Abstract
Brainbow is a genetic cell-labeling technique where hundreds of different hues can be generated by stochastic and combinatorial expression of a few spectrally distinct fluorescent proteins. Unique color profiles can be used as cellular identification tags for multiple applications such as tracing axons through the nervous system, following individual cells during development, or analyzing cell lineage. In recent years, Brainbow and other combinatorial expression strategies have expanded from the mouse nervous system to other model organisms and a wide variety of tissues. Particularly exciting is the application of Brainbow in lineage tracing, where this technique has been instrumental in parsing out complex cellular relationships during organogenesis. Here we review recent findings, new technical improvements, and exciting potential genetic and genomic applications for harnessing this colorful technique in anatomical, developmental, and genetic studies.
Keywords: in vivo imaging, lineage tracing, neural circuitry, clonal analysis, fluorescence microscopy
VISION is arguably the most powerful sensory system in humans. Complex quantitative information portrayed in a visual display is made understandable to the brain by a highly precise visual system, which is accustomed to processing multivariate information present throughout an extremely complex visual field from moment to moment. Visualization tools are therefore particularly useful in the study of dynamic biological systems. In the developing embryo or regenerating tissues for example, cells proliferate, differentiate, and disperse into mature positions. In the nervous system, neurons form complex networks, with thousands of connections potentially overlapping within a small volume (Lichtman and Denk 2011). Analyzing the structure of one of these complex systems through time and/or space is challenging, if not impossible, without a powerful approach for distinguishing among many different individual cellular components. Perhaps the most useful visual modality for tracking gene function and individual cell behavior within these contexts is color.
Following the isolation of green fluorescent protein (GFP) from Aequorea victoria in 1962 (Shimomura et al. 1962), fluorescent proteins have been utilized in a wide array of biological systems to label tissues, cells, organelles, or individual proteins (pioneered by Chalfie et al. 1994). Modifications to GFP have changed its excitation and emission spectra such that new colors could be added to the biological fluorescence palette (e.g., Tsien 1998; Campbell et al. 2002; Shaner et al. 2005; Ai et al. 2007; Goedhart et al. 2012), while unique fluorescent proteins have been identified in other organisms (Matz et al. 1999; Shaner et al. 2004, 2007; Merzlyak et al. 2007). These developments have allowed for genetic targeting of multiple fluorescent proteins (FPs) to visualize different cell types or proteins that interact with one another.
A major limitation in labeling studies has been that cells belonging to one cell type (as defined by a common gene expression pattern) are typically labeled by the same color. Since like cells are often in close proximity with one another, it is difficult to resolve morphology or movement of individual cells. In anatomically complex tissue such as the nervous system, tracking cellular movement and neuronal connections is particularly challenging. This problem can be solved in part by labeling very sparsely (e.g., Luskin et al. 1988; Walsh and Cepko 1988; Lee and Luo 2001; Noctor et al. 2001; Zong et al. 2005), but the scarcity of labeled cells makes it difficult to study interactions between cells or neurites (Luo 2007; Jefferis and Livet 2012). The lack of unique cellular identifiers is also limiting for lineage tracing in developmental studies, which depends on the ability to assign large pool of cells to a common progenitor. As a potential solution to these difficulties, the Brainbow multicolor labeling approach was designed and implemented to generate a wide array of fluorescent colors that serve as unique identification tags in living cells (Livet et al. 2007).
Basic Principle—How to Get Many Colors and What They Mean
The Brainbow strategy capitalizes on the fact that the three primary colors, red, green, and blue, can combine to generate all colors in the visual spectrum. For example, a television screen combines only red (R), green (G), and blue (B) into a multicolor “RGB” display. Brainbow achieves the same effect by combining three or four distinctly colored FPs and expressing them in different ratios within each cell. The resulting color combinations are unique to each Brainbow-expressing cell and can therefore serve as cellular identification tags that can be visualized by the light microscope (Livet et al. 2007; Lichtman et al. 2008).
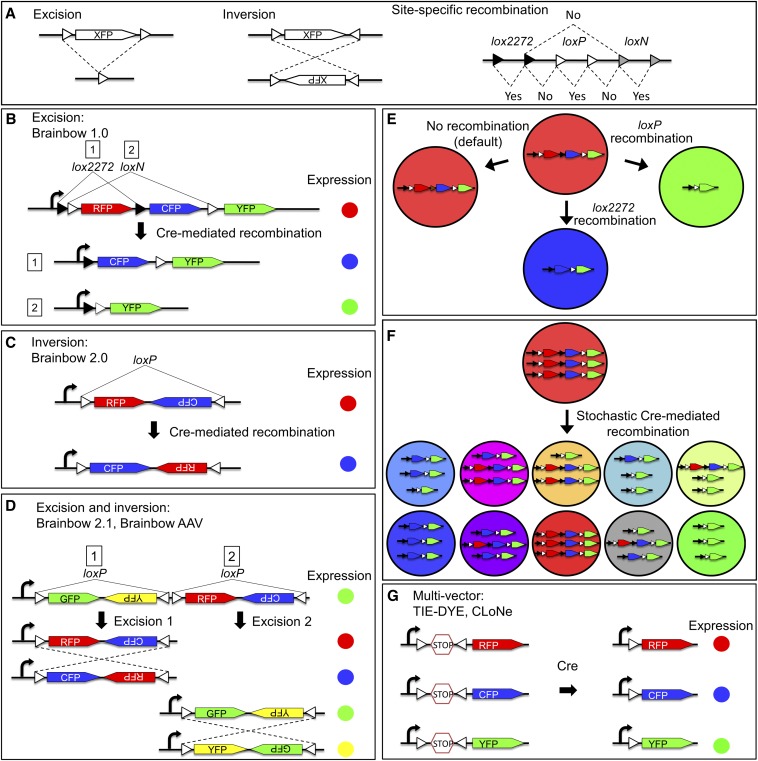
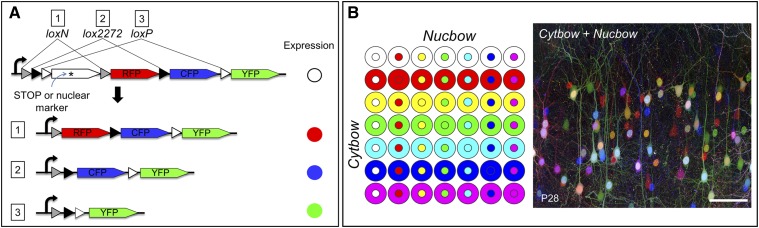
Many different Brainbow and Brainbow-like strategies are now used, utilizing recombinase-mediated DNA excision or DNA inversion (Figure 1A). In DNA excision-based Brainbow (e.g., Brainbow 1.0), three separate FPs are arranged sequentially in the transgene along with two pairs of Cre recombinase recognition sites (Lox sites) that flank the first and second FPs (Figure 1B). The two pairs of Lox sites (loxP and lox2272) can be recognized by Cre only in identical pairs (i.e., loxP with loxP and lox2272 with lox2272). Before recombination, only the first color in the array is expressed (termed the “default” color). Following Cre recombination, one of the three FPs will be exclusively expressed by that copy of the cassette. This strategy can be expanded to four FPs by utilizing a third pair of Lox sites (Livet et al. 2007).
Figure 1.
Principles of Brainbow labeling. (A) Cre recombinase can perform excision or inversion of DNA flanked by Lox sites (triangles), depending on the orientation of the Lox sites. Different lox sites such as lox2272 (black triangle), loxP (white triangle), and loxN (gray triangle) function identically but are incompatible with each other. (B) Excision-based Brainbow. Fluorescent proteins (FPs) are flanked by two pairs of mutually incompatible Lox sites. In the absence of recombination, RFP is expressed. Recombination results in excision expression of either CFP (event 1) or YFP (event 2). (C) Inversion-based Brainbow. FP expression can be changed between RFP and CFP by DNA inversion. (D) In Brainbow 2.1, DNA excision leads to selection of either the GFP/YFP pair or the RFP/CFP pair. DNA inversion then decides which FP of the pair is expressed. Brainbow AAV works similarly. (E) For each copy of Brainbow, only the first FP in the array is expressed. Therefore in a cell population with a single Brainbow transgene, cells can be RFP+ (no recombination, i.e., “default”), CFP+, or YFP+. (F) When multiple copies of Brainbow are present in a cell, each copy recombines independently. Three copies of Brainbow can generate 10 distinct colors and more copies will generate even greater color diversity. (G) Combinatorial multicolor labeling can also be achieved by using multiple vectors, each carrying a single FP. As the expression of each FP is stochastic, the color profile within each cell is different. B and F are modified from Pan et al. (2013).
In DNA inversion-based Brainbow (e.g., Brainbow 2.0; Figure 1C), two matching Lox sites are positioned such that they face each other. Cre inverts (or “flips”) the interspaced DNA as opposed to excising it. In this strategy, two FPs are aligned in head-to-head orientation such that Cre-mediated inversion leads to expression of one of those two colors. By combining excision and inversion, it is also possible to utilize four FPs (e.g., Brainbow 2.1; Figure 1D) (Livet et al. 2007).
Combinatorial expression of multiple FPs requires multiple copies of the Brainbow cassette (Livet et al. 2007; Lichtman et al. 2008). Brainbow is designed to express only one randomly selected FP from each copy of the cassette. For example, if each cell contains only one copy of a three-color construct (e.g., Brainbow 1.0), it would result in a three-color cell population overall (Figure 1E). More complex multicolor expression results when multiple copies of the Brainbow cassette are present in each cell—either via multiple insertions into the genome or through techniques that introduce many copies as extrachromosomal elements (e.g., microinjection, viral transduction, transfection). When more than one copy of the cassette is present in the nucleus, each can act as the generator of a given “pigment” for that cell. Cre acts randomly on each copy, and thus multiple pigments may be present within each cell, and they mix together to create combinatorial hues (Figure 1F). In practice, up to ∼100 colors have been distinguished using various models (Livet et al. 2007; Loulier et al. 2014). The large number of potential colors provides each cell with a specific color barcode and reduces the chance that two cells will randomly become the same color. This is particularly important for cell tracing (where color is used to follow movement or neurites) and lineage analysis (where color is used to distinguish cell populations derived from different progenitors).
