Abstract
The homodimer NGF (nerve growth factor) exerts its neuronal activity upon binding to either or both distinct transmembrane receptors TrkA and p75NTR. Functionally relevant interactions between NGF and these receptors have been proposed, on the basis of binding and signaling experiments. Namely, a ternary TrkA/NGF/p75NTR complex is assumed to be crucial for the formation of the so-called high-affinity NGF binding sites. However, the existence, on the cell surface, of direct extracellular interactions is still a matter of controversy. Here, supported by a small-angle x-ray scattering solution study of human NGF, we propose that it is the oligomerization state of the secreted NGF that may drive the formation of the ternary heterocomplex. Our data demonstrate the occurrence in solution of a concentration-dependent distribution of dimers and dimer of dimers. A head-to-head molecular assembly configuration of the NGF dimer of dimers has been validated. Overall, these findings prompted us to suggest a new, to our knowledge, model for the transient ternary heterocomplex, i.e., a TrkA/NGF/p75NTR ligand/receptors molecular assembly with a (2:4:2) stoichiometry. This model would neatly solve the problem posed by the unconventional orientation of p75NTR with respect to TrkA, as being found in the crystal structures of the TrkA/NGF and p75NTR/NGF complexes.
Introduction
Nerve growth factor (NGF), the first neurotrophin (NT) to be discovered, plays a key role in determining survival, differentiation, and maintenance of specific neuronal populations during development (1–4). Like other members of the NTs family, NGF is a functional homodimer composed of two noncovalently bound chains (5) that exerts its actions through two structurally unrelated transmembrane receptors (6–8), the TrkA receptor tyrosine-kinase (9,10), which is specific for NGF, and the p75NTR receptor (11), a tumor necrosis factor receptor family member that instead is shared among all NTs.
Each receptor binds independently to NGF with predominantly low affinity, (KD = 10−9 M (9,12–14)), but they produce high-affinity binding sites (KD = 10−11 M) upon receptors coexpression (12,15).
Such high-affinity binding sites are generally believed to be crucial for the overall NTs physiology. In fact, induction of high-affinity binding sites has been characterized on embryonic neurons, not only in the presence of NGF (16) but also of brain-derived neurotrophic factor (BDNF) and NT-3 (14,17). Moreover, these sites, in contrast to the low-affinity binding sites, correlate well with some of the best known physiological properties of the NTs. Indeed, not only is the half-maximal neuronal survival typically obtained with NTs concentrations of 5 × 10−12 M, but they also trigger/mediate special functional and pharmacological properties (18–20), ligand selectivity (21,22), and the differential cross-regulation of TrkA with p75NTR (23).
Although it is widely accepted that the biologically active forms of both receptors are the stable and functional symmetric complexes with NT homodimers (24–26), characterized by a 2:2 stoichiometry, the cross talk mechanism between TrkA and p75NTR, which originates the formation of the high-affinity NT binding sites on neuronal cells and so altering the signaling properties of both partners, is still poorly understood. To this regard several models of TrkA/p75NTR complexes have been proposed to produce a site with high affinity and selectivity for NGF, including: 1) the ligand passing model, where p75NTR binds NGF and passes it to TrkA (27); 2) the allosteric model, where interactions between transmembrane and cytoplasmic domains of both TrkA and p75NTR enhance NGF presentation to the TrkA and accelerate the rate of association (15,28); 3) the formation of a transient ternary complex of TrkA/NGF/p75NTR (12); and 4) a ligand-independent TrkA/p75NTR heterocomplex, in which a ligand-induced alteration preludes to the homodimerization of the receptors required for signaling (29). Even if elusive, the in vivo formation of a ternary heterocomplex has been supported by experimental evidences: in addition to the reconstitution binding experiments (12), it has been confirmed by affinity cross-linking and immunoprecipitation studies (30,31). Nonetheless, the unequivocal functional dissection of the domains involved in the formation of the high-affinity binding sites on p75NTR or on TrkA has led to contrasting conclusions (32–37).
In addition, three recent x-ray crystallographic studies reported evidences against the likely formation of a transient ternary complex of TrkA/NGF/p75NTR, i.e., the NGF/TrkA (38,39), NGF/p75NTR (40), and NT-3/p75NTR (26) binary complexes. The superposition of the structures shows the ligand binding sites for either receptor are mutually exclusive and therefore not in favor of a ternary heterocomplex model with a 2:2:2 stoichiometry.
Moreover, the observed opposite orientation of NGF bound to TrkA as compared to p75NTR cannot be easily reconciled even with the existence of a 1:2:1 ternary heterocomplex. Several ad hoc explanations have been proposed, based on 1) possible multiple proteolytic cleavages that would trigger the release of the ectodomain of p75NTR (41,42); 2) likely conformational changes in the mutual orientations of the different modules of the TrkA and p75NTR ectodomains (43); and 3) one receptor handing over the NGF ligand to the other receptor (44).
To solve this conundrum, we now postulate that the existence in solution of oligomeric forms of NGF may support an alternative mechanism that would likely accommodate a transient ternary heterocomplex. Indeed, NGF oligomers have been previously reported to form not only in solution, by equilibrium sedimentation studies (45), but also in the solid state, namely in the crystal structure of the mouse bis-des-octa beta NGF (mNGF) bound to zinc Protein Data Bank (PDB) ID 1BTG (46). In the latter, the potential biological relevance of the observed dimer of dimers (DD) arrangement was not considered, ascribing it as a result of artifacts associated with lattice crystal packing. We therefore sought to further examine and structurally characterize the oligomerization state of hNGF in solution by small-angle x-ray scattering (SAXS) experiments. The analysis of our SAXS data demonstrates a concentration-dependent equilibrium between NGF homodimers and a DD form, and, in addition, provides structural evidence for a head-to-head DD assembly of hNGF in solution. This oligomeric arrangement offers intriguing biological implications.
Indeed, these findings lead us to hypothesize a transient ternary heterocomplex with a 2:4:2 stoichiometry (i.e., TrkA/NGF/NGF/p75NTR), that would reconcile the observed unconventional orientation of the p75NTR and TrkA receptors with respect to NGF as well as their mutual steric obstruction as a result of a simultaneous binding to the same face of NGF (25,38–40).
Materials and Methods
SAXS experiment and data processing
Synchrotron radiation x-ray solution scattering data were collected on the X33 beamline (47,48) of the EMBL at DORIS III storage ring, DESY, Hamburg. Solutions of hNGF, obtained as previously reported (49), in 50 mM Na-phosphate pH 7.0, 1 mM EDTA at 0.43, 1.98, 2.75, 3.77, and 5.5 mg/mL concentrations, were measured using a MAR345 Image Plate at a wavelength λ = 1.5 Å, covering the momentum transfer range 0.012 < s < 0.45 Å−1. To check for radiation damage two 2-min exposures were compared; no radiation effects were observed upon addition of 2 mM DTT to the sample immediately before the measurements. The data were processed and the scattering of the buffer was subtracted. All data manipulations were performed using the program package PRIMUS (50).
The forward scattering I(0) and the radius of gyration Rg were evaluated using the Guinier approximation (51) and the program GNOM (52) that also provides the maximum particle dimension Dmax and the distance distribution function p(r). The molecular masses of the solutes were evaluated by calibration against the reference solutions of bovine serum albumin.
The scattering curves from the crystallographic models of hNGF were calculated using the program CRYSOL (53). For the mixtures of hNGF the experimental intensity Iexp(s) was represented as Iexp(s) = vd Id (s) + vdd Idd (s), where vd and vdd are the volume fractions of hNGF dimers and DD, respectively Id (s) and Idd (s) are the intensities of hNGF dimers and DD, respectively. These volume fractions were determined by the program OLIGOMER (50) to best fit the experimental data. The shape of hNGF was reconstructed by the ab initio program DAMMIN (54) employing simulated annealing to provide a compact interconnected bead model. The results of multiple runs (15 runs) using the program DAMMIN (53), were aligned, averaged, and superposed to determine common structural features using the programs DAMAVER (55) and SUPCOMB (56), respectively.
