Abstract
For many viruses, assembly and budding occur simultaneously during virion formation. Understanding the mechanisms underlying this process could promote biomedical efforts to block viral propagation and enable use of capsids in nanomaterials applications. To this end, we have performed molecular dynamics simulations on a coarse-grained model that describes virus assembly on a fluctuating lipid membrane. Our simulations show that the membrane can promote association of adsorbed subunits through dimensional reduction, but it also introduces thermodynamic and kinetic effects that can inhibit complete assembly. We find several mechanisms by which membrane microdomains, such as lipid rafts, reduce these effects, and thus, enhance assembly. We show how these predicted mechanisms can be experimentally tested. Furthermore, the simulations demonstrate that assembly and budding depend crucially on the system dynamics via multiple timescales related to membrane deformation, protein diffusion, association, and adsorption onto the membrane.
Introduction
Processes in which proteins assemble on membranes to drive topology changes are ubiquitous in biology. Despite extensive experimental and theoretical investigations (e.g., Baumgart et al. (1) and Krauss and Haucke (2)), how assembly-driven membrane deformation depends on protein properties, membrane properties, and membrane compositional inhomogeneity, remains incompletely understood. An important example of this phenomenon occurs during the formation of an enveloped virus, when the virion acquires a membrane envelope by budding from its host cell. Budding is typically driven at least in part by assembly of capsid proteins or viral membrane proteins (3–8), and many enveloped viruses, including HIV and influenza, preferentially bud from membrane microdomains (e.g., lipid rafts) (5,9,10). Understanding how viruses exploit membrane domain structures to facilitate budding would reveal fundamental aspects of the viral lifecycle, and could focus efforts to identify targets for new antiviral drugs that interfere with budding.
There is much interest in developing enveloped viral nanoparticles as targeted transport vehicles equipped to cross cell membranes through fusion (11–13). More generally, identifying the factors that make viral budding robust will shed light on other biological processes in which high-order complexes assemble to reshape membranes. Toward this goal, we perform dynamical simulations in which capsids simultaneously assemble and bud from model lipid membranes. We identify mechanisms by which membrane adsorption either promotes or impedes assembly, and we find multiple mechanisms by which a membrane microdomain significantly enhances assembly and budding.
Enveloped viruses can be divided into two groups based on how they acquire their lipid membrane envelope. For the first group, which includes influenza and type C retroviruses (e.g., HIV), the (immature) nucleocapsid core assembles on the membrane concomitant with budding. In the second group, a core assembles in the cytoplasm before envelopment (reviewed in Sunquist and Kräusslich (3), Hurley et al. (4), and Welsch et al. (5)). In many families from this group, e.g., alphavirus, envelopment is driven by assembly of viral transmembrane glycoproteins around the core (14). For all enveloped viruses, membrane deformation is driven at least in part by a combination of weak protein-protein and protein-lipid interactions. Thus, properties of the membrane should substantially affect budding and assembly timescales. In support of this hypothesis, many viruses from both groups preferentially bud from membrane microdomains 10–100 nm in size that are concentrated with cholesterol and/or sphingolipids (5,9,10). A critical question is whether viruses utilize microdomains primarily to concentrate capsid proteins or other molecules, or if the geometric and physical properties of domains facilitate budding. Answering these questions through experiments alone has been challenging (3–5).
Extensive previous theoretical investigations have studied budding by preassembled cores or nanoparticles (e.g., Ruiz-Herrero et al. (15), Chaudhuri et al. (16), Deserno and Gelbart (17), Deserno and Bickel (18), Deserno (19), Fošnarič et al. (20), Ginzburg and Balijepalli (21), Jiang et al. (22), Li and Xing (23), Li and Gu (24), Smith et al. (25), Tzlil et al. (26), Vácha et al. (27), Yang and Ma (28), and Dasgupta et al. (29,30)); or have budding triggered by nonassembling subunits (31); or have used continuum models to study assembly and budding (32,33). Most closely related to our work, Matthews and Likos (34–36) recently performed simulations on a coarse-grained model of patchy colloidal particles assembling on a membrane represented as a triangulated surface. These elegant simulations provided a first look at the process of simultaneous assembly and budding, and showed that subunit adsorption onto a membrane facilitates assembly through dimensional reduction. Here, we perform dynamical simulations on a model that more closely captures the geometric features of capsid subunits and lipid bilayers, and we explore how the presence of a microdomain within the membrane can influence assembly and budding.
Our simulations show that, while the membrane can promote assembly of partial capsids, free energy penalties and impeded diffusion of adsorbed subunits associated with membrane deformations can inhibit completion of assembly. We find that a microdomain within a certain size range favors membrane geometries that diminish these impediments, and thus can play a key role in enabling complete assembly and budding. Furthermore, our simulations suggest that assembly morphologies depend crucially on multiple timescales, including those of protein-protein association, membrane deformation, and protein adsorption onto the membrane. Finally, we discuss potential effects of simplifications in our coarse-grained model, and how a key prediction from the simulations can be tested in an in vitro assay.
Materials and Methods
Due to the large length- and timescales associated with assembly of a capsid, simulating the process with an all-atom model is beyond the capabilities of contemporary computers (37). Therefore, in this article we aim to elucidate the principles underlying simultaneous assembly and budding by considering a simplified geometric model for capsid proteins, inspired by previous simulations of empty capsid assembly (38–53) and assembly around nucleic acids (51,54–57). Similarly, we consider a simplified model for lipids (31,58) that recapitulates the material properties of biological membranes. Complete details of the model are provided in Supporting Materials and Methods in the Supporting Material; we summarize the model here.
Membrane model
The membrane is represented by the model from Cooke and Deserno (58), in which each amphiphile is represented by one head bead and two tail beads connected by FENE bonds (Fig. 1 c). This is an implicit solvent model; hydrophobic forces responsible for the formation of bilayers are mimicked by attractive interactions between tail beads with interaction strength ε0. This model enables computational feasibility while allowing the formation of bilayers with physical properties such as fluidity, diffusivity, and rigidity that are easily tuned across the range of values measured in biological membranes (15,58). The bead diameter is set to σ = 0.9 nm to obtain bilayers with widths of 5 nm and the lipid-lipid interaction strength is set to kBT/ε0 = 1.1 and ωc = 1.5σ to obtain fluid membranes with bending modulus κ = 8.25 kBT.
Figure 1.
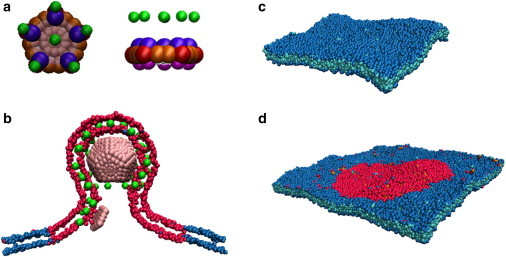
Capsomer and membrane models. (A) Top and side view of the capsomer. (Red and orange) Attractive sites; (blue and purple) top and bottom repulsive sites; (pink) excluders; and (green) capsomer-lipid interaction sites, with the pseudo-atom types defined in Methods and in The Capsid Subunit Model in the Supporting Material. (B) A slice of the membrane and the entire capsid are shown during budding. (Green) Capsomer-lipid interaction sites; (red) domain lipids. (C) Homogeneous membrane patch. (Blue and cyan beads) Lipid heads and lipid tails, respectively. (D) A two-phase membrane. (Red and blue beads) Domain and bulk lipid headgroups, respectively. To see this figure in color, go online.
When studying the effect of a domain, we consider two types of lipids, with M and D referring, respectively, to the lipids outside and inside of the domain, and tail-tail interaction parameters εij (see Eq. S6 in the Supporting Material) set to εDD = εMM = ε0, while εDM is a variable parameter that controls the domain line tension, γ. Varying εDM from 0 to ε0 tunes γ from its maximum value to 0 (see The Membrane Model in the Supporting Material). Within the parameter range studied, the line tension can be approximated by γσ/kBT ≈ 22.9–24.7εDM/ε0 (see Fig. S3 in the Supporting Material).