In addition to Cre-Lox excision and inversion strategies, other approaches have been developed to create colorful cellular identification tags. One alternative is to use Flp recombinase and FRT recognition sites, which are functionally equivalent to Cre recombinase and Lox sites, respectively (e.g., Flybow and Flpbow) (Hadjieconomou et al. 2011; Cai et al. 2013). Another strategy is to utilize multiple single-FP vectors simultaneously (Figure 1G). Each vector is stochastically expressed to create combinatorial and diverse hues (Boldogkoi et al. 2009; Weber et al. 2011; Worley et al. 2013; Garcia-Marques et al. 2014; Garcia-Moreno et al. 2014). This approach is generally more suitable for somatic labeling, as generating and maintaining transgenic lines carrying multiple single-FP transgenes is more challenging. For example, TIE-DYE in Drosophila requires two balancer chromosomes to maintain four transgenes (see below) (Worley et al. 2013).
The use of multiple colors within one cell population allows for a shift in the types of questions that can be asked using standard reporter constructs. Labeling strategies often use a given promoter to drive one-color expression for all members of that particular cell type, which distinguishes that cell type from others (i.e., cell type 1 is a given color and cell type 2 is a different color). While this strategy is ideal for questions that investigate the behavior of cells at the population level, it essentially homogenizes a given cell population, obscuring differences or interactions between like cells. Brainbow labeling, however, is fundamentally different in that it distinguishes among like cells (i.e., individual cells of a given cell type are now many different colors). This approach allows one to address a very different type of question regarding the function of individual cells within the population (as opposed to population behavior) and is ideal for following individual cells over time and space, as well as for tracing projections in the nervous system. Another important property of Brainbow is that it is a genetic labeling technique, and the result of stochastic DNA recombination is inheritable. Therefore an initial pool of progenitor cells that is labeled in specific colors produces labeled progeny that reflect their cellular lineage (Figure 2). In other words, all cells within a clone will have the same color (Snippert et al. 2010; Hadjieconomou et al. 2011; Hampel et al. 2011; Rinkevich et al. 2011; Blanpain and Simons 2013; Pan et al. 2013; Worley et al. 2013; Loulier et al. 2014). Applications of Brainbow generally make use of colors as cellular identification tags or as markers of parentage (Loulier et al. 2014; Roy et al. 2014). While Brainbow can help to make sense of a densely labeled tissue, it also is useful in sparsely labeled regions where two or more cells need to be distinguished.
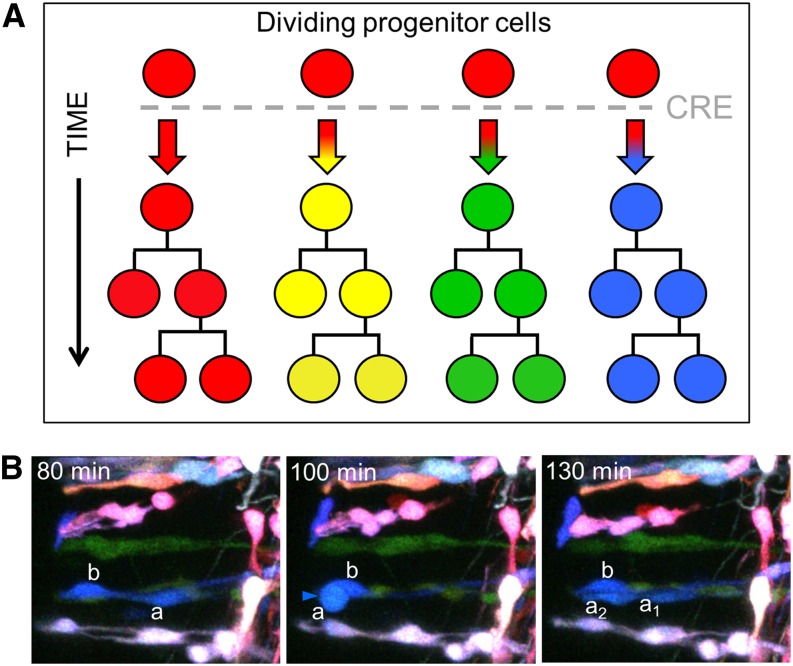
Figure 2.
Brainbow for clonal analysis. (A) A uniform population of dividing progenitor cells becomes multicolor upon Cre recombination. Following recombination, each dividing cell produces progeny that share its unique color, thus color coding its resulting clone. (B) This type of Brainbow labeling was used in vivo to follow dividing radial progenitor cells in the chick spinal cord over time. Over a period of 50 min shown here, one member of the blue clone (cell a) divides, producing two daughter cells (a1 and a2). Panels in B are reprinted from Loulier et al. (2014) with permission from Elsevier.
Brainbow Resources: Mouse, Fly, Fish, and Beyond
One of Brainbow’s strengths is its broad applicability. A number of Brainbow adaptations have been developed in recent years for different tissues and model organisms such as mouse, rat, chick, zebrafish, fruit fly, and plants. These include both germline transgenic approaches and somatic labeling approaches (e.g., nongermline transgenic). We summarize these two approaches below and in Table 1 and Table 2.
Table 1. Transgenic lines.
| Organism | Latin name | Promoter | Transgenic lines |
|---|---|---|---|
| Mouse | Mus musculus | Neuronal | Brainbow 1.0/1.1/2.0/2.1 (Livet et al. 2007), Brainbow 3.0/3.1a/3.2a, Flpbow 1/3a, Autobowb (Cai et al. 2013) |
| Ubiquitous | R26-Confettib (Snippert et al. 2010) | ||
| R26-Rainbow (Rinkevich et al. 2011) | |||
| Rainbow (Tabansky et al. 2013) | |||
| MAGICc (Loulier et al. 2014) | |||
| Ubow (Ghigo et al. 2013) | |||
| Zebrafish | Danio rerio | Gal4 inducible | Brainbow (Robles et al. 2013) |
| Zebrabow (Pan et al. 2013) | |||
| Ubiquitous | PriZm (Gupta and Poss 2012) | ||
| Zebrabow (Pan et al. 2013) | |||
| Fruit fly | Drosophila melanogaster | Gal4 inducible | dBrainbowb (Hampel et al. 2011) |
| Flybow1.0/1.1/2.0a (Hadjieconomou et al. 2011) | |||
| LOLLibow (Boulina et al. 2013) | |||
| Ubiquitous | TIE-DYEb (Worley et al. 2013) | ||
| Plant | Arabidopsis thaliana | Ubiquitous | Brother of Brainbow (Wachsman et al. 2011) |
Default nonfluorescent nuclear marker expression.
No default fluorophore expression in the absence of CRE.
Default nuclear-EBFP2 (equivalent to DAPI-labeling) expression.
Table 2. Somatic expression.
| Transgenesis method | Transgene name | Organism applied | Genome integration | Applications |
|---|---|---|---|---|
| DNA injection in embryo | Brainbow (Pan et al. 2011) | Zebrafish | No | Cell and axon labeling |
| Electroporation | Brainbow (Egawa et al. 2013) | Mouse, chick | Yes (except for Egawa et al. 2013) | Cell and axon labeling, lineage analysis |
| CLoNea (García-Moreno et al. 2014) | ||||
| MAGICb (Loulier et al. 2014) | ||||
| Star Track (García-Marques et al. 2014) | ||||
| Lentivirus | RGB LeGO (Weber et al. 2011) | Mouse, culture cells | Yes | Lineage analysis |
| LeGO with DNA barcode (Cornils et al. 2014) | ||||
| AAV | Brainbow AAVa (Cai et al. 2013) | Mouse | No | Cell and axon labeling |
| Pseudorabies virus | Rainbow PRV (Boldogkoi et al. 2009) | Mouse, rat | No | Axon tracing, brain mapping |
| PRV-263 (Card et al. 2011a,b) |
No default fluorophore expression in the absence of CRE.
Default nuclear-EBFP2 (equivalent to DAPI-labeling) expression.
Germline approaches—Brainbow transgenic lines
Mouse:
A number of transgenic Brainbow mouse lines have been generated, under the control of neuronal (Thy1.2) or ubiquitous promoters (Table 1). Neuronal lines include the original Brainbow lines (Livet et al. 2007), which are now available at The Jackson Laboratory (Bar Harbor, ME). Additional mouse lines were generated recently to expand upon the techniques used in the original neuronal lines (Cai et al. 2013) (see New Improvements to Brainbow). For ubiquitous expression, the R26-Confetti and R26-Rainbow lines were generated by knocking in Brainbow constructs into the ROSA26 locus (Figure 3B) (Red-Horse et al. 2010; Snippert et al. 2010; Rinkevich et al. 2011). The ROSA26 locus is well suited for constitutive and ubiquitous expression, and both lines have been used extensively for lineage studies (R26-Confetti line is available at The Jackson Laboratory). It is worth noting that for these lines, there is only one copy per haploid genome and it is therefore limited to four (R26-Confetti) or three (R26-Rainbow) distinct colors after recombination. Non-knock-in lines include “Rainbow” (Tabansky et al. 2013), “Ubow” (Ghigo et al. 2013), “Cytbow”, and “Nucbow” (Loulier et al. 2014). Transgenesis with pronuclear injection results in single- as well as multicopy genomic insertions and therefore has greater potential for color diversity (Livet et al. 2007).