Modeling, docking, and molecular dynamics simulations
To obtain an overall molecular model of hNGF, as much as complete, the crystallographic structure of hNGF from hNGF/TrkA complex (PDB ID 2IFG) (38), missing some residues (underlined in Fig. S1 in the Supporting Material), was used as template. Its C-term part (A116-A120) was extended by molecular modeling on the basis of the crystal structure of mNGF (PDB ID 1BTG) (46) and its missing loop III (P61-S66) was modeled on the basis of the llama NGF structure (PDB ID 4EFV) (57); the multiple alignment of hNGF sequence with the two templates is shown in Fig. S1. This model of hNGF (G10-A120), devoid of the N-terminal region (S1-R9), and the crystal structure of mNGF (G10-R118) were energy minimized and used to generate the molecular DD assemblies by self-docking with the program SymmDock (58).
All molecular dynamics (MD) simulations were performed in explicit water using the GROMACS 4.5 (Groningen Machine for Chemical Simulation) package (59) in conjunction with the Amber99SB force field. The simple point charge model was used to represent the water. The protonation state of the ionizable groups of either proteins was chosen according to pH 7.0 and an appropriate number of counter ions were added to achieve charge neutrality in the simulation system. After energy minimization by using a steepest descent algorithm, during the equilibration dynamic period the system was thermostated to a temperature of 300 K and maintained at a pressure of 1 bar. Starting from these equilibrated structures, MD production runs of 10 ns in length were performed. All simulations were performed at 1 atm and 300 K by coupling to an external heat and an isotropic pressure bath. The MD simulations were aimed 1) at inspecting the structural fluctuations of the reconstructed regions of the hNGF dimer and 2) at investigating the molecular assembly stability association of the selected solution from the hNGF DD docking.
The dynamical behavior of the DD structure was monitored by calculating 1) the distance between the centers of mass (COMs) of the two dimers and 2) the root mean-square derivation (RMSD) using, as the reference, the starting energy minimized DD assembly.
The ΔASApolar and ΔASAapolar upon the hNGF DD formation were calculated by the program Surface Racer 5.0 (60) with a probe radius of 1.4Å. The heat capacity change ΔCp and burial of solvent-accessible surface (ΔASAapolar and ΔASApolar) can be correlated according to the following equations (61,62):
ΔH(333 K) is defined as the binding enthalpy at 333 K. The latter is the median unfolding temperature of the set of proteins being used for the parameterization of the previous relationships, for which thermodynamic and high-resolution structural information are available (61,62).
All the figures have been created by the program Pymol (63).
Biochemical experiments
For analytical gelfiltration experiments, increasing concentration of hNGF were passed through a Superdex 75 10/300 GL column equilibrated in 50 mM Na Phosphate (pH 7), 1 mM EDTA, 150 mM NaCl at a flow rate of 0.7 mL/min.
In cross-linking experiments, hNGF stock solution (2.5 mg/mL) was serially diluted 1:2 and incubated with Bis(sulfosuccinimidyl)suberate (BS3) at protein/BS3 ratio (mol/mol) of 1/27. These samples were incubated for 24 h at room temperature to allow cross-linking reactions to proceed. The reactions were terminated by adding sodium dodecylsulfate polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer. To monitor the reaction, the samples were analyzed by 15% SDS-PAGE in reduced conditions followed by Coomassie staining.
Results
SAXS experiments on hNGF
A pilot set of SAXS measurements was carried out on a solution of hNGF at a concentration of 5.5 mg/mL. The experimental pattern and the computed p(r) function are shown in Fig. S2 A. The derived parameters of molecular mass of 54 ± 5 kDa, Rg = 29.0 ± 0.5 Å and Dmax = 120 ± 10 Å point to a rather extended structure, suggestive of a DD assembly of hNGF in solution. Remarkably, DD arrangements were observed in the crystal structure of mNGF (PDB ID 1BTG) (46). The asymmetric unit consists of three protomers. Two are related by a twofold noncrystallographic symmetry forming a dimer, whereas the third protomer forms a dimer by applying a twofold crystallographic symmetry operation. A close analysis of the crystal packing clearly showed that the two types of dimers were arranged as DD. Among the eight DD interfaces, present in the crystal and assessed with the Protein interfaces, surfaces, and assemblies service PISA at the European Bioinformatics Institute (64), the head-to-head DD with the wider interface area (549.0 Å2) has been chosen to calculate the scattering curve, yielding a good fit to our experimental data (χ = 1.88) and confirm the existence of a DD assembly of hNGF in solution. Moreover the ab initio shape of hNGF yielded a good superimposition with the previous DD crystallographic assembly of mNGF (46), as shown in Fig. S2 B.
Molecular modeling, self-docking, and MD on hNGF and its assembly
It is worth noting that in addition to differences in the source of the NGF analyzed by SAXS, human instead of mouse, the exploited crystallographic structure of mNGF PDB ID 1BTG (46) actually refers to the bis-des-octa (1–8) form of mNGF, the first eight N-terminal residues being removed by proteolysis. Therefore, we resort to obtain a model of hNGF DD to analyze SAXS data. A structure of partner free hNGF is however still lacking. To date crystal structures have been reported for the partner free NGF from mouse PDB ID 1BET (65), 4EAX (66) and 1BTG (46), from cobra PDB ID 4EC7 (66), and from llama PDB ID 4EFV (57), whereas hNGF crystal structures have been determined only in complex with the ligand binding domain (d5) PDB ID 1WWW (39) or the full length of the TrkA receptor PDB ID 2IFG (38) and of the ectodomain of the unrelated cellular receptor p75NTR PDB ID 1SG1 (40). A complex of hNGF and the ectodomain of p75NTR has also been characterized by solution x-ray scattering (25). Interestingly, a six residues long loop (P61-S66), namely loop III, as well as a few residues at the N- and C-terminals are not defined in the crystal structures PDB ID 1WWW (39), PDB ID 2IFG (38), and PDB ID 1SG1 (40). The loop III of llama NGF PDB ID 4EFV (57), whose sequence is nearly identical to hNGF (except for residue 65 that in hNGF is Asp, whereas in llama NGF is Ala), on the contrary is in one of the two protomers well-defined. Therefore, we have been able to reconstruct a rather comprehensive model of hNGF on the basis of the crystal structures PDB ID 2IFG (38), PDB ID 1BTG (46), and PDB ID 4EFV (57). The obtained hNGF model however still lacks the first N-terminal and the last two C-terminal residues, respectively. Energy minimization and subsequent MD simulations using GROMACS (59) were performed on the resulting model to validate the structures of the modeled regions and to relax the main and side-chain conformations of those residues involved in intermolecular contacts with the TrkA receptor. As expected, the most flexible regions resulted to be the loops and the N- and C-termini. However, the 10 ns long simulation showed that the local structure of the modeled loop III is not significantly more flexible than the other loops. The RMS fluctuations, during the 10 ns of MD simulation, in the Cα position are plotted against the residues of the hNGF modeled. The N-terminal region of hNGF in either protomers resulted to be extremely flexible (Fig. 1 A). In fact, with the only exception of the complex of hNGF with TrkA (an α-helix is formed upon binding) in all of the remaining complexed and uncomplexed NGF known crystal structures, the N-terminal residues are not structurally defined.
Figure 1.
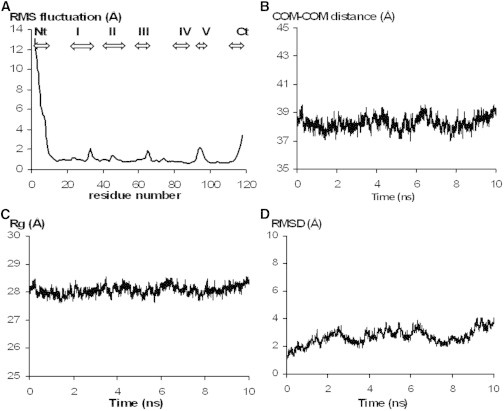
MD simulations of hNGF model. (A) Cα RMS fluctuations over the 10 ns MD simulation (for clarity only a representative protomer is reported. Nt: N-terminal; I: loopI; II: loopII; III: loopIII; IV: loopIV; V: loopV; Ct: C-terminal); and of hNGF DD model: (B) distance between the COMs of the two dimers, (C) radius of gyration and (D) RMSD with respect to the energy minimized starting assembly during a 10 ns simulation.
