dataset was used for multivariate analyses. PCA clustered microbiotas into two groups that incorporated 28% of the data variability (Fig. 1). There was no overlap in the (bivariate normal) ellipses that incorporated 50% of either the pulp or the caries microbiotas. Pulp bacteria centered closely to the 50% ellipse, whereas the caries biofilms were widely dispersed.
Fig. 1.
Principal component analysis based on detection frequencies of species in samples. Most of the pulp samples clustered together, whereas caries samples were widely dispersed. PC1 (x-axis) represents 15.7% of the variability in the data and PC2 (y-axis) represents 12.5% of the variability of the data. The ellipses encompass 50% of the samples in either caries/plaque and are grouped by sample type (caries C1–C8 blue; pulp P1–P9 red).
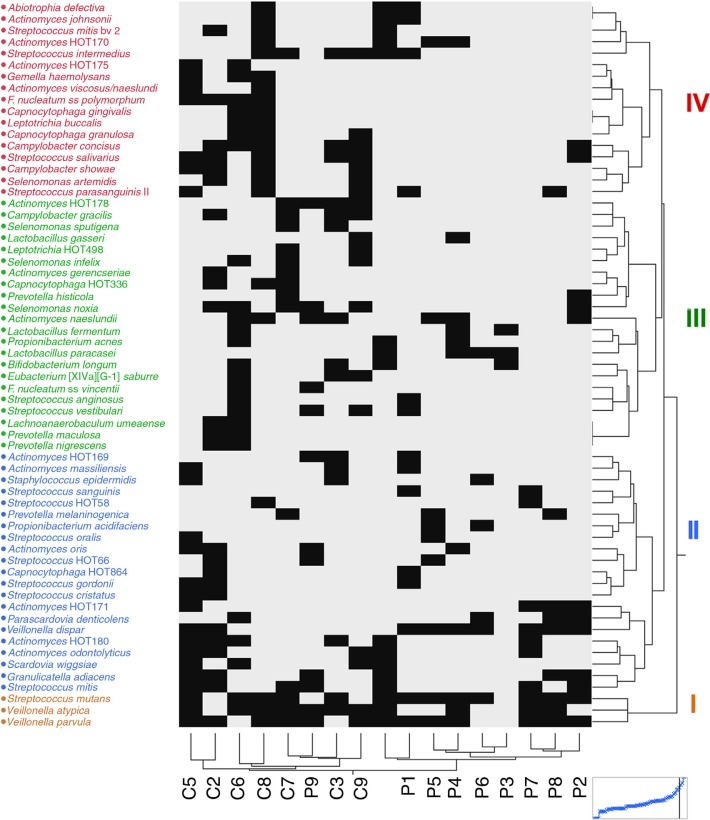
The groups formed from hierarchical cluster analysis of species/taxa in samples were similar to those from the PCA analysis. The species grouped into four levels (brown, blue, green, and magenta, Fig. 2) based on their contribution in cluster formation. Cluster 1 (brown) species were the most influential and comprised Streptococcus mutans, Veillonella atypica, and Veillonella parvula. These species were detected in most of the samples. Cluster 2 species (blue) contained named and unnamed species (designated by HOT groups). Cluster 2 species included Streptococcus mitis, Streptococcus cristatus, Streptococcus gordonii, two unnamed Streptococcus species, S. wiggsiae, Parascardovia denticolens, Actinomyces odontolyticus, A. oris, Actinomyces massiliensis, three unnamed Actinomyces species, Veillonella dispar, and Prevotella melaninogenica. The third (green) and fourth (magenta) species groups were detected less frequently in the pulp than in caries biofilms. Species of interest observed as putative cariogenic pathogens included Bifidobacterium longum, Lactobacillus paracasei, Lactobacillus fermentum, Lactobacillus gasseri, and several Actinomyces species.
Fig. 2.