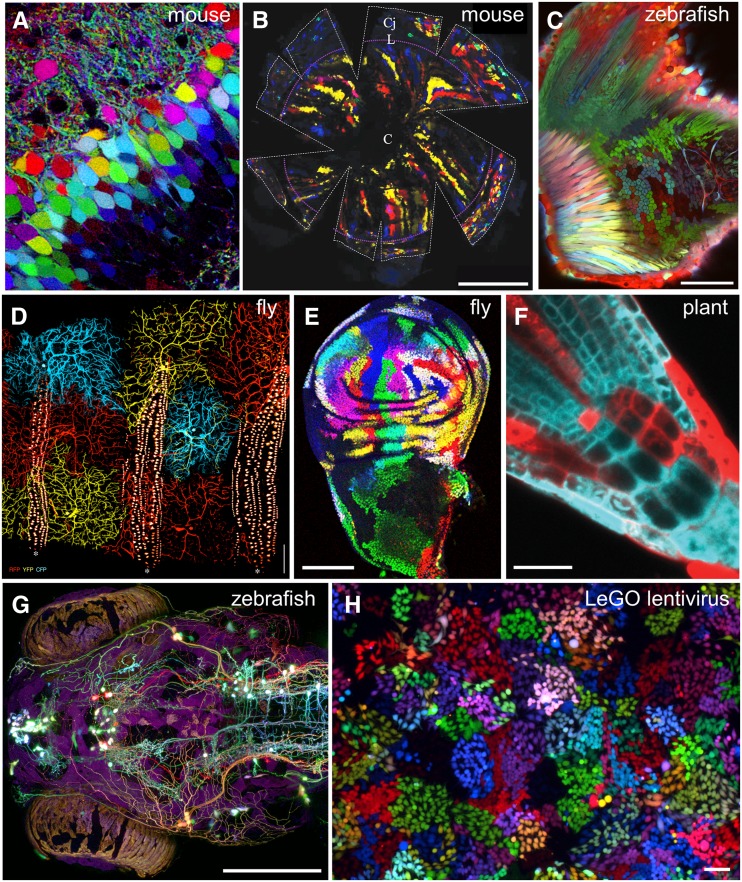
Figure 3.
Brainbow transgenic lines and other approaches. (A) Neurons within the dentate gyrus of the Brainbow mouse hippocampus (line L; Image by T. Weissman and J. Lichtman). (B) Radial clones of cells in the mouse cornea from Di Girolamo et al. (2014), included with permission from Wiley, Copyright © 2014 AlphaMed Press. (C) Pectoral fin in “zebrabow” zebrafish, from Pan et al. (2013). (D) Sensory neurons in the ventrolateral body wall of a Drosophila LOLLIbow larva, adapted from Boulina et al. (2013) with permission from Elsevier. (E) Wing-imaginal disc in TIE-DYE Drosophila, adapted from Worley et al. (2013) with permission from Elsevier. (F) Cells in Arabidopsis thaliana root meristem labeled using the Brother of Brainbow system from Wachsman et al. (2011). Image is copyrighted by the American Society of Plant Biologists and is reprinted with permission. (G) Dorsal view of larval zebrafish injected with Brainbow plasmid DNA. (H) Human embryonic kidney (HEK) cells transduced by LeGO lentivirus. Image is adapted by permission from Macmillan Publishers (Weber et al. 2012). Bars: B, 1 mm; C, E, G, and H, 100 µm; D, 200 µm; and F, 20 µm.
Zebrafish:
In zebrafish, our colleagues and we developed a set of zebrafish Brainbow tools (named “Zebrabow”; Figure 3C) (Pan et al. 2013), which include ubiquitous and Gal4-inducible Brainbow transgenic lines, and established parameters for optimal color diversity. We also showed that the combinatorial color profiles remain constant after cellular growth and division, an important prerequisite for color-based lineage tracing. Additional zebrafish Brainbow lines have been generated by other groups for either broad (“PriZm”) (Gupta and Poss 2012) or Gal4-inducible expression (Robles et al. 2013). The broadly expressed Brainbow lines have been used to follow cell migration, proliferation, and growth of individual clones in the cornea, heart, and brain (Gupta and Poss 2012; Pan et al. 2013; Dirian et al. 2014). The Gal4/UAS system has been particularly useful for tracing densely fasciculated axons in the somatosensory and visual systems (Pan et al. 2011, 2013; Robles et al. 2013). These lines are readily available for public use, and the Zebrabow lines have thus far been distributed to >130 laboratories around the world.
Drosophila:
Several groups have adapted Brainbow 1.0 (Hampel et al. 2011), Brainbow 1.1 (Forster and Luschnig 2012), and Brainbow 2.0 (“Flybow”) (Hadjieconomou et al. 2011) for use with the Gal4-UAS system, enabling expression in specific cell types defined by any Gal4 driver line. Forster and Luschnig (2012), for example, expressed Brainbow in the tracheal tube to reconstruct and quantify the shape and orientation of individual tracheal cells during development, which helped demonstrate a role for the tyrosine kinase Src42A in regulating the expansion of a cylindrical epithelium during development. Boulina et al. (2013) have adapted Brainbow specifically for live imaging in Drosophila, using a photo-inducible form of Cre to activate recombination in vivo (“LOLLIbow”; Figure 3D). Brainbow has been used further in conjunction with the manipulation of gene expression to study gene function. Worley et al. (2013) used a multivector, multicolor approach (“TIE-DYE”; Figure 1G and Figure 3E) to follow multiple cell lineages in the wing imaginal disc, simultaneously interfering with expression of UAS-regulated constructs (e.g., yorkie, cubitus interruptus, or ras) in a subset of labeled cells. Unique color expression in each cell clone allowed for clear visualization of the boundary between mutant and normal cells, revealing differences in cell–cell interactions based on perturbations in gene expression.
Plants:
Brainbow has also been applied for genetic studies in Arabidopsis thaliana, called “Brother of Brainbow” (BOB) (Figure 3F) (Wachsman et al. 2011). One useful feature of this elegant approach is that expression of a gene of interest can be coupled with the default FP (e.g., nuclear YFP), thus extending Brainbow to allow for manipulation and visual determination of gene expression. The authors showed that the retinoblastoma-related (RBR) gene could be coexpressed with BOB, which trans-complements the RBR homozygous mutant background. Cre induction ubiquitously or in cell type-specific manners then results in clones that lose the complementing RBR transgene. This is a powerful approach for testing cell autonomous vs. nonautonomous effects and can also be applied in animals (e.g., Loulier et al. 2014).
Somatic Brainbow labeling approaches
In addition to transgenic lines, Brainbow can be delivered to somatic cells via DNA injection, electroporation, or viral transduction (Table 2). These nongermline approaches can be applied to a wide range of models, allowing for direct cross-species comparisons and applications in organisms for which it is difficult to generate transgenic lines. Furthermore, these methods do not require the time and costs required for generating and maintaining Brainbow transgenic lines.
DNA injection and electroporation:
DNA injection and electroporation are widely used for somatic gene expression and can be applied to most model organisms. DNA (plasmid or BAC) is first injected adjacent to the cells of interest, and then an electrical current is applied to transfer DNA into the cell. DNA can also be injected directly into the cytoplasm of early embryos, as is done in zebrafish (e.g., Pan et al. 2011) (Figure 3G). Brainbow expression with this approach is often very robust and can lead to many color combinations (due to high copy number of the transgene), but diminishes over time due to dilution of nonintegrated transgene through cell divisions. Several recent articles (Garcia-Marques et al. 2014; Garcia-Moreno et al. 2014; Loulier et al. 2014) have overcome this limitation by utilizing genome-integrating transposases such as PiggyBAC and Tol2, allowing long-term cell labeling and lineage tracing. Similarly, transposases also facilitate integration of DNA injected into oocytes (Kawakami 2004; Kikuta and Kawakami 2009).
Viral vectors:
Viral vectors such as lentivirus, adeno-associated virus (AAV), and pseudorabies virus can carry different combinations of FPs to generate diverse color profiles in infected cells (Table 2). These vectors can all infect both dividing and quiescent cells, but have very distinct properties and applications (see below).
Lentivirus:
Lentiviral vectors [e.g., lentiviral gene ontology (LeGO) vectors] are HIV-based, replication-incompetent vectors that have been modified for gene delivery without expression of viral components or alteration of cellular metabolism (Wiznerowicz and Trono 2005). The vectors can be integrated into the host genome for stable expression and are inherited after cell division, making them suitable for clonal studies of tissue regeneration and tumorigenesis. To enable multicolor clonal labeling, Weber et al. (2011) made use of three different-colored LeGO vectors that express FPs in the three primary colors, red, green, and blue (Figure 3H). In this RGB LeGO system, color diversity is generated by stochastic viral insertion and FP expression in each cell. This approach has been applied in vitro and in vivo for tracking transplanted liver, bone, and blood stem cells and tumorigenic cells. Direct injection of RGB LeGO may be a potentially powerful method for in vivo cell labeling for long-term developmental or morphological analysis, although multicolor labeling would be restricted to the injection site, where the viral transduction rate is high (Weber et al. 2012; Gomez-Nicola et al. 2014).
AAV:
AAV is suitable for long-term transgene expression and can be applied to a wide variety of species and tissues. Unlike lentivirus, AAV persists as an extrachromosomal element and does not integrate into the genome, minimizing the threat of mutagenesis. The Brainbow AAV system consists of two types of AAV vectors that, in conjunction, can express four different FPs (Cai et al. 2013). Infection of multiple AAV virions in one cell is very common and results in high diversity of color. In addition, expression of FPs is Cre dependent, allowing cell type-specific Brainbow labeling. This makes it ideal for high-resolution, single-cell anatomical analyses such as defining the connections of genetically defined cell types in the brain (Cai et al. 2013). AAV vectors are not suitable for lineage labeling, however, because AAV episomal DNA is not replicated during mitosis and will therefore be lost after cell division (McCarty et al. 2004).
