SUMMARY
Individual posttranslational modifications (PTMs) of p53 mediate diverse p53-dependent responses, however much less is known about the combinatorial action of adjacent modifications. Here, we describe crosstalk between the early DNA damage response mark p53K382me2 and the surrounding PTMs that modulate binding of p53 co-factors, including 53BP1 and p300. The 1.8 Å resolution crystal structure of the tandem Tudor domain (TTD) of 53BP1 in complex with p53 peptide acetylated at K381 and dimethylated at K382 (p53K381acK382me2) reveals that the dual PTM induces a conformational change in p53. The α-helical fold of p53K381acK382me2 positions the side chains of R379, K381ac, and K382me2 to interact with TTD concurrently, reinforcing a modular design of double PTM mimetics. Biochemical and NMR analyses show that other surrounding PTMs, including phosphorylation of serine/threonine residues of p53, affect association with TTD. Our findings suggest a novel PTM-driven conformation switch-like mechanism that may regulate p53 interactions with binding partners.
INTRODUCTION
p53 undergoes numerous posttranslational modifications (PTMs) that mediate function, stability, and subcellular localization of this tumor suppressor. Recent mass-spectrometry analysis has identified 222 PTMs present on 99 residues of endogenous p53 (DeHart et al., 2014). The PTMs are spread throughout the protein, however are particularly enriched in the C-terminal regulatory domain (CTD) of p53. Among the most common PTMs are phosphorylation of serine and threonine residues, methylation of arginine residues, and acetylation, methylation, ubiquitination, sumoylation and neddylation of lysine residues (Berger, 2010; Dai and Gu, 2010). Generally, phosphorylation and acetylation are thought to activate or stabilize p53, whereas polyubiquitination targets p53 for proteasomal degradation, and methylation can be either an activating or repressive mark. Although some individual PTMs are linked to a particular p53 response, growing evidence suggests widespread crosstalk between the PTMs, which could be either synergistic or antagonistic in nature.
The p53 CTD contains six lysines within a span of 17 residues, including two pairs of contiguous lysine residues, K372K373 and K381K382. As individual lysine residues can be posttranslationally modified in a variety of ways, crosstalk between these PTMs can provide a mechanism for fine-tuning p53 activities. For example, in response to DNA damage, SET7/9-dependent monomethylation of K372 can promote acetylation of nearby lysine residues, including K373 and K382, and enhance the stability and activity of p53, ultimately upregulating p21 and triggering cell cycle arrest (Ivanov et al., 2007). Methylation of K369 in mouse p53 (K372 in human ortholog) is important for the recruitment of the Tip60 lysine acetyltransferase (KAT) complex to p53 and for the subsequent acetylation of K370 and K379 (K373 and K382 in human p53) (Kurash et al., 2008). In the absence of sustained damage, repressive methylation marks have been proposed to keep p53 in an inactive form, however upon DNA damage, acetylation can replace methylation, promoting p53 transcriptional activity (Berger, 2010; Loewer et al., 2010). In agreement, activities of SMYD2 and SET8 lysine methyltransferases (KMTs) responsible for the deposition of the repressive marks p53K370me1 (p53 monomethylated at K370) and p53K382me1 (p53 monomethylated at K382) are reduced following DNA damage, while activity of the CBP/p300 KAT is increased (Huang et al., 2006; Huang et al., 2007; Ivanov et al., 2007; Loewer et al., 2010; Shi et al., 2007; West et al., 2010). The acetylation-methylation interplay can function as a switch, allowing for distinctly different p53 responses to severe DNA damage as opposed to transient low-level DNA breaks that occur during normal cell processes (Berger, 2010; Loewer et al., 2010).
A number of spatial and temporal correlations have been reported within the p53 methylation or acetylation pathways. The SET7/9-produced activating mark p53K372me1 (p53 monomethylated at K372) prevents repressive monomethylation of K370 by inhibiting SMYD2 priming at p53 (Huang et al., 2006; Huang et al., 2007). Acetylation of the CTD lysine residues stimulates p53 transactivation through the recruitment of co-factors, inhibits ubiquitin ligase MDM2-mediated ubiquitination, and is essential for p53 tetramer formation (Barlev et al., 2001; Itahana et al., 2009; Kawaguchi et al., 2006; Luo et al., 2004; Mujtaba et al., 2004; Yamaguchi et al., 2009). Binding of p53 to the transcriptional co-activator PC4 is augmented when K381 and K382 are acetylated (Debnath et al., 2011). Acetylation of p53 by Tip60 selectively impedes MDM2-dependent neddylation, whereas FBXO11-facilitated neddylation suppresses p53 transcriptional activity, possibly via preventing acetylation (Abida et al., 2007; Dohmesen et al., 2008). Sumoylation at K386 blocks consecutive acetylation of p53 by p300, while acetylation shows no antagonistic effect on sumoylation at K386 and alleviates sumoylation-induced inhibition of DNA binding. Genotoxic stress triggers phosphorylation of S366, S378 and T387, which differentially affect p53 acetylation (Ou et al., 2005). Phosphorylation of S378 decreases acetylation of K373 and K382, whereas phosphorylation of S366 and T387 increases it (Ou et al., 2005).
Three histone readers, including the malignant brain tumor (MBT) module of the L3MBTL1 protein, the tandem Tudor domain (TTD) of the PHF20 protein, and the TTD of p53-binding protein 1 (53BP1), have been shown to recognize methylated K382 of p53 (Cui et al., 2012; Kachirskaia et al., 2008; Roy et al., 2010; Shi et al., 2007; West et al., 2010). p53 monomethylated at K382 is generated by the SET8 KMT, which is active in resting cells but is repressed following DNA damage (Shi et al., 2007). In the absence of stress, the MBT module of L3MBTL1 binds to p53K382me1 at promoters of the highly responsive p53 targets, p21 and PUMA, repressing these genes and compacting chromatin (Shi et al., 2007; West et al., 2010). The concomitant decrease in p53K382me1 levels and the increase in p53K382me2 (p53 dimethylated at K382) levels upon DNA damage cause disassociation of L3MBTL1. The dimethylated p53K382 species is recognized by the TTD of 53BP1, a DNA damage response protein and a p53 co-factor that also binds to H4K20me2 (histone 4 dimethylated at K20) (Botuyan et al., 2006; Huang et al., 2007; Huyen et al., 2004; Kachirskaia et al., 2008). 53BP1 association with p53K382me2 has been shown to facilitate p53 accumulation and stabilization at DNA double-strand break (DSB) sites and play a role in DNA repair (Kachirskaia et al., 2008; Roy et al., 2010).