Hence, even though the N-terminal region of hNGF might play a role in the DD formation, it would be affected by a local conformational flexibility that is unlikely to be foreseen by the clustering methods used to analyze the MD trajectories. Therefore, we opted to remove residues S1-R9 from the hNGF modeled structure aiming to increase the likelihood of native-like poses in the subsequent self-docking simulations. The energy minimized model of hNGF, devoid of nine N-terminal residues (S1-R9) was then subjected to the self-docking simulations to generate poses of the DD. For validation purposes, the same protocol was applied to obtain self-docking poses of the mNGF DD model by using the energy minimized crystal structure of the bis-des-octa (1–8) mNGF (PDB ID 1BTG) (46), devoid of 1 N-terminal residue (M9). Furthermore, we aimed to compare the two self-docking simulations by applying the same energy and similarity filtering criteria by assuming that the DD assembly/interface should be conserved across evolution. The resulting top 100 solutions of each self docking simulation, performed with SymmDock (58), ranked according to a scoring function that includes the geometric fit, the atomic desolvation energy and the interface area size, were compared to identify similar poses, and then analyzed to select those whose shapes were the closest to the head-to-head assembly observed in the crystal structure of mNGF (PDB ID 1BTG) (46), that well fitted the experimental SAXS curve.
The hNGF DD pose ranking 13 among the top 100 solutions (with a geometric score of 10,276, an interface area size of 1310.40 Å2, and a desolvation energy of 23.86 kcal/mol), showed the lowest RMSD (15.00 Å), calculated on Cα atoms, from the reference head-to-head crystallographic assembly of the mNGF. In addition, this hNGF DD pose showed to be similar (RMSD 7.04 Å) to the third best scored pose among the top 100 solutions of the mNGF self-docking simulation (with a geometric score of 11,510, an interface area size of 2138.40 Å2, and a desolvation energy of −59.37 kcal/mol).
The hNGF DD interface was analyzed by the 2P2I Inspector web-based tool (67). The calculated interface parameters are within the means and the standard deviations of interfaces parameters calculated for a nonredundant representative data set of homodimeric complexes (for details, see below).
To assess DD stability, this model was subjected to MD simulations using GROMACS (59). The distance between the COMs of each dimer was monitored along the 10 ns long simulation. The COM-COM distance (Fig. 1 B) resulted to be rather stable from the beginning of the simulation and small fluctuations were observed only for the radius of gyration (Fig. 1 C). The computed average Rg (along the 10 ns simulation time) of 28.1 ± 0.2 Å for the hNGF DD model is very close to the experimental Rg value of 29.0 ± 0.5 Å (Guinier plot). These results confirm the stability of the modeled hNGF DD molecular assembly. Moreover, the analysis of the RMSD calculated on Cα atoms over time, computed using as reference the energy minimized starting hNGF DD model (Fig. 1 D), showed that after the initial stabilization, in the first 2 ns, the assembly did not considerably deviate from the starting model. Therefore, the described in silico studies provided comprehensive models of both hNGF dimers and DD to be used for further analysis of the SAXS data.
SAXS studies on concentration-dependent assembly of hNGF
SAXS data at 5.5 mg/mL (curve 1 in Fig. 2 A) were analyzed on the basis of the modeled hNGF DD: the p(r) computed from the atomistic model of DD (dash line in Fig. 2 B) well overlaps with the p(r) obtained from the SAXS profile. The molecular assembly fits well with the shape of hNGF as determined ab initio from the solution scattering data by using the DAMMIN program (54) (see Fig. 2 D).
Figure 2.
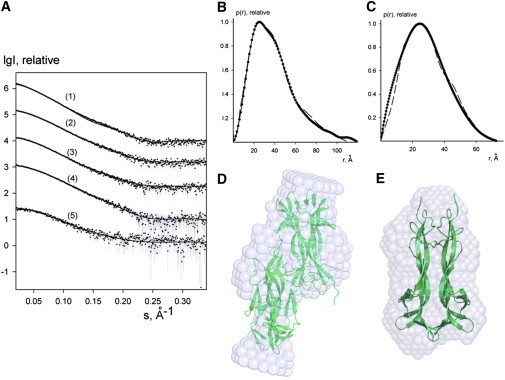
SAXS experiments on the concentration dependence of hNGF DD assembly in solution. (A) The experimental solution x-ray scattering pattern from hNGF curves (1–5) correspond to solute concentrations (mg/mL) of 5.50, 3.77, 2.75, 1.98, and 0.43, respectively. Dots with error bars represent the experimental data; solid lines represent the fits of the experimental scattering curve from a multicomponent mixture (hNGF dimers and DD) by the program OLIGOMER (50). The volume fractions are reported in Table S1. All the plots display the logarithm of the scattering intensity I as a function of momentum transfer s = [4πsin(θ/2)]/λ (Å−1) where θ is the scattering angle and λ = 1.5 Å is the x-ray wavelength. The curves are displaced down by one logarithmic unit for clarity. The p(r) distance distribution functions calculated from the experimental SAXS data (dotted line) corresponding to 5.50 mg/mL (curve 1) and to 0.43 mg/mL (curve 5) are displayed in (B) and in (C), respectively. The p(r) from the hNGF DD and dimer models (dashed line) are also shown in (B) and in (C), respectively. The ab initio envelope bead models for the shapes determination of hNGF (D) at 5.50 mg/mL (curve 1) and (E) at 0.43 mg/mL (curve 5). The cartoon (Cα) representations of the hNGF DD (D) and hNGF dimer (E) models, respectively, are superimposed to the envelope bead models (light blue spheres) obtained by DAMMIN (54). Figure created by the program Pymol (63).
Further SAXS measurements performed on solutions of hNGF at different concentrations (ranging between 0.43 and 5.5 mg/mL, i.e., 17.2–220.0 μM), showed a significant concentration dependence of the measured solution scattering curves. Given the intensities from the components (dimers and DD) OLIGOMER (50) finds the volume fractions by solving a system of linear equations using the algorithm of nonnegative or unconstrained least-squares to minimize the discrepancy between the experimental and calculated scattering curves. Details are reported in Table S1 and in Fig. 2 A (fits from OLIGOMER are displayed as solid lines). The modeled hNGF dimer provides a good fit to the data at 0.43 mg/ml (curve 1 in Fig. 2, A and C), it also well overlaps with the averaged ab initio shape (Fig. 2 E). The singular value decomposition analysis of the scattering curves found only two significant singular values corresponding to two nonrandomly oscillating singular vectors (Fig. S3).
The results, plotted in Fig. 3, show that the hNGF DD molecular assembly appears to dissociate into individual hNGF dimers at low concentrations (1–2 mg/mL, i.e., 40–80 μM).
Figure 3.
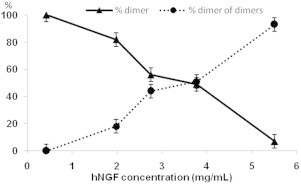
Concentration dependence distribution between hNGF dimers and DD, as assessed by SAXS experiments.
These results are in accordance with previously reported experiments performed on hNGF (∼0.3 mg/mL, i.e., 12 μM) by analytical ultracentrifugation (25,26,45). Moreover, the existence of hNGF DD has also been confirmed by cross-linking experiments (Fig. 4 A) and gel filtration analysis. Indeed, in Fig. 4 B, the appearance of a second peak with a lower retention time corresponds to the DD assembly (highlighted by an arrow).
Figure 4.