Capsid subunit model
We modified and extended a model for assembly of nonenveloped capsids (45,54,59,60) to describe assembly on a membrane. A complete listing of the interaction potentials is provided in The Capsid Subunit Model in the Supporting Material; we summarize them here. The capsid subunit is a rigid body with a pentagonal base and radius of rpentamer = 5σ formed by 15 attractive and 10 repulsive interaction sites (Fig. 1 a and see Fig. S4). Subunit assembly is mediated through a Morse potential between attractor pseudoatoms located in the pentagon plane, with one located at each subunit vertex and two along each edge. Attractions occur between like attractors only, meaning that there are vertex-vertex and edge-edge attractions, but no vertex-edge attractor interactions. The 10 repulsive interaction sites are arranged symmetrically above and below the pentagon plane, so as to favor a subunit-subunit angle consistent with a dodecahedron (116°). Further details are in The Capsid Subunit Model in the Supporting Material.
Membrane-capsomer interaction
The potential between capsomers and lipids accounts for attractive interactions and excluded-volume. We add to the capsomer body six attractor pseudoatoms that have attractive interactions with lipid tail beads. When simulating a phase-separated membrane, the attractors interact only with the domain lipid tails, except for simulations presented in Fig. S1. The attractors are placed one above each vertex and one above the center of the pentagon, each located a distance of 6σ above the pentagon plane (Fig. 1 a). These are motivated by, e.g., the myristate group on retrovirus GAG proteins that promotes subunit adsorption by inserting into the lipid bilayer (61). While in many cases the electrostatic interactions between capsid proteins and membrane polar headgroups also drive membrane adsorption, we did not explicitly include such an effect in order to reduce the number of model parameters. The attractor-tail interaction is the same form as the lipid tail-tail interaction except that there is no repulsive component (see Eq. S14 in the Supporting Material)). It is parameterized by the interaction strength, εad, which tunes the adhesion free energy according to ead = αεad with α = 2.276σ−2 (see Adhesion Energy in the Supporting Material).
To account for capsomer-lipid excluded-volume interactions, a layer of 35 excluder beads, each with diameter 1.25σ, is placed in the pentagon plane (Fig. 1 a). Excluders experience repulsive interactions with all lipid beads. Because the mean location of the attractive interaction sites on adsorbed subunits is near the membrane midplane, the effective radius of the assembled capsid (not including the lipid coat) can be estimated from the distance between the attractors and the capsomer plane plus the capsid inradius (the radius of a sphere inscribed in a dodecahedron), which gives Rcapsid ≈ 15.3σ. As discussed below, this is smaller than any enveloped virus, and thus our results are qualitative.
In this work we are motivated by viruses such as HIV, where expression of the capsid protein (GAG) alone is sufficient for the formation of budded particles (62). Therefore, we consider a model that does not include viral transmembrane proteins (spike proteins). We also do not consider how some viruses use cellular machinery to drive scission (63), because this process is virus-specific and depends on detailed properties of cellular proteins. For those viruses our model may elucidate the mechanisms leading up to the point of scission.
Simulations
Simulations were performed on GPUs with a modified version HOOMD 0.10.1 (64,65). We modified the Andersen barostat (66) implementation to simulate the membrane at constant temperature and constant frame tension (31) and to couple the barostat to rigid-body dynamics. The membrane was coupled to the thermostat and barostat with characteristic times τT = 0.4τ0 and τP = 0.5τ0, respectively, with τ0 the characteristic diffusion time for a lipid bead (defined below). The imposed frame tension was set to zero. Simulations with an alternative method to control tension (36) led to the same behavior.
Each capsomer was simulated as a rigid body using the Brownian dynamics algorithm, which uses the (non-overdamped) Langevin equation to evolve positions and rigid body orientations in time (64,65). To approximate the rotational dynamics of globular proteins, we modified the rigid-body algorithm in HOOMD so that forces and torques arising from drag and random buffeting were applied separately and isotropically. Finally, the code was modified to update rigid-body positions according to changes in the box size generated by the barostat at each time step.
Matthews and Likos (36) showed that hydrodynamic interactions (HI) between lipid particles can increase the rate of membrane deformation. However, since the mechanisms of assembly and budding appeared to be similar in simulations that did not include HI, the timescales for protein diffusion and association are only qualitative in a coarse-grained model, and the computational cost required to include HI is large in our more detailed model, we neglect HI in our simulations.
Units
We set the units of energy, length, and time in our simulations equal to the characteristic energy, size, and diffusion time for a lipid bead of ε0, σ, and τ0, respectively. The remaining parameters can be assigned physical values by setting the system to room temperature, T = 300 K, and noting that the typical width of a lipid bilayer is ∼5 nm, and the mass of a typical phospholipid is ∼660 g/mol. The units of our system can then be assigned as follows:
For each set of parameters, the results from four or more independent simulations were averaged to estimate the mean behavior of the system.
Timescales
The diffusion coefficient of capsomers in solution is D ≈ 4.2σ2/τ0, while for capsomers adsorbed on the membrane, D ∈ [0.004, 0.02] for ead/αkBT ranging from 0.6 to 0.2. Thus timescales to diffuse by one capsomer diameter (10σ) are τd ≈ 25τ0 for capsomers in solution and τD ∈[500, 2500]τ0 on the membrane. We note that these timescales are qualitative, because the reduction in degrees of freedom associated with coarse-grained models reduces the ruggedness of the underlying free energy landscape (67). For example, comparison between the lipid model employed here with real values of lipid diffusion indicates a speed-up factor of ∼103 (58).
System
To simulate an infinite membrane, periodic boundary conditions were employed for the lateral dimensions and a wall was placed at the bottom of the box. Thus, the capsomers remained below the membrane unless they budded through it. To maintain a constant and equal ideal gas pressure above and below the membrane (despite the imbalance of capsomer concentrations), phantom particles were added to the system. These particles experienced excluded-volume interactions with the lipid head beads, and no other interactions.
For most simulations of inhomogeneous membranes the membrane contained n = 16,200 lipids, including those belonging to the domain. An initial bilayer configuration was equilibrated and then placed with its normal along the z axis in a cubic box of side-length Lx = Ly = 90σ and Lz = 100σ. For large domains (rdomain > 40) the membrane contained n = 28,800 lipids and the initial box size was 130 × 130 × 100σ3. For most simulations of homogeneous membranes, the bilayer contained n = 7,164 lipids and the initial box size was 63.5 × 63.5 × 100σ3; additional simulations on larger membranes were performed to rule out finite size effects.
The capsomers were introduced in the box in two different ways, to understand how the rate of subunit translation and/or targeting to the membrane affects assembly. The first set of simulations considered budding via quasi-equilibrium states, meaning that capsid proteins adsorb onto the membrane slowly in comparison to assembly and membrane deformation timescales. This scenario corresponds to the limit of low subunit concentration and a rate of subunit protein translation or targeting of subunits to the membrane, which is slow in comparison to assembly. Specifically, each capsomer was injected at ∼50σ below the membrane midplane once all previously injected subunits were part of the same cluster. For other simulations, capsomers were injected one by one with an interval τinject until reaching a predefined maximum number of subunits. In the limit of τinject = 0, all capsomers were placed randomly at distances between 30 and 50σ below the membrane at the beginning of the simulation. For all simulations, the initial configuration had three free capsomers placed at 30σ below the membrane. Images were generated using the program VMD (68).