In this study, we characterize crosstalk between PTMs in the K381K382 region and the surrounding residues of p53. We demonstrate that dimethylation of p53K382 is rapidly induced following DNA damage, whereas acetylation of p53K381 remains relatively unchanged. The TTD domain of 53BP1 recognizes the p53K381acK382me2 (p53 acetylated at K381 and dimethylated at K382) modification. This dual PTM causes a significant conformational change in the p53 structure, converting a U-shaped p53K382me2 peptide into α-helical p53K381acK382me2. Other PTMs, including phosphorylation of serine and threonine residues of p53 further influence the interaction with 53BP1 TTD. Our data suggest a novel p53 regulatory mechanism in which different combinations of PTMs trigger distinct p53 conformations when bound to specific interactors.
RESULTS AND DISCUSSION
p53K382me2 is an early DNA damage response PTM
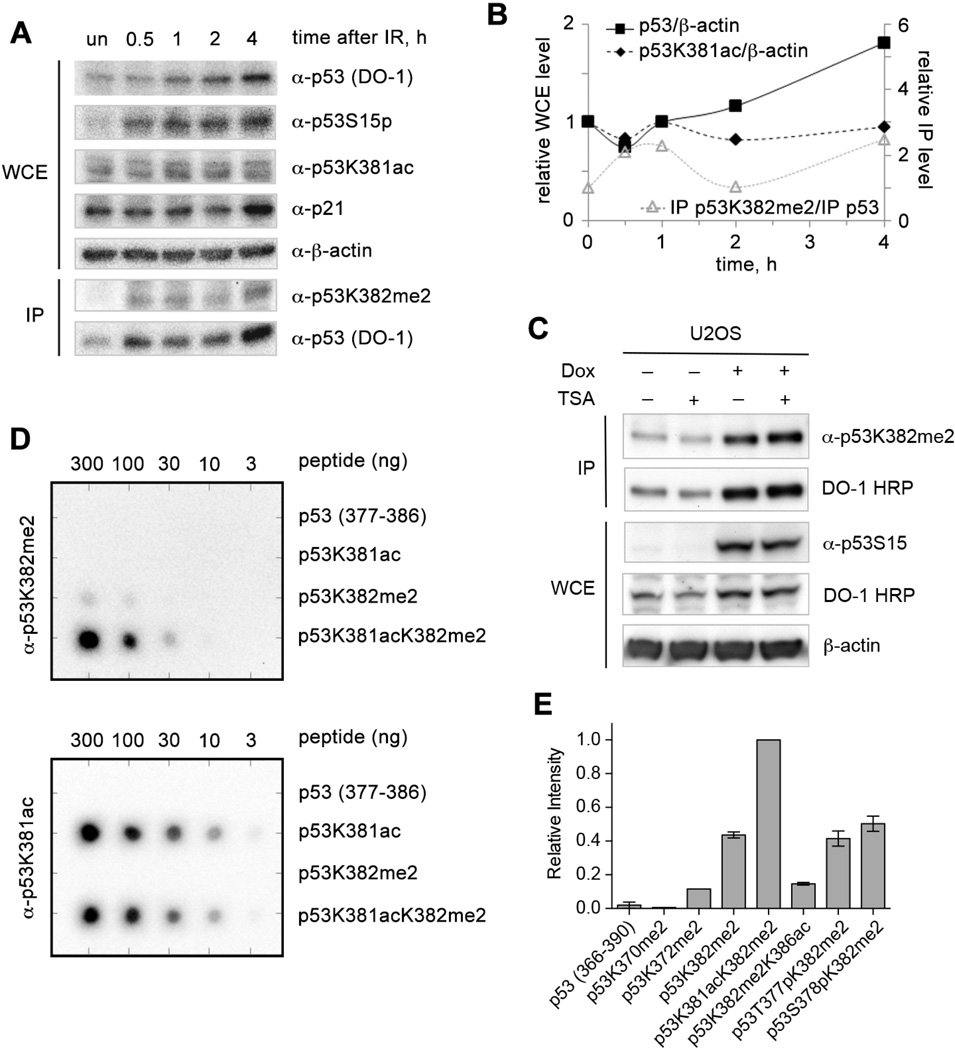
Dimethylated K382 has been identified as a DNA damage-associated PTM of p53 (Kachirskaia et al., 2008), however the dynamics of this PTM had not been investigated. We used immunoblot analysis to examine the occurrence of p53K382me2 in human U2OS osteosarcoma cells in the context of the well-characterized response to ionizing radiation (IR). As expected, the levels of total and phospho-Ser15 p53 increased during the four hours following exposure to 5 Gy IR, indicating increased stability and activation of p53 (Figure 1A). The level of the cyclin-dependent kinase inhibitor, p21 (CDKN1A), a transcriptional target of p53, was sharply elevated at 4 h after exposure to IR. In agreement with previous reports (Kachirskaia et al., 2008), p53K382me2 was undetectable in untreated cells, however its level increased considerably 0.5 h after DNA damage, indicating that dimethylation of K382 is a PTM elicited early in the response to DNA damage (Figure 1B). The p53K382me2 level continued rising 1 h after irradiation, reduced at 2 h, and increased again at 4 hours, revealing that this modification is subject to regulation. The dynamic nature of p53K382me2 levels following exposure to IR resembles oscillations in the activity of Set7/9 after DNA damage (Ivanov et al., 2007), as well as ATM activity, p53 levels, and p53 transcriptional target levels that result from re-activation of DNA damage response signaling by the presence of persistent DNA damage (Batchelor et al., 2008).
Figure 1. p53K382me2 is an early DNA damage response PTM.
(A) Immunoblot analysis of whole cell extract (WCE) or immunoprecipitated (IP) proteins from U2OS cells following exposure to IR using the indicated antibodies is shown. Cells were left untreated or were exposed to 5 Gy IR and harvested at the indicated times. p53 proteins were immunoprecipitated by agarose-conjugated DO1-antibodies. The lower of the two bands detected by the antip53K381ac antibody co-migrates with the bands detected by the anti-p53 (DO-1) and antip53S15p antibodies. (B) A graph of the relative levels of total p53 and p53 bearing K381ac and K382me2 modifications following exposure to IR is shown. Relative protein levels are based on immunoblot analysis. The relative levels of total p53 (DO-1 reactivity) and p53K381ac in WCE are given relative to β-actin levels (left axis); the relative levels of anti-p53K382me2 reactivity in the IP material are given relative to the total p53 (DO-1) level recovered in the IP material (right axis). Protein level ratios were normalized to the value in the untreated samples. Quantitation of p53K381ac levels was based on the integrated signal intensity for the lower band only. (C) Immunoblot analysis of WCE or IP proteins from U2OS cells using the indicated antibodies is shown. Cells were treated without or with 5 µg/mL doxorubicin (Dox) for 4 h, either in the absence or presence of 400 nM Trichostatin A (TSA) for the last 1 h before harvest. p53 proteins were immunoprecipitated by agarose-conjugated DO1-antibodies. (D) Dot blot analysis of anti-p53K382me2 antibody and anti-p53K381ac antibody reactivities toward p53 peptides containing K381ac and K382me2 combinations. (E) Microarray analysis of anti-p53K382me2 antibody reactivity for the indicated p53 peptides. Data is presented as normalized mean intensities from six individual spots per peptide. Error is presented as s.e.m., see also Figure S1.