Two-way hierarchical cluster of binary levels for 17 samples across 63 species/taxa. Sample groupings were similar to those of the principal component analysis (Fig. 1). Species grouped in four clusters (color coded). The figure illustrates the fewer species detected in pulp than in caries samples of the same children although S. mutans, V. parvula, and V. atypica were detected in most samples.
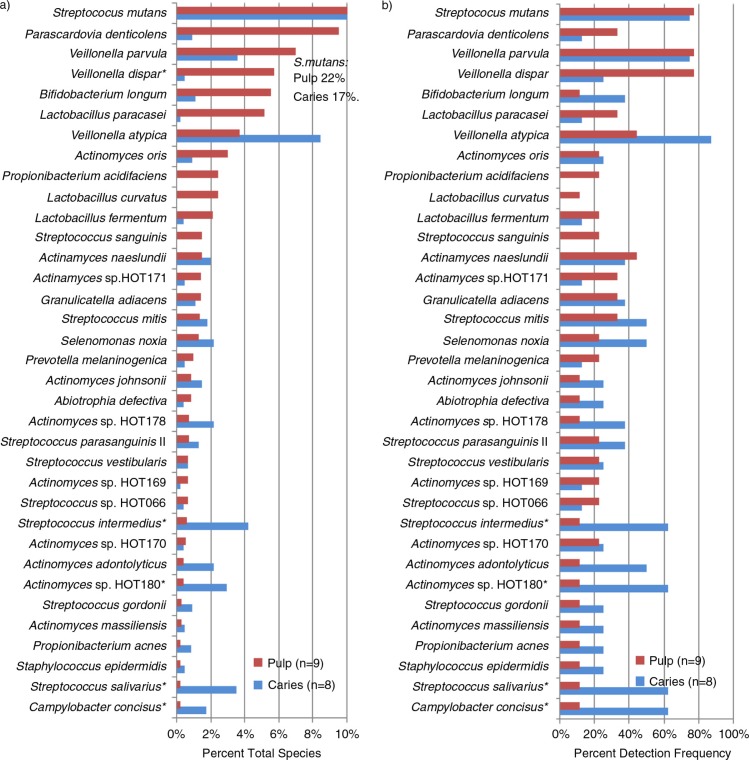
Figure 3 summarizes the species detected in pulps and their detection in carious lesions. The principal pulp species included S. mutans, P. denticolens, B. longum, L. paracasei, and several additional Lactobacillus and Actinomyces species. Only V. dispar (p=0.035) was at higher proportions in the pulp than caries biofilms. Species detected at higher proportions or more frequently in caries than pulps included Streptococcus intermedius (p=0.05), Actinomyces sp. HOT 180 (p=0.05), Streptococcus salivarius (p=0.05), and Campylobacter concisus (p=0.05) (Fig. 3b).
Fig. 3.
Taxa in exposed pulps and their presence in caries. (a) Percent of total taxa identified; (b) percent detection frequency. *p≤0.05. Species are in descending order of percent of all species detected/site in pulps of deciduous teeth and were detected in at least two pulp or caries samples (except for Lactobacillus curvatus which was included as it comprised 10 isolates in one pulp sample). Species at highest mean proportions in the pulps were S. mutans, Bifidobacteriaceae (Parascardovia denticolens and Bifidobacterium longum), and Veillonella species followed by Lactobacillus paracasei and several Actinomyces species including A. oris and A. naeslundii.
Several additional species were detected more frequently in caries but were not detected in the pulp microbiotas: Streptococcus mitis biovar 2 (p=0.05), Campylobacter gracilis (p=0.03), Campylobacter showae (p=0.03), F. nucleatum subsp. polymorphum (p=0.03), and S. wiggsiae (p=0.03).