Results
To simulate capsid protein and membrane dynamics on time- and length-scales relevant to assembly and budding, we use the models illustrated in Fig. 1, a and b, and Fig. S4. The physical mechanisms that control the formation and size of domains (with a typical size of 10–100 nm) in cell membranes are poorly understood (69–71). To focus on the effect of a domain on assembly rather than its formation, we simulate a minimal heterogeneous membrane comprised of two lipid species, with interaction strengths that lead to phase separation within the membrane, with the minor species forming a circular domain (Fig. 1 d). The bulk membrane and domain have the same bending coefficient and area per lipid (to focus on mechanisms other than curvature- or bending stiffness-sorting (1)), but protein subunits preferentially partition into the domain (see Fig. S1; note that a complete listing of the interaction potentials is provided in Materials and Methods and see also The Capsid Subunit Model in the Supporting Material).
We performed simulations for a range of subunit-membrane interaction strengths ead, microdomain sizes rdomain, microdomain line tensions γ, and timescales for subunit association to the membrane τinject. All simulations were performed with subunit-subunit interaction strengths of and between vertex and edge attractors respectively (see Supporting Materials and Methods in the Supporting Material). While assembly can proceed readily in bulk (into the same capsid geometry) under these conditions, in all simulations that we performed (for all values of τinject) subunits adsorbed onto the membrane before assembling into any oligomer larger than a trimer. This behavior is consistent with enveloped viruses for which assembly in the cytosol is limited to small oligomers (e.g., HIV (72)). The results presented here correspond to long but finite simulation times, at which point assembly outcomes appeared roughly independent of increasing simulation time. Although these results need not necessarily correspond to equilibrium configurations, note that capsid assembly must proceed within finite timescales in in vivo or in vitro settings as well (73).
The homogeneous membrane inhibits complete assembly
Given that capsid proteins may be targeted to the membrane rather than arriving by diffusion (74), we have considered several modes of introducing subunits into our simulated system, as described in Materials and Methods. We began by simulating assembly on a homogeneous membrane (a single species of lipid) (Fig. 1 c) via quasi-equilibrium states, meaning that free subunits were injected into the system far from the membrane one by one, each after all previously injected subunits were assembled (see Materials and Methods). This scenario corresponds to the limit of low subunit concentration and a rate of subunit protein translation or targeting of subunits to the membrane that is slow in comparison to assembly.
We found that assembly of membrane-absorbed subunits required large subunit-subunit interactions (as compared to those required for assembly in bulk solution), but that such subunits could undergo rapid nucleation on the membrane. However, we found no parameter sets for which our model undergoes complete assembly and budding on a homogeneous membrane. In most simulations, assembly slows dramatically after formation of a half-capsid (six subunits). The nature of subsequent assembly depends on the adhesion strength. For low adhesion strengths (ead < 0.2α), assembly beyond a half-capsid occurs when particles detach from the membrane, sometimes leading to nearly completely assembled but partially wrapped capsids (Fig. 2, a and b). At intermediate adhesion strengths (0.2 ≤ ead/α ≤ 0.4), particles do not readily dissociate from the membrane and assembly typically stalls at a half-capsid. Higher adhesion strengths (ead > 0.4α) yield deformed, open structures that cannot drive complete budding (Fig. 2 d).
Figure 2.
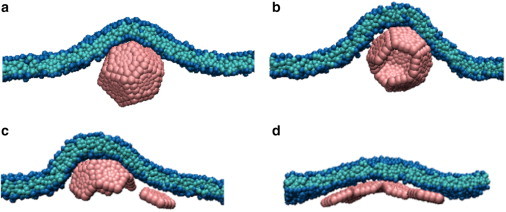
Typical end-products for assembly on a homogeneous membrane as a function of subunit-membrane adhesion strength ead. (A and B) Assembled but partially wrapped capsids for (A) ead = 0.1α and (B) ead = 0.15α. (C) Assembly stalls at a half capsid for ead = 0.2α. (D) A deformed, open structure forms for ead = 0.4α. To see this figure in color, go online.
These results reveal that adsorption to a membrane has mixed effects on assembly. Through dimensional reduction, membrane adsorption reduces the search space and can reduce nucleation barriers by generating a high local subunit concentration (34,36). Similar effects occur during assembly on a polymer (54–56,73,75,76). However, assembly on the membrane also introduces new impediments to assembly.
Formation of a completely enveloped capsid incurs a membrane bending free energy cost of 8πκ, independent of capsid size (77). This free energy penalty must be compensated by subunit-subunit and subunit-membrane interactions. In our model the subunit-membrane interactions do not promote membrane curvature, and thus large subunit-subunit interactions are required for assembly on the membrane. For these parameters, nucleation also occurs in bulk solution if there is no membrane present (nucleation did not occur in bulk solution with a membrane present for any value of τinject because subunits adsorbed onto the membrane before undergoing nucleation). We also considered a model in which the surface of the subunit is curved (see Fig. S10), so that subunit-membrane adsorption does promote local curvature. Interestingly, this model did not lead to improved assembly as compared to the flat subunits.
This result and the frustrated assembly dynamics of half-capsid intermediates illustrate the fact that the geometry and energetics of membranes affect the assembly of membrane-associated proteins in multiple ways, altering both the probability of binding for subunits in the vicinity of an assemblage and the apparent diffusion-limited flux of subunits to the assemblage. Once intermediates reach one-half the capsid size, additional subunits approach with orientations that are not conducive to association. Addition of such a subunit requires a large membrane deformation, which is energetically unfavorable for physically relevant values of the membrane bending rigidity and thus rare (see Fig. S6 b). Assembly therefore stalls or, in the case of weak adhesion energy, proceeds by detachment of subunits from the membrane leading to assembled but partially wrapped capsids. The stalled assembly states resemble the partially assembled states predicted theoretically (32,33), while the partially wrapped capsids are consistent with the metastable partially wrapped states found for a preassembled particle in our previous simulations (15). A second impediment to assembly arises because subunit-membrane attractions are reduced in regions where the membrane curvature is large on the length scale of the rigid subunit (see Fig. S7). This effect hinders subunit diffusion across the neck (see Movies S1 and S2), therefore decreasing the flux of subunits to the assembling capsid.
As discussed below, the large magnitude of the membrane-induced barrier to assembly arises in part due to the small capsid size and relatively large subunits of our model. However, the barrier is intrinsic to assembly of a spherical or convex polygonal structure on a deformable two-dimensional manifold, and thus will exist for any such model.
Assembly and budding from a membrane microdomain
We next simulated assembly in the presence of a phase-separated membrane (Fig. 1 d) to understand the effects of a membrane domain on assembly and budding. While the mechanisms by which rafts form are incompletely understood, we focus on the effect that the presence of a domain can exert on assembly and budding. We emphasize that we consider lipid-lipid interaction parameters and domain sizes for which the domain is flat and stable in the absence of capsid subunits (see Fig. S3 b); i.e., the domain line tension is insufficient to drive budding. We first consider budding in the quasi-equilibrium limit.
Effect of line tension and adhesion energy
Fig. 3 (left) shows the predominant final system configurations as a function of ead and line tension for fixed domain size rdomain = 35σ, which corresponds to 1.3 times the area required to wrap the capsid. Moderate adhesion strengths and small line tensions lead to complete assembly and budding (Fig. 4), meaning that: 12 subunits form a complete capsid, the capsid is completely wrapped by the membrane, and the membrane undergoes scission through spontaneous fusion of the neck to release the membrane-enveloped capsid. Because it requires a relatively large thermal fluctuation, scission is characterized by long timescales. After scission, the portion of the domain not enveloping the capsid remains within the membrane.
Figure 3.