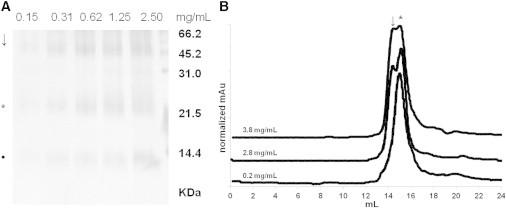
Distribution between hNGF dimers (star) and DD (arrow), as assessed by biochemical experiments. (A) Cross-linking of serial 1:2 dilutions of hNGF; (B) Gel filtration profiles of hNGF at different concentrations; mAu values have been normalized setting 100 the maximum of each peak for a clearer comparison. (↓, DD; ∗, dimers; •, protomers).
When this chromatographic peak has been subjected to a second run of gel filtration, it dissociates into a homogeneous peak of purely dimeric hNGF (highlighted by a star), because of the dilution of the sample. This further confirms that the DD assembly is concentration dependent in the exploited range of 0.2–3.8 mg/mL.
The concentration-dependent DD assembly of hNGF is not shared by its precursor form
It is worth noting that the observed hNGF DD arrangement is restricted to the mature form of hNGF. Indeed, a parallel SAXS study carried out on the mouse precursor form of NGF, m-proNGF (amino acid sequence identity 85.06% proNGF; 89.17% NGF; 80.99% pro-peptide with the human ortholog), in the concentration range of 3.2–13.0 mg/mL, i.e., 64.0–260.0 μM, pointed to the only existence in solution of a dimeric assembly (68,69).
The resulting representative m-proNGF model that was obtained by the ensemble optimization method according to the experimental SAXS scattering pattern (69) and exploiting a structural constraint available from previously reported H/D exchange experiments (70), explains the observed different oligomerization behavior between the mature NGF and its precursor (Fig. 5).
Figure 5.
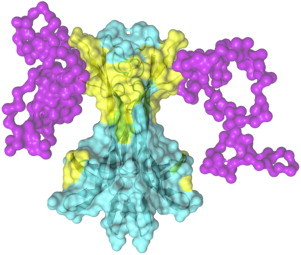
Mapping the NGF DD interaction surface on the proNGF model. The prodomains (magenta) hinder the NGF DD interacting surfaces (yellow) on mature mNGF (cyan). Figure created by the program Pymol (63).
This is likely caused by the direct interaction of residue W21 with the propeptide region of the m-proNGF (70). W21 is also part of the interface of the NGF/receptor complex in both TrkA and p75NTR receptors. Remarkably, W21 is engaged in an hydrogen bond mediating DD assembly (Table 1).
Table 1.
Salt bridges and hydrogen bond interactions across the interface of the selected model of the hNGF DD molecular assembly
| Dimer AB | Dimer CD | Distance (Å) |
|---|---|---|
| Salt bridges | ||
| A: R69 [NH2] | C: D16 [OD1] | 2.80 |
| A: R69 [NH1] | C: D16 [OD2] | 3.10 |
| Hydrogen bonds | ||
| A: S19 [N] | D: R114 [NH2] | 2.80 |
| A: W21 [NE1] | D: V117 [O] | 2.00 |
| A: R59 [NH1] | C: G70 [N] | 2.30 |
| A: G70 [NH1] | C: R59 [NH1] | 2.30 |
| B: R114 [NH2] | C: S19 [O] | 2.30 |
| B: V117 [O] | C: W21 [NE1] | 2.00 |
In the proposed model of the hNGF DD, the surfaces engaged in dimer/dimer interactions encompass residues located near the N- and C-termini, the loop III and the residues on the surrounding β-sheet regions of the hNGF dimer (Fig. 2 A): the contribution of the interacting residues to the stability of the DD assembly was analyzed by PISA (64) and by the 2P2I Inspector (67). The total interface area is 2248.70 Å2. The ΔASApolar and ΔASAapolar of 1084.75 Å2 and 1166.71 Å2, respectively, represent the changes in the polar and apolar solvent-accessible surfaces upon the hNGF DD formation. The energetic calculations relative to the model of the hNGF DD head-to-head assembly, due to the changes in the solvent-accessible surfaces, resulted in a ΔCp and ΔH(333K) of −0.77 J·K−1·mol−1 and −100.95 kJ·mol−1, respectively.
Overall, 12 stretches of residues are engaged at the dimer/dimer interface resulting in 254 nonbonded contacts, 6 hydrogen bonds, and 2 salt bridges (Table 1), that involve either protomers of the two dimers (namely AD and BC). The secondary structure at the dimer/dimer interface is mainly β-sheet and it is characterized by 25.0% of charged residues. The two interacting protein dimers enclose a volume of 5474.25 Å3 (gap volume).
A multiple sequence alignment in UniProt (71) showed that residues whose side chains are involved in hydrogen bonds and salt bridges are in NGF from different species highly conserved across evolution (except for R69 in distantly related organisms like Danio rerio and Xiphophorus maculatus), suggesting their crucial role in this NGF/NGF interaction and in NGF activity (data not shown).
Our antiparallel head-to-head hNGF DD model led us to put forward, to our knowledge, a novel three-dimensional arrangement for the transient ternary heterocomplex TrkA/NGF/p75NTR that would reconcile the unexpected incongruity in the crystallographic-derived heterocomplex with a 2:2:2 stoichiometry. In fact, the crystal structures of NGF/TrkA and NGF/p75NTR (38–40) clearly show not only that both receptors share a mutually exclusive NGF binding site but, additionally, that the relative orientations of the p75NTR and TrkA receptors are opposite with respect to NGF (Fig. 6 A).
Figure 6.
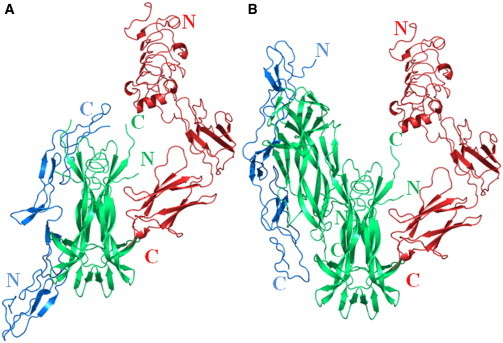
Models of the ternary p75NTR/NGF/TrkA heterocomplex. (A) Model obtained by merging the individual crystal structures of hNGF in complex with TrkA (red), PDB ID 2IFG (38), and p75NTR (blue), PDB ID 1SG1 (40). (B) Model obtained by superimposing the crystal structures of the individual hNGF complexes with its receptors on the hNGF DD model validated by SAXS. Figure created by the program Pymol (63).
Indeed, the existence of NGF as an antiparallel DD in solution, with the two NGF dimers oriented in a head-to-head fashion would instead pave the way for a 2:4:2 TrkA/NGF/NGF/p75NTR heterocomplex (Fig. 6 B), whereby an NGF DD could neatly be involved in a simultaneous transient binding to the ectodomains of the TrkA and p75NTR receptors in their native orientations on the cellular surface.
Discussion
The initial focus of this study was the characterization of the oligomerization state of hNGF in solution. SAXS experiments proved that, in solution, hNGF appears mainly as dimers in equilibrium with tetramers (self-associated DD), whose distributions are concentration dependent. Furthermore, the three-dimensional envelope generated from the one-dimensional SAXS curves, well supported the occurrence of a head-to-head DD assembly in solution. This unexpected finding posed the question of the biological function and implications, if any, of the observed hNGF DD, namely in the context of TrkA/p75NTR receptors cross talk mechanism and interactions that have been well documented in the creation of high-affinity NGF binding sites and signaling modulation (72). Although it is widely accepted that NGF high-affinity binding sites are generated by p75NTR and TrkA coexpression, the existence of heterocomplexes between these receptors being assessed by in vitro and in vivo bioassays, contrasting evidences have been reported about which of the receptor domains, i.e., the extracellular, the transmembrane, or the intracellular, participate with the formation of the NGF high-affinity sites. A recent fluorescence resonance energy transfer study (73) highlighted a direct physical interaction of TrkA and p75NTR intracellular domains and suggested that, in a basal condition, there is a pool of interacting TrkA and p75NTR. This interaction diminishes as NGF binds and internalizes TrkA with a faster kinetics than p75NTR. Furthermore, given that the superimposition of the NGF/p75NTR (25,40), NT-3/p75NTR (26), and NGF/TrkA (38,39) structures showed that the binding sites on the ligands were mutually exclusive, the possibility of a physical interaction of p75NTR and TrkA through their extracellular domains was discarded.
