SUMMARY
Skeletal development and invasion by tumor cells depends on proteolysis of collagen by the pericellular metalloproteinase MT1-MMP. Its hemopexin-like (HPX) domain binds to collagen substrates to facilitate their digestion. Spin labeling and paramagnetic NMR detection have revealed how the HPX domain docks to collagen I-derived triple-helix. Mutations impairing triple-helical peptidase activity corroborate the interface. Saturation transfer difference NMR suggests rotational averaging around the longitudinal axis of the triple-helical peptide. Part of the interface emerges as unique and potentially targetable for selective inhibition. The triple-helix crosses the junction of blades I and II at a 45° angle to the symmetry axis of the HPX domain, placing the scissile Gly~Ile bond near the HPX domain and shifted ~25 Å from MMP-1 complexes. This raises the question of the MT1-MMP catalytic domain folding over the triple-helix during catalysis, a possibility accommodated by the flexibility between domains suggested by AFM images.
Keywords: collagenolysis, collagen triple-helical peptide, triple-helical peptidase, NMR paramagnetic relaxation enhancement, spin labeling, atomic force microscopy, invasion
INTRODUCTION
Collagen degradation by membrane type 1 matrix metalloproteinase (MT1-MMP or MMP-14) contributes to several important pathophysiological processes. Without MT1-MMP, development of the skeleton is deranged (Holmbeck et al., 1999), and osteocytes fail to digest collagen (Holmbeck et al., 2005). Invasion of 3D collagen matrices by endothelial cells, tumor cells, and rheumatoid synovial fibroblasts is conferred by MT1-MMP at their leading edges (Galvez et al., 2001; Miller et al., 2009; Sabeh et al., 2004; Stratman et al., 2009). Formation of new blood vessels requires MT1-MMP (Chun et al., 2004) and endothelial cells form vascular guidance tunnels via MT1-MMP activity (Stratman et al., 2009). Tumor cells enmeshed in 3D collagen are restricted to spheroid morphology without growing or dividing, whereas MT1-MMP expression frees the tumor cells to grow and proliferate (Hotary et al., 2003). After myocardial infarction, increased cardiac expression of MT1-MMP decreases both left ventricular function and survival of mice (Zavadzkas et al., 2011). MT1-MMP is also over-expressed in atherosclerotic plaques, possibly destabilizing the plaques (Johnson et al., 2008) by degrading collagen (Schneider et al., 2008).
In invasive cell pseudopodia, the collagenase activity of MT1-MMP depends upon interacting with palladin which connects it to the actin cytoskeleton (von Nandelstadh et al., 2014) and upon dynamic homodimerization regulated by the reorganization of the actin cytoskeleton by Rho GTPases (Itoh et al., 2011). This self-association that can occur at the leading but not trailing cell surfaces appears to depend on the hemopexin-like (HPX) (Itoh et al., 2001), transmembrane, and cytoplasmic domains (Itoh et al., 2008; Lehti et al., 2002; Rozanov et al., 2001). Collagen degradation was disrupted efficiently by over-expression of MT1-MMP without its catalytic domain and less efficiently when the transmembrane anchor was also deleted (Itoh et al., 2006; Miller et al., 2009). These truncations disrupted assembly of MT1-MMP into a collagenolytic form in the plasma membrane. This was attributed to defective homodimerization (Itoh et al., 2006), but the truncations may also have disturbed other interactions that facilitate its collagenase activity, such as with cell adhesion complexes which contain β1-integrins (Woskowicz et al., 2013) and with the cytoskeleton via palladin (von Nandelstadh et al., 2014). The soluble ectodomain of MT1-MMP is monomeric and very active in collagen degradation, propelling it in one direction along the collagen fibril, analogous to MMP-1 (Collier et al., 2011).
The soluble ectodomain of MT1-MMP comprises the catalytic and HPX domains but not the C-terminal transmembrane anchor. The ectodomain digests fibrillar collagens I, II, and III into the ¾ and ¼ fragments characteristic of collagenases (Ohuchi et al., 1997). The bonds hydrolyzed are Gly775-Ile776 of α1(I) and Gly775-Leu776 and Gly781-Ile782 of α2(I) chains (Ohuchi et al., 1997). The HPX domain of MT1-MMP binds collagen (Tam et al., 2004), but not in a cellular assay (Itoh et al., 2006). Effects of deleting the HPX domain from MT1-MMP included elimination of collagen invasion by cells (Wang et al., 2004), diminution of collagenolysis at the cell surface, retention of half of collagen-invasive activity of cells (Li et al., 2008), and prevention of tumor growth in vivo (Remacle et al., 2012). Single chain antibodies against the HPX domain of MT1-MMP inhibit collagen degradation and cell invasion and sprouting through type I collagen, suggesting the therapeutic potential of this approach (Basu et al., 2012).
The HPX domain has the fold of a four-bladed β-propeller (Tochowicz et al., 2011) (Figure 1A) of 23 kDa, with sequence features repeated among the blades. Its migration with apparent MW of 32 to 40 kDa by gel permeation was interpreted as dimerization in solution (Itoh et al., 2006; Itoh et al., 2001). This view, however, was revised by quantitative evidence from analytical ultracentrifugation for 62% monomer and 38% dimer in solution (Tochowicz et al., 2011). A crystal structure suggests a tilted dimer with a symmetric interface about blades II and III. Mutations at this interface indicate its relevance to collagen degradation, migration, and proposed “biological dimerization” (Tochowicz et al., 2011). The isolated HPX domain was found to be monomeric in solution (Basu et al., 2012; Overall et al., 2000).
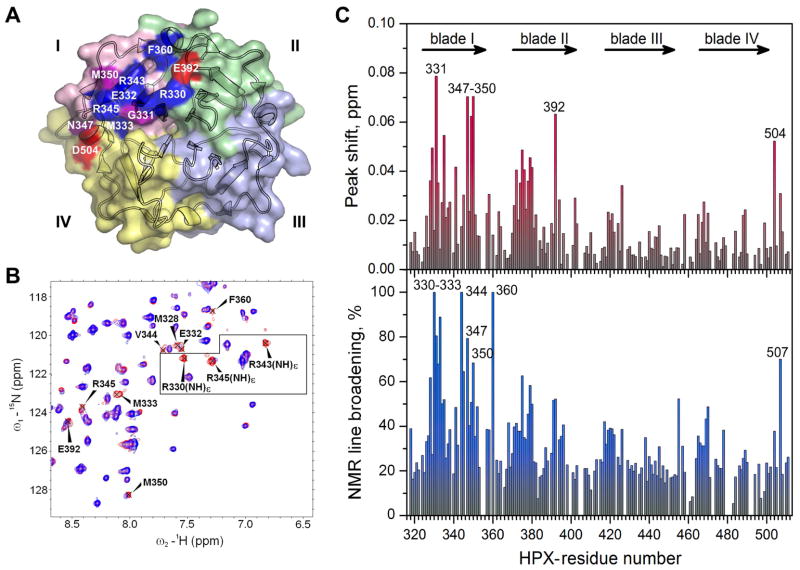
Figure 1. Perturbations of NMR spectra suggest the THP binding site on the HPX domain of MT1-MMP.
(A) The crystal structure of the HPX domain (pdb: 3c7x) (Tochowicz et al., 2011) colors blades I – IV pink, green, light blue, and yellow, respectively. THP-induced perturbations of NMR peaks are colored red where shifted, blue where broadened, and purple for both.
(B) NMR peaks in the 15N TROSY spectrum of the HPX domain affected by addition of α1(I)772-786 THP are labeled. The spectral contours of the free state are red and the bound state blue (1.5-fold molar excess of THP, pH 5, 25 °C). The box encloses arginine side chain NHε peaks, which are assigned in Figure S1D.
(C) THP-induced chemical shift perturbations (upper panel) and NMR line broadening (lower panel) are plotted versus sequence. The chemical shift perturbation is quantified as ΔωHN = (ΔωH2 + (ΔωN/5)2)1/2, where ΔωH and ΔωN are the peak shifts in ppm in the 1H and 15N dimensions. The NMR line-broadening is quantified as 1−ITHP/I0, where ITHP and I0 are the peak heights with and without one equivalent of THP.
In collagenolysis by MMPs, the HPX and catalytic domains working in tandem have been expected to loosen the collagen triple-helix to expose a single strand for digestion (Bode et al., 1994). Due to the size and insolubility of intact collagen, investigations of this question and its details have relied heavily on self-assembling triple-helical peptides (THP) as collagen mimetics (Fields, 2010). THPs have long served as excellent substrates to study the triple-helical peptidase activity and specificity of MMPs (Lauer-Fields et al., 2009; Minond et al., 2007; Robichaud et al., 2011). Most exosites of THP binding to the HPX domain of MMP-1 were mapped to blade I (Arnold et al., 2011; Lauer-Fields et al., 2009). Recent structural models of complexes of MMP-1 with THPs have been interpreted to suggest significant destabilization of the triple-helix (Bertini et al., 2012) or a small degree of bending and shift of the triple-helix (Manka et al., 2012).
Despite the similarity of ¾ and ¼ fragments generated, collagenolytic MMPs may differ partly in collagen sequence recognized and in mode of engagement of the triple-helix. MT1-MMP joins MT2-MMP, MMP-8, and MMP-13 in hydrolyzing more thermally stable triple-helices and in preferring an aromatic side chain at P1′ (Minond et al., 2007). Though MMP-1 prefers extension of native collagen sequence C-terminal to the scissile bond, MMP-8, MMP-13, and MT1-MMP prefer N-terminal extension of native collagen sequence (Robichaud et al., 2011). MT1-MMP appears to bind collagen at a site different from that of MMP-1 and -8 (Tam et al., 2004). The promise and need for selective inhibitors (Basu et al., 2012; Remacle et al., 2012) and insight into mechanistic differences among collagenolytic MMPs demands structural study of collagen triple-helix interactions with MT1-MMP.