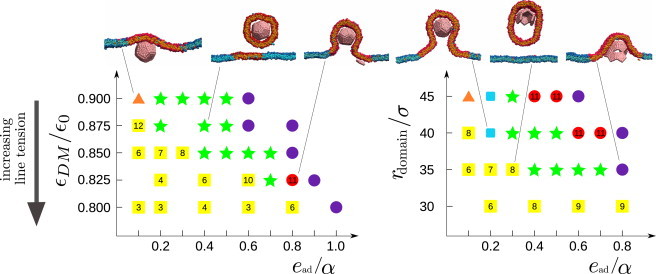
Predominant end-products from assembly simulations via quasi-equilibrium states with a membrane microdomain, as a function of adhesion strength ead and (left) line tension (γσ/kBT ≈ 22.9–24.7εDM/ε0) with fixed domain radius rdomain = 35σ and (right) varying rdomain with fixed line tension εDM/ε0 = 0.85 (γ ≈ 1.9 kBT/σ) and α = 2.276σ−2. The most frequent outcomes are indicated as complete assembly and budding (green solid stars); budding of the entire domain before assembly completes, with the number indicating the typical partial capsid size upon budding (yellow solid squares); complete assembly but incomplete wrapping (orange solid triangles); stalled assembly with wrapping (red solid circles); complete assembly and wrapping without fusion of the neck (blue squares); and malformed assembly (purple circles). Snapshots from simulations for the corresponding parameter sets are also shown. The complete distribution of outcome frequencies and assembly times are shown for some parameter sets in Fig. S11 and Fig. S12. To see this figure in color, go online.
Figure 4.
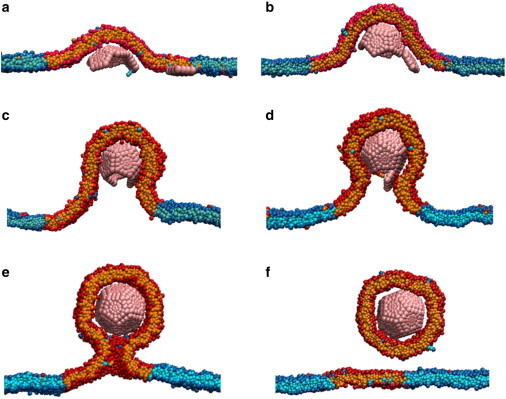
Capsid assembly and budding from a domain. Two-dimensional slices of configurations at different times extracted from MD simulations for ead = 0.4α, rdomain = 35σ, and εDM/ε0 = 0.875 (γ ≈ 1.3 kBT/σ). The membrane wraps the growing capsid (A–D) until the complete, enveloped capsid is connected to the rest of the membrane by a narrow neck (E). Finally, thermal fluctuations lead to fusion of the neck and the encapsulated capsid escapes from the membrane (F). To see this figure in color, go online.
Analysis of simulation trajectories identified three mechanisms by which the domain facilitates assembly. First, partitioning of adsorbed proteins into the domain generates a high local subunit concentration, and thus, promotes nucleation. However, in simulations in which the degree of subunit partitioning into the domain was varied (see Fig. S1), the outcome was insensitive to this parameter. This insensitivity arises because our system requires strong subunit-subunit interactions to drive membrane bending and thus nucleation occurs readily. Second, as noted in the case of spherical particle budding through a raft (25), the domain line tension promotes membrane curvature (buckling), because buckling reduces the length of the domain interface (78). While a positive membrane tension inhibits budding (19), the effective compressive force arising due to line tension promotes budding. Modeling the partial capsid as a hemispherical cap (79), and neglecting curvature outside the vicinity of the capsid (see Theoretical Analysis on the Effect of the Domain on Neck Geometry in the Supporting Material, and Foret (33)), the length of the interface as a function of number of subunits n in a partial capsid is given by
| (1) |
with N = 12 the number of subunits in a capsid and Rcapsid ≈ 15.3σ as the capsid radius. The change in interfacial energy between a flat domain and a completely wrapped capsid is then given by
| (2) |
For rdomain = 35σ and γ = 1.9 kBT/σ, we have ΔEint ≈ −215 kBT, which is comparable to the total bending energy associated with a wrapped capsid, Ebend = 8πκ = 207 kBT. The significance of this effect and consequently the ability of a domain to promote budding diminishes as the domain size becomes large in comparison to the capsid (see Fig. S2).
The simulations also identified a third effect of the domain that promotes complete assembly—the presence of the domain interface changes the geometry of the membrane in the vicinity of the capsid intermediate, promoting a long shallow neck. While curvature energy favors capsid assembly in the domain interior, the line tension is minimized by a neck that extends to the domain interface. The relatively shallow curvature of such a neck reduces the impediments to assembly on a membrane discussed in the previous section. Subunits diffuse readily across a long neck, and subsequent attachment to the assembling capsid incurs relatively small membrane deformation energies. The influence of the neck in subunit diffusion and association is illustrated by animations from assembly trajectories in Movie S1 and Movie S2. Formation of a long neck is governed by a competition between the loss of subunit-membrane adhesion energy associated with partial detachment from the capsid and the reduction in interfacial energy caused by the extended neck. The free energy difference between tightly wrapped capsids and extended necks is approximately calculated in Fig. S9, which indicates that moderate adhesion energies and small domains favor extended necks.
Outside of optimal parameter values, we observe five classes of alternative end-products, as follows:
-
1.
For large values of γ or small domains, formation of a partial capsid triggers budding of the entire domain before assembly completes. Budding of the whole domain is driven by the interfacial energy, Eint(n,rdomain) = γlint(n,rdomain). In the absence of assembly (n = 0), this driving force is insufficient to overcome the deformation free energy barrier to domain budding for the parameters we simulate. However, assembly of a partial capsid intermediate stabilizes curvature and thus reduces the bending energy required for budding of the whole domain. A simple estimate of this effect is obtained by assuming that the partial capsid compensates for the bending energy of a budded domain by an amount proportional to its wrapped area:
| (3) |
Following Lipowsky (78), budding of the whole domain is favorable when Eint(n,rdomain) > Ebend(n,rdomain). However, larger intermediates are required for budding to be spontaneous, thus enabling assembly to complete. Furthermore, for rdomain > 2Rcapsid and intermediates beyond a half-capsid (n > N/2), wrapping of the intermediate produces curvature incommensurate with whole-domain budding. Higher values of ead promote tight wrapping with strong curvature of the capsid, and thus disfavor whole-domain budding (Fig. 4 B).
-
2.
For small γ and ead, the capsid assembles but wrapping is incomplete. Here the subunit-membrane adhesion energy is insufficient to compensate for the membrane bending energy cost associated with wrapping.
-
3.
For larger-than-optimal adhesion strengths, the membrane wraps the assembling capsid tightly with a short neck. As discussed in the previous section, the high curvature associated with a short neck inhibits association of the final subunit leading to stalled, incomplete assembly.
-
4.
For large ead, subunit-membrane adhesion energy dominates over subunit-subunit interactions leading to misassembled structures.
-
5.
At other domain sizes (Fig. 4, right) we observe configurations in which the capsid is completely wrapped, but the neck does not undergo scission. To illustrate the timescales, interactions, and coupling between assembly and membrane configurations, the total subunit-subunit attractive interaction energy and the magnitude of membrane deformation are plotted as a function of time for a trajectory leading to each type of outcome in Fig. 5.
Figure 5.
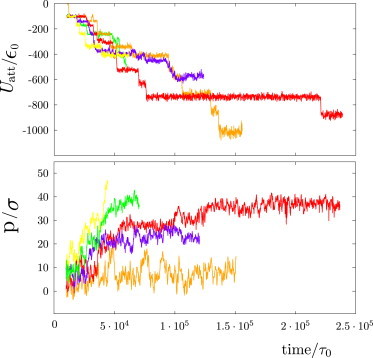
Total subunit-subunit attractive interaction energy (top) and amplitude of membrane deformation, measured by the capsid penetration, p (bottom), as a function of time for a trajectory leading to each type of outcome described in the main text. The capsid penetration p is measured as the distance between the top of the capsid and the center of mass of the membrane. The color code represents the outcome type and follows the same format as in Fig. 3: successful assembly (green), budding of a partial capsid (yellow), complete assembly but incomplete wrapping (orange), stalled assembly with wrapping (red) and malformed assembly (violet). To see this figure in color, go online.