Discussion
The advanced stage of dental caries in the sampled children in this pilot project allowed sampling of bacteria associated with dental caries and with pulps that were exposed during caries removal before restorative therapy. While fewer species were detected in the deciduous pulps than in carious lesion biofilms, the overall similarity of species between sample sites suggests that the vital pulp microbiota reflected those species that had recently invaded from deep dentinal lesions. This extends the previous observation of similarity between isolates from carious lesions and root canals in ECC using typing by repetitive sequence-based PCR fingerprinting methods (24). The literature study compared caries and root canal detection of several Streptococcus and Lactobacillus species, but did not report detection of Bifidobacterium, Parascardovia, or Actinomyces species as in the current report. In another report, species cultured from primary teeth with necrotic pulps were principally Bifidobacterium subsp2 and S. intermedius (18). The sequence of Bifidobacterium subsp. 2 (GenBank: GQ900831.1) was identified to S. wiggsiae in the HOMD database (100% identity, 0/781 mismatch) indicating that they had cultured S. wiggsiae from necrotic pulps. They also identified Actinomyces species so our data were more comparable to this study, although fewer species were identified in the literature study (18). In the current study, samples were incubated longer than the literature study, which probably accounted for the greater diversity of species detected. The pulp microbiotas detected in the current report comprised mainly of S. mutans, P. denticolens, Lactobacillus, and Actinomyces species. Our findings together with those of Ledezma-Rastillo et al. (18) suggest that studies focusing on selected gram-negative taxa are underestimating the species diversity present in exposed pulps of deciduous teeth. The frequent detection of Lactobacillus, Bifidobacterium, Propionibacterium, and Actinomyces species from root canal infections of permanent teeth (25) suggests that presence of gram-positive rod species extends beyond the primary to the permanent dentition.
Overall, the microbiota detected in severe ECC and exposed pulps was heterogeneous including highly acidic and/or acid-tolerant gram-positive species in combination with weak or non-acidogenic, but proteolytic gram-negative species. The presence of a complex microbiota in dentinal caries was revealed using open-ended, non-selective methods including anaerobic culture allowing for the growth of fastidious taxa (9, 26) and molecular cloning/sequencing and pyrosequencing approaches (10, 27–29). Recent reports have indicated that different components of the microbiota play different roles in initial enamel lesions compared to caries extension into dentin (30–33). The hydroxyapatite-rich enamel likely requires a more acidic microbiota for demineralization compared with dentin. The highly acidogenic species would include S. mutans, acidogenic non-mutans streptococci and Actinomyces species, and Bifidobacterium/Scardovia species. The mechanism of caries progression into dentin may include proteolysis by Prevotella species of proteins denatured by the acidic species (32). Whether acidic or proteolytic mechanisms, or a combination of methods, initiate pulpitis, it seems likely that the proteolytic component would lead to pulp tissue necrosis considering the frequent detection of gram-negative taxa in root canal infections, including in deciduous teeth (12–15). The caries biofilms analyzed could have included interproximal lesions, so some of the gram-negative species detected in the current report could reflect a gingivitis-associated microbiota as we observed in the microbiota of white spot lesions associated with fixed orthodontic appliances (34).
Fewer species were detected in the pulp than in caries biofilms as has been observed for root canal infections in deciduous teeth (13, 18). Detection of fewer species in pulp than caries microbiotas may reflect that the pulp samples were smaller than the caries biofilms, and the smaller biomass could reduce the likelihood of detecting individual species. This might explain the lack of detection of S. wiggsiae in pulps of the current study, whereas S. wiggsiae was the most frequently detected species cultured from pulp samples of deciduous teeth (as Bifidobacterium two species) (18). The relatively lower species detection in pulps compared to caries was the likely basis for the clustering of samples by clinical site. The lower pulp microbiota diversity could reflect the confined nutritionally restricted environment of deciduous pulps compared with carious biofilms that are exposed to the oral cavity. The detection of lactobacilli in pulp is consistent with finding lactobacilli in initial pulpal infections of permanent teeth (35) and root canal infections (25).