This work investigates the mode of collagen I-based THP binding to the HPX domain of MT1-MMP. Solution NMR and enzymatic assay of mutations mapped the interface between the HPX domain and THP. Distances were measured between spin-labeled THP samples and the HPX domain of MT1-MMP by NMR detection of paramagnetic relaxation enhancements (PREs). The PREs have been used in calculating the solution structural model of the predominant orientation of this transient complex of modest affinity. The interfacial contacts, distinctive translational position, a potential site for selective inhibition, interdomain flexibility, and implications for engagement of the collagen triple-helix by the ectodomain of MT1-MMP are discussed.
RESULTS AND DISCUSSION
NMR Mapping of the THP Binding Site in the HPX Domain
For investigating the site of collagen triple-helix binding in the HPX domain, we assigned 95% of its backbone amide resonances at pH 5 by triple resonance NMR. The methyl and carbon resonances of the Leu/Val/Ile side chains of the HPX domain were assigned by interpreting HMCBCA spectra together with HMQC, triple resonance, and 13C and 15N-edited NOESY spectra (Figure S1A,B). To assign peaks of arginine side chains in the interface, each candidate was replaced by an alanine (Figure S1C).
Changes introduced to 15N TROSY NMR spectra of the HPX domain by several additions of α1(I)772-786 THP include both shifts and broadening. The association fails to saturate with additions up to four-fold molar excess. The chemical shift perturbations are generally small, but larger where the peaks also undergo severe line broadening. The side chain Nε-Hε peaks of Arg330, Arg343, and Arg345 are considerably broadened and shifted by the THP (Figure 1B). The backbone peaks most shifted by the THP center on blade I, but span from blade II to blade IV, namely Gly331, Asn347-Met350, Glu392, and Asp504 (red or purple in Figure 1A,C). The THP induces locally extreme line broadening in blade I at Arg330 – Met333, Val344, Arg345, Asn347, and Met350, which are located in the loops connecting β-strand 1 with 2, β-strand 3 with 4a, and the bulge in the outermost strand (Figure 1A,D). The broadening is also very evident at Phe360 of the helix connecting blades I and II and Gly507 at the C-terminus of blade IV (Figure 1D). The progressive THP-induced shifts and broadenings of the NMR peaks indicate the titrations to be intermediate on the time scale of NMR chemical shift differences between free and bound states. This suggests that the association with the THP is a dynamic equilibrium in which the interfacial residues (Figure 1A) experience fluctuation between the free and THP-bound states.
Impact of THP-Binding Patch on Triple-Helical Peptidase Activity
We evaluated the contribution of this THP-binding patch to processing of a collagen triple-helix by the ectodomain. Steady-state kinetic parameters of triple-helical peptidase activity were fitted to progress curves of fluorescence increase with hydrolysis (Palmier and Van Doren, 2007). The HPX domain in the ectodomain enhances the catalytic efficiency kcat/Km towards the fTHP-15 substrate over that of the catalytic domain alone, through a clearly more favorable Km (Table 1). In proteolysis of the linear peptide substrate FS-6, however, the ectodomain experiences decreases relative to the catalytic domain of two-fold in kcat/Km, three-fold in kcat, and 1.5-fold in Km (Table 1). The catalytic turnover rate constant kcat for triple-helical peptide substrates is also three-fold higher by the catalytic domain than the ectodomain. These observations suggest that the HPX domain may modestly hinder substrates from entering or cleaved products from leaving the catalytic cleft.
Table 1.
Steady-state kinetic parameters for hydrolysis of fTHP-15 and FS-6 by MT1-MMP variants at 27 °C, pH 7.5.
| Enzyme variant | fTHP-15 | FS-6 | ||||
|---|---|---|---|---|---|---|
| kcat/Km, M−1s−1 | kcat, s−1 | Km, nM | kcat/Km, M−1s−1 | kcat, s−1 | Km, nM | |
|
Wild type CATA |
7660 ± 30 | 0.068 | 8900 ±200 | 1,180,000 ± 80000 | 1.90 | 1600 ± 200 |
|
Wild type ECDB |
26700 ± 100 | 0.022 | 820 ± 10 | 589,000 ± 10000 | 0.62 | 1100 ± 30 |
|
ECD R330A |
18500 ± 100 (70%) | 0.018 | 980 ± 10 | 597,000 ± 5000 | 0.80 | 1300 ± 20 |
|
ECD R343A |
14900 ± 100 (56%) | 0.064 | 4300 ± 100 | 575,000 ±17000 | 0.59 | 1000 ± 50 |
|
ECD R345A |
17900 ± 100 (67%) | 0.030 | 1700 ± 20 | 819,000 ±18,000 | 0.69 | 850 ± 30 |
|
ECD N347A |
28400 ± 100 (106%) | 0.022 | 790 ±10 | 564,000 ±10000 | 0.58 | 1000 ± 30 |
|
ECD F360G |
16100 ± 100 (60%) | 0.027 | 1700± 20 | 535,000 ± 5000 | 0.74 | 1400 ± 20 |
|
ECD E392A |
15500 ± 100 (58%) | 0.024 | 1600 ± 20 | 514,000 ± 9000 | 0.53 | 1000 ± 30 |
CAT, catalytic domain
ECD, ectodoman
Since THP perturbations of amide NMR peaks of the HPX domain might occur not only at the interface but potentially elsewhere through indirect effects, site-directed mutagenesis was used to examine residues of the HPX domain suggested by NMR to interact with THPs. Surface residues Arg330, Arg343, Arg345, Asn347, and Glu392 (Figure 2A) were selected for alanine substitution from among the residues with NMR peaks most affected by the α1(I)772-786 THP. Arg330 and Glu392 form a salt bridge across the interface between blades I and II (Figures 1A, 2A, 5B). Phe360 with its side chain exposed was mutated to glycine for full removal of a hydrophobic side chain, but it may also enhance backbone flexibility. The variants exhibit similar catalytic efficiency upon the linear peptide substrate FS–6, suggesting that the mutations in the HPX domain do not affect the catalytic domain, but that the R345A mutation appears to enhance Km slightly for the linear peptide (Table 1).
Figure 2. Triple-helical peptidase activity of the ectodomain of MT1-MMP impaired by alanine substitutions at the THP binding site of its HPX domain.
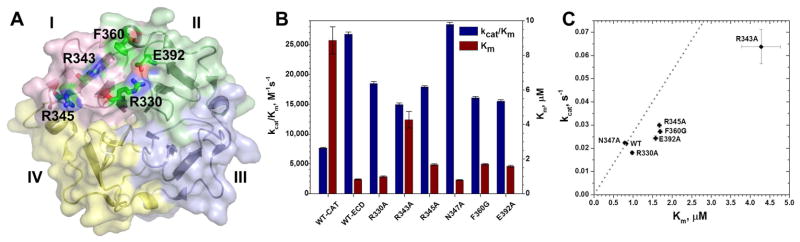
(A) Residues that impair triple-helical peptidase activity when mutated are shown with sticks and labels.
(B) The catalytic efficiency kcat/Km and Michaelis constant Km of the triple-helical peptidase activity towards fTHP-15 by the variants of the ectodomain (catalytic through HPX domains) of MT1-MMP at 27 °C, pH 7.5 are from fits of two progress curves (Palmier and Van Doren, 2007). The uncertainties plotted are three-fold the fitting errors.
(C) Catalytic turnover kcat is plotted versus Km for THP hydrolysis. Variants falling to the right of the trend line through the WT point are decreased in kcat/Km of the triple-helical peptidase activity.
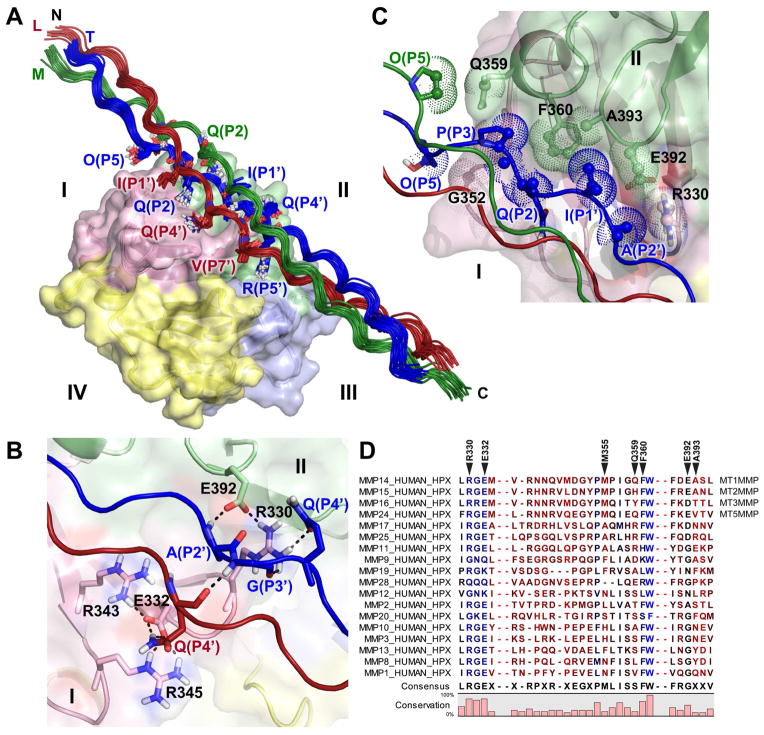
Figure 5. Structural ensemble of the complex of THP with HPX domain of MT1-MMP and interface between scissile bond region and blades I and II.
