Abstract
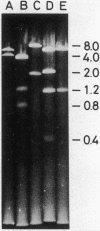
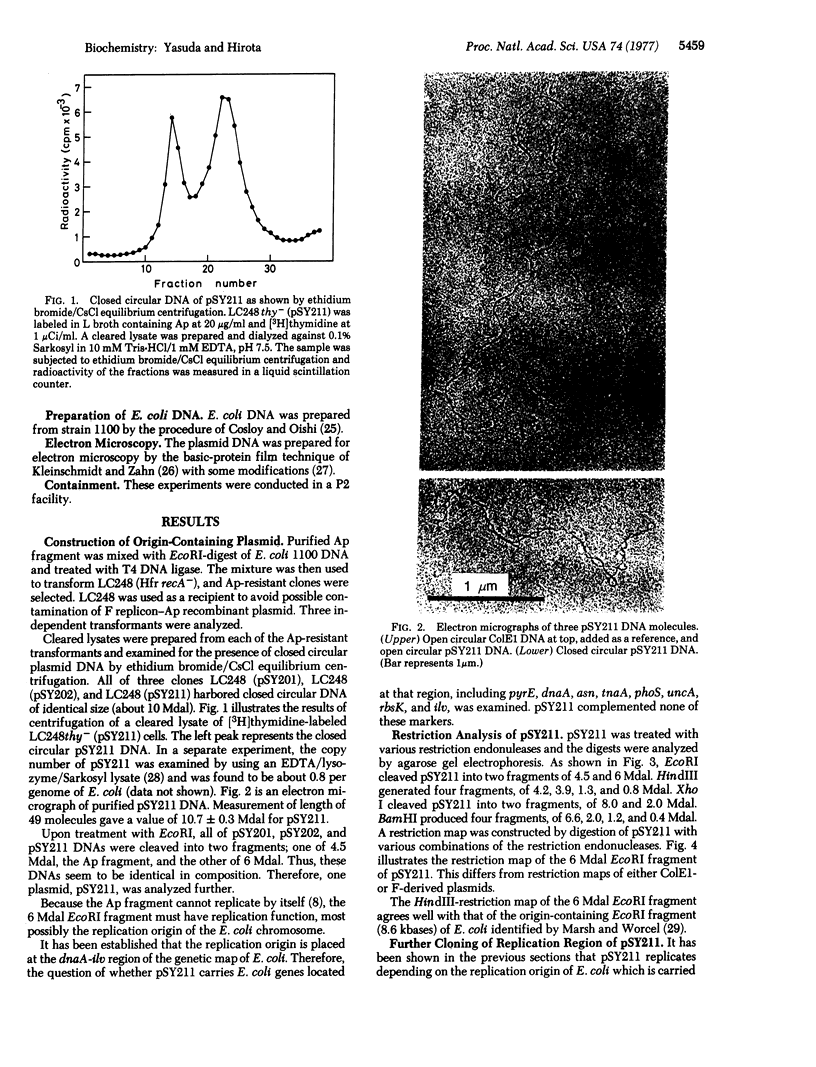
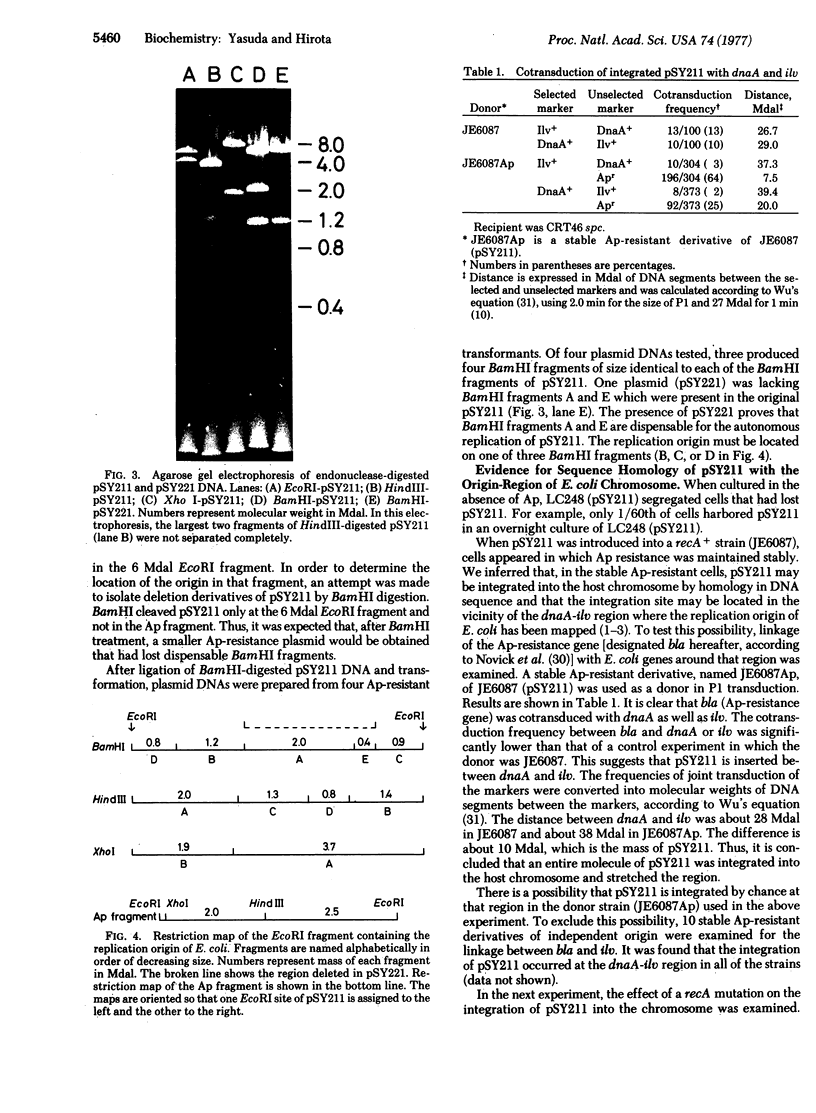
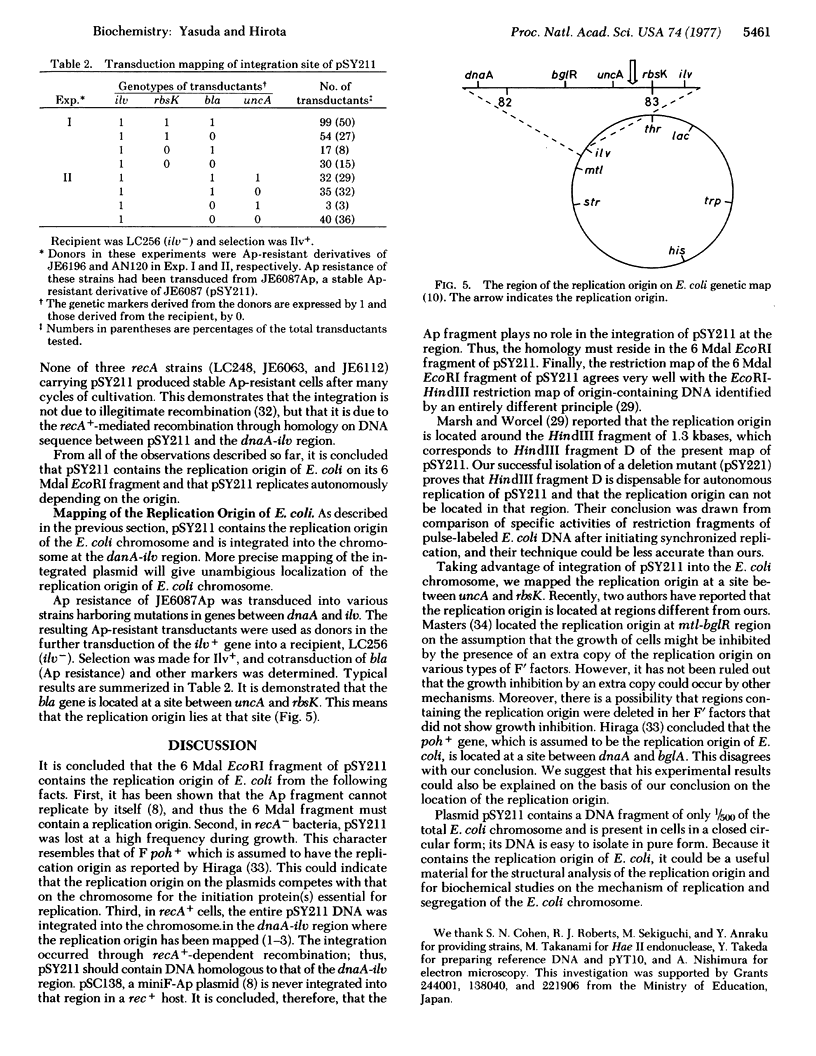
The replication origin of Escherichia coli has been cloned on a nonreplicating DNA fragment coding for ampicillin resistance. This recombinant DNA, named pSY211, replicates depending on the presence of the replication origin and can be recovered as a closed circular plasmid DNA of 10.7 megadaltons (Mdal). A restriction map has been constructed. EcoRI cleaves pSY211 into two fragments: one is the ampicillin fragment of 4.5 Mdal and the other is a chromosomal fragment of 6 Mdal and contains the origin. The 6 Mdal EcoRI fragment has four BamHI sites, three HindIII sites, and one Xho I site. A mutant of pSY211 has been isolated which is lacking two BamHI fragments of the chromosomal fragment. In recA hosts, pSY211 is lost at a high frequency. In recA+ hosts, pSY211 is integrated into the chromosome due to nucleotide sequence homology between pSY211 and the replication origin of the E. coli chromosome. The integration site has been mapped. We conclude that the replication origin is located at a site between uncA and rbsK, at about 83 min on the genetic map of E. coli.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bachmann B. J., Low K. B., Taylor A. L. Recalibrated linkage map of Escherichia coli K-12. Bacteriol Rev. 1976 Mar;40(1):116–167. doi: 10.1128/br.40.1.116-167.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazaral M., Helinski D. R. Circular DNA forms of colicinogenic factors E1, E2 and E3 from Escherichia coli. J Mol Biol. 1968 Sep 14;36(2):185–194. doi: 10.1016/0022-2836(68)90374-4. [DOI] [PubMed] [Google Scholar]
- Bird R. E., Louarn J., Martuscelli J., Caro L. Origin and sequence of chromosome replication in Escherichia coli. J Mol Biol. 1972 Oct 14;70(3):549–566. doi: 10.1016/0022-2836(72)90559-1. [DOI] [PubMed] [Google Scholar]