To demonstrate that the increase in p53K382me2 levels is a general response to DNA damage, we treated U2OS cells with doxorubicin, a chemotherapeutic agent that induces DSBs in DNA, and with Trichostatin A (TSA), a broad-spectrum deacetylase inhibitor. As shown in Figures 1C and S1A, little p53K382me2 was present in cells under basal conditions, either with or without TSA treatment. However the p53K382me2 level increased substantially in p53 immunoprecipitated from cells treated with doxorubicin. Unexpectedly, we found that anti-p53K382me2 antibodies showed higher reactivity toward p53 immunoprecipitated from cells treated with both doxorubicin and TSA, even though the total amount of p53, as detected by DO-1 antibodies, remained essentially the same. These results suggest that acetylation of p53 may affect anti-p53K382me2 antibody reactivity.
Generally, acetylation of p53 has been shown to increase in response to DNA damage and correlate with p53 activation and stabilization (Barlev et al., 2001; Luo et al., 2004; Tang et al., 2008), however DNA damage-dependent behavior of p53K381ac has not been characterized. We found that in contrast to p53K382me2, the level of p53K381ac in U2OS cells remained relatively unaffected by the exposure to IR (Figure 1A and B). As expected, treatment of U2OS cells with TSA in the presence or absence of doxorubicin significantly elevated the p53K381ac level (Figure S1B). Together, the DNA damage assays reveal that the presence of acetylated K381 may influence the recognition of dimethylated K382 by anti-p53K382me2 antibodies.
Acetylation of K381 enhances reactivity of anti-p53K382me2 antibodies
We note that the anti-p53K382me2 antibodies were raised in rabbits immunized with p53K382me2 peptide containing residues 377–387 of human p53 and were positively selected for binding to p53K382me2 peptide and negatively selected for binding to unmodified p53 peptide (Kachirskaia et al., 2008), however the effect of acetylation of the adjacent K381 residue on the antibody reactivity had not been examined. To investigate the role of K381 acetylation, we synthesized unmodified p53 peptide (residues 377–386), and p53 peptides carrying single modifications, p53K381ac (p53 acetylated at K381) or p53K382me2, and the double modification p53K381acK382me2 and tested them in dot-blot assays. As shown in Figure 1D, the generated anti-p53 antibodies strongly recognized the p53K381acK382me2 peptide, reacted 16-fold more weakly with the p53K382me2 peptide, and did not associate with unmodified p53 or p53K381ac peptides. In contrast, the anti-p53K381ac antibodies recognized p53K381ac and p53K381acK382me2 peptides almost equally well.
To verify the anti-p53K382me2 antibody selectivity, we synthesized an expanded series of biotinylated p53 CTD peptides (residues 366–390) and printed them on streptavidin-coated glass slides for microarray analysis. In agreement with the dot-blot results, the anti-p53K382me2 antibodies reacted preferentially with the p53K381acK382me2 peptide over unmodified, p53K382me2, p53K370me2, and p53K372me2 peptides (Figure 1E). Importantly, acetylation at K386 or phosphorylation at T377 or S378 did not enhance recognition of K382me2. The higher reactivity of the generated anti-p53 antibodies toward peptides containing both K381ac and K382me2 modifications suggest that acetylation at K381 alters the p53 conformation such that it enhances antibody binding, although we cannot rule out the possibility that the immunogen was subjected to acetylation at K381 in vivo.
DNA damage promotes co-localization of p53 and p300
p53 is acetylated by p300/CBP at multiple lysine residues including K381 in the CTD (Luo et al., 2004; Tang et al., 2008) (Figure 2A). To establish whether DNA damage promotes colocalization of p53 and p300, we analyzed changes in genome-wide association of these proteins using publically available datasets. The p53 binding sites before and after DNA damage have recently been identified (Lee et al., 2010; Li et al., 2012). p53 binds to ~7,800 regions in untreated mouse embryonic stem (ES) cells, however under stress, the number of p53-bound regions increases to ~52,000 (Li et al., 2012). Using the p300/CBP ChIP-seq data from mouse ES cells generated by ENCODE consortium, we examined whether p53 colocalizes with the regions occupied by p300/CBP (Consortium et al., 2012). Under basal conditions, p53 associated with only ~3% (914 regions out of 23654) of p300-bound regions (Figure 2B). In response to stress, p53 and p300 co-localization increased substantially, with p53 occupying ~23% of p300-bound regions (5,506 out of 23,654, p < 0.0002). In agreement with these results, meta-analysis of p53 target regions showed enrichment of p300/CBP in cells treated with adriamycin (a trade name for doxorubicin) (Figure 2C). Gene ontology analysis revealed that p53/p300 co-localization occurs at genes involved in response to oxidative stress, as well as regulation and development (Figure 2D). Together, these analyses suggest a synergistic activity of p53 and p300/CBP in modulating cellular processes upon DNA damage.
Figure 2. DNA damage promotes co-localization of p53 and p300.
(A) Scheme of p53 K381 and K382 modifications in the context of the DNA damage response. (B) Pie chart displaying p53 overlap on p300-bound regions in mouse ES cells before and after DNA damage. p value < 0.0002 Fisher’s exact test. (C) Normalized p300 ChIP-seq tag density surrounding the center of p53 bound regions. Graphs include 2 Kb upstream and downstream from the center of p53-bound regions. (D) Gene ontology (GO) of p53/p300 regulated genes upon DNA damage. Biological processes include responses to oxidative stress, which may correlate with dimethylation of K382, as well as regulatory and development processes, which may correlate with p53 activating PTMs, including acetylation of multiple lysine residues and dimethylation of K370. DAVID generated GO processes with p values lower than 0.05 were re-analyzed using REVIGO. (E) Superimposed 1H,15N HSQC spectra of BD of p300 collected during titration with p53K381ac peptide (left) and p53K381acK382me2 peptide (right). Spectra are color coded according to the protein:peptide molar ratio. (F) Representative binding curves used to determine the Kd values by NMR.