(A) The ensemble of 15 best low-energy structures are plotted with the leading, middle, and trailing chains of the THP colored red, green, and blue, respectively, and blades I through IV colored according to Figure 1A. THP residues in contact with the HPX domain are plotted with sticks. See also Figure S6.
(B) Hydrogen bonds are shown as dashed lines across the interface and to their neighbors, in the structure with best HADDOCK score.
(C) Hydrophobic interactions in the interface are shown. Carbon atoms with distances < 4 Å across the interface are shown with dots and small spheres.
(D) Sites of THP contact are marked on the alignment of human MMP sequences. Bright blue indicates the most conserved positions in sequence and bright red the least conserved. Darker shades denote intermediate levels of conservation. Sequence omissions of 10 and 30 residues are indicated by two dashes.
Assays of the catalytic domain only suggest the maximal effects of removal of the HPX domain on triple-helical peptidase activity are the 3.5-fold decrease of kcat/Km and 10.8-fold increase of Km (Table 1, Figure 2B). The R343A mutation realizes half this much impairment of Km, by 5.2-fold that is, and 44% loss of catalytic efficiency. The R345A, F360G, E392A lesions increase Km by ~2-fold and decrease catalytic efficiency by about 33%, 40%, and 42%, respectively (Figure 2B). Consequently, these four residues may contribute to productive binding and processing of the THP substrate. The 30% loss of kcat/Km and increase of Km of the R330A mutation is more subtle. The lack of effect of the N347A mutation suggests this position plays no direct role. The muted mutational effects on catalytic efficiency appear to be related to the trend of kcat increasing with Km (Figure 2C), similar to the positive correlation of kcat with Km as the length of the triple-helical substrate was varied (Robichaud et al., 2011). Less affinity for a substrate may correlate with faster off-rate of the cleaved product to hasten kcat and account for partial correlation of kcat with Km (Figure 2C).
Side Chains of the THP that Contact the HPX Domain
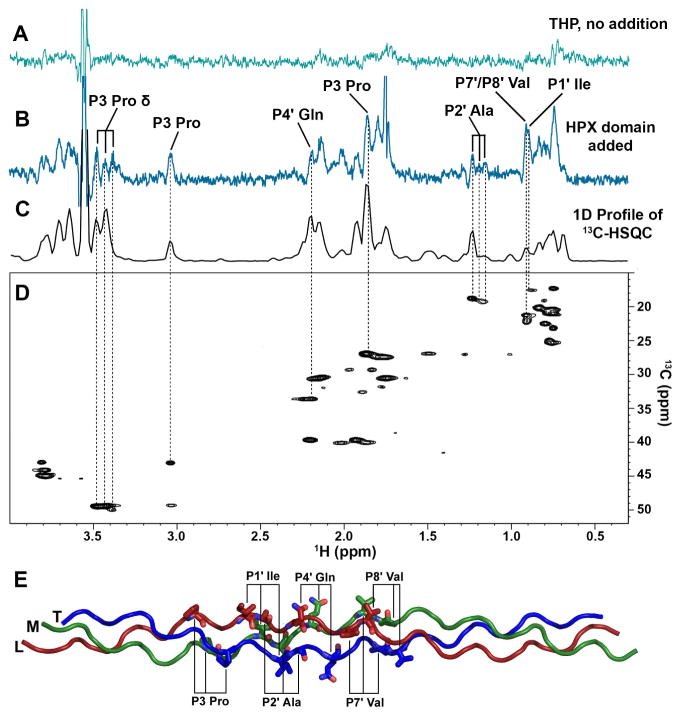
Saturation transfer difference (STD) NMR identified groups in α1(I)772-786 THP that contact the HPX domain. Saturation of an upfield methyl peak unique to the spectrum of the HPX domain spread across the domain and across its interface to several aliphatic groups of the THP in contact (Figure 3B). The side chains of the THP in STD contact and sufficiently resolved in 1H NMR spectra with the HPX domain include the Pro γ and δ at the P3 position in the substrate sequence, as well as Ile γ at P1′, Ala β at P2′, Gln γ at P4′, and Val γ at either of the spectrally overlapped P7′/P8′ positions (Figure 3B,E). The peaks from all three chains of the THP at the P3 Pro δ and P2′ Ala β positions received saturation from the HPX domain (Figure 3B–D). This indicates that the THP binds as an average of rotational orientations. The rotational averaging, fast-intermediate exchange regime, and the limited effects of the mutations suggest modest affinity and dynamic averaging in this complex.
Figure 3. Side chains of the THP in contact with the HPX domain of MT1-MMP revealed by saturation transfer difference NMR.
(A) STD spectrum of 400 μM α1(I)772-786 THP (pH 5, 25 °C) without addition of HPX domain.
(B) STD spectrum of the THP after addition of 10 μM HPX domain. Side chains of the THP receiving saturation from the HPX domain and assigned without ambiguity are labeled.
(C) 1D projection of and (D) 2D 13C HSQC of the same THP are aligned for comparison.
(E) THP side chains in contact with the HPX domain are labeled on the homology model using the nomenclature of protease substrates relative to the scissile bond at P1–P1′. Leading, middle, and trailing chains are colored dark red, green, and blue, respectively.
Monomer in Solution
The quaternary structure of the HPX domain is relevant to its recognition of collagen-mimicking THPs. The HPX domain was characterized by hydrodynamic methods, with an interest in any self-association or dependence on [CaCl2], which is present in the extracellular matrix at 2 to 3 mM and in several crystal structures of HPX domains, beginning with MMP-2 (Libson et al., 1995). However, a sodium ion appears at the corresponding site where the four blades meet in the crystal structure of the HPX domain of MT1-MMP (Tochowicz et al., 2011). Migration of the HPX domain (folded with EDTA present) on an analytical gel filtration column is very consistent with a theoretical monomeric MW of 23 kDa and is unchanged from 50 to 200 μM, suggesting lack of self-association (Figure S2A). After addition of 5 mM CaCl2 or 5 mM EDTA, the rate of migration is retained within the uncertainties (Figure S2A). This suggests the monomeric behavior to be unaffected by calcium ions. Dynamic light scattering (DLS) indicates the HPX domain to be monodisperse and suggests Ca2+-free samples to have hydrodynamic radius of 2.3 nm with apparent MW of ~22 kDa (Figure S2B). DLS of the fraction eluting from the gel permeation column in 5 mM CaCl2 suggests an apparent hydrodynamic radius of 2.05 nm and apparent MW of ~17 kDa (Figure S2C). Addition of 5 mM CaCl2 fails to perturb the 15N TROSY NMR spectrum (Figure S2D), further suggesting a lack of interaction of calcium ions with the HPX domain of MT1-MMP. 15N NMR cross-correlated relaxation (Figure S2E) was used to compare rotational correlation times, τc, at 37°C for samples free of Ca2+ (pH 5), in 5 mM Ca2+ (pH 7), and in 5 mM EDTA with 2.5 mM Ca2+ (pH 7) and suggested them to be 10.8 ± 0.2 ns, 11.75 ± 0.5 ns, and 12.2 ± 1.2, respectively (Table S1). These conditions without and with Ca2+ ions available are all much closer to the theoretically predicted τc values of a monomer of 9.1 ± 0.6 ns than to the symmetric dimer described (Tochowicz et al., 2011), which has a theoretically predicted τc of almost 24 ns (Table S1). Thus, the samples of HPX domain of MT1-MMP studied are predominantly monomeric, consistently across a range of relevant conditions.
Close Approaches of Spin-Labeled THPs with the HPX Domain
We sought to dock α1(I)772-786 THP with the monomer from the crystal structure of the HPX domain of MT1-MMP (Tochowicz et al., 2011). We prepared a homology model of this THP (Figure 3E) from the crystal structure of a homologous THP (Manka et al., 2012) as template. We measured intermolecular distances using a method effective for complexes of modest affinity in dynamic equilibrium. The approach places spin labels in one partner in order to introduce distance-dependent NMR line broadenings known as PREs (Clore and Iwahara, 2009). We introduced a nitroxide spin label at the δ position of the six-membered side chain ring of the artificial amino acid TOAC, incorporated into the proline-poor region of α1(I)772-786 THP. The THP samples with TOAC incorporated at either P4′ or P7′ formed a triple-helix, as evident from the positive feature at λ = 225 nm in the CD spectra at 22 °C (Figure S3).
We monitored the effects of TOAC substitution upon folding stability of the THP and the binding interface of the HPX domain. TOAC substitution at P4′ decreased Tm to 23 °C and at P7′ to 27.5 °C (Figure S3), each below the Tm of 41 °C of the native sequence. To stay well below the melting transitions, NMR experiments were performed at 12.5 °C using TOAC substitution at P4′ or at 17 °C using TOAC substitution at P7′. When added to the HPX domain, the TOAC-substituted THPs (reduced with ascorbate to avoid paramagnetic broadening) induced similar patterns of line broadening as the unmodified α1(I)772-786 THP (Figure S4). α1(I)772-786 THP broadened the peaks of Phe360 and Ser470 more than the TOAC-containing variants. The TOAC substitution at P7′ heightened line broadenings for residues of blades II to IV in and near the interface identified using unmodified α1(I)772-786 THP (Figures 1C, S4), as well as at five more distant residues of blade III, suggesting a subtle perturbation of part of blade III (Figure S4). Nonetheless, the shared locations of line broadening introduced by all THPs indicate the same binding region on the HPX domain.