Pseudorabies virus:
Pseudorabies virus preferentially infects neurons and can spread across synaptic junctions to label both downstream and upstream neurons. It is therefore a powerful tool to trace the functional neuronal connectivity in the developing and mature nervous system. Multicolor pseudorabies virus has been developed to further facilitate visualization of neuronal morphology and map connectivity of intersecting brain pathways (Boldogkoi et al. 2009; Kobiler et al. 2010; Card et al. 2011a, b). Pseudorabies virus is best suited for anatomical studies and short-term neurophysiological studies. It is not suitable for long-term cell labeling, as chronic infection leads to changes to cellular physiology and eventually death.
In summary, both germline and somatic approaches are suitable for short-term cell labeling experiments and long-term lineage analyses, but they have different strengths and weaknesses. Germline approaches have the advantage of consistent transgene copy number and more homogenous expression. It is easier to produce consistent labeling density and color diversity across different animals. In contrast, labeling density and color diversity are often more variable with somatic labeling, which allows for flexibility in terms of titrating each parameter. Brainbow transgene copy number is usually higher at the injection site and decays with distance, resulting in variable color diversity within an injected individual. The primary strength of the somatic approach is speed and flexibility. Brainbow labeling can be directly applied to strains of interest, even in animals where germline transgenics are less common (e.g., rat and chick).
New Improvements to Brainbow
Since its invention in 2007, some limitations of Brainbow have been recognized (Weissman et al. 2011; Cai et al. 2013; Roy et al. 2014), and several groups have modified the original approach to adapt to different species and improve performance. Here we summarize some of the most notable improvements to Brainbow that make multicolor labeling more robust and more easily applicable.
Improving color balance
The first wave of Brainbow constructs work by switching expression from one (default) FP to another (alternative) FP (Figure 1E). This ensures that all cells express at least one FP (default or alternative) for easy screening of transgenic animals. The caveat of this strategy is that color balance is dependent on recombinase activity (Figure 4). Another limitation is the perdurance of the default FP after recombination; when recombination occurs after the onset of Brainbow expression, there is accumulation of the default FP that needs to be degraded for the cell to display its appropriate genome-specified hue. This can be a potential issue for lineage tracing, as color may change over time within the same lineage (Pan et al. 2013; Loulier et al. 2014).
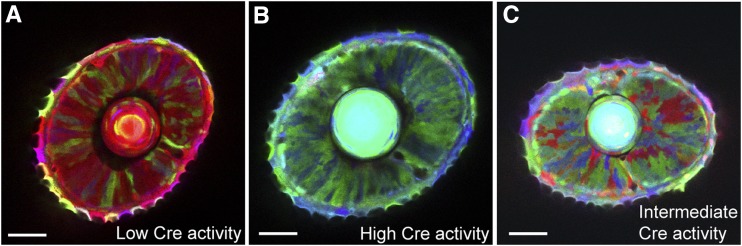
Figure 4.
Tuning Cre activity to maximize color diversity. Images show larval zebrafish eyes expressing Brainbow 1.0 (Zebrabow). (A) When Cre activity is low, most copies of Brainbow express the default FP (RFP). (B) When Cre activity is high, all of the Brainbow copies are recombined, resulting in only the nondefault FPs (CFP and YFP). (C) An intermediate level of Cre activity results in much greater color diversity. Bar, 50 μm. Modified from Pan et al. (2013).
Such limitations can be circumvented by modulating the timing and strength of recombinase activity, but Brainbow without default expression is desirable for lineage tracing or when analyses are done soon after the onset of recombination. Several groups have now generated transgenic lines and somatic labeling tools that drive multicolor labeling in recombined cells while eliminating the default FP expression (with a transcriptional stop signal) or utilize a nuclear localized FP that can be clearly distinguished from the FP expressed after recombination (Figure 5A) (Snippert et al. 2010; Hampel et al. 2011; Cai et al. 2013; Loulier et al. 2014). In these configurations, all alternative FPs have equal chances of being expressed. Constructs with or without default expression are noted in Table 1 and Table 2.
Figure 5.
Improvements in color balance and discrimination. (A) A Brainbow construct with no default FP expression. After recombination, there is an equal probability of expressing RFP, CFP, or YFP, and thus color balance is not dependent on Cre activity. (B) Combining Brainbow constructs with different subcellular localization improves color discrimination. Shown in the left diagram, each cell can express both a nuclear color from Nucbow and a cytoplasmic color from Cytbow. When there are two copies of each, 49 total color combinations can be generated. Fluorescence image on the right shows a mouse cortex labeled by the Cytbow and Nucbow combination. Bar, 100 μm. B was modified from Loulier et al. (2014) with permission from Elsevier.
Antibody amplification
Many commonly used FPs are derived from jellyfish (A. victoria) (e.g., GFP, YFP, BFP, and CFP) (Tsien 1998), coral (Discosoma sp.) (e.g., dsRed, dTomato, mOrange, and mCherry) (Shaner et al. 2004), or anemone (Entacmaea quadricolor) (e.g., TagRFP, TagBFP, and mKate2) (Merzlyak et al. 2007). FPs derived from the same species can have distinct endogenous fluorescence spectra, but are too similar antigenically to be distinguished by antibodies. This poses a challenge for histological analyses, where endogenous fluorescence often becomes too weak after fixation. To overcome this limitation, Hampel et al. (2011) (dBrainbow) added unique epitope tags to each FP, so that fluorescence from each can be independently amplified via antibody labeling. An alternative approach was developed by Cai et al. (2013), which utilizes FPs derived from different species (PhiYFP from Phialidium sp., mOrange from Discosoma sp., GFP from A. victoria, and mKate2 from E. quadricolor) so that each FP can be recognized by antibodies specific to each FP. Both of these approaches allow for the boosting of fluorescence intensity for analysis in fixed tissue.
Improving color discrimination
From 3 to ∼100 colors can be generated by Brainbow (Livet et al. 2007). Dividing a finite color space into increasing numbers of colors, however, requires the investigator to distinguish between closely related hues (e.g., between different shades of yellow). It is therefore of great concern whether perceived differences in color (by eye or digital quantification) represent true differences in cellular identity/lineage or simply experimental variability.
An elegant solution is to increase the dimension of labeling by targeting different FPs to different subcellular compartments (Garcia-Moreno et al. 2014; Loulier et al. 2014). Consider a cell with two copies of cytoplasmic Brainbow (Cytbow) and two copies of nuclear Brainbow (Nucbow). Each copy of Cytbow and Nucbow recombines independently, resulting in 7 possible cytoplasmic and 7 possible nuclear colors (including unlabeled for each) and thus 7 × 7 = 49 different possible color combinations overall (Figure 5B). Moreover, these 49 possibilities would be relatively straightforward to visualize and quantify, because it is technically easier to cluster 7 distinct hues for two different subcellular compartments than to cluster 49 distinct hues in one compartment. Loulier et al. (2014) showed that this approach can be used to distinguish different topological adjacent and intermixed clones. Similar logic potentially can be applied to trace cellular projections or specific structures. For example, axons are robustly labeled when FPs are targeted to the plasma membrane; such labeling could be combined with cytoplasmic- and mitochondria-targeted Brainbow to increase dynamic range and distinguish among axons within a more complex population of projections (e.g., Livet et al. 2007; Cai et al. 2013; Loulier et al. 2014).
Current and Emerging Brainbow Applications
Brainbow has had significant impact on a number of diverse disciplines, including neurobiology, developmental biology, cancer, and stem cell biology. Recent developments in imaging, computation, and genomics can potentially synergize with Brainbow to allow more multifaceted and comprehensive analyses of biological systems. Here we highlight several current applications and discuss emerging avenues for applying Brainbow.