On the other hand, the existence in solution of a head-to-head NGF DD arrangement, as described here, would make a ternary heterocomplex TrkA/NGF/NGF/p75NTR feasible, reconciling the available experimental evidences in one unifying scheme where the extracellular domains of the receptors play a crucial role in the generation of the high-affinity binding sites. In this scenario such a transient heterocomplex could take place involving the basal preformed homodimers of TrkA and p75NTR receptors, leading to the observed quick dissociation of the homodimers of the receptors themselves. Indeed, the interaction surface between the NGF DD is characterized by a less favored binding enthalpy (−100.95 kJ·mol−1) compared to TrkA/NGF (−68.69 kJ·mol−1) and p75NTR/NGF (−61.99 kJ·mol−1) binding enthalpies, that in turn could easily lead to the dissociation of the transient ternary heterocomplex characterized by a 2:4:2 stoichiometry, leaving each NGF dimer bound to one of the two engaged cognate receptors and therefore available for the formation of TrkA/NGF and p75NTR/NGF symmetric complexes with a 2:2 stoichiometry embedding distinct functional and signaling capabilities.
Interestingly, 63% of the residues encompassing the oligomeric interface of the hNGF DD model, are conserved among the human members of the NT superfamily, being 42% identical, 18% conserved, and 3% semiconserved. In comparison, the overall amino acid sequences of the human members of the NT superfamily indicate residues to be 35% identical, 21% conserved, and 4.5% semiconserved.
It is interesting to note that the less conserved residues mainly cluster on NGF loop III. This suggests that whether other members of the NT superfamily would, alike NGF, form as suggested by Narhi et al. (45) self-associated oligomers, the respective DD assembly may not be necessarily identical but rather be shaped by the conformation of loop III and its physicochemical properties (charge, hydrophobicity-hydrophilicity). Thus, upon p75NTR binding, a distinctive binding site would be exposed for the recognition of the NT by the cognate Trk receptor. This scenario would indeed take into account the fact that p75NTR enhances Trk specificities for their cognate NTs (21,22,74).
It is worth noting that, although unconventional, the NGF DD arrangement closely resembles that being observed in the crystal structure of FGF1 (fibroblast growth factor) in complex with its high affinity receptor (FGFR2). The main difference being a central heparin molecule bound to two FGF1 ligands recruiting two FGF receptors (75). The thermodynamics and stoichiometry of the ternary complex suggested that in solution FGF1 binds to heparin in a transdimeric manner before FGFR recruitment (76).
Even if NGF DDs are observed in solution as a result of direct protein-protein contacts, as we have showed here, we cannot exclude that in vivo this interaction might even be strengthened by means of a matrix component, hence closely resembling the FGF1 binding to heparin.
A recent carbohydrate microarray study reported compelling evidences on the interactions between a specific sulfated epitope on chondroitin sulfate, CS-E, and the NTs, showing that CS is capable of assembling multimeric signaling complexes and of modulating neurotrophin signaling pathways (77). In this study, all of the NTs show concentration-dependent binding to CS-E tetrasaccharides, with NGF displaying the greatest specificity. Furthermore, NGF and BDNF enhance the binding of their cognate Trk receptor with a selectivity that is greater for the complex than for the NT alone, whereas NT-3 and NT-4/5 do not have the same effect on TrkC and TrkB, respectively. Besides the proposed role of CS polysaccharides in the assembly of NT/Trk receptor complexes, these molecules might also be involved in an additional level of regulation by acting on NT oligomerization at the cell surface, suggesting that the spatiotemporal expression of CS-E in vivo might differentially regulate specific NT signaling pathways by several different mechanisms.
Finally, it is worth noting that the observed NGF DD arrangement is only restricted to the mature form of NGF, whereas the precursor proNGF maintains a dimeric assembly (68,69) in the range of the exploited concentrations. These data point to another significant difference between the two forms of NGF, which may have profound consequences on their downstream signaling abilities and account for their distinctive selectivity for the p75NTR coreceptor. Indeed, on the sole basis of in vitro binding studies (78), there would be no clear rationale for TrkA not to be a preferred receptor for proNGF as it is p75NTR, by considering the in vitro affinities for proNGF of either receptor being between 15 and 20 nM.
The relevance of the described NGF DD for the in vivo biological functions of NGF (and, by extension, of NTs) remains to be ascertained, though such an arrangement is likely to play a critical role in the sorting process of NGF for regulatory secretion pathway. Indeed, secretory granules contain very high (90–150 mg/ml) concentrations of their secretory products (79–81). In addition, aggregation, condensation, and loop-mediated homodimerization (82–85) are known to be crucial for specific segregation of soluble proteins for the regulated secretory pathway. In particular, the protein cores of growth hormone and prolactin granules disperse slowly (86,87), insulin crystallizes in β cell granules (88) and peptide and protein hormones in secretory granules of the endocrine system aggregate selectively into amyloid-like cross-β-sheet rich conformation upon prohormone processing (89). Therefore, one could speculate that NGF oligomerization might be involved in the sorting process for regulated secretory pathway being stored in secretory granules as oligomer, i.e., DD form. Upon secretion the concentration gradient between the DD and the dimer forms would be affected by the rate of diffusion and/or by the interactions with matrix component at the level of the cell surface, such as CS polysaccharides.
Different oligomeric forms of NTs, including the DD, may also be involved as modulators in the recruitment of different molecules for synaptic tagging. NTs could be transported anterogradely from the soma to presynaptic terminals, released as a consequence of neuronal spiking, and received by the postsynaptic neuron. Conversely, neural or synaptic activity could led to secretion of NTs from dendrites, and the secreted NTs might act as retrograde factors on the presynaptic neuron (90,91).
Because NGF is overexpressed in the oxidative environment linked to neurodegenerative processes (92), this protein could also be a target for peroxynitrite-mediated oxidative modifications. Interestingly, the oxidation of NGF by peroxynitrite in vitro causes nitration (tyrosine and tryptophan residues) and induces the formation of stable high-molecular weight oligomers of NGF (93).
In conclusion, we have provided new, to our knowledge, molecular details on the NGF concentration-dependent oligomerization that has not been observed for the unprocessed proNGF species. The observation that in solution a head-to-head DD NGF molecular assembly has been detected, likely to be found also for other NTs, prompts a straightforward answer to the puzzle of the formation of a transient ternary heterocomplex between TrkA and p75NTR receptors and NGF. The proposed model shall pave the way to further biochemical and biological in vitro as well in vivo experiments that will ultimately prove it or disprove it.
Acknowledgments
We are grateful to the late Prof. R. Rudolph (Martin-Luther University Halle-Wittenberg, Halle, Germany) for providing a first sample of hNGF. Annalisa Pastore (Department of Clinical Neurosciences, King's College London, UK) is thanked for her thoughtful discussions and insights.
We acknowledge the support of the EMBL EU funding for recording at EMBL Hamburg SAXS beamline. D. Svergun and P. Koranev acknowledge support from the German Ministry of Education and Science (BMBF) project BIOSCAT, grant 05K20912. A. Cattaneo acknowledges funding from MIUR (PRIN-2005057070 and FIRB RBLA03FLJC) and Telethon. This work was in part supported by the FP7-PAINCAGE integrative project.
Contributor Information
Doriano Lamba, Email: doriano.lamba@ts.ic.cnr.it.
Antonino Cattaneo, Email: antonino.cattaneo@sns.it.