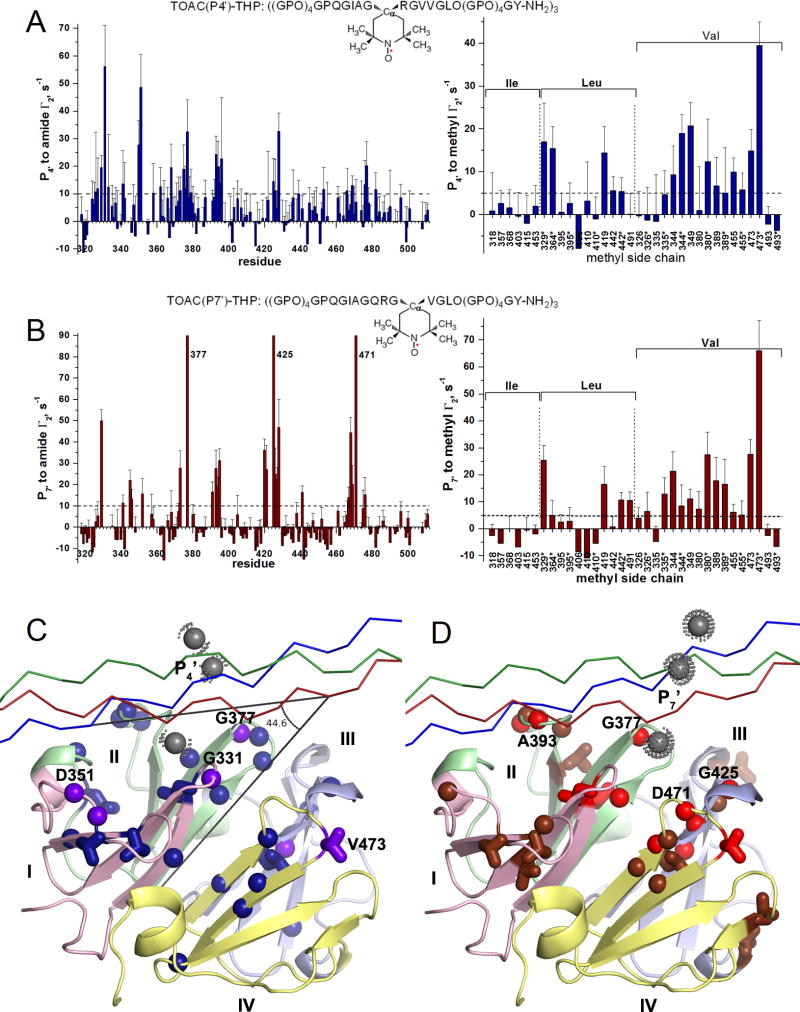
PREs were measured as Γ2 differences between the relaxation rate constants in the paramagnetic and diamagnetic states (the latter via ascorbate reduction of the nitroxide) (Figure S5). The highly distance-dependent PREs emanating from the TOAC placed in the P4′ or P7′ position of the THP sequence are plotted in Figure 4. The spin labeling of the P4′ position of the THP introduced many strong PREs to blades I and II (Figure 4A,C), resembling the spatial pattern of peak perturbations by the unmodified THP (Figure 1). The largest of these PREs, i.e., 56 s−1 to Gly331 (Figure 4A), suggests this to be the closest approach of the HPX domain to the P4′ position in the THP. This point in blade I lies in the β-hairpin loop nearest the central axis of pseudo four-fold symmetry and is represented by the purple sphere nearest a gray sphere in Figure 4C. The next largest PREs (Figure 4A) and next closest approaches radiate outward from Gly331 to Asp351 and Met350 of the neighboring loop in blade I, to a Val473 methyl group in the innermost β-hairpin loop of blade IV, and to Gly377 of the innermost β-hairpin loop of blade II (purple in Figure 4C). The PREs together suggest that the P4′ region of the THP passes near blades I and II and the central loop of blade IV of the HPX domain.
Figure 4. Proximity of the THP to the HPX domain revealed by PREs from nitroxide spin labels.
(A) PREs from TOAC at the P4′ position of the THP (see inset) were detected on amide groups (left, blue) and methyl groups (right, blue) of the HPX domain. The methyl groups are organized by residue type.
(B) PREs from TOAC at the P7′ position (see inset) to the amide and methyl groups are plotted with dull red. PREs are quantified as Γ2 = R2,para − R2,dia. The errors of Γ2 are propagated from the fitting uncertainties of R2,para and R2,dia. Each asterisk (*) of the X-axis refers to the proS methyl group; the other methyl group is proR.
(C) Significant PREs from TOAC at P4′ to amide or methyl groups are marked on the crystal structure of the HPX domain as spheres or sticks, respectively: Purple designates PREs > 25 s−1 while dark blue designates smaller PREs > 10 s−1 to amide groups and > 5 s−1 to methyl groups. The chains of the THP are plotted with Cα traces (red, green, and blue).
(D) PREs from TOAC at P7′ are plotted likewise but with PREs > 25 s−1 colored bright red and the smaller PREs colored dull red. In (C,D), the gray dotted spheres mark the approximate positions of the nitroxide group at P4′ (C) or at P7′ (D) of the THP. See also Figure S5.
PREs from the TOAC at the P7′ position appear in all four blades of the HPX domain at almost periodically repeating intervals (Figure 4B, left panel), suggestive of similar distances of TOAC at P7′ to all four blades. Extreme broadening of amide peaks of Gly377, Gly425, and Asp471 (Figure 4B) suggests them to make the closest approaches to the TOAC at P7′ of the THP, likely within ~13 Å. These three residues are located in the first and innermost β-hairpin loops of blades II, III, and IV (three red spheres nearest a nitroxide in Figure 4D). The large PRE from the P7′ TOAC to Leu329 indicates the first β-hairpin loop of blade I to be nearby as well. These PREs suggest positioning of the P7′ TOAC adjacent to these four inner loops of the HPX domain, with smaller PREs radiating outward (Figure 4D).
Structure Reveals Mode of THP Binding and Contacts with Blades I and II
Significant PREs with Γ2 values above 10 s−1 for amide groups or above 5 s−1 for methyl groups were converted using eq. 1 into distance restraints for docking the crystal structure of the HPX domain of MT1-MMP with the homology model of α1(I)772-786 THP. 108 distance restraints from the HPX domain to the spin label at P4′ and 111 to the spin label at P7′ were obtained (Table 2). Ambiguous distance restraints at the interface supplemented the docking calculations, i.e. five residues highlighted in Figure 1A and twelve THP residues highlighted in Figure 3E. The ensemble of 15 lowest energy structures converged with a backbone RMSD of 0.31 Å to the mean (Figure 5A, Table 2). Comparison of PRE-estimated distances with the distances in the best-scoring structure and a Q-factor of 0.13 indicate good agreement (Figure S6, Table 2).
Table 2.
Structural statistics of PRE-based structural ensembles.
| PDB: 2MQS | |
| BMRB: 25048 | |
| Restraints to TOAC at P4′ of THP | |
| HN 25 aaa *3 TOAC = 75 | |
| HCH3 11aab *3 TOAC = 33 | |
| Restraints to TOAC at P7′ of THP | |
| HN 23 aac * 3 TOAC = 69 | |
| HCH3 14 aad *3 TOAC = 42 | |
| Total PRE-based distance restraints = 219 | |
| Ambiguous restraints | |
| 5 aae of HPX domain and 12 aaf of THP | |
| RMS deviations | |
| All backbone atoms (Å)g | 0.31 |
| All heavy atoms (Å)g | 0.36 |
| Backbone to crystal structure of HPX domain (3C7X, Å)h | 0.62 |
| Q-factor for PRE-based distance restraintsh | 0.13 |
| Number of violations larger than 2 or 1 Å | 1i, 3j |
| Statistics of overall structural quality | Mean Score |
| Procheck G-factor (all dihedrals) | 0.01 |
| Molprobityk | 1.44 |
| Residues in most favored regions of Ramachandran plot l | 84.5 % |
| Residues in additional allowed regions | 14.5 % |
A327, L329, G331, F341, M350, D351, R362, I368, E373, K375, D376, G377, F379, A393 – E396, G425, Y428, N439, S466, G469, Y476, F477, K482
L329(δ2), L364(δ2), L419(δ1), L442 (δ1, δ2), V344(γ1, γ2), V349(γ1), V380(γ2), V473(γ1, γ2)
L329, F341, R345, N346, G352, E373, G377, D391, A393 – L395, F420, W421, G425 – Y428, E441, F467 – G469, D471, Y476
L329(δ2), L419(δ1), L442 (δ2), L491(δ1), V335(γ2), V344(γ1, γ2), V349(γ1), V380(γ1, γ2), V389 (γ1, γ2), V473(γ1, γ2)
R330, R343, R345, F360, E392
P(P3), I(P1′), A(P2′), Q(P4′) x three chains of homotrimer
Calculated based on the 15 lowest energy structures.
Calculated based on the best structure ranked by HADDOCK score.
L442(δ2)
L395(HN), Y428(HN), G352(HN)
Molprobity score combines the clash score, rotamers and Ramachandran evaluation into a single score.
Ramachandran plot analysis (%) from Procheck-NMR.
The N-terminal half of the α1(I)772-786 THP traverses the interface between blades I and II (Figure 5A,C), covering the pit in this interface that was designated the S10′ subsite in MMP-1 (Manka et al., 2012). The THP passes across the pseudo-four-fold axis of symmetry among the blades and makes a 45° angle with this axis (Figure 4C). The structures of the ensemble bury an average of 1100 Å2 of surface area in the interface between the THP and blades I and II of the HPX domain.