Binding of bromodomain (BD) of CBP to p53K382ac (p53 acetylated at K382) has been found to contribute to stabilization of the CBP-p53 complex at chromatin, leading to acetylation of histones and p53 transactivation (Mujtaba et al., 2004; Zeng et al., 2008). Interestingly, BD of CBP displays selectivity for K382 and does not recognize p53 acetylated at K320 or K373 (Mujtaba et al., 2004). To determine whether p53K381acK382me2 can recruit BD-containing cofactors, we examined interaction of the p300 BD with acetylated p53 peptides by NMR. Titration of either p53K381acK382me2 peptide or p53K381ac peptide into 15N-labeled BD of p300 led to substantial resonance changes in the 1H,15N HSQC spectra of BD, implying that BD recognizes both peptides (Figure 2E). Binding affinities of the p300 BD, measured via chemical shift perturbation analysis, were found to be 241 µM and 282 µM for p53K381ac and p53K381acK382me2, respectively (Figure 2F). These affinities were in the same range as the binding affinity of CBP BD for p53K382ac (dissociation constant (Kd) of 187 µM) (Zeng et al., 2008). These results indicate that p53K381ac can serve as a docking site for BD and that dimethylation of K382 slightly diminishes binding of BD to p53K381ac. Much like interaction of CBP BD with p53K382ac (Mujtaba et al., 2004; Zeng et al., 2008), the p300 BD-p53K381ac association can stabilize the p300-p53 complex, though the BD can be displaced from p53K381acK382me2 by the TTD module of 53BP1, which binds to this dual PTM more strongly (see below).
K381acK382me2 induces an α-helical structure in p53
The tandem Tudor domain of 53BP1 has been shown to bind to p53K382me2 in response to DNA damage (Kachirskaia et al., 2008), therefore we examined the effect of K381 acetylation on recognition of this PTM by TTD. Although acetylation causes significant changes in the size and electrostatic properties of the lysine side chain, we found that K381ac has only a small effect on the binding free energy (Figure 3). The Kd value measured by fluorescence spectroscopy was estimated to be 760 nM for the interaction of the TTD with p53K381acK382me2 peptide and was comparable to the Kd of 900 nM determined for the interaction with p53K382me2 (Figure 3A and B) (Roy et al., 2010).
Figure 3. Molecular basis for the recognition of p53K381acK382me2 by TTD.
(A) Binding affinities of wild type (WT) and mutated 53BP1 TTD for the indicated histone peptides measured by tryptophan fluorescence. (*) Measured by NMR; (**) taken from (Roy et al., 2010). (B) Representative binding curves used to determine the Kd values by fluorescence spectroscopy. (C) Superimposed 1H,15N HSQC spectra of 53BP1 TTD recorded as p53K381acK382me2 peptide was titrated in. Spectra are color coded according to the protein:peptide molar ratio. (D) The normalized chemical shift changes in 1H,15N HSQC spectra of TTD induced by p53K381acK382me2 peptide as a function of residue. Residues showing large chemical shift differences are labeled. Differences greater than the average plus one standard deviation (SD), the average plus one-half SD, and the average are shown in red, orange and yellow, respectively, see also Figures S2 and S3.
To obtain molecular insight into the association with the dual PTM, we co-crystallized the TTD with p53K381acK382me2 peptide and determined a 1.8 Å resolution structure of the complex (Figure 4 and Table 1). The structure reveals a canonical TTD fold containing a pair of β-barrels closely coupled through a C-terminal α-helix (Figure 4A and B). The p53K381acK382me2 peptide folds in an α-helix and is positioned near the top of the first β-barrel. The α-helix is stabilized through the characteristic pattern of backbone carbonyl-amide hydrogen bonds involving R379, K381ac, K382me2, L383 and F385 residues of the peptide.
Figure 4. The crystal structure of the 53BP1 TTD-p53K381acK382me2 complex.
(A, B) TTD is shown as a ribbon diagram (A) and a solid surface (B) with the peptide depicted as a yellow ribbon. Residues of TTD forming the binding sites for R379, K381ac and K382me2 of the p53 peptide are colored green, salmon and blue, respectively. (C) A close view of the R379-, K381ac- and K382me2-binding pockets. Dashed lines represent intermolecular hydrogen bonds. (D) Structural overlay of the p53K381acK382me2 peptide (yellow) bound to TTD and p53K382me2 peptide (gray) bound to TTD (described in the accompanying paper). TTD is not depicted for clarity.
Table1.
Data collection and refinement statistics for the 53BP1 TTD-p53K381acK382me2 complex
| TTD-p53K381acK382me2 | ||
|---|---|---|
| Data collection | ||
| Wavelength(Å) | 1.000 | |
| Space group | C2221 | |
| Resolution (Å) | 50–1.8(1.84–1.80) | |
| Cell dimensions (Å) | a=58.89 b=110.94 c=96.92 | α=β=γ=90° |
| Number of measured reflections | 616,822 | |
| Number of unique reflections | 29571 | |
| Completeness(%) | 100(99.8) | |
| Redundancy | 7.3(6.2) | |
| I/sigma (I) | 25.7 (2.4) | |
| Rmerge(%) | 7.7(64.1) | |
| Refinement | ||
| Resolution (Å) | 48.5–1.8(1.87–1.8) | |
| Number of reflections | 28055 | |
| R-factor (%) | 18.68(25.5) | |
| R-free (%) | 21.84(30.0) | |
| Number of protein atoms | 2085 | |
| Number of heterogen atoms | 270 | |
| Number of water molecules | 270 | |
| R.m.s.d. from ideal values | ||
| Bond lengths (Å) | 0.007 | |
| Bond angles (°) | 1.156 | |
| Average B-values (Å2) | ||
| Protein chain A / B | 20.6/29.9 | |
| Peptide chain C / D | 28.9/56.6 | |
| Water | 34.0 | |
| Ramachandran Plot analysis | ||
| Residues in most favored regions | 92.7% | |
| Residues in additional allowed regions | 7.3% | |
| Residues in generously allowed regions | 0 | |
| Residues in disallowed regions | 0 | |
Values in parentheses refer to data in the highest resolution shell
R-free is calculated basing on 5% of the reflections
The α-helical structure of the p53K381acK382me2 peptide places R379, K381ac and K382me2 on the same face of the α-helix, thereby allowing the side chains of these residues to interact with the TTD simultaneously in a tentacle-like manner. The three side chains adopt an extended conformation and are inserted deeply into separate binding pockets (Figure 4B and C). The guanidinium moiety of R379 is restrained by two hydrogen bonds, one formed with the carboxylic group of E1551 and another formed with the carboxylic group of D1521. The dimethylated K382 occupies an aromatic pocket consisting of four aromatic residues and an aspartate. The aromatic rings of W1495, Y1502, F1519 and Y1523 are arranged almost perpendicular to each other and are involved in cation-π interactions with the positively charged dimethylammonium group of K382. The hydrogen bond seen between this group and the carboxylate of D1521 is likely to account for the preference of TTD for the dimethyllysine substrate. The neutral side chain of K381ac is bound in an adjacent aromatic/hydrophobic pocket formed by Y1500, F1553 and I1587. The side chain amide of K381ac is hydrogen bonded to the hydroxyl group of Y1502. Additionally, the N-terminus of the peptide is restrained through the transient polar interaction between the hydroxyl group of S378 and the carboxyl of E1551.