- Borck K., Beggs J. D., Brammar W. J., Hopkins A. S., Murray N. E. The construction in vitro of transducing derivatives of phage lambda. Mol Gen Genet. 1976 Jul 23;146(2):199–207. doi: 10.1007/BF00268089. [DOI] [PubMed] [Google Scholar]
- Butlin J. D., Cox G. B., Gibson F. Oxidative phosphorylation in Escherichia coli K12. Mutations affecting magnesium ion- or calcium ion-stimulated adenosine triphosphatase. Biochem J. 1971 Aug;124(1):75–81. doi: 10.1042/bj1240075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clewell D. B., Helinski D. R. Properties of a supercoiled deoxyribonucleic acid-protein relaxation complex and strand specificity of the relaxation event. Biochemistry. 1970 Oct 27;9(22):4428–4440. doi: 10.1021/bi00824a026. [DOI] [PubMed] [Google Scholar]
- Cosloy S. D., Oishi M. The nature of the transformation process in Escherichia coli K12. Mol Gen Genet. 1973 Jul 31;124(1):1–10. doi: 10.1007/BF00267159. [DOI] [PubMed] [Google Scholar]
- Covey C., Richardson D., Carbon J. A method for the deletion of restriction sites in bacterial plasmid deoxyribonucleic acid. Mol Gen Genet. 1976 May 7;145(2):155–158. doi: 10.1007/BF00269587. [DOI] [PubMed] [Google Scholar]
- Dürwald H., Hoffmann-Berling H. Endonuclease-I-deficient and ribonuclease I-deficient Escherichia coli mutants. J Mol Biol. 1968 Jul 14;34(2):331–346. doi: 10.1016/0022-2836(68)90257-x. [DOI] [PubMed] [Google Scholar]
- Helling R. B., Goodman H. M., Boyer H. W. Analysis of endonuclease R-EcoRI fragments of DNA from lambdoid bacteriophages and other viruses by agarose-gel electrophoresis. J Virol. 1974 Nov;14(5):1235–1244. doi: 10.1128/jvi.14.5.1235-1244.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmstetter C., Cooper S., Pierucci O., Revelas E. On the bacterial life sequence. Cold Spring Harb Symp Quant Biol. 1968;33:809–822. doi: 10.1101/sqb.1968.033.01.093. [DOI] [PubMed] [Google Scholar]