While the ensemble represents the most probable structure in solution in a single rotational orientation (Figure 5A), the contacts with all three chains at the P3 and P2′ positions (Figure 3) imply an average of rotational orientations actually present in solution. In the most probable orientation represented by the NMR-elucidated complex, the trailing (T) chain of α1(I)772-786 THP contributes the majority of the contacts with the HPX domain: Hyp at P5, Pro at P3, Gln at P2, Ile at P1′, Ala at P2′, Gln at P4′, and Arg at P5′ (Figure 5). The leading (L) chain places the Ile at P1′, Gln at P4′, and Val at P7′ in contact with blade I (Figure 5A). Both Hyp at P5 and Gln at P2 from the middle (M) chain make contact with blade II. The side chain of Gln at P4′ in the L chain donates hydrogen bonds to Glu332 in blade I (Figure 5B). In the T chain, Gly at P3′ and the side chain of Gln at P4′ accept hydrogen bonds donated by Arg330 from blade I, while Ala at P2′ donates a hydrogen bond to Glu392 (Figure 5B). Alanine substitutions of Arg330 or Glu392, engaged in a salt bridge, impaired triple-helical peptidase activity (Figure 2A, Table 1). The R343A and R345A mutations may have impaired activity indirectly by disrupting the hydrogen bonding of Glu332, which forms salt bridges with both arginine residues (Figure 5B). Gln359 approaches the Hyp at P5 in the M chain (Figure 5C). The rim of the pocket between blades I and II makes hydrophobic contacts with the T chain (Figure 5C): A methylene group of Arg330 appears to touch Ala at P2′. Gly352 contacts Hyp at P5. Phe360 meets the methylene groups of Pro at P3 and Gln at P2. Ala393 and the methylene groups of Glu392 contact Ile at P1′.
Conserved vs. Protease-Specific Interfacial Contacts
Arg330 and Glu332 of MT1-MMP, which hydrogen bond the THP in the coordinates, are conserved among most human MMPs (Figure 5B,D). Phe360 that presents a hydrophobic surface to the THP is conserved among the membrane-type and collagenolytic MMPs (Figure 5C,D). In contrast, THP-binding residues Glu392 and Ala393 are shared only between MT1-MMP and MT2-MMP (Figure 5B–D). While Gln359 is only shared with MT5-MMP (Figure 5C,D). These latter three interfacial residues could potentially be targeted in development of therapeutic or diagnostic agents with selectivity for membrane-type MMPs such as MT1-MMP.
A tumor-suppressing inhibitor of MT1-MMP was proposed to bind in the central pocket between Arg330, Met422, and Ser470 where the four blades meet (Remacle et al., 2012). This binding site partially overlaps the THP-binding interface in Figure 5A,C, raising the question of the inhibitor competing with binding of collagen.
Positioning of THP with Respect to Digestion
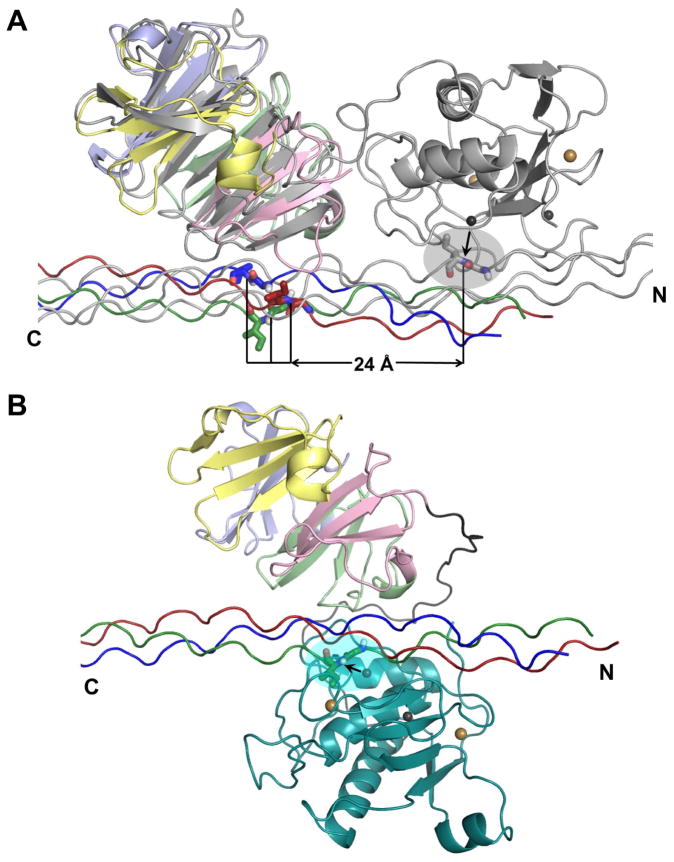
The NMR-based structural model (Figure 5) could represent a partial or initial association of MT1-MMP with the collagen triple-helix, in which the HPX domain enhances the affinity (Figure 2, Table 1). Structural superposition of the α1(I)772-786 THP complexes with MT1-MMP and full-length MMP-1 highlights the similarity in the angle and surface at which the THP crosses the two different HPX domains (Figure 6A). However, in the MT1-MMP complex the THP is translated by nine residues from its position in the MMP-1 complex. That is, Ile at P1′ at the scissile bond lies over the pocket of MT1-MMP corresponding to the S10′ subsite of MMP-1 (Figure 6A). In the MMP-1 complex, Leu at P10′ of the same THP instead lies over S10′. Independent NMR analysis of interactions between α1(I)772-786 THP and the MT1-MMP HPX domain also shows shifted binding of the THP (C. Luchinat, M. Fragai, L. Cerofolini, S. Amar, S. Pahwa, and G.B. Fields, manuscript in preparation), using the same approach that did not show shifted binding of the THP to MMP-1 (Bertini et al., 2012). The translational shift seems consistent with the reports of differences in collagen binding by MMP-1 and MT1-MMP and their HPX domains (Minond et al., 2007; Robichaud et al., 2011; Tam et al., 2004). However, the NMR structure (Figure 5A) suggests that if the catalytic domain is positioned like in MMP-1, it would be too far to reach the scissile bond (Figure 6A). A change in arrangement seems necessary for the collagen triple-helix to interact with both soluble domains of MT1-MMP during collagenolysis. The large shift and transient nature of the MT1-MMP complex suggests that more than one rearrangement could be possible.
Figure 6. Collagen triple-helix positioning across MT1-MMP HPX domain and fulllength MMP-1.
(A) The complex of THP with HPX domain of MT1-MMP (colored as in Figure 5) is superimposed via the HPX domain with the NMR model of the THP complex with full-length MMP-1 (Bertini et al., 2012) colored gray. The scissile Gly-Ile peptide bond in the MMP-1 complex is shown with sticks within the gray oval. The horizontal line marks potential distances of translation between the scissile bond positions in the MT1-MMP complex and the MMP-1 complex.
(B) Illustration of the hypothesis that the catalytic domain (cyan) of MT1-MMP folds over the collagen triple-helix to form a sandwich with the HPX domain. The scissile bond is highlighted by an arrow and a cyan oval at the active site. Calcium and zinc ions are indicated by gold and gray spheres, respectively. Hypothetical paths of the interdomain linker are colored gray. See Movie S1 for a simulated, speculative trajectory of reorientation between the hypothetical modes of binding of panels A and B.
Two potential modes of binding of the triple-helix for productive hydrolysis by MT1-MMP are considered: a sliding model or a clasping model. If the side-by-side positioning of MMP-1 domains in THP complexes (Bertini et al., 2012; Manka et al., 2012) (gray in Figure 6A) could represent MT1-MMP complexes, that would suggest shifting of the triple-helix by nine residues and around 24 to 28 Å in order to place the scissile Gly~Ile/Leu peptide bond at the active site. Greater affinity of the catalytic domain for the proline-poor region of the triple-helix might facilitate the hypothetical shift. The modest affinity and transient binding of the THP to the HPX domain might permit the THP to move across MT1-MMP to place the scissile bond in the active site and shift Leu at P10′ into the S10′ subsite like in MMP-1.
Consider if, however, the THP bound to the HPX domain of MT1-MMP (Figure 5) does represent the final translational positioning of collagen triple-helix bound to full-length MT1-MMP. The hypothesis that the HPX domain folds back upon the collagen triple-helix at the active site (Bode, 1995) would then offer the needed proximity of both domains to the scissile bond region (Figure 6B). The ease of rotating the orientations between the catalytic and HPX domains is suggested by the tremendous range of fluctuating orientations between the corresponding domains of MMP-1 (Cerofolini et al., 2013), which was proposed to generalize to all MMPs due to the greater length of their flexible linkers (Bertini et al., 2008). We used atomic force microscopy (AFM) of the ectodomain to investigate this hypothesis of loose tethering of the catalytic to HPX domain of MT1-MMP, as well as the range of their relative positions. The features in AFM images are monodisperse. In some features, the domains appear to be stacked vertically above the mica surface. In many other features, the catalytic and HPX domains appear to be laterally adjacent and distinct (Figure 7A–C). The distances between the highest points of the two domains mostly range from four to seven nm, with greater extensions to more than 10 nm also observed (Figure 7). The variable distance between the domains suggests the extensibility and flexibility of the 35-residue linker between the domains. This provides freedom of diffusional positioning of the domains. Such repositioning might aid the diffusive movement of MT1-MMP ectodomain on collagen (Collier et al., 2011; Overall and Butler, 2007). The long, flexible linker and variable separation between the domains suggests that they are potentially free enough to reorient between the side-by-side orientations observed for MMP-1 and an arrangement to sandwich the collagen triple-helix between the two domains. Simulation of a hypothetical progression of domain orientations suggests the linker between the domains is long enough for a clasping arrangement (Movie S1), as hypothesized for binding a collagen monomer (Bode, 1995).
Figure 7. AFM images reveal variable distances between the catalytic and HPX domains of MT1-MMP, suggesting flexible tethering and diffusion of the domains.