Effect of domain size
The dependence of assembly and budding on the domain radius rdomain for constant line tension γ = 1.9 kBT/σ is shown in Fig. 3 (right). There is an optimal domain size ∼1–2 times the area of a wrapped capsid (35σ ≲ rdomain ≲ 40σ) that leads to robust assembly and budding over a broad range of adhesion energies ead. For smaller domains, low values of adhesion lead to budding of the entire domain before assembly completes. In the absence of protein assembly, line tension triggers budding above a threshold domain size; smaller domains are stable because bending energy dominates over interfacial energy (78). However, we find here that partial capsid intermediates stabilize membrane deformation over an area proportional to their size, and thus drive budding within domains below a threshold size (Eq. 3). On the other hand, for larger-than-optimal domains the assembling capsid only deforms a fraction of the domain; thus, the domain interface does not promote a long neck (see Fig. S9), and it provides a smaller driving force due to interfacial energy (Eq. 3). The behavior of such domains is therefore comparable to that in a homogeneous membrane.
Effect of subunit adsorption timescale
In the quasi-equilibrium simulations discussed so far, the assembly outcomes were determined by the relative timescales of membrane deformation and partial capsid annealing. To determine the effect of the subunit adsorption timescale, we characterized the system behavior for subunit injection timescales τeject (see Materials and Methods) between the quasi-equilibrium limit and 0, where all subunits were introduced at the inception of the simulation (Fig. 6). We set rdomain = 35σ.
Figure 6.
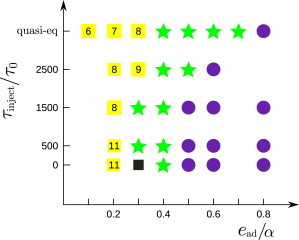
Predominant end-products as a function of the subunit injection timescale τinject and the adhesion strength ead are shown for a domain with rdomain = 25σ and γ = 1.9 kBT/σ. The most frequent outcome is shown for every set of parameters. (Symbols are defined as in Fig. 3 except for black solid square symbols, which denote budding of the whole domain with a malformed capsid inside.) Alternative outcomes observed at some parameter sets are documented in Fig. S15. To see this figure in color, go online.
The predominant end-products are shown as a function of the adhesion strength and the subunit injection timescale in Fig. 6. We see that the qualitative behavior is independent of the injection timescale; for all injection rates there is range of intermediate adhesion strengths at ∼ead = 0.4α, for which complete assembly and budding is observed. However, as the injection timescale decreases, both the lower and upper bounds of this optimal range shift to weaker adhesion energies. Weak adhesion energies avoid malformed assemblages and also increase the timescale for budding of the entire domain (because domain curvature is less stabilized), thus increasing the probability of complete well-formed capsids. However, overly weak adhesion energies lead to longer necks (see Fig. S9) that reduce the timescale for budding of the entire domain, so that, even at high injection rates, budding precedes capsid completion. Stronger-than-optimal adhesion energies tend to result in malformed assemblages (see Fig. S13 b) at the lower injection timescales.
This result can be understood from previous studies of assembly into empty capsids or around polymers (39,41,51,55,73,80,81)—higher adhesion energies lead to an exponential increase in the timescale for annealing of partial capsid configurations; kinetic traps occur when annealing timescales exceed the subunit binding timescale. The ultimate fate of these large aggregates depends on the adhesion energy ead. For smaller-than-optimal adhesion energies, assemblages are loosely wrapped and the entire domain undergoes budding once the assemblage reaches a threshold size (e.g., Fig. S13 b). For larger ead, malformed aggregates are tightly wrapped by the membrane and remain attached by a neck (e.g., Fig. S13 a). The shortest injection timescales and largest adhesion energies we investigated lead to large flat aggregates that do not bend the membrane (see Fig. S13 c), or partial capsids emerging from a flat aggregate (see Fig. S13 d). Finally, we note that as the subunit injection timescale is decreased, the diversity of outcomes at a given parameter set increases and thus the yield of budded, well-formed capsids decreases (see Fig. S15).
Effect of subunit copy number
We found that the dynamics is qualitatively similar when excess subunits are included in the simulation. For example, we performed simulations on systems with 19 capsomers, ∼60% more than needed for capsid formation. For an injection timescale of τeject = 500τ0, the behavior is similar to the small τeject results discussed above, except that subunits on the periphery of an assembling capsid typically form flat aggregates that can hinder budding (see Fig. S14). For adhesion strengths between 0.3 and 0.4 ε0, budding is observed (see Fig. S14), whereas larger values of ead lead to the forms of kinetic traps discussed above.
Discussion
Our simulations demonstrate that, while a fluctuating membrane can promote assembly through dimensional reduction, it also can inhibit complete assembly by limiting the orientational fluctuations and diffusion of adsorbed subunits. These effects, which are not present for assembly in bulk solution (73), can engender metastable partially assembled or partially budded structures. While the degree of inhibition may depend on the specific membrane and protein properties (see below), it is generic to the assembly of a curved structure on a deformable surface. We find that assembly from a membrane microdomain can substantially diminish these effects, which could partly account for the prevalence of enveloped viruses that preferentially bud from lipid rafts or other membrane microdomains.
As an initial exploration of the relationship between membrane domain structure and budding, we considered a minimal model for a microdomain, which accounts only for preferential partitioning or targeting of capsid proteins within the domain. Our simulations identified three effects by which such a domain can promote assembly and budding.
-
1.
Generating a high local concentration of adsorbed subunits,
-
2.
Generating a buckling pressure that promotes budding, and
-
3.
Enhancing the diffusive flux of subunits to the assembling capsid by lengthening the neck around the budding capsid.
Importantly, the predicted effects are sensitive to the domain size (Fig. 3), with an optimal domain size of ∼1–2 times the area of a wrapped capsid. Smaller domains lead to budding before completion of assembly, whereas facilitation of budding becomes ineffective when the domain radius becomes large in comparison to the capsid size. These predictions could be tested by in vitro experiments in which capsid proteins assemble and bud from multicomponent artificial phospholipid vesicles as studied for other membrane-associated proteins (e.g., Becalska et al. (82), Thiam et al. (83), and Manneville et al. (84)) with phase-separated or heterogeneous vesicles (e.g., Manneville et al. (84)).
Finally, we consider the limitations of the model studied here. The effective diameter of our enveloped T = 1 capsid is ∼28 nm, while the smallest enveloped viruses found in nature have diameters of 40–50 nm (e.g., Jones et al. (85)). Although the relationship between particle size and budding has been explored in detail for preassembled nucleocapsids or nanoparticles (e.g., Ruiz-Herrero et al. (15), Ginzburg and Balijepalli (21), and Yue and Zhang (86)), to our knowledge, our simulations here have identified new factors that control simultaneous assembly and budding. During assembly of a larger capsid, each subunit would individually comprise a smaller fraction of the total capsid area and thus would incur a smaller increment of membrane deformation energy when associating with the capsid. Similarly, intrasubunit degrees of freedom could allow subunit distortions that would facilitate diffusion across the neck. However, note that such distortions would themselves involve free energy penalties and thus would still hinder diffusion.
In principle, other forms of excluded-volume interaction with the membrane could improve subunit diffusion, but changing the softness of the repulsive excluded volume interaction between our subunits in the membrane did not change the results. We also note that the potential used for the subunit-membrane interaction in this work does not represent local distortions of the lipid hydrophobic tails resulting from insertion of a hydrophobic group. Such insertions could lead to local membrane curvatures and membrane-mediated subunit interactions that could either facilitate or hinder assembly and budding; the study of these phenomena is an open and active field (e.g., Weikl et al. (87), Semrau et al. (88), Reynwar and Deserno (89), Goulian et al. (90), and Deserno (67)). Given the qualitative nature of subunit-subunit interactions in our model, we do not expect these effects to qualitatively change the results.