Mapping neuronal connectivity with Brainbow
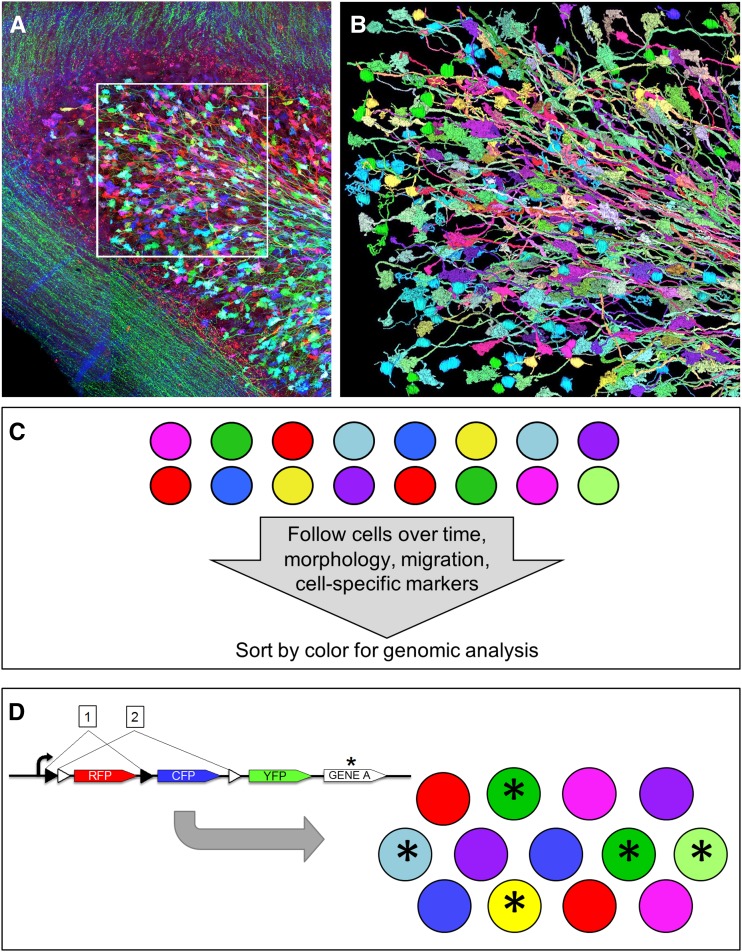
Mapping the connectivity patterns between diverse neuron types within the brain is one of the major challenges in neuroscience. Brainbow’s color diversity provides a unique way to unambiguously trace axons and identify neuronal connections over long distances. Livet et al. (2007) utilized Brainbow to decipher the connectivity between mossy fiber axons (originating in the brainstem and cerebral cortex) and granule neurons within the cerebellum. In total, 341 axons and 93 granule neurons in a small three-dimensional volume (160 μm2 × 65 μm) were digitally reconstructed to demonstrate the convergence of multiple presynaptic neurons onto individual granule cells (Figure 6, A and B). More recently, Kang and Lichtman (2013) used Brainbow expression to distinguish among multiple axons reinnervating the neuromuscular junction following peripheral nerve injury, showing that regenerating axons avoid other axonal branches only if they arise from the same parent neuron. Multicolor tracing with Brainbow has also been applied in Drosophila (Hampel et al. 2011; Hadjieconomou et al. 2011; Boulina et al. 2013), zebrafish (Pan et al. 2011; Heap et al. 2013; Robles et al. 2013), and chick (Egawa et al. 2013). Notably, Egawa et al. (2013) used Brainbow to detect the refinement of multiple inputs into the ciliary ganglion during embryonic chick development, identifying the precise time point suitable for optogenetic manipulation of neural activity.
Figure 6.
Current and emerging applications. (A) A cerebellar folium from the Brainbow mouse line H was imaged using confocal microscopy. Three-dimensional volume (160 μm2 × 65 μm) indicated in the box was segmented using semiautomated methods and reconstructed digitally, as shown in B. (B) Digital reconstruction of 341 axons and 93 granule neurons from volume marked in A. A and B are modified from Livet et al. (2007). (C) Multicolor cells can be followed over time in living tissue and then sorted by color (e.g., FACS) for sequencing or gene expression analysis. (D) In this schematized construct, a particular gene (gene A*) is coexpressed with YFP (following excision at loxP site 2). In the resulting cell population at right, only cells expressing any level of YFP will also express the gene of interest.
The present challenge is to expand brain mapping analyses from small three-dimensional volumes to the whole brain (Lichtman and Denk 2011), and recent technical advancements may pave the way. Notably, computational methods use machine learning and visualization tools for complex brain-wiring data sets (Kim et al. 2014; Oh et al. 2014), tissue-clearing techniques make the brain optically transparent (Dodt et al. 2007; Hama et al. 2011; Chung et al. 2013), and imaging techniques correct for light distortion in thick samples (C. Wang et al. 2014; K. Wang et al. 2014). Coupled with multicolor Brainbow labeling, these methods could allow whole brain neuronal imaging while retaining the ability to identify neurites from many individual cells. This would complement current brain mapping approaches, which either are restricted to small monochromatic volumes (e.g., serial electron microscopy) (Bock et al. 2011; Helmstaedter et al. 2013; Takemura et al. 2013) or do not have single-cell resolution (e.g., anterograde and retrograde viral tracers) (Osten and Margrie 2013). Furthermore, Brainbow-powered light microscopy can be done within intact and living tissues, allowing one to track cellular dynamics and function in real time (e.g., multiple time-point assays and genomic analysis of imaged cells). It will be exciting to see what the field can achieve by combining Brainbow with the rapidly advancing suite of new imaging techniques.
Cellular dynamics and lineage tracing
While Brainbow was originally developed for use in brain mapping and connectivity studies, its application to the field of development has been particularly impactful. During development, orderly proliferation and cellular migration determine tissue size and the correct assembly of component cells. Conventional viral or single FP lineage labeling methods are often unable to determine whether cells are derived from a single clone, especially for cells that undergo extensive migration. Viral vectors with genetic barcodes provide additional proof of lineage, but cannot be utilized when multiple clones are intermingled, restricting labeling to a very low density (one to two clones per animal) (Luskin et al. 1988; Walsh and Cepko 1988).
The diversity of Brainbow color provides an ideal method to unambiguously label multiple clones in close proximity (Figure 2). Multicolor labeling can be targeted to specific cell populations and developmental stages by utilizing a cell-type-specific promoter driving the Brainbow construct, Cre, or driving CreER, a chemically inducible form of Cre. In a series of elegant experiments, Clevers and colleagues combined intestinal stem cell-specific expression of CreER (Lgr5-CreER) and Brainbow (R26-Confetti) to investigate the dynamics of stem cell proliferation and homeostasis within the intestinal crypt (Snippert et al. 2010, 2014; Schepers et al. 2012; Ritsma et al. 2014). In combination with quantitative analysis, these studies suggest that stem cell homeostasis is regulated by neutral competition between dividing stem cells for a spatially limited proliferative niche and that adenoma cells are derived from Lgr5+ intestinal stem cells. Some other notable applications include the analysis of lineage in astrocytes and neurons (Dirian et al. 2014; García-Marques et al. 2014; Garcia-Moreno et al. 2014; Loulier et al. 2014), coronary arteries (Red-Horse et al. 2010), corneal epithelial cells (Pan et al. 2013; Amitai-Lange et al. 2014; Di Girolamo et al. 2014), germline progenitor cells (Zhang et al. 2012; Komai et al. 2014), cleavage stage blastomeres (Tabansky et al. 2013), Langerhans cells (Ghigo et al. 2013), papillae of the tongue (Tanaka et al. 2013), radial glial cells (Pilz et al. 2013), developing nephrons (Barker et al. 2012), hematopoietic cells (Wang et al. 2013), distal digit (Rinkevich et al. 2011), and cardiomyocytes (Gupta and Poss 2012). Rapid developments in high-speed light microscopy such as selective plane illumination microscopy (also called light sheet) (Keller et al. 2008; Amat et al. 2014), as well as multicolor volume microscopy (Mahou et al. 2012, 2014) will further improve the resolution and duration of lineage tracing experiments.
Moving beyond just color—genomic and genetic analyses
Current applications for utilizing Brainbow in anatomical, developmental, or lineage studies to date have focused mostly at the cellular level. The genomic profiles and cellular identities that determine specific neuronal connectivity or clonal behavior largely remain mysteries. We believe future studies will aim to combine in vivo observations with genetic analysis. The study of blood cells has led the way in this respect, where distinctly colored clones can be identified in vivo, isolated by flow cytometry, and then analyzed by RT-PCR and sequencing (Figure 6C). Different approaches would be suited for different tissue types. Less adherent cells, for example, would be more suitable for flow cytometry, whereas large adherent clones would be more suitable for laser capture microdissection. An attractive possibility is to combine multicolor lineage tracing with single-cell sequencing and utilize the relative amounts of different FPs to determine the cellular origin.
Another approach for using Brainbow to manipulate gene activity is to link color to the expression of specific transgenes (Figure 6D) (Wachsman et al. 2011; Worley et al. 2013; Loulier et al. 2014). If a gene is fused with (or expressed in tandem with) one particular FP, then cells with detectable levels of the FP can then be followed to measure the effect of gene expression and to test for cell autonomous vs. noncell autonomous effects. For example, Loulier et al. (2014) designed a Brainbow construct in which one of the FPs (CFP) is coexpressed with a gene that regulates mitotic spindle orientation (dominant-negative LGN). Four days after electroporation of the dominant-negative LGN–Brainbow construct into mouse embryonic cortex, it was found that cells expressing CFP (and thus dnLGN) were less likely than CFP-negative cells to be located in the ventricular zone and more likely to have moved to the cortical plate, consistent with predicted roles for LGN in development (reviewed in Pevre and Morin 2012). Expanding upon this approach, FP intensity could even be quantified to assay dosing and combinatorial effects of gene expression. Theoretically, each FP could be fused with a separate gene (e.g., gene 1 with YFP, gene 2 with RFP, and gene 3 with CFP). In this case, the hue of each cell would represent a specific ratio of gene expression. Such techniques will expand Brainbow’s potential in genetic studies.
Practical Tips for Brainbow Optimization and Analysis
There are several useful step-by-step guides for Brainbow labeling and we encourage readers to consult them for specific hardware requirements and protocols (Pan et al. 2011; Weissman et al. 2011; Mahou et al. 2012; Shimosako et al. 2014). Here we highlight several challenges and key issues that we and other users have encountered.
Maximizing color diversity
Two main factors are important for determining color diversity: (1) the copy number of the Brainbow transgene and (2) the timing and level of Cre activity. In general, as more copies of the Brainbow DNA construct are present in cells, higher expression levels and more color combinations result (mixture of more pigments); however, too many copies per cell can result in reduced color diversity (since all cells have all pigments). For example, Cai et al. (2013) observed reduced color diversity at the Brainbow AAV injection site (likely due to too many copies present in each cell) with maximal color diversity farther away from the injection site (fewer copies per cell). In transgenic lines that have incorporated multiple copies in a tandem array, there is some evidence that recombination between matching pairs of Lox sites extends across insertions, leading to a decrease in copy number and reduced color diversity following Cre activity (Loulier et al. 2014; J. Livet, personal communication). The ideal number of colors is not the same for every experiment. Some studies that investigate regions with many overlapping cells may require a large number of distinct colors, while a handful of colors may be appropriate in studies that consider less dense regions.