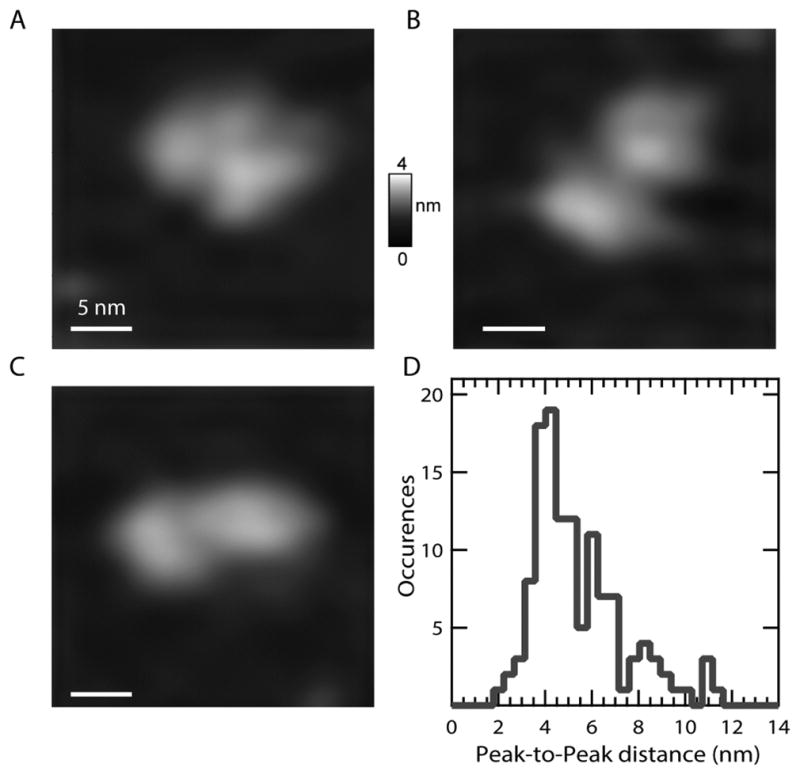
(A–C) Features with distances between high points of the domains of 4.2 nm (A), 6.1 nm.(B), and 8 nm (C). The vertical scale rises to 4 nm and the lateral scale bar represents 5 nm.
(D) The histogram shows the distribution of peak-to-peak distances (N = 134 features) between high points of adjoining domains in 150 mM NaCl, 10 mM CaCl2, at pH 7.5. The bin size is 0.45 nm.
CONCLUSIONS
The structure of the complex of the HPX domain of MT1-MMP with a collagen I-derived triple-helical peptide has been captured by NMR detection of intermolecular PREs to spin labels, despite the modest affinity and transiency of the complex. Part of the interface is conserved and part is unique to membrane-type MMPs and could be exploited for selective inhibition to increase therapeutic potential of inhibiting tumor cell invasion and preserving the left ventricle after heart attack. There is enough freedom between the catalytic and HPX domains not only to bind adjacent sites on the collagen triple-helix but also to reorient for movement on collagen and possibly to embrace a collagen monomer between the domains, a potentially distinctive mode of action.
EXPERIMENTAL PROCEDURES
Folding of Domains
Domains of MT1-MMP were expressed in E. coli, labeled with isotopes for NMR, folded from inclusion bodies, and purified in denaturant as described in Supplemental Experimental Procedures. The HPX domain was folded at pH 5 in the absence of Ca2+, or using 10 mM CaCl2 at pH 7 for comparison. Folding of the catalytic domain and ectodomain (C127S-substituted) and its variants used 10 mM CaCl2 at pH 7 – 7.5.
α1(I)772-786 THP
The peptide ((Gly-Pro-Hyp)4-Gly-Pro-Gln-Gly-Ile-Ala-Gly-Gln-Arg-Gly-Val-Val-Gly-Leu-Hyp-(Gly-Pro-Hyp)4-Gly-Tyr-NH2)3 was synthesized and characterized as described (Lauer-Fields et al., 2009). Underlining marks residues 772-786 of the type I collagen α1 chain. The NMR peak assignments of this THP are available (Bertini et al., 2012).
NMR Spectroscopy and Titrations to Map THP Binding Site on HPX Domain
NMR spectra of 15N/2H- or 15N/13C/2H-labeled HPX domain at 0.3 – 0.4 mM (pH 5 or 7) were acquired on a Bruker Avance III 800 MHz spectrometer with TCI cryoprobe. NMR peak assignments are described in Supplemental Experimental Procedures. Methyl peak assignments are depicted in Figure S1.
Stock solutions of α1(I) 772-786 THP were prepared at 8 to 9 mM in folded trimer in 10 mM sodium acetate (pH 5), 0.02% NaN3, and 7% D2O, as determined by A280 and extinction coefficient of 3591 M−1 cm−1. 15N BEST-TROSY(Lescop et al., 2010) spectra of 15N-labeled HPX domain were collected at 25 °C with p rogressive additions of small volume aliquots of the α1(I) 772-786 THP.
Site-Directed Mutagenesis of Prospective THP Binding Site
Mutations in the HPX domain were generated in the ectodomain construct (which also contains the catalytic domain and intervening linker) using QuikChange mutagenesis (Stratagene), and confirmed by DNA sequencing.
Enzyme Kinetic Assays
The triple-helical peptidase activity of WT and mutant ectodomains of MT1-MMP was detected as the increased fluorescence of fTHP-15 with sequence of ((Gly-Pro-Hyp)5-Gly-Pro-Lys(Mca)-Gly-Pro-Gln-Gly~Leu-Arg-Gly-Gln-Lys(Dnp)-Gly-Val-Arg-(Gly-Pro-Hyp)5-NH2)3 with Mca as fluorophore and Dnp as quencher (Robichaud et al., 2011). The linear peptide substrate FS-6 (Mca-Lys-Pro-Leu-Gly~Leu-Dpa-Ala-Arg-NH2) (Neumann et al., 2004) served as a control. Hydrolysis of these substrates at 27 °C in 20 mM Tris·HCl (pH 7.5), 10 mM CaCl2, 10 μM ZnCl2 was monitored with a BioTek Synergy MX plate reader as fluorescence emission at λ=393 nm using excitation of fTHP-15 at λ=324 nm and FS–6 at 328 nm. Km and kcat were estimated by fitting progress curves at two substrate concentrations (Palmier and Van Doren, 2007). The concentration of intact enzyme active sites was quantified by titration with the inhibitor galardin (GM6001, EMD).
Homology Model of the THP
The crystal structure of α1(II) THP (Manka et al., 2012) (PDB ID 4auo) provided the template for creating the homology model of α1(I) 772-786 THP, with modifications to sequence made in PyMol. The homology model was refined by energy minimization followed by molecular dynamics to equilibrate it in water solvent using GROMACS (Pronk et al., 2013).
STD NMR to Map Binding Sites on THP
STD relied on transfer of selective saturation from a dilute protein (0, 3, 10, or 30 μM HPX domain herein) to the ligand, which was 0.4 mM α1(I)772-786 THP at 25 °C. The HPX domain was selectively saturated in its methyl region at −0.5 ppm. The reference spectra utilized dummy saturation off-resonance at −20.7 ppm.
Measurement of PREs
To measure distances between α1(I)772-786 THP and the HPX domain, PREs were introduced to the complex by the nitroxide spin label of the TOAC substitution for either Gln at P4′ or Val at P7′ (Figure 4A,B). Each TOAC-labeled THP was present at two-fold molar excess over the 2H/15N/13C-1H3-labeled HPX domain. The diamagnetic reference state of each complex was prepared by reducing the nitroxide with 20 mM sodium ascorbate for at least 2 h. Amide and methyl proton relaxation was measured before and after ascorbate reduction using a CPMG sequence that suppresses 1H-1H J-couplings (Aguilar et al., 2012). Detection of amide groups employed CPMG relaxation periods of 0, 4, 8, 12, 16 and 20 ms added to the 15N HSQC. Methyl groups were detected by HMQC joined with CPMG relaxation periods of 0, 4, 8, 12, 16, 24, and 32 ms. R2, para and R2, dia refer to the respective paramagnetic and diamagnetic states. Each PRE is the difference: Γ2 = R2,para − R2,dia.
Distance restraints from PREs
PRE-based distances r from the nitroxide of the TOAC to each amide or methyl group of the HPX domain were calibrated using eq. 1 (Battiste and Wagner, 2000):
| eq. 1 |
with τc as rotational correlation time (at 12.5 or 17 °C), with K a collection of physical constants of 1.23×10−32 cm6s−2, and ωh the proton Larmor frequency of 800 MHz · 2π radians. Amide PREs with Γ2 > 10 s−1 and methyl PREs with Γ2 > 5 s−1 (provided Γ2 > uncertainties) were interpreted with eq. 1. The lower bounds of distance restraints were 10 Å below and upper bounds 3 Å above these target distances. For the strongest PREs (at residues 377, 425 and 471), the severe paramagnetic line broadening implies target distances < ~13 Å, and an upper bound of 15 Å was assigned to their distance restraints.
Atomic Force Microscopy
Following recent work (Fulcher et al., 2014), WT MT1-MMP ectodomain was deposited at 100 nM in imaging buffer of 20 mM Tris·HCl (pH 7.5), 10 mM CaCl2, and 150 mM NaCl on freshly cleaved muscovite mica surface (V-1 grade, SPI Supplies), incubated for 1 min, rinsed three times with ~100 μL of the imaging buffer, and incubated another 5 min. Images were acquired at room temperature in the imaging buffer using tapping mode and a Cypher instrument (Asylum Research). Biolever mini tips (BL-AC40TS, Olympus) with spring constants ~0.06 N/m were used. Images were recorded with an estimated tip-sample force <100 pN for minimal distortion of proteins.
Experimental Docking of THP and HPX Domain
Monomeric coordinates from the crystal structure of the HPX domain (Tochowicz et al., 2011) (PDB ID 3c7x) and the homology model of α1(I)772-786 THP were docked using explicit PRE-based distance restraints and ambiguous distance restraints to represent residues at the interface (Table 2). The HADDOCK webserver used for the calculations treated the proteins initially as rigid bodies, subsequently allowed the side chains to be flexible and backbones to be flexible within 5 Å of the interface, and finally refined in water solvent (de Vries et al., 2007; Dominguez et al., 2003). PRE-based restraints to the TOAC nitroxide were pointed at NE2 of the Gln at P4′ or at CB of the Val at P7′ in the THP sequence.