To minimize the number of model parameters, we have set the material constants (e.g., bending modulus and fluidity) equal for the domain and background lipid species. Although this study demonstrates three contributions by which such a domain can promote membrane deformation, it will be interesting to explore additional effects that may arise due to varying material properties within microdomains (1). For example, it is believed that biological rafts have higher bending moduli than nonraft membrane environments. Similarly, for some viruses important roles are played by recruitment of additional viral proteins (6), other cellular factors that create or support membrane curvature (4,91,92), and cytoskeletal machinery that actively drives budding (e.g., Welsch et al. (5), Balasubramaniam and Freed (74), Taylor et al. (93), and Gladnikoff et al. (94)).
While these results can be systematically incorporated into the model, our simulations provide a starting point to understand how microdomains facilitate budding and, through comparison with experiments, to identify the critical steps in budding.
Acknowledgments
Computational resources were provided by the National Science Foundation through XSEDE computing resources (Longhorn, Keeneland, and Maverick) and the Brandeis HPCC, which is partially supported by the Brandeis University NSF MRSEC DMR-1420382 (see below).
This work was supported by award No. R01GM108021 from the National Institute of General Medical Sciencesthe Brandeis University NSF MRSEC DMR-1420382, MODELICO grant No. S2009/ESP-1691 from Comunidad Autónoma de Madrid, and grant No. FIS2010-22047-C05-01 from Ministerio de Ciencia e Innovación de España.
Footnotes
Teresa Ruiz-Herrero’s present address is School of Engineering and Applied Sciences, Harvard University, Cambridge, Massachusetts.
Supporting Material
Supporting Citations
References (95–99) appear in the Supporting Material.
References
- 1.Baumgart T., Capraro B.R., Das S.L. Thermodynamics and mechanics of membrane curvature generation and sensing by proteins and lipids. Annu. Rev. Phys. Chem. 2011;62:483–506. doi: 10.1146/annurev.physchem.012809.103450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Krauss M., Haucke V. Shaping membranes for endocytosis. In: Amara S.G., Bamberg E., Fleischmann B.K., Gudermann T., Jahn R., Lederer W.J., Lill R., Nilius B., Offermanns S., editors. Vol. 161. Springer; Berlin, Germany: 2011. pp. 45–66. (Reviews of Physiology, Biochemistry and Pharmacology). [DOI] [PubMed] [Google Scholar]
- 3.Sundquist W.I., Kräusslich H.-G. HIV-1 assembly, budding, and maturation. Cold Spring Harb. Perspect. Med. 2012;2:a006924. doi: 10.1101/cshperspect.a006924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hurley J.H., Boura E., Różycki B. Membrane budding. Cell. 2010;143:875–887. doi: 10.1016/j.cell.2010.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Welsch S., Müller B., Kräusslich H.-G. More than one door—budding of enveloped viruses through cellular membranes. FEBS Lett. 2007;581:2089–2097. doi: 10.1016/j.febslet.2007.03.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Solon J., Gareil O., Gaudin Y. Membrane deformations induced by the matrix protein of vesicular stomatitis virus in a minimal system. J. Gen. Virol. 2005;86:3357–3363. doi: 10.1099/vir.0.81129-0. [DOI] [PubMed] [Google Scholar]
- 7.Vennema H., Godeke G.J., Rottier P.J. Nucleocapsid-independent assembly of coronavirus-like particles by co-expression of viral envelope protein genes. EMBO J. 1996;15:2020–2028. doi: 10.1002/j.1460-2075.1996.tb00553.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garoff H., Hewson R., Opstelten D.-J.E. Virus maturation by budding. Microbiol. Mol. Biol. Rev. 1998;62:1171–1190. doi: 10.1128/mmbr.62.4.1171-1190.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Waheed A.A., Freed E.O. The role of lipids in retrovirus replication. Viruses. 2010;2:1146–1180. doi: 10.3390/v2051146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rossman J.S., Lamb R.A. Influenza virus assembly and budding. Virology. 2011;411:229–236. doi: 10.1016/j.virol.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lundstrom K. Alphaviruses in gene therapy. Viruses. 2009;1:13–25. doi: 10.3390/v1010013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheng F., Tsvetkova I.B., Mukhopadhyay S. The packaging of different cargo into enveloped viral nanoparticles. Mol. Pharm. 2013;10:51–58. doi: 10.1021/mp3002667. [DOI] [PubMed] [Google Scholar]
- 13.Rowan K. Oncolytic viruses move forward in clinical trials. J. Natl. Cancer Inst. 2010;102:590–595. doi: 10.1093/jnci/djq165. [DOI] [PubMed] [Google Scholar]
- 14.Garoff H., Sjöberg M., Cheng R.H. Budding of alphaviruses. Virus Res. 2004;106:103–116. doi: 10.1016/j.virusres.2004.08.008. [DOI] [PubMed] [Google Scholar]
- 15.Ruiz-Herrero T., Velasco E., Hagan M.F. Mechanisms of budding of nanoscale particles through lipid bilayers. J. Phys. Chem. B. 2012;116:9595–9603. doi: 10.1021/jp301601g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chaudhuri A., Battaglia G., Golestanian R. The effect of interactions on the cellular uptake of nanoparticles. Phys. Biol. 2011;8:04600. doi: 10.1088/1478-3975/8/4/046002. [DOI] [PubMed] [Google Scholar]
- 17.Deserno M., Gelbart W.M. Adhesion and wrapping in colloid-vesicle complexes. J. Phys. Chem. B. 2002;106:5543–5552. [Google Scholar]
- 18.Deserno M., Bickel T. Wrapping of a spherical colloid by a fluid membrane. Europhys. Lett. 2003;62:767. [Google Scholar]
- 19.Deserno M. Elastic deformation of a fluid membrane upon colloid binding. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 2004;69:031903. doi: 10.1103/PhysRevE.69.031903. [DOI] [PubMed] [Google Scholar]
- 20.Fošnarič M., Iglič A., May S. Monte Carlo simulations of complex formation between a mixed fluid vesicle and a charged colloid. J. Chem. Phys. 2009;131:105103. [Google Scholar]
- 21.Ginzburg V.V., Balijepalli S. Modeling the thermodynamics of the interaction of nanoparticles with cell membranes. Nano Lett. 2007;7:3716–3722. doi: 10.1021/nl072053l. [DOI] [PubMed] [Google Scholar]
- 22.Jiang W., Kim B.Y.S., Chan W.C.W. Nanoparticle-mediated cellular response is size-dependent. Nat. Nanotechnol. 2008;3:145–150. doi: 10.1038/nnano.2008.30. [DOI] [PubMed] [Google Scholar]
- 23.Li X., Xing D. A simple method to evaluate the optimal size of nanoparticles for endocytosis based on kinetic diffusion of receptors. Appl. Phys. Lett. 2010;97:153704. [Google Scholar]
- 24.Li Y., Gu N. Thermodynamics of charged nanoparticle adsorption on charge-neutral membranes: a simulation study. J. Phys. Chem. B. 2010;114:2749–2754. doi: 10.1021/jp904550b. [DOI] [PubMed] [Google Scholar]
- 25.Smith K.A., Jasnow D., Balazs A.C. Designing synthetic vesicles that engulf nanoscopic particles. J. Chem. Phys. 2007;127:084703. doi: 10.1063/1.2766953. [DOI] [PubMed] [Google Scholar]
- 26.Tzlil S., Deserno M., Ben-Shaul A. A statistical-thermodynamic model of viral budding. Biophys. J. 2004;86:2037–2048. doi: 10.1016/S0006-3495(04)74265-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vácha R., Martinez-Veracoechea F.J., Frenkel D. Receptor-mediated endocytosis of nanoparticles of various shapes. Nano Lett. 2011;11:5391–5395. doi: 10.1021/nl2030213. [DOI] [PubMed] [Google Scholar]