We compared the structures of TTD bound to the p53K381acK382me2 and p53K382me2 peptides and found that 53BP1 uses different mechanisms to recognize each of these PTM-containing p53 sequences (Figure 4D). In the p53K382me2 complex, the p53 peptide is bound in a U-shape conformation with its N-terminus positioned near the rim of the second β-barrel and the C-terminus traced between the β-barrels. In contrast, the p53K381acK382me2 sequence is bound in a different orientation and lays across the open end of the first β-barrel. Apart from recognition of the dimethylammonium group of K382 through the aromatic cage, no similarity in coordination of any other p53 residue is seen in the p53K381acK382me2-TTD and p53K382me2-TTD complexes. The fact that K381acK382me2 induces such a dramatic conformational change in the p53 CTD structure, transforming a loop into an α-helix, suggests a PTM-driven conformation switch that may regulate p53.
Molecular mechanism of the p53K381acK382me2-TTD interaction
To assess the roles of the separate binding pockets in recognition of the K381ac and K382me2 modifications, we tested interactions of the TTD by NMR and tryptophan fluorescence. Large chemical shift changes in 1H,15N HSQC spectra of 15N-labeled TTD were observed upon addition of p53K381acK382me2 peptide (Figure 3C). Plotting chemical shift changes for each backbone amide of TTD allowed us to identify the protein regions that were most affected due to binding in solution (Figure 3D). In agreement with the crystal structure of the complex, residues of the K382me2-binding aromatic cage, such as W1495, Y1523 and D1521, and adjacent residues were affected to the highest degree. Resonances of the K381ac-binding pocket residues, Y1500 and F1553, were also substantially perturbed.
A close comparison of chemical shift perturbations in the TTD upon binding to p53K381acK382me2 and p53K382me2 revealed that both peptides induce the most significant changes in the K382me2-binding cage (Figure 3D). The essential role of the cage residues was substantiated through mutational analysis. Substitution of Y1502 with alanine reduced binding of TTD to p53K381acK382me2 ~14-fold, whereas mutation of W1495 to alanine completely abrogated this interaction (Figure 3A and Suppl. Figure S2).
NMR resonances of residues in the second β-barrel, including the K381ac-binding site residue F1553, were less perturbed upon binding to p53K381acK382me2 as compared with binding to p53K382me2. By contrast, another K381ac-binding site residue, Y1500, was perturbed to a greater extent upon titration with p53K381acK382me2. Replacement of Y1500 and F1553 individually or in combination weakened the interaction with p53K381acK382me2 by ~6-, ~8- and ~37-fold, respectively, corroborating the significance of the K381ac-binding site (Figure 3A). We note that in the absence of the K382me2 mark, the wild type TTD associates with p53K381ac peptide very weakly (Kd >5 mM) (Figure S3).
The carboxyl group of D1521 forms hydrogen bonds with K382me2 and R379 of the p53K381acK382me2 peptide, tightly locking the side chains of both p53 residues. When D1521 was mutated to a tyrosine to preserve the K382me2-binding aromatic cage but eliminate contact with R379, the binding affinity of TTD decreased ~9 fold. Alanine substitution of D1521 led to a ~83-fold reduction in affinity for p53K381acK382me2, implying that the D1521A mutant is unable to make favorable contacts with either K382me2 or R379 of the peptide. Together, these data indicate that proper coordination of not only K382me2 but also K381ac and R379 is necessary for strong binding of TTD.
Acetyl-methyllysine switches mediate binding of the 53BP1 TTD
Tip60-dependent acetylation of K16 of histone H4 has been shown to reduce binding of the 53BP1 TTD to H4K20me2, whereas H4 deacetylation facilities 53BP1 foci formation (Hsiao and Mizzen, 2013; Tang et al., 2013). To compare the acetyl-methyllysine switches in histone H4 and p53, we superimposed structures of the TTD bound to p53K381acK382me2 and H4K20me2 peptides (Tang et al., 2013) (Figure 5A). The overlay reveals distinct mechanisms for the recognition of these sequences. Unlike the p53 peptide, which interacts primarily with the first Tudor of TTD, the histone peptide forms a long loop that spreads over both Tudors. 1H,15N HSQC titration experiments confirmed the more extensive binding site for the H4K20me2 peptide, with residues in the second Tudor exhibiting substantial chemical shift changes upon binding to H4K20me2 (Figure 5B). Furthermore, K16 in the TTD-H4K20me2 complex occupies a binding pocket positioned far from the binding pocket for acetylated K381 of p53, suggesting that the histone and p53 acetyl-methyllysine switches can modulate distinct 53BP1 activities and could also be selectively targeted. The rigid structure of p53K381acK382me2 particularly allows for modular design of small molecules to selectively disrupt the interaction. We employed fixed distances between αC atoms of R379, K381ac and K382me2 and the helical turn as restrains to develop three-point pharmacophores with high complementarity in charge, shape, and hydrophobicity to the p53 peptide (see Suppl. Info). While the lead compound pyrrolinoquinolinol (PQ) binds to the TTD more weakly than does the p53 peptide, at high concentrations it can displace the peptide, and the pattern of chemical shift perturbations indicates that it occupies the same binding pocket (Figure 5C and D and Figures S4 and S5). We expect to increase affinity through mitigating the entropic factor by incorporating shorter tethers in the succeeding generation of dual PTM mimetics.
Figure 5. Comparison of the acetyl-methyllysine switches.
(A) Superimposition of the structures of the TTD in complex with the p53K381acK382me2 (yellow) and H4K20me2 (dark red) (PDB: 2LVM) peptides. The TTD from the p53K381acK382me2 complex is shown with the K381ac- and K382me2-binding pockets colored as in Figure 4. The TTD from the H4K20me2 complex is omitted for clarity. (B) Normalized chemical shift changes observed in 1H,15N HSQC spectra of TTD after addition of the H4K20me2 peptide. The TTD:peptide ratio is 1:10. (C) 1H,15N HSQC overlays of TTD in the free state (black) and in the presence of 2 mM of PQ (pink). Residues that exhibit significant PQ-induced resonance perturbations in (C) are mapped onto the surface of the 53BP1 TTD and labeled. The PQ compound is shown as brown sticks and is docked to TTD. PQ synthesis is described in Figure S4, see also Figure S5.