A mixture of antibiotics is routinely used for disinfection of immature teeth in regenerative endodontic procedures (36) and has been used successfully to treat primary teeth with pulp infections (37). Recognition of the diversity and nature of early bacterial invaders into the pulp of patients with severe ECC is important for the development of anti-bacterial medicaments for vital pulp therapy in such patients.
We conclude that the microbiota of vital, infected pulps in primary teeth with severe ECC is quite similar to that of advanced carious lesion biofilms. While the dominant species detected was S. mutans, the detection of Parascardovia, Lactobacillus, and Veillonella species in pulps suggests that these taxa may play direct or supporting roles in pulp pathology in children. Knowing the dominant species may be important for the development of vital pulp therapy for severe ECC patients.
Acknowledgements
This study was performed with grants from NIDCR at the National Institutes of Health DE-015847, DE016937, and T32-DE-007327.
Conflict of interests and funding
There are no conflicts of interests or funding related to this project by any author.
References
- 1.Fisher-Owens SA, Isong IA, Soobader MJ, Gansky SA, Weintraub JA, Platt LJ, et al. An examination of racial/ethnic disparities in children's oral health in the United States. J Public Health Dent. 2013;73:166–74. doi: 10.1111/j.1752-7325.2012.00367.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dietrich T, Culler C, Garcia RI, Henshaw MM. Racial and ethnic disparities in children's oral health: the National Survey of Children's Health. J Am Dent Assoc. 2008;139:1507–17. doi: 10.14219/jada.archive.2008.0077. [DOI] [PubMed] [Google Scholar]
- 3.Kassa D, Day P, High A, Duggal M. Histological comparison of pulpal inflammation in primary teeth with occlusal or proximal caries. Int J Paediatr Dent. 2009;19:26–33. doi: 10.1111/j.1365-263X.2008.00962.x. [DOI] [PubMed] [Google Scholar]
- 4.Berkowitz RJ, Amante A, Kopycka-Kedzierawski DT, Billings RJ, Feng C. Dental caries recurrence following clinical treatment for severe early childhood caries. Pediatr Dent. 2011;33:510–14. [PubMed] [Google Scholar]
- 5.Dulgergil CT, Colak H. Do the more caries in early primary dentition indicate the more caries in permanent dentition? Results of a 5-years follow-up study in rural-district. J Int Soc Prev Community Dent. 2012;2:48–52. doi: 10.4103/2231-0762.109364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ng MW, Ramos-Gomez F, Lieberman M, Lee JY, Scoville R, Hannon C, et al. Disease management of early childhood caries: ECC Collaborative Project. Int J Dent. 2014;2014:327801. doi: 10.1155/2014/327801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rashewsky S, Parameswaran A, Sloane C, Ferguson F, Epstein R. Time and cost analysis: pediatric dental rehabilitation with general anesthesia in the office and the hospital settings. Anesth Prog. 2012;59:147–53. doi: 10.2344/0003-3006-59.4.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marchant S, Brailsford SR, Twomey AC, Roberts GJ, Beighton D. The predominant microflora of nursing caries lesions. Caries Res. 2001;35:397–406. doi: 10.1159/000047482. [DOI] [PubMed] [Google Scholar]