Supplementary Material
HIGHLIGHTS.
Complex of cancer-critical MT1-MMP domain with collagen mimic is captured by PRE NMR
The collagen triple-helix is translated ~25 Å from MMP-1 complexes
Part of interface distinguishes MT-MMPs and is attractive for therapeutic development
The domains of MT1-MMP are separated enough for reorientation on collagen
Acknowledgments
This work was supported by NIH grants R01 CA098799 and R01 GM057289. GMK acknowledges support from the Burroughs Wellcome Fund Career Award at the Scientific Interface. The 800 MHz NMR was purchased with funds from NIH grant S10 RR022341 and the University of Missouri. We thank Dorota Tokmina-Roszyk for synthesizing samples of unlabeled THP and THP substrate.
Abbreviations
- AFM
atomic force microscopy
- DLS
dynamic light scattering
- HPX
hemopexin-like
- Hyp or O
4-hydroxyproline
- MT1-MMP
membrane-type 1 matrix metalloproteinase
- STD
saturation transfer difference
- THP
triple-helical peptide
- TOAC
2,2,6,6-tetramethylpiperidine-1oxyl-4-amino-4-carboxylic acid
- WT
wild-type
Footnotes
ACCESSION NUMBERS
The ensemble of 15 structural models has been deposited under Protein Data Bank ID 2mqs. The NMR peak assignments have been deposited under BioMagResBank code 25048.
Supplemental information includes six figures, a table, and Supplemental Experimental procedures with this article online.
AUTHOR CONTRIBUTIONS
YZ and SRV planned the research and prepared the manuscript. YZ conducted most of the experiments and calculations. TCM contributed the STD-NMR and methyl NMR peak assignments and RRSG, BPM, and GMK the AFM analysis. RS and GBF prepared and characterized the THP samples. GBF edited the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aguilar JA, Nilsson M, Bodenhausen G, Morris GA. Spin echo NMR spectra without J modulation. Chem Commun. 2012;48:811–813. doi: 10.1039/c1cc16699a. [DOI] [PubMed] [Google Scholar]
- Arnold LH, Butt LE, Prior SH, Read CM, Fields GB, Pickford AR. The Interface between Catalytic and Hemopexin Domains in Matrix Metalloproteinase-1 Conceals a Collagen Binding Exosite. J Biol Chem. 2011;286:45073–45082. doi: 10.1074/jbc.M111.285213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu B, Correa de Sampaio P, Mohammed H, Fogarasi M, Corrie P, Watkins NA, Smethurst PA, English WR, Ouwehand WH, Murphy G. Inhibition of MT1-MMP activity using functional antibody fragments selected against its hemopexin domain. Int J Biochem Cell Biol. 2012;44:393–403. doi: 10.1016/j.biocel.2011.11.015. [DOI] [PubMed] [Google Scholar]
- Battiste JL, Wagner G. Utilization of site-directed spin labeling and high-resolution heteronuclear nuclear magnetic resonance for global fold determination of large proteins with limited nuclear overhauser effect data. Biochemistry. 2000;39:5355–5365. doi: 10.1021/bi000060h. [DOI] [PubMed] [Google Scholar]
- Bertini I, Calderone V, Fragai M, Jaiswal R, Luchinat C, Melikian M, Mylonas E, Svergun DI. Evidence of reciprocal reorientation of the catalytic and hemopexin-like domains of full-length MMP-12. J Am Chem Soc. 2008;130:7011–7021. doi: 10.1021/ja710491y. [DOI] [PubMed] [Google Scholar]
- Bertini I, Fragai M, Luchinat C, Melikian M, Toccafondi M, Lauer JL, Fields GB. Structural Basis for Matrix Metalloproteinase 1-Catalyzed Collagenolysis. J Am Chem Soc. 2012;134:2100–2110. doi: 10.1021/ja208338j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bode W. A helping hand for collagenases: the haemopexin-like domain. Structure. 1995;3:527–530. doi: 10.1016/s0969-2126(01)00185-x. [DOI] [PubMed] [Google Scholar]
- Bode W, Reinemer P, Huber R, Kleine T, Schnierer S, Tschesche H. The X-ray crystal structure of the catalytic domain of human neutrophil collagenase inhibited by a substrate analogue reveals the essentials for catalysis and specificity. EMBO J. 1994;13:1263–1269. doi: 10.1002/j.1460-2075.1994.tb06378.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerofolini L, Fields GB, Fragai M, Geraldes CF, Luchinat C, Parigi G, Ravera E, Svergun DI, Teixeira JM. Examination of matrix metalloproteinase-1 in solution: a preference for the pre-collagenolysis state. J Biol Chem. 2013;288:30659–30671. doi: 10.1074/jbc.M113.477240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun TH, Sabeh F, Ota I, Murphy H, McDonagh KT, Holmbeck K, Birkedal-Hansen H, Allen ED, Weiss SJ. MT1-MMP-dependent neovessel formation within the confines of the three-dimensional extracellular matrix. J Cell Biol. 2004;167:757–767. doi: 10.1083/jcb.200405001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clore GM, Iwahara J. Theory, practice, and applications of paramagnetic relaxation enhancement for the characterization of transient low-population states of biological macromolecules and their complexes. Chem Rev. 2009;109:4108–4139. doi: 10.1021/cr900033p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collier IE, Legant W, Marmer B, Lubman O, Saffarian S, Wakatsuki T, Elson E, Goldberg GI. Diffusion of MMPs on the Surface of Collagen Fibrils: The Mobile Cell Surface – Collagen Substratum Interface. PLoS ONE. 2011;6:e24029. doi: 10.1371/journal.pone.0024029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries SJ, van Dijk AD, Krzeminski M, van Dijk M, Thureau A, Hsu V, Wassenaar T, Bonvin AM. HADDOCK versus HADDOCK: new features and performance of HADDOCK2.0 on the CAPRI targets. Proteins. 2007;69:726–733. doi: 10.1002/prot.21723. [DOI] [PubMed] [Google Scholar]
- Dominguez C, Boelens R, Bonvin AM. HADDOCK: a protein-protein docking approach based on biochemical or biophysical information. J Am Chem Soc. 2003;125:1731–1737. doi: 10.1021/ja026939x. [DOI] [PubMed] [Google Scholar]
- Fields GB. Synthesis and biological applications of collagen-model triple-helical peptides. Organic & Biomolecular Chemistry. 2010;8:1237–1258. doi: 10.1039/b920670a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulcher YG, Sanganna Gari RR, Frey NC, Zhang F, Linhardt RJ, King GM, Van Doren SR. Heparinoids activate a protease, secreted by mucosa and tumors, via tethering supplemented by allostery. ACS Chem Biol. 2014;9:957–966. doi: 10.1021/cb400898t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galvez BG, Matias-Roman S, Albar JP, Sanchez-Madrid F, Arroyo AG. Membrane type 1-matrix metalloproteinase is activated during migration of human endothelial cells and modulates endothelial motility and matrix remodeling. J Biol Chem. 2001;276:37491–37500. doi: 10.1074/jbc.M104094200. [DOI] [PubMed] [Google Scholar]
- Holmbeck K, Bianco P, Caterina J, Yamada S, Kromer M, Kuznetsov SA, Mankani M, Robey PG, Poole AR, Pidoux I, et al. MT1-MMP-deficient mice develop dwarfism, osteopenia, arthritis, and connective tissue disease due to inadequate collagen turnover. Cell. 1999;99:81–92. doi: 10.1016/s0092-8674(00)80064-1. [DOI] [PubMed] [Google Scholar]
- Holmbeck K, Bianco P, Pidoux I, Inoue S, Billinghurst RC, Wu W, Chrysovergis K, Yamada S, Birkedal-Hansen H, Poole AR. The metalloproteinase MT1-MMP is required for normal development and maintenance of osteocyte processes in bone. J Cell Sci. 2005;118:147–156. doi: 10.1242/jcs.01581. [DOI] [PubMed] [Google Scholar]
- Hotary KB, Allen ED, Brooks PC, Datta NS, Long MW, Weiss SJ. Membrane type I matrix metalloproteinase usurps tumor growth control imposed by the three-dimensional extracellular matrix. Cell. 2003;114:33–45. doi: 10.1016/s0092-8674(03)00513-0. [DOI] [PubMed] [Google Scholar]
- Itoh Y, Ito N, Nagase H, Evans RD, Bird SA, Seiki M. Cell surface collagenolysis requires homodimerization of the membrane-bound collagenase MT1-MMP. Mol Biol Cell. 2006;17:5390–5399. doi: 10.1091/mbc.E06-08-0740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh Y, Ito N, Nagase H, Seiki M. The second dimer interface of MT1-MMP, the transmembrane domain, is essential for ProMMP-2 activation on the cell surface. J Biol Chem. 2008;283:13053–13062. doi: 10.1074/jbc.M709327200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh Y, Palmisano R, Anilkumar N, Nagase H, Miyawaki A, Seiki M. Dimerization of MT1-MMP during cellular invasion detected by fluorescence resonance energy transfer. Biochem J. 2011;440:319–326. doi: 10.1042/BJ20110424. [DOI] [PubMed] [Google Scholar]