Supporting Material
References
- 1.Levi-Montalcini R., Hamburger V. Selective growth stimulating effects of mouse sarcoma on the sensory and sympathetic nervous system of the chick embryo. J. Exp. Zool. 1951;116:321–361. doi: 10.1002/jez.1401160206. [DOI] [PubMed] [Google Scholar]
- 2.Levi-Montalcini R. The nerve growth factor 35 years later. Science. 1987;237:1154–1162. doi: 10.1126/science.3306916. [DOI] [PubMed] [Google Scholar]
- 3.Bothwell M. Functional interactions of neurotrophins and neurotrophin receptors. Annu. Rev. Neurosci. 1995;18:223–253. doi: 10.1146/annurev.ne.18.030195.001255. [DOI] [PubMed] [Google Scholar]
- 4.Chao M.V. Neurotrophins and their receptors: a convergence point for many signalling pathways. Nat. Rev. Neurosci. 2003;4:299–309. doi: 10.1038/nrn1078. [DOI] [PubMed] [Google Scholar]
- 5.Angeletti R.H., Bradshaw R.A. Nerve growth factor from mouse submaxillary gland: amino acid sequence. Proc. Natl. Acad. Sci. USA. 1971;68:2417–2420. doi: 10.1073/pnas.68.10.2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lewin G.R., Barde Y.A. Physiology of the neurotrophins. Annu. Rev. Neurosci. 1996;19:289–317. doi: 10.1146/annurev.ne.19.030196.001445. [DOI] [PubMed] [Google Scholar]
- 7.Dechant G., Barde Y.A. The neurotrophin receptor p75(NTR): novel functions and implications for diseases of the nervous system. Nat. Neurosci. 2002;5:1131–1136. doi: 10.1038/nn1102-1131. [DOI] [PubMed] [Google Scholar]
- 8.Roux P.P., Barker P.A. Neurotrophin signaling through the p75 neurotrophin receptor. Prog. Neurobiol. 2002;67:203–233. doi: 10.1016/s0301-0082(02)00016-3. [DOI] [PubMed] [Google Scholar]
- 9.Kaplan D.R., Hempstead B.L., Parada L.F. The trk proto-oncogene product: a signal transducing receptor for nerve growth factor. Science. 1991;252:554–558. doi: 10.1126/science.1850549. [DOI] [PubMed] [Google Scholar]
- 10.Klein R., Jing S.Q., Barbacid M. The trk proto-oncogene encodes a receptor for nerve growth factor. Cell. 1991;65:189–197. doi: 10.1016/0092-8674(91)90419-y. [DOI] [PubMed] [Google Scholar]
- 11.Johnson D., Lanahan A., Chao M. Expression and structure of the human NGF receptor. Cell. 1986;47:545–554. doi: 10.1016/0092-8674(86)90619-7. [DOI] [PubMed] [Google Scholar]
- 12.Hempstead B.L., Martin-Zanca D., Chao M.V. High-affinity NGF binding requires coexpression of the trk proto-oncogene and the low-affinity NGF receptor. Nature. 1991;350:678–683. doi: 10.1038/350678a0. [DOI] [PubMed] [Google Scholar]
- 13.Rodriguez-Tébar A., Dechant G., Barde Y.A. Binding of brain-derived neurotrophic factor to the nerve growth factor receptor. Neuron. 1990;4:487–492. doi: 10.1016/0896-6273(90)90107-q. [DOI] [PubMed] [Google Scholar]
- 14.Rodríguez-Tébar A., Dechant G., Barde Y.A. Binding of neurotrophin-3 to its neuronal receptors and interactions with nerve growth factor and brain-derived neurotrophic factor. EMBO J. 1992;11:917–922. doi: 10.1002/j.1460-2075.1992.tb05130.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mahadeo D., Kaplan L., Hempstead B.L. High affinity nerve growth factor binding displays a faster rate of association than p140trk binding. Implications for multi-subunit polypeptide receptors. J. Biol. Chem. 1994;269:6884–6891. [PubMed] [Google Scholar]
- 16.Sutter A., Riopelle R.J., Shooter E.M. Nerve growth factor receptors. Characterization of two distinct classes of binding sites on chick embryo sensory ganglia cells. J. Biol. Chem. 1979;254:5972–5982. [PubMed] [Google Scholar]
- 17.Rodriguez-Tébar A., Barde Y.A. Binding characteristics of brain-derived neurotrophic factor to its receptors on neurons from the chick embryo. J. Neurosci. 1988;8:3337–3342. doi: 10.1523/JNEUROSCI.08-09-03337.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maliartchouk S., Saragovi H.U. Optimal nerve growth factor trophic signals mediated by synergy of TrkA and p75 receptor-specific ligands. J. Neurosci. 1997;17:6031–6037. doi: 10.1523/JNEUROSCI.17-16-06031.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.MacPhee I.J., Barker P.A. Brain-derived neurotrophic factor binding to the p75 neurotrophin receptor reduces TrkA signaling while increasing serine phosphorylation in the TrkA intracellular domain. J. Biol. Chem. 1997;272:23547–23551. doi: 10.1074/jbc.272.38.23547. [DOI] [PubMed] [Google Scholar]
- 20.Lad S.P., Peterson D.A., Neet K.E. Individual and combined effects of TrkA and p75NTR nerve growth factor receptors. A role for the high affinity receptor site. J. Biol. Chem. 2003;278:24808–24817. doi: 10.1074/jbc.M212270200. [DOI] [PubMed] [Google Scholar]
- 21.Brennan C., Rivas-Plata K., Landis S.C. The p75 neurotrophin receptor influences NT-3 responsiveness of sympathetic neurons in vivo. Nat. Neurosci. 1999;2:699–705. doi: 10.1038/11158. [DOI] [PubMed] [Google Scholar]
- 22.Kuruvilla R., Zweifel L.S., Ginty D.D. A neurotrophin signaling cascade coordinates sympathetic neuron development through differential control of TrkA trafficking and retrograde signaling. Cell. 2004;118:243–255. doi: 10.1016/j.cell.2004.06.021. [DOI] [PubMed] [Google Scholar]
- 23.Maliartchouk S., Debeir T., Saragovi H.U. Genuine monovalent ligands of TrkA nerve growth factor receptors reveal a novel pharmacological mechanism of action. J. Biol. Chem. 2000;275:9946–9956. doi: 10.1074/jbc.275.14.9946. [DOI] [PubMed] [Google Scholar]
- 24.Kaplan D.R., Stephens R.M. Neurotrophin signal transduction by the Trk receptor. J. Neurobiol. 1994;25:1404–1417. doi: 10.1002/neu.480251108. [DOI] [PubMed] [Google Scholar]
- 25.Aurikko J.P., Ruotolo B.T., Blundell T.L. Characterization of symmetric complexes of nerve growth factor and the ectodomain of the pan-neurotrophin receptor, p75NTR. J. Biol. Chem. 2005;280:33453–33460. doi: 10.1074/jbc.M503189200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gong Y., Cao P., Jiang T. Crystal structure of the neurotrophin-3 and p75NTR symmetrical complex. Nature. 2008;454:789–793. doi: 10.1038/nature07089. [DOI] [PubMed] [Google Scholar]
- 27.Barker P.A., Shooter E.M. Disruption of NGF binding to the low affinity neurotrophin receptor p75LNTR reduces NGF binding to TrkA on PC12 cells. Neuron. 1994;13:203–215. doi: 10.1016/0896-6273(94)90470-7. [DOI] [PubMed] [Google Scholar]
- 28.Mehta H.M., Woo S.B., Neet K.E. Comparison of nerve growth factor receptor binding models using heterodimeric muteins. J. Neurosci. Res. 2012;90:2259–2271. doi: 10.1002/jnr.23116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ross G.M., Shamovsky I.L., Riopelle R.J. Reciprocal modulation of TrkA and p75NTR affinity states is mediated by direct receptor interactions. Eur. J. Neurosci. 1998;10:890–898. doi: 10.1046/j.1460-9568.1998.00094.x. [DOI] [PubMed] [Google Scholar]
- 30.Huber L.J., Chao M.V. A potential interaction of p75 and trkA NGF receptors revealed by affinity cross-linking and immunoprecipitation. J. Neurosci. Res. 1995;40:557–563. doi: 10.1002/jnr.490400415. [DOI] [PubMed] [Google Scholar]