The timing and amount of recombination also determine clone size and color balance and need to be titrated appropriately for each system. For Brainbow vectors with default expression (i.e., expression of a FP in the absence of Cre, such as Brainbow 1.0), tunable Cre expression is necessary to adjust color balance (Figure 4). For Brainbow constructs with no default expression (e.g., Brainbow 3.0), color balance is not dependent on the level of Cre activity. Furthermore, the timing of Cre activity determines when progenitor cells are labeled: if recombination occurs very early in a lineage when only a few progenitor cells are present, the entire resulting cell population may inherit only those few same colors. Delaying recombination allows for a larger progenitor pool to recombine separately, generating more colors for the resulting cell population.
Color constancy
Since recombination is random, color will vary from one animal to the next. For a given promoter, however, the same cell populations should be labeled. If individual cells are being followed over time or space, it is crucial to keep constant the image acquisition settings, since small changes from one imaging session to the next can change color appearance significantly. If tracking development, the growth of the organism can send the cells of interest deeper into the tissue, affecting the relative scatter of each FP’s emitted wavelength. In this case, the use of nearby contextual landmarks to ensure cellular identity is useful. If relying only upon color to assign neuronal identity (for example, concluding that a light pink cell body in one part of brain corresponds to a light pink axon in a different region, without tracing it there), a rigorous controlled approach must be used for acquiring images and quantifying color (Livet et al. 2007; Weissman et al. 2011; Cai et al. 2013).
In general, any factor that affects the FPs differentially can lead to an undesired change of a cell’s overall hue. Short wavelengths (e.g., blue light) scatter more readily than longer wavelengths (e.g., red), and thus the same cell may appear different when it is located superficially as opposed to deep (i.e., it may appear more blue toward the surface). Bleaching can also affect each FP to different extents, since each has its own photostability. In general, in vivo preparations are less vulnerable to bleaching of fluorescence expression.
Color quantification
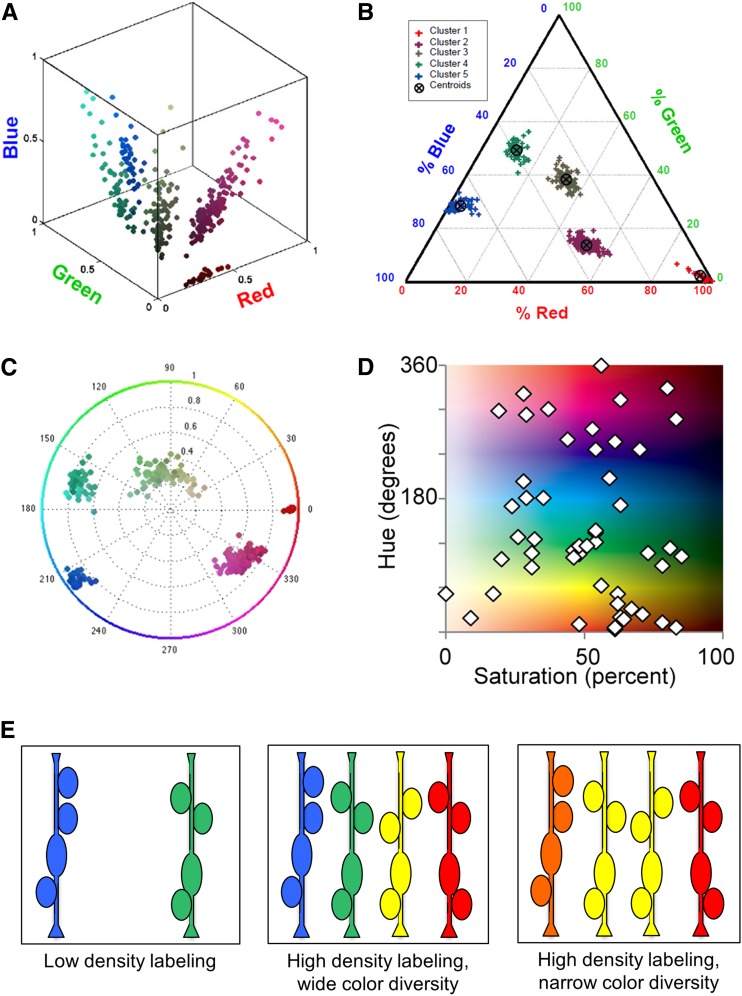
In most cases it is necessary to quantify the diverse colors observed by eye, particularly when conclusions are drawn based on similarity or dissimilarity of color, e.g., assigning lineage relationships, or as readout of gene expression. In image processing software such as Image J and Photoshop, colors are displayed as three channels: red, green, and blue. This is known as the RGB color model. With three-FP Brainbow, RFP is usually designated as red, YFP as green, and CFP as blue. When values from each channel are directly plotted on a three-dimensional graph, there is substantial variability along a diagonal intersecting zero (Figure 7A). This reflects variability in brightness, which depends on cellular topography, imaging depth, and promoter activity. It is therefore preferable to normalize brightness level and measure the relative proportions of each FP. This can be done with a ternary graph that has three axes, each representing the percentage of a color (red, green, or blue; Figure 7B) (Loulier et al. 2014). Another approach is to convert RGB values to hue, saturation, and brightness (also known as the HSB or HSV color model). Color values can be plotted as a two-dimensional hue vs. saturation graph, which is independent of brightness (Figure 7, C and D). Ratiometric color quantification has been extended to up to five FPs (Malide et al. 2012).
Figure 7.
Quantification of color. A–C compare the same data set with different quantification methods. (A) When color is plotted in a three-dimensional RGB graph without correction for brightness, fluorescence values of different cells are widely scattered along the diagonal. (B) When colors are expressed as a ratio along three color axes, color distribution becomes more clustered. (C and D) Color can also be normalized by plotting hue (0°–360°) against saturation (0–100%). This can be displayed either as a circular plot (C) or as an XY distribution (D). A–C are modified from Loulier et al. (2014) with permission from Elsevier. D was modified from Pan et al. (2013). (E) Schematized clones of related cells are shown, labeled at low and high densities. If the average number of cells per clone is four in low-density labeling (left), this average number should also be four in high-density labeling (center). Narrow color diversity (right) could result in labeling of neighboring clones the same hue (two yellow clones in the center). In this case the average clone size would appear larger.
For lineage analysis, cells with the same color are likely derived from the same progenitor. It is important to keep in mind, however, that it is still possible for two cells with a distinct lineage to arrive at the same hue by chance (Figure 7E). Wider color diversity greatly reduces the likelihood that unrelated cells will have the same color, but additional verification is necessary when color combinations are few (e.g., single genomic insertion of Brainbow or suboptimal Cre activity) or when cells from different clones are intermingled (see Blanpain and Simons 2013). One powerful way to test whether single-color cells belong to the same clone is to compare the average number of cells per single-color clone (clone size) at different labeling densities (Figure 7E): if cells with the same color are clonally related, the average clone size should be the same regardless of labeling density, similar to what has been done in retroviral clonal analysis (Galileo et al. 1990).
Conclusions
Brainbow is a tremendously powerful tool for visualizing the dynamics of large numbers of cells, unraveling neural circuits, and piecing together lineage relationships. Emerging applications can now combine visual observations with genetic and genomic analyses. Particularly intriguing are approaches that use color to first visualize and identify a given cellular population and then quantify gene expression in those cells, in addition to approaches that manipulate gene function by linking expression of FPs with given transgenes, thus genetically targeting a subset of clearly identifiable cells.
The Brainbow toolbox has greatly expanded in recent years. A variety of transgenic Brainbow lines are available for use in common model systems such as mouse, Drosophila, and zebrafish. Somatic labeling approaches such as microinjection and viral vectors further extend Brainbow to model systems that are less amenable to transgenesis. New improvements to the original Brainbow constructs have increased its practical use significantly, especially in terms of color detection and discrimination.
While Brainbow initially gained attention for its beauty, it has now proved to be a wellspring of biological insights, for example into neuronal connectivity patterns (Livet et al. 2007; Egawa et al. 2013; Heap et al. 2013; Kang and Lichtman 2013; Robles et al. 2013), dynamics of stem cell proliferation and organ homeostasis (Snippert et al. 2010; Rinkevich et al. 2011; Gupta and Poss 2012; Schepers et al. 2012; Tabansky et al. 2013), and genetic regulation of single cells in vivo (Wachsman et al. 2011; Forster and Luschnig 2012; Loulier et al. 2014). We believe the new resources and applications will make Brainbow an increasingly valuable research tool, and we look forward to seeing exciting (and beautiful) Brainbow genetic studies in the future.
Acknowledgments
We thank Jean Livet, Karine Loulier, Emmanual Beaurepaire, Maria Boulina, Akira Chiba, Nick Di Girolamo, Iswar Hariharan, Jeff Lichtman, Xavier Morin, Kristoffer Riecken, Ben Scheres, Guy Wachsman, and Melanie Worley for sharing their Brainbow images, and Jean Livet and Joshua Sanes for helpful comments on this manuscript. Our work was supported by grant R01HD067140 from the National Institute of Child Health and Development of the National Institutes of Health, as well as by the National Science Foundation and the M. J. Murdock Charitable Trust.