- 9.Tanner AC, Mathney JM, Kent RL, Chalmers NI, Hughes CV, Loo CY, et al. Cultivable anaerobic microbiota of severe early childhood caries. J Clin Microbiol. 2011;49:1464–74. doi: 10.1128/JCM.02427-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aas JA, Griffen AL, Dardis SR, Lee AM, Olsen I, Dewhirst FE, et al. Bacteria of dental caries in primary and permanent teeth in children and young adults. J Clin Microbiol. 2008;46:1407–17. doi: 10.1128/JCM.01410-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Corby PM, Lyons-Weiler J, Bretz WA, Hart TC, Aas JA, Boumenna T, et al. Microbial risk indicators of early childhood caries. J Clin Microbiol. 2005;43:5753–9. doi: 10.1128/JCM.43.11.5753-5759.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tavares WL, Neves de Brito LC, Teles RP, Massara ML, Ribeiro Sobrinho AP, Haffajee AD, et al. Microbiota of deciduous endodontic infections analysed by MDA and checkerboard DNA-DNA hybridization. Int Endod J. 2011;44:225–35. doi: 10.1111/j.1365-2591.2010.01805.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ruviere DB, Leonardo MR, da Silva LA, Ito IY, Nelson-Filho P. Assessment of the microbiota in root canals of human primary teeth by checkerboard DNA-DNA hybridization. J Dent Child (Chic) 2007;74:118–23. [PubMed] [Google Scholar]
- 14.Sato T, Hoshino E, Uematsu H, Noda T. Predominant obligate anaerobes in necrotic pulps of human deciduous teeth. Microbial Ecol Health Dis. 1993;6:269–75. [Google Scholar]
- 15.Triches TC, de Figueiredo LC, Feres M, de Freitas SF, Zimmermann GS, Cordeiro MM. Microbial profile of root canals of primary teeth with pulp necrosis and periradicular lesion. J Dent Child (Chic) 2014;81:14–19. [PubMed] [Google Scholar]
- 16.Topcuoglu N, Bozdogan E, Kulekci G. Presence of oral bacterial species in primary endodontic infections of primary teeth. J Clin Pediatr Dent. 2013;38:155–60. doi: 10.17796/jcpd.38.2.5252712533082gt0. [DOI] [PubMed] [Google Scholar]
- 17.Gomes GB, Sarkis-Onofre R, Bonow ML, Etges A, Jacinto RC. An investigation of the presence of specific anaerobic species in necrotic primary teeth. Braz Oral Res. 2013;27:149–55. doi: 10.1590/s1806-83242013000100020. [DOI] [PubMed] [Google Scholar]
- 18.Ledezma-Rasillo G, Flores-Reyes H, Gonzalez-Amaro AM, Garrocho-Rangel A, Ruiz-Rodriguez Mdel S, Pozos-Guillen AJ. Identification of cultivable microorganisms from primary teeth with necrotic pulps. J Clin Pediatr Dent. 2010;34:329–33. doi: 10.17796/jcpd.34.4.20124lu111544377. [DOI] [PubMed] [Google Scholar]
- 19.Kanasi E, Dewhirst FE, Chalmers NI, Kent R, Jr., Moore A, Hughes CV, et al. Clonal analysis of the microbiota of severe early childhood caries. Caries Res. 2010;44:485–97. doi: 10.1159/000320158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Palmer CA, Kent R, Jr., Loo CY, Hughes CV, Stutius E, Pradhan N, et al. Diet and caries-associated bacteria in severe early childhood caries. J Dent Res. 2010;89:1224–9. doi: 10.1177/0022034510376543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hughes CV, Dahlan M, Papadopolou E, Loo CY, Pradhan NS, Lu SC, et al. Aciduric microbiota and mutans streptococci in severe and recurrent severe early childhood caries. Pediatr Dent. 2012;34:16–23. [PMC free article] [PubMed] [Google Scholar]
- 22.Drury TF, Horowitz AM, Ismail AI, Maertens MP, Rozier RG, Selwitz RH. Diagnosing and reporting early childhood caries for research purposes. A report of a workshop sponsored by the National Institute of Dental and Craniofacial Research, the Health Resources and Services Administration, and the Health Care Financing Administration. J Public Health Dent. 1999;59:192–7. doi: 10.1111/j.1752-7325.1999.tb03268.x. [DOI] [PubMed] [Google Scholar]