- 31.Bibel M., Hoppe E., Barde Y.A. Biochemical and functional interactions between the neurotrophin receptors trk and p75NTR. EMBO J. 1999;18:616–622. doi: 10.1093/emboj/18.3.616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Battleman D.S., Geller A.I., Chao M.V. HSV-1 vector-mediated gene transfer of the human nerve growth factor receptor p75hNGFR defines high-affinity NGF binding. J. Neurosci. 1993;13:941–951. doi: 10.1523/JNEUROSCI.13-03-00941.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bilderback T.R., Gazula V.R., Dobrowsky R.T. Phosphoinositide 3-kinase regulates cross talk between Trk A tyrosine kinase and p75(NTR)-dependent sphingolipid signaling pathways. J. Neurochem. 2001;76:1540–1551. doi: 10.1046/j.1471-4159.2001.00171.x. [DOI] [PubMed] [Google Scholar]
- 34.Esposito D., Patel P., Hempstead B.L. The cytoplasmic and transmembrane domains of the p75 and Trk A receptors regulate high affinity binding to nerve growth factor. J. Biol. Chem. 2001;276:32687–32695. doi: 10.1074/jbc.M011674200. [DOI] [PubMed] [Google Scholar]
- 35.Hempstead B.L., Patil N., Chao M.V. Deletion of cytoplasmic sequences of the nerve growth factor receptor leads to loss of high affinity ligand binding. J. Biol. Chem. 1990;265:9595–9598. [PubMed] [Google Scholar]
- 36.Mischel P.S., Smith S.G., Reichardt L.F. The extracellular domain of p75NTR is necessary to inhibit neurotrophin-3 signaling through TrkA. J. Biol. Chem. 2001;276:11294–11301. doi: 10.1074/jbc.M005132200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Matusica D., Skeldal S., Coulson E.J. An intracellular domain fragment of the p75 neurotrophin receptor (p75(NTR)) enhances tropomyosin receptor kinase A (TrkA) receptor function. J. Biol. Chem. 2013;288:11144–11154. doi: 10.1074/jbc.M112.436469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wehrman T., He X., Garcia K.C. Structural and mechanistic insights into nerve growth factor interactions with the TrkA and p75 receptors. Neuron. 2007;53:25–38. doi: 10.1016/j.neuron.2006.09.034. [DOI] [PubMed] [Google Scholar]
- 39.Wiesmann C., Ultsch M.H., de Vos A.M. Crystal structure of nerve growth factor in complex with the ligand-binding domain of the TrkA receptor. Nature. 1999;401:184–188. doi: 10.1038/43705. [DOI] [PubMed] [Google Scholar]
- 40.He X.L., Garcia K.C. Structure of nerve growth factor complexed with the shared neurotrophin receptor p75. Science. 2004;304:870–875. doi: 10.1126/science.1095190. [DOI] [PubMed] [Google Scholar]
- 41.Urra S., Escudero C.A., Bronfman F.C. TrkA receptor activation by nerve growth factor induces shedding of the p75 neurotrophin receptor followed by endosomal gamma-secretase-mediated release of the p75 intracellular domain. J. Biol. Chem. 2007;282:7606–7615. doi: 10.1074/jbc.M610458200. [DOI] [PubMed] [Google Scholar]
- 42.Jung K.M., Tan S., Kim T.W. Regulated intramembrane proteolysis of the p75 neurotrophin receptor modulates its association with the TrkA receptor. J. Biol. Chem. 2003;278:42161–42169. doi: 10.1074/jbc.M306028200. [DOI] [PubMed] [Google Scholar]
- 43.Zampieri N., Chao M.V. Structural biology. The p75 NGF receptor exposed. Science. 2004;304:833–834. doi: 10.1126/science.1098110. [DOI] [PubMed] [Google Scholar]
- 44.Barker P.A. High affinity not in the vicinity? Neuron. 2007;53:1–4. doi: 10.1016/j.neuron.2006.12.018. [DOI] [PubMed] [Google Scholar]
- 45.Narhi L.O., Rosenfeld R., Yphantis D.A. Comparison of the biophysical characteristics of human brain-derived neurotrophic factor, neurotrophin-3, and nerve growth factor. J. Biol. Chem. 1993;268:13309–13317. [PubMed] [Google Scholar]
- 46.Holland D.R., Cousens L.S., Matthews B.W. Nerve growth factor in different crystal forms displays structural flexibility and reveals zinc binding sites. J. Mol. Biol. 1994;239:385–400. doi: 10.1006/jmbi.1994.1380. [DOI] [PubMed] [Google Scholar]
- 47.Boulin C.J., Kemof R., Koch M.H.J. Data acquisition systems for linear and area X-ray detectors using delay line readout. Nucl. Instrum. Methods Sec. A. 1988;269:312–320. [Google Scholar]
- 48.Roessle M.K., Ristau R., Svergun D.I. Upgrade of the small-angle X-ray scattering beamline X33 at the European Molecular Biology Laboratory, Hamburg. J. Appl. Cryst. 2007;40:s190–s194. [Google Scholar]
- 49.Covaceuszach S., Capsoni S., Cattaneo A. Development of a non invasive NGF-based therapy for Alzheimer’s disease. Curr. Alzheimer Res. 2009;6:158–170. doi: 10.2174/156720509787602870. [DOI] [PubMed] [Google Scholar]
- 50.Konarev P.V., Volkov V.V., Svergun D.I. PRIMUS: a Windows PC-based system for small-angle scattering data analysis. J. Appl. Cryst. 2003;36:1277–1282. [Google Scholar]
- 51.Guinier A. La diffraction des rayons X aux très faibles angles: applications à l’etude des phénomènes ultra-microscopiques. Ann. Phys. (Paris) 1939;12:161–237. [Google Scholar]
- 52.Svergun D.I., Semenyuk A. Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J. Appl. Cryst. 1992;25:495–503. [Google Scholar]
- 53.Svergun D.I., Barberato C., Koch M.H.J. CRYSOL - a program to evaluate X-ray solution scattering of biological macromolecules from atomic coordinates. J. Appl. Cryst. 1995;28:768–773. [Google Scholar]
- 54.Svergun D.I. Restoring low resolution structure of biological macromolecules from solution scattering using simulated annealing. Biophys. J. 1999;76:2879–2886. doi: 10.1016/S0006-3495(99)77443-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Volkov V.V., Svergun D.I. Uniqueness of ab-initio shape determination in small-angle scattering. J. Appl. Cryst. 2003;36:860–864. doi: 10.1107/S0021889809000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kozin M., Svergun D.I. Automated matching of high- and low-resolution structural models. J. Appl. Cryst. 2001;34:33–41. [Google Scholar]
- 57.Ratto M.H., Leduc Y.A., Adams G.P. The nerve of ovulation-inducing factor in semen. Proc. Natl. Acad. Sci. USA. 2012;109:15042–15047. doi: 10.1073/pnas.1206273109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schneidman-Duhovny D., Inbar Y., Wolfson H.J. PatchDock and SymmDock: servers for rigid and symmetric docking. Nucleic Acids Res. 2005;33:W363–W367. doi: 10.1093/nar/gki481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hess B., Kutzner C., Lindahl E. GROMACS 4: algorithms for highly efficient, load-balanced, and scalable molecular simulation. J. Chem. Theory Comput. 2008;4:435–447. doi: 10.1021/ct700301q. [DOI] [PubMed] [Google Scholar]
- 60.Tsodikov O.V., Record M.T., Jr., Sergeev Y.V. Novel computer program for fast exact calculation of accessible and molecular surface areas and average surface curvature. J. Comput. Chem. 2002;23:600–609. doi: 10.1002/jcc.10061. [DOI] [PubMed] [Google Scholar]
- 61.Murphy K.P., Freire E. Thermodynamics of structural stability and cooperative folding behavior in proteins. Adv. Protein Chem. 1992;43:313–361. doi: 10.1016/s0065-3233(08)60556-2. [DOI] [PubMed] [Google Scholar]
- 62.Xie D., Freire E. Molecular basis of cooperativity in protein folding. V. Thermodynamic and structural conditions for the stabilization of compact denatured states. Proteins. 1994;19:291–301. doi: 10.1002/prot.340190404. [DOI] [PubMed] [Google Scholar]