Footnotes
Communicating editor: P. Sengupta
Literature Cited
- Ai H. W., Shaner N. C., Cheng Z., Tsien R. Y., Campbell R. E., 2007. Exploration of new chromophore structures leads to the identification of improved blue fluorescent proteins. Biochemistry 46: 5904–5910. [DOI] [PubMed] [Google Scholar]
- Amat F., Lemon W., Mossing D. P., McDole K., Wan Y., et al. , 2014. Fast, accurate reconstruction of cell lineages from large-scale fluorescence microscopy data. Nat. Methods 11: 951–958. [DOI] [PubMed] [Google Scholar]
- Amitai-Lange A., Altshuler A., Bubley J., Dbayat N., Tiosano B., et al. , 2014. Lineage tracing of stem and progenitor cells of the murine corneal epithelium. Stem Cells 33: 230–239. [DOI] [PubMed] [Google Scholar]
- Barker N., Rookmaaker M. B., Kujala P., Ng A., Leushacke M., et al. , 2012. Lgr5(+ve) stem/progenitor cells contribute to nephron formation during kidney development. Cell Rep. 2: 540–552. [DOI] [PubMed] [Google Scholar]
- Blanpain C., Simons B. D., 2013. Unravelling stem cell dynamics by lineage tracing. Nat. Rev. Mol. Cell Biol. 14: 489–502. [DOI] [PubMed] [Google Scholar]
- Bock D. D., Lee W. C., Kerlin A. M., Andermann M. L., Hood G., et al. , 2011. Network anatomy and in vivo physiology of visual cortical neurons. Nature 471: 177–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boldogkoi Z., Balint K., Awatramani G. B., Balya D., Busskamp V., et al. , 2009. Genetically timed, activity-sensor and rainbow transsynaptic viral tools. Nat. Methods 6: 127–130. [DOI] [PubMed] [Google Scholar]
- Boulina M., Samarajeewa H., Baker J. D., Kim M. D., Chiba A., 2013. Live imaging of multicolor-labeled cells in drosophila. Development 140: 1605–1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai D., Cohen K. B., Luo T., Lichtman J. W., Sanes J. R., 2013. Improved tools for the brainbow toolbox. Nat. Methods 10: 540–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell R. E., Tour O., Palmer A. E., Steinbach P. A., Baird G. S., et al. , 2002. A monomeric red fluorescent protein. Proc. Natl. Acad. Sci. USA 99: 7877–7882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Card J. P., Kobiler O., Ludmir E. B., Desai V., Sved A. F., et al. , 2011a A dual infection pseudorabies virus conditional reporter approach to identify projections to collateralized neurons in complex neural circuits. PLoS ONE 6: e21141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Card J. P., Kobiler O., McCambridge J., Ebdlahad S., Shan Z., et al. , 2011b Microdissection of neural networks by conditional reporter expression from a brainbow herpesvirus. Proc. Natl. Acad. Sci. USA 108: 3377–3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalfie M., Tu Y., Euskirchen G., Ward W. W., Prasher D. C., 1994. Green fluorescent protein as a marker for gene expression. Science 263: 802–805. [DOI] [PubMed] [Google Scholar]
- Chung K., Wallace J., Kim S. Y., Kalyanasundaram S., Andalman A. S., et al. , 2013. Structural and molecular interrogation of intact biological systems. Nature 497: 332–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornils K., Thielecke L., Huser S., Forgber M., Thomaschewski M., et al. , 2014. Multiplexing clonality: combining RGB marking and genetic barcoding. Nucleic Acids Res. 42(7):e56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Girolamo N., Bobba S., Raviraj V., Delic N. C., Slapetova I., et al. , 2014. Tracing the fate of limbal epithelial progenitor cells in the murine cornea. Stem Cells . 10.1002/stem.1769 [DOI] [PubMed] [Google Scholar]
- Dirian L., Galant S., Coolen M., Chen W., Bedu S., et al. , 2014. Spatial regionalization and heterochrony in the formation of adult pallial neural stem cells. Dev. Cell 30: 123–136. [DOI] [PubMed] [Google Scholar]
- Dodt H. U., Leischner U., Schierloh A., Jahrling N., Mauch C. P., et al. , 2007. Ultramicroscopy: three-dimensional visualization of neuronal networks in the whole mouse brain. Nat. Methods 4: 331–336. [DOI] [PubMed] [Google Scholar]
- Egawa R., Hososhima S., Hou X., Katow H., Ishizuka T., et al. , 2013. Optogenetic probing and manipulation of the calyx-type presynaptic terminal in the embryonic chick ciliary ganglion. PLoS ONE 8: e59179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forster D., Luschnig S., 2012. Src42A-dependent polarized cell shape changes mediate epithelial tube elongation in drosophila. Nat. Cell Biol. 14: 526–534. [DOI] [PubMed] [Google Scholar]
- Galileo D. S., Gray G. E., Owens G. C., Majors J., Sanes J. R., 1990. Neurons and glia arise from a common progenitor in chicken optic tectum: demonstration with two retroviruses and cell type-specific antibodies. Proc. Natl. Acad. Sci. USA 87: 458–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Marques J., Nunez-Llaves R., Lopez-Mascaraque L., 2014. NG2-glia from pallial progenitors produce the largest clonal clusters of the brain: time frame of clonal generation in cortex and olfactory bulb. J. Neurosci. 34: 2305–2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Moreno F., Vasistha N. A., Begbie J., Molnar Z., 2014. CLoNe is a new method to target single progenitors and study their progeny in mouse and chick. Development 141: 1589–1598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghigo C., Mondor I., Jorquera A., Nowak J., Wienert S., et al. , 2013. Multicolor fate mapping of Langerhans cell homeostasis. J. Exp. Med. 210: 1657–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goedhart J., von Stetten D., Noirclerc-Savoye M., Lelimousin M., Joosen L., et al. , 2012. Structure-guided evolution of cyan fluorescent proteins towards a quantum yield of 93%. Nat. Commun. 3: 751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Nicola D., Riecken K., Fehse B., Perry V. H., 2014. In-vivo RGB marking and multicolour single-cell tracking in the adult brain. Sci. Rep. 4: 7520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta V., Poss K. D., 2012. Clonally dominant cardiomyocytes direct heart morphogenesis. Nature 484: 479–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadjieconomou D., Rotkopf S., Alexandre C., Bell D. M., Dickson B. J., et al. , 2011. Flybow: genetic multicolor cell labeling for neural circuit analysis in drosophila melanogaster. Nat. Methods 8: 260–266. [DOI] [PubMed] [Google Scholar]
- Hama H., Kurokawa H., Kawano H., Ando R., Shimogori T., et al. , 2011. Scale: a chemical approach for fluorescence imaging and reconstruction of transparent mouse brain. Nat. Neurosci. 14: 1481–1488. [DOI] [PubMed] [Google Scholar]
- Hampel S., Chung P., McKellar C. E., Hall D., Looger L. L., et al. , 2011. Drosophila brainbow: a recombinase-based fluorescence labeling technique to subdivide neural expression patterns. Nat. Methods 8: 253–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heap L. A., Goh C. C., Kassahn K. S., Scott E. K., 2013. Cerebellar output in zebrafish: an analysis of spatial patterns and topography in eurydendroid cell projections. Front. Neural Circuits 7: 53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmstaedter M., Briggman K. L., Turaga S. C., Jain V., Seung H. S., et al. , 2013. Connectomic reconstruction of the inner plexiform layer in the mouse retina. Nature 500: 168–174. [DOI] [PubMed] [Google Scholar]
- Jefferis G. S., Livet J., 2012. Sparse and combinatorial neuron labelling. Curr. Opin. Neurobiol. 22: 101–110. [DOI] [PubMed] [Google Scholar]
- Kang H., Lichtman J. W., 2013. Motor axon regeneration and muscle reinnervation in young adult and aged animals. J. Neurosci. 33: 19480–19491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami K., 2004. Transgenesis and gene trap methods in zebrafish by using the Tol2 transposable element. Methods Cell Biol. 77: 201–222. [DOI] [PubMed] [Google Scholar]
- Keller P. J., Schmidt A. D., Wittbrodt J., Stelzer E. H., 2008. Reconstruction of zebrafish early embryonic development by scanned light sheet microscopy. Science 322: 1065–1069. [DOI] [PubMed] [Google Scholar]
- Kikuta H., Kawakami K., 2009. Transient and stable transgenesis using tol2 transposon vectors. Methods Mol. Biol. 546: 69–84. [DOI] [PubMed] [Google Scholar]
- Kim J. S., Greene M. J., Zlateski A., Lee K., Richardson M., et al. , 2014. Space-time wiring specificity supports direction selectivity in the retina. Nature 509: 331–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobiler O., Lipman Y., Therkelsen K., Daubechies I., Enquist L. W., 2010. Herpesviruses carrying a brainbow cassette reveal replication and expression of limited numbers of incoming genomes. Nat. Commun. 1: 146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komai Y., Tanaka T., Tokuyama Y., Yanai H., Ohe S., et al. , 2014. Bmi1 expression in long-term germ stem cells. Sci. Rep. 4: 6175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T., Luo L., 2001. Mosaic analysis with a repressible cell marker (MARCM) for drosophila neural development. Trends Neurosci. 24: 251–254. [DOI] [PubMed] [Google Scholar]
- Lichtman J. W., Denk W., 2011. The big and the small: challenges of imaging the brain’s circuits. Science 334: 618–623. [DOI] [PubMed] [Google Scholar]
- Lichtman J. W., Livet J., Sanes J. R., 2008. A technicolour approach to the connectome. Nat. Rev. Neurosci. 9: 417–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livet J., Weissman T. A., Kang H., Draft R. W., Lu J., et al. , 2007. Transgenic strategies for combinatorial expression of fluorescent proteins in the nervous system. Nature 450: 56–62. [DOI] [PubMed] [Google Scholar]