Phosphorylation of p53 affects binding of 53BP1 TTD
The p53 sequence around K382 contains a number of PTMs that are proposed to modulate binding of co-factors and/or prevent deposition of other PTMs. These include phosphorylation of T377 and S378 and acetylation of K386. To determine whether the neighboring PTMs affect binding of 53BP1 to p53K382me2, we tested a GST-tagged TTD construct using a biotinylated p53 peptide microarray (Table S1). As shown in Figure 6, the GST-TTD recognizes p53K382me2, p53K381acK382me2 and p53K382me2K386ac peptides, however binds more weakly to p53T377pK382me2 and p53S378pK382me2 peptides. The structure of the TTDp53K381acK382me2 complex provides insight into the inhibitory effects of phosphorylation. The side chain of S378 is fully buried in the groove formed by the D1521, M1584 and E1551 residues of TTD, with the hydroxyl group of S378 donating a transient hydrogen bond to the carboxyl group of E1551 (Figure 6C). Phosphorylation of S378 would eliminate the hydrogen bond as well as increase steric hindrance and repulsion between the negatively charged phosphate group and the two negatively charged carboxyl groups of E1551 and D1521. Phosphorylation of T377 would be unfavorable for similar reasons. Although we did not observe a clear electron density for T377 in the complex, its position adjacent to S378 implies that T377 is bound in the same well-defined groove (Figure 6C). While the contributions of these individual PTMs to the p53 activity is still not fully understood, the peptide microarray results suggest that phosphorylation of T377 and S378 may provide a mechanism for releasing 53BP1 from p53. The peptide microarray also demonstrated that the TTD binds p53K370me2 and p53K372me2 peptides more weakly (Kds of 20 µM) than p53K382me2, corroborating previous observations that 53BP1 TTD selects for dimethylated p53K382 (Roy et al., 2010).
Figure 6. The effect of neighboring PTMs on binding of TTD.
(A) Scatterplot of duplicate peptide microarray binding assays for GST 53BP1-TTD. The correlation coefficient was calculated by linear regression analysis using GraphPad Prism v5, (B) top, Diagram of the p53 CTD and modifications studied. M; methylation, P; phosphorylation, A; acetylation. bottom, Heat maps depicting the results of four replicate array binding experiments are presented as normalized mean intensities on a color scale from 0 (black; undetectable binding) to 1 (yellow; strong binding). (C) The zoom-in view of the S378-binding site.
CONCLUDING REMARKS
Single PTMs of p53 have been shown to have an impact on transcriptional and transcription-independent functions of p53 (Berger, 2010; Chuikov et al., 2004; Huang and Berger, 2008; Huang et al., 2010; Huang et al., 2006; Huang et al., 2007; Kachirskaia et al., 2008; Shi et al., 2007). However, characterization of cross-regulation or combinatorial action of multiple dynamic marks on p53 remains a daunting challenge. Multiple modifications can act synergistically to enhance the affinity and specificity of p53-binding effectors or antagonistically to inhibit interactions. The first evidence of a crosstalk between methylated and acetylated lysine residues in the p53 CTD was reported in 2007. It was found that DNA damage-induced monomethylation of p53K372 promotes acetylation of nearby K373 and K382, and prior acetylation of K373 and K382 inhibits subsequent methylation of p53K372 by SET7/9 (Ivanov et al., 2007; Kurash et al., 2008).
In this work, we examined the relationship between PTMs in the K381K382 region and the surrounding residues of p53. Our results demonstrate that dimethylation of p53K382 increases notably within the first hour after DNA damage and is subject to further regulation, whereas the level of p53K381ac does not change significantly. The dual p53K381acK382me2 mark is robustly recognized by the TTD domain of 53BP1, and weakly by BD of p300. We found that the K381acK382me2 combination of PTMs induces a large conformational change in the p53 CTD. Unlike p53K382me2 peptide, which adopts a U-shape conformation, the p53K381acK382me2 peptide folds into an α-helix when bound to TTD, and the two binding sites in TTD only partially overlap. These data suggest a possible mechanism for the regulation of p53 activity, i.e. different combinations of PTMs could promote distinct conformations of p53. The PTM-triggered changes in the CTD structure can mediate p53 ability to selectively interact with binding partners, facilitating p53 commitment to specific biological processes, such as DNA repair.
EXPERIMENTAL PROCEDURES
Dot blot analysis of antibody specificity
The generation and purification of the rabbit polyclonal antibody specific for p53 dimethylated at K382 was described previously (Kachirskaia et al., 2008). The anti-p53K381ac antibody was obtained from Abcam (ab61241, Abcam, Cambridge, MA). Peptides were dissolved in water to yield 1 or 2 mg/mL stock solutions. The peptides were further diluted into 0.1% BSA in PBS. One microliter of the peptide dilutions were spotted onto a Whatman Protran nitrocellulose membrane (0.45 µm pore size, GE Healthcare Life Sciences, Pittsburgh, PA), allowed to air dry for 1h at room temperature and stored at 4 °C overn ight. Membranes were blocked with 1% BSA in PBS-T (PBS with 0.1% Tween-20) for 1h, incubated with a 1:100 dilution of antip53K382me2 for 1h at 4 °C, then washed 5 times with PBS-T. The membranes were incubated with horse radish peroxidase (HRP)-conjugated goat anti-rabbit secondary antibody (Thermo Scientific, Rockford, IL) for 1h at room temperature with shaking, then washed 5 times with PBS-T. Images were developed with ECL Plus substrate (Thermo Scientific, Rockford, IL) and exposed to Hyperfilm ECL (GE Healthcare Life Sciences, Pittsburgh, PA) or imaged using a ChemiDoc MP imaging system (BioRad).
Cell culture, immunoprecipitation and immunoblotting
The U2OS cell line was cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum. Sub-confluent cultures were used for experiments. Where indicated, cells were exposed to 5 or 10 Gy of IR from a 137Cs source using a Shepart Mark II irradiator, treated with 0.5 µg/mL doxorubicin (Selleck Chemicals) for 4h or 400 nM TSA (Cell Signaling Technologies) for 1h. For cells treated with both agents, TSA was added 3 h after addition of doxorubicin and cells were harvested 1 h later. Cells were rinsed with ice-cold PBS and harvested by scraping, with the addition of protease inhibitors to the last of three ice-cold PBS rinses. Protein extracts were prepared by suspending cell pellets in lysis buffer (50 mM Tris·HCl (pH 7.4), 250 mM NaCl, 0.5% Triton X-100, 10% glycerol, 1 mM dithiothreitol, 1 mM phenylmethanesulfonylfluoride (Sigma), protease inhibitors (Roche), and phosphatase inhibitors), incubating on ice for 20 m, then centrifuged at 14,000 × g at 4 °C for 10 m. Total protein was determined using a modified Bradford method (Pierce). Lysates containing 1 mg of total protein were incubated with anti-p53 DO1-conjugated agarose beads (Santa Cruz Biotechnology) on a rotating mixer overnight at 4 °C. Agarose beads were washed three times with lysis buffer and suspended in 20 µl of 2 × LDS sample buffer (Life Technologies) without reducing agents and heated for 10 min at 70 °C. El uted proteins were separated by denaturing gel electrophoresis and transferred to PVDF membranes. Membranes were blocked as described above and incubated with anti-p53K382me2 overnight at 4 °C. Potential interference by immunoglobulin heavy chains was avoided by use of an anti-rabbit light chain specific secondary antibody (Jackson ImmunoResearch) for detection of p53 K382me2 or horse radish peroxidase-conjugated DO-1 anti-p53 (Santa Cruz Biotechnology) for detection of total p53. Other reagents used include anti-p21 (Santa Cruz Biotechnologies) and anti-p53 Ser15p (Cell Signaling Technologies). Specific bands were detected by Western Lightning (PerkinElmer) or ECL Plus (Thermo Scientific) chemiluminescence reagents.