- Hershfield V., Boyer H. W., Chow L., Helinski D. R. Characterization of a mini-ColC1 plasmid. J Bacteriol. 1976 Apr;126(1):447–453. doi: 10.1128/jb.126.1.447-453.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiraga S. Novel F prime factors able to replicate in Escherichia coli Hfr strains. Proc Natl Acad Sci U S A. 1976 Jan;73(1):198–202. doi: 10.1073/pnas.73.1.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohlfeld R., Vielmetter W. Bidirectional growth of the E. coli chromosome. Nat New Biol. 1973 Apr 4;242(118):130–132. doi: 10.1038/newbio242130a0. [DOI] [PubMed] [Google Scholar]
- LENNOX E. S. Transduction of linked genetic characters of the host by bacteriophage P1. Virology. 1955 Jul;1(2):190–206. doi: 10.1016/0042-6822(55)90016-7. [DOI] [PubMed] [Google Scholar]
- Lederberg E. M., Cohen S. N. Transformation of Salmonella typhimurium by plasmid deoxyribonucleic acid. J Bacteriol. 1974 Sep;119(3):1072–1074. doi: 10.1128/jb.119.3.1072-1074.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louarn J., Funderburgh M., Bird R. E. More precise mapping of the replication origin in Escherichia coli K-12. J Bacteriol. 1974 Oct;120(1):1–5. doi: 10.1128/jb.120.1.1-5.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovett M. A., Helinski D. R. Method for the isolation of the replication region of a bacterial replicon: construction of a mini-F'kn plasmid. J Bacteriol. 1976 Aug;127(2):982–987. doi: 10.1128/jb.127.2.982-987.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh R. C., Worcel A. A DNA fragment containing the origin of replication of the Escherichia coli chromosome. Proc Natl Acad Sci U S A. 1977 Jul;74(7):2720–2724. doi: 10.1073/pnas.74.7.2720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masters M. Strains of Escherichia coli diploid for the chromosomal origin of DNA replication. Mol Gen Genet. 1975 Dec 30;143(1):105–111. doi: 10.1007/BF00269427. [DOI] [PubMed] [Google Scholar]