- 28.Yang K., Ma Y.-q. Wrapping and internalization of nanoparticles by lipid bilayers: a computer simulation study. Aust. J. Chem. 2011;64:894. [Google Scholar]
- 29.Dasgupta S., Auth T., Gompper G. Shape and orientation matter for the cellular uptake of nonspherical particles. Nano Lett. 2014;14:687–693. doi: 10.1021/nl403949h. [DOI] [PubMed] [Google Scholar]
- 30.Dasgupta S., Auth T., Gompper G. Wrapping of ellipsoidal nano-particles by fluid membranes. Soft Matter. 2013;9:5473–5482. [Google Scholar]
- 31.Reynwar B.J., Illya G., Deserno M. Aggregation and vesiculation of membrane proteins by curvature-mediated interactions. Nature. 2007;447:461–464. doi: 10.1038/nature05840. [DOI] [PubMed] [Google Scholar]
- 32.Zhang R., Nguyen T.T. Model of human immunodeficiency virus budding and self-assembly: role of the cell membrane. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 2008;78:051903. doi: 10.1103/PhysRevE.78.051903. [DOI] [PubMed] [Google Scholar]
- 33.Foret L. Shape and energy of a membrane bud induced by protein coats or viral protein assembly. Eur. Phys. J. E Soft Matter. 2014;37:42. doi: 10.1140/epje/i2014-14042-1. [DOI] [PubMed] [Google Scholar]
- 34.Matthews R., Likos C. Influence of fluctuating membranes on self-assembly of patchy colloids. Phys. Rev. Lett. 2012;109:178302. doi: 10.1103/PhysRevLett.109.178302. [DOI] [PubMed] [Google Scholar]
- 35.Matthews R., Likos C.N. Structures and pathways for clathrin self-assembly in the bulk and on membranes. Soft Matter. 2013;9:5794–5806. [Google Scholar]
- 36.Matthews R., Likos C.N. Dynamics of self-assembly of model viral capsids in the presence of a fluctuating membrane. J. Phys. Chem. B. 2013;117:8283–8292. doi: 10.1021/jp4037099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Freddolino P.L., Arkhipov A.S., Schulten K. Molecular dynamics simulations of the complete satellite tobacco mosaic virus. Structure. 2006;14:437–449. doi: 10.1016/j.str.2005.11.014. [DOI] [PubMed] [Google Scholar]
- 38.Schwartz R., Shor P.W., Berger B. Local rules simulation of the kinetics of virus capsid self-assembly. Biophys. J. 1998;75:2626–2636. doi: 10.1016/S0006-3495(98)77708-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hagan M.F., Chandler D. Dynamic pathways for viral capsid assembly. Biophys. J. 2006;91:42–54. doi: 10.1529/biophysj.105.076851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hicks S.D., Henley C.L. Irreversible growth model for virus capsid assembly. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 2006;74:031912. doi: 10.1103/PhysRevE.74.031912. [DOI] [PubMed] [Google Scholar]
- 41.Nguyen H.D., Reddy V.S., Brooks C.L., 3rd Deciphering the kinetic mechanism of spontaneous self-assembly of icosahedral capsids. Nano Lett. 2007;7:338–344. doi: 10.1021/nl062449h. [DOI] [PubMed] [Google Scholar]
- 42.Wilber A.W., Doye J.P.K., Wong P. Reversible self-assembly of patchy particles into monodisperse icosahedral clusters. J. Chem. Phys. 2007;127:085106. doi: 10.1063/1.2759922. [DOI] [PubMed] [Google Scholar]
- 43.Nguyen H.D., Brooks C.L., 3rd Generalized structural polymorphism in self-assembled viral particles. Nano Lett. 2008;8:4574–4581. doi: 10.1021/nl802828v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nguyen H.D., Reddy V.S., Brooks C.L., 3rd Invariant polymorphism in virus capsid assembly. J. Am. Chem. Soc. 2009;131:2606–2614. doi: 10.1021/ja807730x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Johnston I.G., Louis A.A., Doye J.P.K. Modelling the self-assembly of virus capsids. J. Phys. Condens. Matter. 2010;22:104101. doi: 10.1088/0953-8984/22/10/104101. [DOI] [PubMed] [Google Scholar]
- 46.Wilber A.W., Doye J.P.K., Lewis A.C.F. Monodisperse self-assembly in a model with protein-like interactions. J. Chem. Phys. 2009;131:175102. doi: 10.1063/1.3243581. [DOI] [PubMed] [Google Scholar]
- 47.Wilber A.W., Doye J.P.K., Louis A.A. Self-assembly of monodisperse clusters: dependence on target geometry. J. Chem. Phys. 2009;131:175101. doi: 10.1063/1.3243580. [DOI] [PubMed] [Google Scholar]
- 48.Rapaport D., Johnson J., Skolnick J. Supramolecular self-assembly: molecular dynamics modeling of polyhedral shell formation. Comput. Phys. Commun. 1999;121–122:231–235. [Google Scholar]
- 49.Rapaport D.C. Self-assembly of polyhedral shells: a molecular dynamics study. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 2004;70:051905. doi: 10.1103/PhysRevE.70.051905. [DOI] [PubMed] [Google Scholar]
- 50.Rapaport D.C. Role of reversibility in viral capsid growth: a paradigm for self-assembly. Phys. Rev. Lett. 2008;101:186101. doi: 10.1103/PhysRevLett.101.186101. [DOI] [PubMed] [Google Scholar]
- 51.Hagan M.F., Elrad O.M., Jack R.L. Mechanisms of kinetic trapping in self-assembly and phase transformation. J. Chem. Phys. 2011;135:104115. doi: 10.1063/1.3635775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ayton G.S., Voth G.A. Multiscale computer simulation of the immature HIV-1 virion. Biophys. J. 2010;99:2757–2765. doi: 10.1016/j.bpj.2010.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen B., Tycko R. Simulated self-assembly of the HIV-1 capsid: protein shape and native contacts are sufficient for two-dimensional lattice formation. Biophys. J. 2011;100:3035–3044. doi: 10.1016/j.bpj.2011.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Perlmutter J.D., Qiao C., Hagan M.F. Viral genome structures are optimal for capsid assembly. eLife. 2013;2:e00632. doi: 10.7554/eLife.00632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Elrad O.M., Hagan M.F. Encapsulation of a polymer by an icosahedral virus. Phys. Biol. 2010;7:045003. doi: 10.1088/1478-3975/7/4/045003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mahalik J.P., Muthukumar M. Langevin dynamics simulation of polymer-assisted virus-like assembly. J. Chem. Phys. 2012;136:135101. doi: 10.1063/1.3698408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang R., Linse P. Icosahedral capsid formation by capsomers and short polyions. J. Chem. Phys. 2013;138:154901. doi: 10.1063/1.4799243. [DOI] [PubMed] [Google Scholar]
- 58.Cooke I.R., Deserno M. Solvent-free model for self-assembling fluid bilayer membranes: stabilization of the fluid phase based on broad attractive tail potentials. J. Chem. Phys. 2005;123:224710. doi: 10.1063/1.2135785. [DOI] [PubMed] [Google Scholar]
- 59.Wales D.J. The energy landscape as a unifying theme in molecular science. Philos. Trans. A Math. Phys. Eng. Sci. 2005;363:357–377. doi: 10.1098/rsta.2004.1497. [DOI] [PubMed] [Google Scholar]
- 60.Fejer S.N., James T.R., Wales D.J. Energy landscapes for shells assembled from pentagonal and hexagonal pyramids. Phys. Chem. Chem. Phys. 2009;11:2098–2104. doi: 10.1039/b818062h. [DOI] [PubMed] [Google Scholar]
- 61.Hamard-Peron E., Muriaux D. Retroviral matrix and lipids, the intimate interaction. Retrovirology. 2011;8:15. doi: 10.1186/1742-4690-8-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Johnson M.C., Scobie H.M., Vogt V.M. Nucleic acid-independent retrovirus assembly can be driven by dimerization. J. Virol. 2002;76:11177–11185. doi: 10.1128/JVI.76.22.11177-11185.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Baumgärtel V., Ivanchenko S., Lamb D.C. Live-cell visualization of dynamics of HIV budding site interactions with an ESCRT component. Nat. Cell Biol. 2011;13:469–474. doi: 10.1038/ncb2215. [DOI] [PubMed] [Google Scholar]