- 23.Dewhirst FE, Chen T, Izard J, Paster BJ, Tanner AC, Yu WH, et al. The human oral microbiome. J Bacteriol. 2010;192:5002–17. doi: 10.1128/JB.00542-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Novakova D, Svec P, Kukletova M, Zackova L, Sedlacek I. Evaluation of the strain identity between isolates from caries lesions and root canals in early childhood caries cases. Folia Microbiol (Praha) 2013;58:649–56. doi: 10.1007/s12223-013-0254-6. [DOI] [PubMed] [Google Scholar]
- 25.Chavez De Paz LE, Dahlen G, Molander A, Moller A, Bergenholtz G. Bacteria recovered from teeth with apical periodontitis after antimicrobial endodontic treatment. Int Endod J. 2003;36:500–8. doi: 10.1046/j.1365-2591.2003.00686.x. [DOI] [PubMed] [Google Scholar]
- 26.Munson MA, Banerjee A, Watson TF, Wade WG. Molecular analysis of the microflora associated with dental caries. J Clin Microbiol. 2004;42:3023–9. doi: 10.1128/JCM.42.7.3023-3029.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Becker MR, Paster BJ, Leys EJ, Moeschberger ML, Kenyon SG, Galvin JL, et al. Molecular analysis of bacterial species associated with childhood caries. J Clin Microbiol. 2002;40:1001–9. doi: 10.1128/JCM.40.3.1001-1009.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jiang W, Zhang J, Chen H. Pyrosequencing analysis of oral microbiota in children with severe early childhood dental caries. Curr Microbiol. 2013;67:537–42. doi: 10.1007/s00284-013-0393-7. [DOI] [PubMed] [Google Scholar]
- 29.Crielaard W, Zaura E, Schuller AA, Huse SM, Montijn RC, Keijser BJ. Exploring the oral microbiota of children at various developmental stages of their dentition in the relation to their oral health. BMC Med Genomics. 2011;4:22. doi: 10.1186/1755-8794-4-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Simon-Soro A, Belda-Ferre P, Cabrera-Rubio R, Alcaraz LD, Mira A. A tissue-dependent hypothesis of dental caries. Caries Res. 2013;47:591–600. doi: 10.1159/000351663. [DOI] [PubMed] [Google Scholar]
- 31.Simon-Soro A, Tomas I, Cabrera-Rubio R, Catalan MD, Nyvad B, Mira A. Microbial geography of the oral cavity. J Dent Res. 2013;92:616–21. doi: 10.1177/0022034513488119. [DOI] [PubMed] [Google Scholar]
- 32.Hashimoto K, Sato T, Shimauchi H, Takahashi N. Profiling of dental plaque microflora on root caries lesions and the protein-denaturing activity of these bacteria. Am J Dent. 2011;24:295–9. [PubMed] [Google Scholar]
- 33.Takahashi N, Nyvad B. The role of bacteria in the caries process: ecological perspectives. J Dent Res. 2011;90:294–303. doi: 10.1177/0022034510379602. [DOI] [PubMed] [Google Scholar]
- 34.Tanner AC, Sonis AL, Lif Holgerson P, Starr JR, Nunez Y, Kressirer CA, et al. White-spot lesions and gingivitis microbiotas in orthodontic patients. J Dent Res. 2012;91:853–8. doi: 10.1177/0022034512455031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nadkarni MA, Simonian MR, Harty DW, Zoellner H, Jacques NA, Hunter N. Lactobacilli are prominent in the initial stages of polymicrobial infection of dental pulp. J Clin Microbiol. 2010;48:1732–40. doi: 10.1128/JCM.01912-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Trope M. Regenerative potential of dental pulp. J Endod. 2008;34:S13–17. doi: 10.1016/j.joen.2008.04.001. [DOI] [PubMed] [Google Scholar]
- 37.Takushige T, Cruz EV, Asgor Moral A, Hoshino E. Endodontic treatment of primary teeth using a combination of antibacterial drugs. Int Endod J. 2004;37:132–8. doi: 10.1111/j.0143-2885.2004.00771.x. [DOI] [PubMed] [Google Scholar]