- Itoh Y, Takamura A, Ito N, Maru Y, Sato H, Suenaga N, Aoki T, Seiki M. Homophilic complex formation of MT1-MMP facilitates proMMP-2 activation on the cell surface and promotes tumor cell invasion. EMBO J. 2001;20:4782–4793. doi: 10.1093/emboj/20.17.4782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JL, Sala-Newby GB, Ismail Y, Aguilera CM, Newby AC. Low Tissue Inhibitor of Metalloproteinases 3 and High Matrix Metalloproteinase 14 Levels Defines a Subpopulation of Highly Invasive Foam-Cell Macrophages. Arterioscl Thromb Vasc Biol. 2008;28:1647–1653. doi: 10.1161/ATVBAHA.108.170548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauer-Fields JL, Chalmers MJ, Busby SA, Minond D, Griffin PR, Fields GB. Identification of specific hemopexin-like domain residues that facilitate matrix metalloproteinase collagenolytic activity. J Biol Chem. 2009;284:24017–24024. doi: 10.1074/jbc.M109.016873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehti K, Lohi J, Juntunen MM, Pei D, Keski-Oja J. Oligomerization through Hemopexin and Cytoplasmic Domains Regulates the Activity and Turnover of Membrane-type 1 Matrix Metalloproteinase. J Biol Chem. 2002;277:8440–8448. doi: 10.1074/jbc.M109128200. [DOI] [PubMed] [Google Scholar]
- Lescop E, Kern T, Brutscher B. Guidelines for the use of band-selective radiofrequency pulses in hetero-nuclear NMR: Example of longitudinal-relaxation-enhanced BEST-type 1H-15N correlation experiments. J Magn Reson. 2010;203:190–198. doi: 10.1016/j.jmr.2009.12.001. [DOI] [PubMed] [Google Scholar]
- Li XY, Ota I, Yana I, Sabeh F, Weiss SJ. Molecular dissection of the structural machinery underlying the tissue-invasive activity of membrane type-1 matrix metalloproteinase. Mol Biol Cell. 2008;19:3221–3233. doi: 10.1091/mbc.E08-01-0016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libson AM, Gittis AG, Collier IE, Marmer BL, Goldberg GI, Lattman EE. Crystal structure of the haemopexin-like C-terminal domain of gelatinase A. Nat Struct Biol. 1995;2:938–942. doi: 10.1038/nsb1195-938. [DOI] [PubMed] [Google Scholar]
- Manka SW, Carafoli F, Visse R, Bihan D, Raynal N, Farndale RW, Murphy G, Enghild JJ, Hohenester E, Nagase H. Structural insights into triplehelical collagen cleavage by matrix metalloproteinase 1. Proc Natl Acad Sci U S A. 2012 doi: 10.1073/pnas.1204991109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MC, Manning HB, Jain A, Troeberg L, Dudhia J, Essex D, Sandison A, Seiki M, Nanchahal J, Nagase H, et al. Membrane type 1 matrix metalloproteinase is a crucial promoter of synovial invasion in human rheumatoid arthritis. Arthritis Rheum. 2009;60:686–697. doi: 10.1002/art.24331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minond D, Lauer-Fields JL, Cudic M, Overall CM, Pei D, Brew K, Moss ML, Fields GB. Differentiation of secreted and membrane-type matrix metalloproteinase activities based on substitutions and interruptions of triple-helical sequences. Biochemistry. 2007;46:3724–3733. doi: 10.1021/bi062199j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann U, Kubota H, Frei K, Ganu V, Leppert D. Characterization of Mca-Lys-Pro-Leu-Gly-Leu-Dpa-Ala-Arg-NH2, a fluorogenic substrate with increased specificity constants for collagenases and tumor necrosis factor converting enzyme. Anal Biochem. 2004;328:166–173. doi: 10.1016/j.ab.2003.12.035. [DOI] [PubMed] [Google Scholar]
- Ohuchi E, Imai K, Fujii Y, Sato H, Seiki M, Okada Y. Membrane type 1 matrix metalloproteinase digests interstitial collagens and other extracellular matrix macromolecules. J Biol Chem. 1997;272:2446–2451. doi: 10.1074/jbc.272.4.2446. [DOI] [PubMed] [Google Scholar]
- Overall CM, Butler GS. Protease yoga: extreme flexibility of a matrix metalloproteinase. Structure. 2007;15:1159–1161. doi: 10.1016/j.str.2007.10.001. [DOI] [PubMed] [Google Scholar]
- Overall CM, Tam E, McQuibban GA, Morrison C, Wallon UM, Bigg HF, King AE, Roberts CR. Domain interactions in the gelatinase A.TIMP-2. MT1-MMP activation complex The ectodomain of the 44-kDa form of membrane type-1 matrix metalloproteinase does not modulate gelatinase A activation. J Biol Chem. 2000;275:39497–39506. doi: 10.1074/jbc.M005932200. [DOI] [PubMed] [Google Scholar]
- Palmier MO, Van Doren SR. Rapid determination of enzyme kinetics from fluorescence: overcoming the inner filter effect. Anal Biochem. 2007;371:43–51. doi: 10.1016/j.ab.2007.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pronk S, Páll S, Schulz R, Larsson P, Bjelkmar P, Apostolov R, Shirts MR, Smith JC, Kasson PM, van der Spoel D, et al. GROMACS 4.5: a high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics. 2013;29:845–854. doi: 10.1093/bioinformatics/btt055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remacle AG, Golubkov VS, Shiryaev SA, Dahl R, Stebbins JL, Chernov AV, Cheltsov AV, Pellecchia M, Strongin AY. Novel MT1-MMP Small-Molecule Inhibitors Based on Insights into Hemopexin Domain Function in Tumor Growth. Cancer Res. 2012;72:2339–2349. doi: 10.1158/0008-5472.CAN-11-4149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robichaud TK, Steffensen B, Fields GB. Exosite Interactions Impact Matrix Metalloproteinase Collagen Specificities. J Biol Chem. 2011;286:37535–37542. doi: 10.1074/jbc.M111.273391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozanov DV, Deryugina EI, Ratnikov BI, Monosov EZ, Marchenko GN, Quigley JP, Strongin AY. Mutation analysis of membrane type-1 matrix metalloproteinase (MT1-MMP). The role of the cytoplasmic tail Cys(574), the active site Glu(240), and furin cleavage motifs in oligomerization, processing, and self-proteolysis of MT1-MMP expressed in breast carcinoma cells. J Biol Chem. 2001;276:25705–25714. doi: 10.1074/jbc.M007921200. [DOI] [PubMed] [Google Scholar]
- Sabeh F, Ota I, Holmbeck K, Birkedal-Hansen H, Soloway P, Balbin M, Lopez-Otin C, Shapiro S, Inada M, Krane S, et al. Tumor cell traffic through the extracellular matrix is controlled by the membrane-anchored collagenase MT1-MMP. J Cell Biol. 2004;167:769–781. doi: 10.1083/jcb.200408028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider F, Sukhova GK, Aikawa M, Canner J, Gerdes N, Tang SM, Shi GP, Apte SS, Libby P. Matrix Metalloproteinase-14 Deficiency in Bone Marrow-Derived Cells Promotes Collagen Accumulation in Mouse Atherosclerotic Plaques. Circulation. 2008 doi: 10.1161/CIRCULATIONAHA.107.707448. [DOI] [PubMed] [Google Scholar]
- Stratman AN, Saunders WB, Sacharidou A, Koh W, Fisher KE, Zawieja DC, Davis MJ, Davis GE. Endothelial cell lumen and vascular guidance tunnel formation requires MT1-MMP-dependent proteolysis in 3D collagen matrices. Blood. 2009 doi: 10.1182/blood-2008-12-196451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam EM, Moore TR, Butler GS, Overall CM. Characterization of the distinct collagen binding, helicase and cleavage mechanisms of matrix metalloproteinase 2 and 14 (gelatinase A and MT1-MMP): the differential roles of the MMP hemopexin c domains and the MMP-2 fibronectin type II modules in collagen triple helicase activities. J Biol Chem. 2004;279:43336–43344. doi: 10.1074/jbc.M407186200. [DOI] [PubMed] [Google Scholar]
- Tochowicz A, Goettig P, Evans R, Visse R, Shitomi Y, Palmisano R, Ito N, Richter K, Maskos K, Franke D, et al. The dimer interface of the membrane type 1 matrix metalloproteinase hemopexin domain: crystal structure and biological functions. J Biol Chem. 2011;286:7587–7600. doi: 10.1074/jbc.M110.178434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Nandelstadh P, Gucciardo E, Lohi J, Li R, Sugiyama N, Carpen O, Lehti K. Actin-associated protein palladin promotes tumor cell invasion by linking extracellular matrix degradation to cell cytoskeleton. Mol Biol Cell. 2014;25:2556–2570. doi: 10.1091/mbc.E13-11-0667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Nie J, Pei D. The hemopexin domain of membrane-type matrix metalloproteinase-1 (MT1-MMP) Is not required for its activation of proMMP2 on cell surface but is essential for MT1-MMP-mediated invasion in three-dimensional type I collagen. J Biol Chem. 2004;279:51148–51155. doi: 10.1074/jbc.M409074200. [DOI] [PubMed] [Google Scholar]
- Woskowicz AM, Weaver SA, Shitomi Y, Ito N, Itoh Y. MT-LOOP-dependent Localization of Membrane Type I Matrix Metalloproteinase (MT1-MMP) to the Cell Adhesion Complexes Promotes Cancer Cell Invasion. J Biol Chem. 2013;288:35126–35137. doi: 10.1074/jbc.M113.496067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zavadzkas JA, Mukherjee R, Rivers WT, Patel RK, Meyer EC, Black LE, McKinney RA, Oelsen JM, Stroud RE, Spinale FG. Direct regulation of membrane type 1 matrix metalloproteinase following myocardial infarction causes changes in survival, cardiac function, and remodeling. Am J Physiol Heart Circ Physiol. 2011;301:H1656–H1666. doi: 10.1152/ajpheart.00141.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.