- Loulier K., Barry R., Mahou P., Le Franc Y., Supatto W., et al. , 2014. Multiplex cell and lineage tracking with combinatorial labels. Neuron 81: 505–520. [DOI] [PubMed] [Google Scholar]
- Luo L., 2007. Fly MARCM and mouse MADM: genetic methods of labeling and manipulating single neurons. Brain Res. Brain Res. Rev. 55: 220–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luskin M. B., Pearlman A. L., Sanes J. R., 1988. Cell lineage in the cerebral cortex of the mouse studied in vivo and in vitro with a recombinant retrovirus. Neuron 1: 635–647. [DOI] [PubMed] [Google Scholar]
- Mahou P., Zimmerley M., Loulier K., Matho K. S., Labroille G., et al. , 2012. Multicolor two-photon tissue imaging by wavelength mixing. Nat. Methods 9: 815–818. [DOI] [PubMed] [Google Scholar]
- Mahou P., Vermot J., Beaurepaire E., Supatto W., 2014. Multicolor two-photon light-sheet microscopy. Nat. Methods 11: 600–601. [DOI] [PubMed] [Google Scholar]
- Malide D., Metais J. Y., Dunbar C. E., 2012. Dynamic clonal analysis of murine hematopoietic stem and progenitor cells marked by 5 fluorescent proteins using confocal and multiphoton microscopy. Blood 120: e105–e116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matz M. V., Fradkov A. F., Labas Y. A., Savitsky A. P., Zaraisky A. G., et al. , 1999. Fluorescent proteins from nonbioluminescent anthozoa species. Nat. Biotechnol. 17: 969–973. [DOI] [PubMed] [Google Scholar]
- McCarty D. M., Young S. M., Jr, Samulski R. J., 2004. Integration of adeno-associated virus (AAV) and recombinant AAV vectors. Annu. Rev. Genet. 38: 819–845. [DOI] [PubMed] [Google Scholar]
- Merzlyak E. M., Goedhart J., Shcherbo D., Bulina M. E., Shcheglov A. S., et al. , 2007. Bright monomeric red fluorescent protein with an extended fluorescence lifetime. Nat. Methods 4: 555–557. [DOI] [PubMed] [Google Scholar]
- Noctor S. C., Flint A. C., Weissman T. A., Dammerman R. S., Kriegstein A. R., 2001. Neurons derived from radial glial cells establish radial units in neocortex. Nature 409: 714–720. [DOI] [PubMed] [Google Scholar]
- Oh S. W., Harris J. A., Ng L., Winslow B., Cain N., et al. , 2014. A mesoscale connectome of the mouse brain. Nature 508: 207–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osten P., Margrie T. W., 2013. Mapping brain circuitry with a light microscope. Nat. Methods 10: 515–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan, Y. A., J. Livet, J. R. Sanes, J. W. Lichtman and A. F. Schier, 2011 Multicolor brainbow imaging in zebrafish. Cold Spring Harb. Protoc. 2011: pdb.prot5546. [DOI] [PMC free article] [PubMed]
- Pan Y. A., Freundlich T., Weissman T. A., Schoppik D., Wang X. C., et al. , 2013. Zebrabow: multispectral cell labeling for cell tracing and lineage analysis in zebrafish. Development 140: 2835–2846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pevre E., Morin X., 2012. An oblique view on the role of spindle orientation in vertebrate neurogenesis. Dev. Growth Differ. 54: 287–305. [DOI] [PubMed] [Google Scholar]
- Pilz G. A., Shitamukai A., Reillo I., Pacary E., Schwausch J., et al. , 2013. Amplification of progenitors in the mammalian telencephalon includes a new radial glial cell type. Nat. Commun. 4: 2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Red-Horse K., Ueno H., Weissman I. L., Krasnow M. A., 2010. Coronary arteries form by developmental reprogramming of venous cells. Nature 464: 549–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinkevich Y., Lindau P., Ueno H., Longaker M. T., Weissman I. L., 2011. Germ and lineage restricted stem/progenitors regenerate the mouse digit tip. Nature 476: 409–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritsma L., Ellenbroek S. I., Zomer A., Snippert H. J., de Sauvage F. J., et al. , 2014. Intestinal crypt homeostasis revealed at single-stem-cell level by in vivo live imaging. Nature 507: 362–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robles E., Filosa A., Baier H., 2013. Precise lamination of retinal axons generates multiple parallel input pathways in the tectum. J. Neurosci. 33: 5027–5039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy E., Neufield Z., Livet J., Khosrotehrani K., 2014. Understanding clonal dynamics in homeostasis and injury through multicolour lineage tracing. Stem Cells 32: 3046–3054. [DOI] [PubMed] [Google Scholar]
- Schepers A. G., Snippert H. J., Stange D. E., van den Born M., van Es J. H., et al. , 2012. Lineage tracing reveals Lgr5+ stem cell activity in mouse intestinal adenomas. Science 337: 730–735. [DOI] [PubMed] [Google Scholar]
- Shaner N. C., Campbell R. E., Steinbach P. A., Giepmans B. N., Palmer A. E., et al. , 2004. Improved monomeric red, orange and yellow fluorescent proteins derived from discosoma sp. red fluorescent protein. Nat. Biotechnol. 22: 1567–1572. [DOI] [PubMed] [Google Scholar]
- Shaner N. C., Steinbach P. A., Tsien R. Y., 2005. A guide to choosing fluorescent proteins. Nat. Methods 2: 905–909. [DOI] [PubMed] [Google Scholar]
- Shaner N. C., Patterson G. H., Davidson M. W., 2007. Advances in fluorescent protein technology. J. Cell Sci. 120: 4247–4260. [DOI] [PubMed] [Google Scholar]
- Shimomura O., Johnson F. H., Saiga Y., 1962. Extraction, purification and properties of aequorin, a bioluminescent protein from the luminous hydromedusan, aequorea. J. Cell. Comp. Physiol. 59: 223–239. [DOI] [PubMed] [Google Scholar]
- Shimosako N., Hadjieconomou D., Salecker I., 2014. Flybow to dissect circuit assembly in the drosophila brain. Methods Mol. Biol. 1082: 57–69. [DOI] [PubMed] [Google Scholar]
- Snippert H. J., van der Flier L. G., Sato T., van Es J. H., van den Born M., et al. , 2010. Intestinal crypt homeostasis results from neutral competition between symmetrically dividing Lgr5 stem cells. Cell 143: 134–144. [DOI] [PubMed] [Google Scholar]
- Snippert H. J., Schepers A. G., van Es J. H., Simons B. D., Clevers H., 2014. Biased competition between Lgr5 intestinal stem cells driven by oncogenic mutation induces clonal expansion. EMBO Rep. 15: 62–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabansky I., Lenarcic A., Draft R. W., Loulier K., Keskin D. B., et al. , 2013. Developmental bias in cleavage-stage mouse blastomeres. Curr. Biol. 23: 21–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T., Komai Y., Tokuyama Y., Yanai H., Ohe S., Okazaki K., Ueno H., 2013. Identification of stem cells that maintain and regenerate lingual keratinized epithelial cells. Nat. Cell Biol. 15(5):511–518. [DOI] [PubMed] [Google Scholar]
- Takemura S. Y., Bharioke A., Lu Z., Nern A., Vitaladevuni S., et al. , 2013. A visual motion detection circuit suggested by drosophila connectomics. Nature 500: 175–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsien R. Y., 1998. The green fluorescent protein. Annu. Rev. Biochem. 67: 509–544. [DOI] [PubMed] [Google Scholar]
- Wachsman G., Heidstra R., Scheres B., 2011. Distinct cell-autonomous functions of RETINOBLASTOMA-RELATED in arabidopsis stem cells revealed by the brother of brainbow clonal analysis system. Plant Cell 23: 2581–2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh C., Cepko C. L., 1988. Clonally related cortical cells show several migration patterns. Science 241: 1342–1345. [DOI] [PubMed] [Google Scholar]
- Wang C., Liu R., Milkie D. E., Sun W., Tan Z., et al. , 2014. Multiplexed aberration measurement for deep tissue imaging in vivo. Nat. Methods 11: 1037–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K., Milkie D. E., Saxena A., Engerer P., Misgeld T., et al. , 2014. Rapid adaptive optical recovery of optimal resolution over large volumes. Nat. Methods 11: 625–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Benedito R., Bixel M. G., Zeuschner D., Stehling M., et al. , 2013. Identification of a clonally expanding haematopoietic compartment in bone marrow. EMBO J. 32: 219–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber K., Thomaschewski M., Warlich M., Volz T., Cornils K., et al. , 2011. RGB marking facilitates multicolor clonal cell tracking. Nat. Med. 17: 504–509. [DOI] [PubMed] [Google Scholar]
- Weber K., Thomaschewski M., Benten D., Fehse B., 2012. RGB marking with lentiviral vectors for multicolor clonal cell tracking. Nat. Protoc. 7: 839–849. [DOI] [PubMed] [Google Scholar]
- Weissman T. A., Sanes J. R., Lichtman J. W., Livet J., 2011. Generating and imaging multicolor brainbow mice. Cold Spring Harb. Protoc. 2011: 763–769. [DOI] [PubMed] [Google Scholar]
- Wiznerowicz M., Trono D., 2005. Harnessing HIV for therapy, basic research and biotechnology. Trends Biotechnol. 23: 42–47. [DOI] [PubMed] [Google Scholar]
- Worley M. I., Setiawan L., Hariharan I. K., 2013. TIE-DYE: a combinatorial marking system to visualize and genetically manipulate clones during development in drosophila melanogaster. Development 140: 3275–3284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Zheng W., Shen Y., Adhikari D., Ueno H., et al. , 2012. Experimental evidence showing that no mitotically active female germline progenitors exist in postnatal mouse ovaries. Proc. Natl. Acad. Sci. USA 109: 12580–12585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zong H., Espinosa J. S., Su H. H., Muzumdar M. D., Luo L., 2005. Mosaic analysis with double markers in mice. Cell 121: 479–492. [DOI] [PubMed] [Google Scholar]