- 64.Anderson J.A., Lorenz C.D., Travesset A. General purpose molecular dynamics simulations fully implemented on graphics processing units. J. Comput. Phys. 2008;227:5342–5359. [Google Scholar]
- 65.Nguyen T.D., Phillips C.L., Glotzer S.C. Rigid body constraints realized in massively-parallel molecular dynamics on graphics processing units. Comput. Phys. Commun. 2011;182:2307–2313. [Google Scholar]
- 66.Andersen H.C. Molecular dynamics simulations at constant pressure and/or temperature. J. Chem. Phys. 1980;72:2384. [Google Scholar]
- 67.Deserno M. Mesoscopic membrane physics: concepts, simulations, and selected applications. Macromol. Rapid Commun. 2009;30:752–771. doi: 10.1002/marc.200900090. [DOI] [PubMed] [Google Scholar]
- 68.Humphrey W., Dalke A., Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14:27–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 69.Lingwood D., Simons K. Lipid rafts as a membrane-organizing principle. Science. 2010;327:46–50. doi: 10.1126/science.1174621. [DOI] [PubMed] [Google Scholar]
- 70.Kerviel A., Thomas A., Muriaux D. Virus assembly and plasma membrane domains: which came first? Virus Res. 2013;171:332–340. doi: 10.1016/j.virusres.2012.08.014. [DOI] [PubMed] [Google Scholar]
- 71.Parton D.L., Tek A., Sansom M.S.P. Formation of raft-like assemblies within clusters of influenza hemagglutinin observed by MD simulations. PLOS Comput. Biol. 2013;9:e1003034. doi: 10.1371/journal.pcbi.1003034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ivanchenko S., Godinez W.J., Lamb D.C. Dynamics of HIV-1 assembly and release. PLoS Pathog. 2009;5:e1000652. doi: 10.1371/journal.ppat.1000652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hagan M.F. Modeling viral capsid assembly. Adv. Chem. Phys. 2014;155:1–68. doi: 10.1002/9781118755815.ch01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Balasubramaniam M., Freed E.O. New insights into HIV assembly and trafficking. Physiology (Bethesda) 2011;26:236–251. doi: 10.1152/physiol.00051.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kivenson A., Hagan M.F. Mechanisms of capsid assembly around a polymer. Biophys. J. 2010;99:619–628. doi: 10.1016/j.bpj.2010.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Perlmutter J.D., Perkett M.R., Hagan M.F. Pathways for virus assembly around nucleic acids. J. Mol. Biol. 2014;426:3148–3165. doi: 10.1016/j.jmb.2014.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Phillips R.B., Kondev J., Garcia H. 2nd Ed. Garland Science; New York: 2013. Physical Biology of the Cell. [Google Scholar]
- 78.Lipowsky R. Domain-induced budding of fluid membranes. Biophys. J. 1993;64:1133–1138. doi: 10.1016/S0006-3495(93)81479-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zandi R., van der Schoot P., Reiss H. Classical nucleation theory of virus capsids. Biophys. J. 2006;90:1939–1948. doi: 10.1529/biophysj.105.072975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Grant J., Jack R.L., Whitelam S. Analyzing mechanisms and microscopic reversibility of self-assembly. J. Chem. Phys. 2011;135:214505. doi: 10.1063/1.3662140. [DOI] [PubMed] [Google Scholar]
- 81.Rapaport D.C. Modeling capsid self-assembly: design and analysis. Phys. Biol. 2010;7:045001. doi: 10.1088/1478-3975/7/4/045001. [DOI] [PubMed] [Google Scholar]
- 82.Becalska A.N., Kelley C.F., Rodal A.A. Formation of membrane ridges and scallops by the F-BAR protein Nervous Wreck. Mol. Biol. Cell. 2013;24:2406–2418. doi: 10.1091/mbc.E13-05-0271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Thiam A.R., Antonny B., Pincet F. COPI buds 60-nm lipid droplets from reconstituted water-phospholipid-triacylglyceride interfaces, suggesting a tension clamp function. Proc. Natl. Acad. Sci. USA. 2013;110:13244–13249. doi: 10.1073/pnas.1307685110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Manneville J.-B., Casella J.-F., Goud B. COPI coat assembly occurs on liquid-disordered domains and the associated membrane deformations are limited by membrane tension. Proc. Natl. Acad. Sci. USA. 2008;105:16946–16951. doi: 10.1073/pnas.0807102105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Jones C.T., Ma L., Kuhn R.J. Flavivirus capsid is a dimeric α-helical protein. J. Virol. 2003;77:7143–7149. doi: 10.1128/JVI.77.12.7143-7149.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yue T., Zhang X. Molecular understanding of receptor-mediated membrane responses to ligand-coated nanoparticles. Soft Matter. 2011;7:9104. [Google Scholar]
- 87.Weikl T.R., Kozlov M.M., Helfrich W. Interaction of conical membrane inclusions: effect of lateral tension. Phys. Rev. E. 1998;57:6988–6995. [Google Scholar]
- 88.Semrau S., Idema T., Storm C. Membrane-mediated interactions measured using membrane domains. Biophys. J. 2009;96:4906–4915. doi: 10.1016/j.bpj.2009.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Reynwar B.J., Deserno M. Membrane-mediated interactions between circular particles in the strongly curved regime. Soft Matter. 2011;7:8567–8575. [Google Scholar]
- 90.Goulian M., Bruinsma R., Pincus P. Long-range forces in heterogeneous fluid membranes. Europhys. Lett. 1993;22:145–150. [Google Scholar]
- 91.McMahon H.T., Gallop J.L. Membrane curvature and mechanisms of dynamic cell membrane remodeling. Nature. 2005;438:590–596. doi: 10.1038/nature04396. [DOI] [PubMed] [Google Scholar]
- 92.Doherty G.J., McMahon H.T. Mechanisms of endocytosis. Annu. Rev. Biochem. 2009;78:857–902. doi: 10.1146/annurev.biochem.78.081307.110540. [DOI] [PubMed] [Google Scholar]
- 93.Taylor M.P., Koyuncu O.O., Enquist L.W. Subversion of the actin cytoskeleton during viral infection. Nat. Rev. Microbiol. 2011;9:427–439. doi: 10.1038/nrmicro2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Gladnikoff M., Shimoni E., Rousso I. Retroviral assembly and budding occur through an actin-driven mechanism. Biophys. J. 2009;97:2419–2428. doi: 10.1016/j.bpj.2009.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Cooke I.R., Kremer K., Deserno M. Tunable generic model for fluid bilayer membranes. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 2005;72:011506. doi: 10.1103/PhysRevE.72.011506. [DOI] [PubMed] [Google Scholar]
- 96.Weeks J.D., Chandler D., Andersen H.C. Role of repulsive forces in determining the equilibrium structure of simple liquids. J. Chem. Phys. 1971;54:5237. [Google Scholar]
- 97.Grest G.S., Kremer K. Molecular dynamics simulation for polymers in the presence of a heat bath. Phys. Rev. A. 1986;33:3628–3631. doi: 10.1103/physreva.33.3628. [DOI] [PubMed] [Google Scholar]
- 98.Reynwar B.J., Deserno M. Membrane composition-mediated protein-protein interactions. Biointerphases. 2008;3:FA117. doi: 10.1116/1.2977492. [DOI] [PubMed] [Google Scholar]
- 99.Bereau T., Wang Z.-J., Deserno M. More than the sum of its parts: coarse-grained peptide-lipid interactions from a simple cross-parametrization. J. Chem. Phys. 2014;140:115101. doi: 10.1063/1.4867465. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