- Novick R. P., Clowes R. C., Cohen S. N., Curtiss R., 3rd, Datta N., Falkow S. Uniform nomenclature for bacterial plasmids: a proposal. Bacteriol Rev. 1976 Mar;40(1):168–189. doi: 10.1128/br.40.1.168-189.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oka A., Takanami M. Cleavage map of colicin E1 plasmid. Nature. 1976 Nov 11;264(5582):193–196. doi: 10.1038/264193a0. [DOI] [PubMed] [Google Scholar]
- Radloff R., Bauer W., Vinograd J. A dye-buoyant-density method for the detection and isolation of closed circular duplex DNA: the closed circular DNA in HeLa cells. Proc Natl Acad Sci U S A. 1967 May;57(5):1514–1521. doi: 10.1073/pnas.57.5.1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- So M., Gill R., Falkow S. The generation of a ColE1-Apr cloning vehicle which allows detection of inserted DNA. Mol Gen Genet. 1975 Dec 30;142(3):239–249. doi: 10.1007/BF00425649. [DOI] [PubMed] [Google Scholar]
- Sompayrac L., Maaloe O. Autorepressor model for control of DNA replication. Nat New Biol. 1973 Jan 31;241(109):133–135. doi: 10.1038/newbio241133a0. [DOI] [PubMed] [Google Scholar]
- Timmis K., Cabello F., Cohen S. N. Cloning, isolation, and characterization of replication regions of complex plasmid genomes. Proc Natl Acad Sci U S A. 1975 Jun;72(6):2242–2246. doi: 10.1073/pnas.72.6.2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vapnek D., Rupp W. D. Identification of individual sex-factor DNA strands and their replication during conjugation in thermosensitive DNA mutants of Escherichia coli. J Mol Biol. 1971 Sep 28;60(3):413–424. doi: 10.1016/0022-2836(71)90178-1. [DOI] [PubMed] [Google Scholar]
- Wu T. T. A model for three-point analysis of random general transduction. Genetics. 1966 Aug;54(2):405–410. doi: 10.1093/genetics/54.2.405. [DOI] [PMC free article] [PubMed] [Google Scholar]