Protein crystallization and structure determination by X-ray crystallography
53BP1 TTD (15 mg/ml) was mixed with p53K381acK382me2 peptide in a 1:1.5 molar ratio on ice for one hour prior to crystallization. Crystals for crystallographic analysis were obtained at 18°C after extensive screening and optimization using the hanging-drop vapor diffusion method. The optimized condition contained 0.1 M sodium chloride and 4.0 M sodium formate in a 0.1 M Tris-HCl pH 7.5 buffer. The data were collected at 100 K on a "NOIR-1" MBC system detector at beamline 4.2.2 at the Advanced Light Source in Berkeley, CA, integrated and scaled using the HKL2000. The structure was obtained by molecular replacement using the TTD structure (PDB: 3LGL) as a search model. The Refmac, Phenix-refine, and Coot programs were used in the refinement. The crystallographic statistics are shown in Table 1.
Supplementary Material
HIGLIGHTS.
Dimethylated K382 of p53 is an early DNA damage response PTM
Dual modification p53K381acK382me2 is recognized by the TTD of 53BP1
The acetyllysine-methyllysine PTM induces substantial conformational changes in p53
PTM-driven conformation switch-like mechanism may regulate p53 interactions
ACKNOWLEDGMENTS
We thank Siddhartha Roy and Catherine Musselman for help with experiments and Jay Nix at beam line 4.2.2 of the ALS in Berkeley for help with X-ray crystallographic data collection. We thank the ENCODE Consortium and the ENCODE production laboratories (Ren Lab-LICRSCSD) that generated datasets used in this study. This research is supported by grants from the NIH, GM101664 (T.G.K.), HL65440 (M.G.), T32CA009657 (H.R-A), GM110058 (B.D.S.), CA181343 (S.B.R.), CA132878 (G.M.) and GM093930 (A.G.K.), and, in part, by the Intramural Research Program of the National Cancer Institute, NIH (EA).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
ACCESSION NUMBER
Atomic coordinates for the structure have been deposited in Protein Data Bank under accession code 4X34.
SUPPLEMENTAL INFORMATION
Supplemental Information includes one text file, one table and five figures.
Author contributions: Q.T., S.J.M., H.R-A., S.B.R., D.M.K., G.C., W.H.L., Y.G. and B.J.K. performed experiments and together with L.J., G.M., A.G.K., B.D.S., M.G., E.A. and T.G.K. analyzed the data. T.G.K. wrote the manuscript with input from all authors.
REFERENCES
- Abida WM, Nikolaev A, Zhao W, Zhang W, Gu W. FBXO11 promotes the Neddylation of p53 and inhibits its transcriptional activity. J Biol Chem. 2007;282:1797–1804. doi: 10.1074/jbc.M609001200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlev NA, Liu L, Chehab NH, Mansfield K, Harris KG, Halazonetis TD, Berger SL. Acetylation of p53 activates transcription through recruitment of coactivators/histone acetyltransferases. Mol Cell. 2001;8:1243–1254. doi: 10.1016/s1097-2765(01)00414-2. [DOI] [PubMed] [Google Scholar]
- Batchelor E, Mock CS, Bhan I, Loewer A, Lahav G. Recurrent initiation: a mechanism for triggering p53 pulses in response to DNA damage. Mol Cell. 2008;30:277–289. doi: 10.1016/j.molcel.2008.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger SL. Keeping p53 in check: a high-stakes balancing act. Cell. 2010;142:17–19. doi: 10.1016/j.cell.2010.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botuyan MV, Lee J, Ward IM, Kim JE, Thompson JR, Chen J, Mer G. Structural basis for the methylation state-specific recognition of histone H4-K20 by 53BP1 and Crb2 in DNA repair. Cell. 2006;127:1361–1373. doi: 10.1016/j.cell.2006.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuikov S, Kurash JK, Wilson JR, Xiao B, Justin N, Ivanov GS, McKinney K, Tempst P, Prives C, Gamblin SJ, et al. Regulation of p53 activity through lysine methylation. Nature. 2004;432:353–360. doi: 10.1038/nature03117. [DOI] [PubMed] [Google Scholar]
- Consortium EP, Bernstein BE, Birney E, Dunham I, Green ED, Gunter C, Snyder M. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui G, Park S, Badeaux AI, Kim D, Lee J, Thompson JR, Yan F, Kaneko S, Yuan Z, Botuyan MV, et al. PHF20 is an effector protein of p53 double lysine methylation that stabilizes and activates p53. Nat Struct Mol Biol. 2012;19:916–924. doi: 10.1038/nsmb.2353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai C, Gu W. p53 post-translational modification: deregulated in tumorigenesis. Trends in molecular medicine. 2010;16:528–536. doi: 10.1016/j.molmed.2010.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debnath S, Chatterjee S, Arif M, Kundu TK, Roy S. Peptide-protein interactions suggest that acetylation of lysines 381 and 382 of p53 is important for positive coactivator 4-p53 interaction. J Biol Chem. 2011;286:25076–25087. doi: 10.1074/jbc.M110.205328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeHart CJ, Chahal JS, Flint SJ, Perlman DH. Extensive post-translational modification of active and inactivated forms of endogenous p53. Molecular & cellular proteomics : MCP. 2014;13:1–17. doi: 10.1074/mcp.M113.030254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohmesen C, Koeppel M, Dobbelstein M. Specific inhibition of Mdm2-mediated neddylation by Tip60. Cell Cycle. 2008;7:222–231. doi: 10.4161/cc.7.2.5185. [DOI] [PubMed] [Google Scholar]
- Hsiao KY, Mizzen CA. Histone H4 deacetylation facilitates 53BP1 DNA damage signaling and double-strand break repair. Journal of molecular cell biology. 2013;5:157–165. doi: 10.1093/jmcb/mjs066. [DOI] [PubMed] [Google Scholar]
- Huang J, Berger SL. The emerging field of dynamic lysine methylation of non-histone proteins. Curr Opin Genet Dev. 2008;18:152–158. doi: 10.1016/j.gde.2008.01.012. [DOI] [PubMed] [Google Scholar]