- 63.DeLano W.L. DeLano Scientific LLC; Palo Alto, CA: 2002. The PyMOL Molecular Graphics System.http://www.pymol.org [Google Scholar]
- 64.Krissinel E., Henrick K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 65.McDonald N.Q., Lapatto R., Blundell T.L. New protein fold revealed by a 2.3-A resolution crystal structure of nerve growth factor. Nature. 1991;354:411–414. doi: 10.1038/354411a0. [DOI] [PubMed] [Google Scholar]
- 66.Tong Q., Wang F., Jiang T. Structural and functional insights into lipid-bound nerve growth factors. FASEB J. 2012;26:3811–3821. doi: 10.1096/fj.12-207316. [DOI] [PubMed] [Google Scholar]
- 67.Basse M.J., Betzi S., Roche P. 2P2Idb: a structural database dedicated to orthosteric modulation of protein-protein interactions. Nucleic Acids Res. 2013;41:D824–D827. doi: 10.1093/nar/gks1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Paoletti F., Konarev P.V., Svergun D.I. Structural and functional properties of mouse proNGF. Biochem. Soc. Trans. 2006;34:605–606. doi: 10.1042/BST0340605. [DOI] [PubMed] [Google Scholar]
- 69.Paoletti F., Covaceuszach S., Lamba D. Intrinsic structural disorder of mouse proNGF. Proteins. 2009;75:990–1009. doi: 10.1002/prot.22311. [DOI] [PubMed] [Google Scholar]
- 70.Kliemannel M., Golbik R., Lilie H. The pro-peptide of proNGF: structure formation and intramolecular association with NGF. Protein Sci. 2007;16:411–419. doi: 10.1110/ps.062376207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.The UniProt Consortium Activities at the Universal Protein Resource (UniProt) Nucleic Acids Res. 2014;42:D191–D198. doi: 10.1093/nar/gkt1140. (UniProt release 2014_01 - Jan 22, 2014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Huang E.J., Reichardt L.F. Trk receptors: roles in neuronal signal transduction. Annu. Rev. Biochem. 2003;72:609–642. doi: 10.1146/annurev.biochem.72.121801.161629. [DOI] [PubMed] [Google Scholar]
- 73.Iacaruso M.F., Galli S., Pietrasanta L.I. Structural model for p75(NTR)-TrkA intracellular domain interaction: a combined FRET and bioinformatics study. J. Mol. Biol. 2011;414:681–698. doi: 10.1016/j.jmb.2011.09.022. [DOI] [PubMed] [Google Scholar]
- 74.Benedetti M., Levi A., Chao M.V. Differential expression of nerve growth factor receptors leads to altered binding affinity and neurotrophin responsiveness. Proc. Natl. Acad. Sci. USA. 1993;90:7859–7863. doi: 10.1073/pnas.90.16.7859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pellegrini L., Burke D.F., Blundell T.L. Crystal structure of fibroblast growth factor receptor ectodomain bound to ligand and heparin. Nature. 2000;407:1029–1034. doi: 10.1038/35039551. [DOI] [PubMed] [Google Scholar]
- 76.Brown A., Robinson C.J., Blundell T.L. Cooperative heparin-mediated oligomerization of fibroblast growth factor-1 (FGF1) precedes recruitment of FGFR2 to ternary complexes. Biophys. J. 2013;104:1720–1730. doi: 10.1016/j.bpj.2013.02.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rogers C.J., Clark P.M., Hsieh-Wilson L.C. Elucidating glycosaminoglycan-protein-protein interactions using carbohydrate microarray and computational approaches. Proc. Natl. Acad. Sci. USA. 2011;108:9747–9752. doi: 10.1073/pnas.1102962108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Nykjaer A., Lee R., Petersen C.M. Sortilin is essential for proNGF-induced neuronal cell death. Nature. 2004;427:843–848. doi: 10.1038/nature02319. [DOI] [PubMed] [Google Scholar]
- 79.Hutton J.C., Penn E.J., Peshavaria M. Low-molecular-weight constituents of isolated insulin-secretory granules. Bivalent cations, adenine nucleotides and inorganic phosphate. Biochem. J. 1983;210:297–305. doi: 10.1042/bj2100297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Oyarce A.M., Hand T.A., Eipper B.A. Dopaminergic regulation of secretory granule-associated proteins in rat intermediate pituitary. J. Neurochem. 1996;67:229–241. doi: 10.1046/j.1471-4159.1996.67010229.x. [DOI] [PubMed] [Google Scholar]
- 81.Dannies P.S. Protein hormone storage in secretory granules: mechanisms for concentration and sorting. Endocr. Rev. 1999;20:3–21. doi: 10.1210/edrv.20.1.0354. [DOI] [PubMed] [Google Scholar]
- 82.Tam W.W., Andreasson K.I., Loh Y.P. The amino-terminal sequence of pro-opiomelanocortin directs intracellular targeting to the regulated secretory pathway. Eur. J. Cell Biol. 1993;62:294–306. [PubMed] [Google Scholar]
- 83.Cool D.R., Fenger M., Loh Y.P. Identification of the sorting signal motif within pro-opiomelanocortin for the regulated secretory pathway. J. Biol. Chem. 1995;270:8723–8729. doi: 10.1074/jbc.270.15.8723. [DOI] [PubMed] [Google Scholar]
- 84.Chanat E., Weiss U., Tooze S.A. Reduction of the disulfide bond of chromogranin B (secretogranin I) in the trans-Golgi network causes its missorting to the constitutive secretory pathways. EMBO J. 1993;12:2159–2168. doi: 10.1002/j.1460-2075.1993.tb05864.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Thiele C., Huttner W.B. The disulfide-bonded loop of chromogranins, which is essential for sorting to secretory granules, mediates homodimerization. J. Biol. Chem. 1998;273:1223–1231. doi: 10.1074/jbc.273.2.1223. [DOI] [PubMed] [Google Scholar]
- 86.Angleson J.K., Cochilla A.J., Betz W.J. Regulation of dense core release from neuroendocrine cells revealed by imaging single exocytic events. Nat. Neurosci. 1999;2:440–446. doi: 10.1038/8107. [DOI] [PubMed] [Google Scholar]
- 87.Dannies P.S. Mechanisms for storage of prolactin and growth hormone in secretory granules. Mol. Genet. Metab. 2002;76:6–13. doi: 10.1016/s1096-7192(02)00011-2. (Review) [DOI] [PubMed] [Google Scholar]
- 88.Michael J., Carroll R., Steiner D.F. Studies on the molecular organization of rat insulin secretory granules. J. Biol. Chem. 1987;262:16531–16535. [PubMed] [Google Scholar]
- 89.Maji S.K., Perrin M.H., Riek R. Functional amyloids as natural storage of peptide hormones in pituitary secretory granules. Science. 2009;325:328–332. doi: 10.1126/science.1173155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Schinder A.F., Poo M. The neurotrophin hypothesis for synaptic plasticity. Trends Neurosci. 2000;23:639–645. doi: 10.1016/s0166-2236(00)01672-6. [DOI] [PubMed] [Google Scholar]
- 91.Poo M.M. Neurotrophins as synaptic modulators. Nat. Rev. Neurosci. 2001;2:24–32. doi: 10.1038/35049004. [DOI] [PubMed] [Google Scholar]
- 92.Fanhnestock M., Scott S.A., Crutcher K.A. Nerve growth factor mRNA and protein levels measured in the same tissue from normal and Alzheimer’s disease partial cortex. Mol. Brain Res. 1996;4:175–178. doi: 10.1016/s0169-328x(96)00193-3. [DOI] [PubMed] [Google Scholar]
- 93.Pehar M., Vargas M.R., Barbeito L. Peroxynitrite transforms nerve growth factor into an apoptotic factor for motor neurons. Free Radic. Biol. Med. 2006;41:1632–1644. doi: 10.1016/j.freeradbiomed.2006.08.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