- Huang J, Dorsey J, Chuikov S, Zhang X, Jenuwein T, Reinberg D, Berger SL. G9a and Glp methylate lysine 373 in the tumor suppressor p53. J Biol Chem. 2010;285:9636–9641. doi: 10.1074/jbc.M109.062588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Perez-Burgos L, Placek BJ, Sengupta R, Richter M, Dorsey JA, Kubicek S, Opravil S, Jenuwein T, Berger SL. Repression of p53 activity by Smyd2-mediated methylation. Nature. 2006;444:629–632. doi: 10.1038/nature05287. [DOI] [PubMed] [Google Scholar]
- Huang J, Sengupta R, Espejo AB, Lee MG, Dorsey JA, Richter M, Opravil S, Shiekhattar R, Bedford MT, Jenuwein T, Berger SL. p53 is regulated by the lysine demethylase LSD1. Nature. 2007;449:105–108. doi: 10.1038/nature06092. [DOI] [PubMed] [Google Scholar]
- Huyen Y, Zgheib O, Ditullio RA, Jr, Gorgoulis VG, Zacharatos P, Petty TJ, Sheston EA, Mellert HS, Stavridi ES, Halazonetis TD. Methylated lysine 79 of histone H3 targets 53BP1 to DNA double-strand breaks. Nature. 2004;432:406–411. doi: 10.1038/nature03114. [DOI] [PubMed] [Google Scholar]
- Itahana Y, Ke H, Zhang Y. p53 Oligomerization is essential for its C-terminal lysine acetylation. J Biol Chem. 2009;284:5158–5164. doi: 10.1074/jbc.M805696200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov GS, Ivanova T, Kurash J, Ivanov A, Chuikov S, Gizatullin F, Herrera-Medina EM, Rauscher F, 3rd, Reinberg D, Barlev NA. Methylation-acetylation interplay activates p53 in response to DNA damage. Mol Cell Biol. 2007;27:6756–6769. doi: 10.1128/MCB.00460-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kachirskaia I, Shi X, Yamaguchi H, Tanoue K, Wen H, Wang EW, Appella E, Gozani O. Role for 53BP1 Tudor domain recognition of p53 dimethylated at lysine 382 in DNA damage signaling. J Biol Chem. 2008;283:34660–34666. doi: 10.1074/jbc.M806020200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi Y, Ito A, Appella E, Yao TP. Charge modification at multiple C-terminal lysine residues regulates p53 oligomerization and its nucleus-cytoplasm trafficking. J Biol Chem. 2006;281:1394–1400. doi: 10.1074/jbc.M505772200. [DOI] [PubMed] [Google Scholar]
- Kurash JK, Lei H, Shen Q, Marston WL, Granda BW, Fan H, Wall D, Li E, Gaudet F. Methylation of p53 by Set7/9 mediates p53 acetylation and activity in vivo. Mol Cell. 2008;29:392–400. doi: 10.1016/j.molcel.2007.12.025. [DOI] [PubMed] [Google Scholar]
- Lee KH, Li M, Michalowski AM, Zhang X, Liao H, Chen L, Xu Y, Wu X, Huang J. A genomewide study identifies the Wnt signaling pathway as a major target of p53 in murine embryonic stem cells. Proc Natl Acad Sci U S A. 2010;107:69–74. doi: 10.1073/pnas.0909734107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, He Y, Dubois W, Wu X, Shi J, Huang J. Distinct regulatory mechanisms and functions for p53-activated and p53-repressed DNA damage response genes in embryonic stem cells. Mol Cell. 2012;46:30–42. doi: 10.1016/j.molcel.2012.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewer A, Batchelor E, Gaglia G, Lahav G. Basal dynamics of p53 reveal transcriptionally attenuated pulses in cycling cells. Cell. 2010;142:89–100. doi: 10.1016/j.cell.2010.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J, Li M, Tang Y, Laszkowska M, Roeder RG, Gu W. Acetylation of p53 augments its site-specific DNA binding both in vitro and in vivo. Proc Natl Acad Sci U S A. 2004;101:2259–2264. doi: 10.1073/pnas.0308762101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mujtaba S, He Y, Zeng L, Yan S, Plotnikova O, Sachchidanand, Sanchez R, Zeleznik-Le NJ, Ronai Z, Zhou MM. Structural mechanism of the bromodomain of the coactivator CBP in p53 transcriptional activation. Mol Cell. 2004;13:251–263. doi: 10.1016/s1097-2765(03)00528-8. [DOI] [PubMed] [Google Scholar]
- Ou YH, Chung PH, Sun TP, Shieh SY. p53 C-terminal phosphorylation by CHK1 and CHK2 participates in the regulation of DNA-damage-induced C-terminal acetylation. Mol Biol Cell. 2005;16:1684–1695. doi: 10.1091/mbc.E04-08-0689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy S, Musselman CA, Kachirskaia I, Hayashi R, Glass KC, Nix JC, Gozani O, Appella E, Kutateladze TG. Structural insight into p53 recognition by the 53BP1 tandem Tudor domain. J Mol Biol. 2010;398:489–496. doi: 10.1016/j.jmb.2010.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi X, Kachirskaia I, Yamaguchi H, West LE, Wen H, Wang EW, Dutta S, Appella E, Gozani O. Modulation of p53 function by SET8-mediated methylation at lysine 382. Mol Cell. 2007;27:636–646. doi: 10.1016/j.molcel.2007.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J, Cho NW, Cui G, Manion EM, Shanbhag NM, Botuyan MV, Mer G, Greenberg RA. Acetylation limits 53BP1 association with damaged chromatin to promote homologous recombination. Nat Struct Mol Biol. 2013;20:317–325. doi: 10.1038/nsmb.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y, Zhao W, Chen Y, Zhao Y, Gu W. Acetylation is indispensable for p53 activation. Cell. 2008;133:612–626. doi: 10.1016/j.cell.2008.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West LE, Roy S, Lachmi-Weiner K, Hayashi R, Shi X, Appella E, Kutateladze TG, Gozani O. The MBT repeats of L3MBTL1 link SET8-mediated p53 methylation at lysine 382 to target gene repression. J Biol Chem. 2010;285:37725–37732. doi: 10.1074/jbc.M110.139527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi H, Woods NT, Piluso LG, Lee HH, Chen J, Bhalla KN, Monteiro A, Liu X, Hung MC, Wang HG. p53 acetylation is crucial for its transcription-independent proapoptotic functions. J Biol Chem. 2009;284:11171–11183. doi: 10.1074/jbc.M809268200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng L, Zhang Q, Gerona-Navarro G, Moshkina N, Zhou MM. Structural basis of site-specific histone recognition by the bromodomains of human coactivators PCAF and CBP/p300. Structure. 2008;16:643–652. doi: 10.1016/j.str.2008.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.