Abstract
Rho signaling regulates a variety of biological processes, but whether it is implicated in aging remains an open question. Here we show that a guanine nucleotide exchange factor of the Dbl family, OSG-1, confers functional aging by dysregulating Rho GTPases activities in C. elegans. Thus, gene reporter analysis revealed widespread OSG-1 expression in muscle and neurons. Loss of OSG-1 gene function was not associated with developmental defects. In contrast, suppression of OSG-1 lessened loss of function (chemotaxis) in ASE sensory neurons subjected to conditions of oxidative stress generated during natural aging, by oxidative challenges, or by genetic mutations. RNAi analysis showed that OSG-1 was specific toward activation of RHO-1 GTPase signaling. RNAi further implicated actin-binding proteins ARX-3 and ARX-5, thus the actin cytoskeleton, as one of the targets of OSG-1/RHO-1 signaling. Taken together these data suggest that OSG-1 is recruited under conditions of oxidative stress, a hallmark of aging, and contributes to promote loss of neuronal function by affecting the actin cytoskeleton via altered RHO-1 activity.
Keywords: Rho GTPase, reactive oxygen species, oxidative stress, chemotaxis
THE Rho guanosine triphosphatases (GTPases) are small signaling G proteins ubiquitously expressed across phyla where they act to regulate actin dynamics and other fundamental biological processes, including cell growth, motility and adhesion, cytokinesis, lipid metabolism, membrane trafficking, and transcription (reviewed in Bustelo et al. 2007). Phylogenetically, Rho genes constitute their own family within the Ras superfamily of GTPases. There are 22 Rho GTPases in mammals with Rho, Rac1, and Cdc42 being the most studied, 11 in Drosophila melanogaster, 6 in Caenorhabditis elegans, and 5 in yeast (reviewed in Jaffe and Hall 2005; Lundquist 2006). Rho GTPases act as molecular switches cycling between an active, GTP-bound state and an inactive, GDP-bound state. As such, they are endowed by three major types of ancillary proteins: guanine nucleotide exchange factors (GEFs) that activate the GTPase by catalyzing exchange of GDP for GTP; GTPase-activating proteins (GAPs) that switch the GTPase off by facilitating its intrinsic GTP hydrolysis and guanine nucleotide dissociation inhibitors (GDIs), which act as anchors and block spontaneous activation (reviewed in Schmidt and Hall 2002). Thus, a complex array of upstream regulatory proteins and downstream effector proteins converge around relatively few handyman proteins, the GTPases, to control a number of cellular activities. The organization of this network is such that signaling specificity and spatiotemporal control are not under the command of the GTPases, but are rather delegated to the GEFs. Accordingly, there are ∼6 GEFs in yeast, 18 in C. elegans, 23 in Drosophila, and 60 in Homo sapiens that endow GTPases of the Rho family (Schmidt and Hall 2002). These GEFs form the Dbl family, a name that originates from the catalytic Dbl homology (DH) domain present in these proteins. Despite the fact that DH domains share little sequence homology to each other, crystallographic and NMR evidence reveals highly conserved DH three-dimensional structure (Jaffe and Hall 2005). Generally, the DH domain is coupled to the Pleckstrin homology (PH) domain, which is invariably located next to it. Since PH domains are known to bind to phosphorylated phosphoinositides it is assumed that the PH domain has a role in targeting GEFs to their intracellular locations. The PH domain also appears to increase catalytic efficiency even though it is not absolutely required for catalysis of nucleotide exchange. In most cases the DH–PH tandem is sufficient for catalytic function but typically GEFs exhibit additional structural domains in their primary sequences (Jaffe and Hall 2005).
The multifaceted nature of Rho signaling argues that the GTPases and their associated genes may play a role in the aging process. In fact, there is growing evidence that the actin cytoskeleton may be a regulator of aging (Gourlay and Ayscough 2005b), but whether Rho signaling is involved in this process remains an open question. In this study we shed light on the issue by reporting the first example of a GEF that confers aging susceptibility to C. elegans neurons. This gene, christened oxidative-stress-susceptible GEF1 or OSG-1 appears to be specifically recruited under conditions of oxidative stress, a key aspect of aging. The protein promotes loss of neuronal function by acting to dysregulate Rho signaling, which presumably affects actin dynamics.
Methods and Materials
Buffers and chemicals
The buffers and chemicals used are listed and described here:
M9 buffer: 22 mM KH2PO4, 22 mM NaH2PO4, 85 mM NaCl, 1 mM MgSO4
NMG plates: 17 g/liter agar, 3 g/liter NaCl, 2.5 g/liter bacto peptone, 1 mM CaCl2, 1 mM MgSO4, 5 mg/liter cholesterol, 25 mM potassium phosphate buffer (132 mM K2HPO4, 868 mM KH2PO4)
DNA extraction buffer: 50 mM KCl, 10 mM Tris-HCl pH 8.3, 2.5 mM MgCl2, 0.45% NP-40, 0.45% Tween-20, 0.01% gelatin, 0.2 mg/ml Proteinase K.
All chemicals were purchased from Sigma Aldrich (St. Louis, MO) if not otherwise stated.
Molecular biology
To create Posg-1::GFP, a ∼2.3-kb genomic fragment upstream from the OSG-1 gene containing the open reading frame was amplified by PCR and subcloned into pPD95.75 Fire vector using PstI and BamHI restriction sites. Primers sequences were:
AACTGCAGTGTGACTTGGGATCATCGGA
CGGGATCCTTGAGCCACAGGGGAGCCAATG
from 5′ to 3′. The forward primer is listed first.
Strains and transgenic animals
We used the strains listed below, which had been previously constructed in our lab or obtained from the CGC:
N2: Bristol
FDX25: sesIs25(Pflp-6::Aβ; Pgcy-5::GFP; rol-6) (micro-injection; Cotella et al. 2012).
We constructed the strains listed below, by crossing, by micro-injection as previously described (integrated transgenic lines were outcrossed four times to remove undesired mutations) (Duan and Sesti 2013) or by ethyl methanesulfonate (EMS) mutagenesis:
EMS mutagenesis
FDX25 worms were incubated in M9 buffer + 50 mM EMS for 4 hr with moderate shaking. Then they were washed five times with M9 buffer and placed on growth media, and actively moving L4 animals were transferred to a fresh plate where they were allowed to recover and develop into young adults. Single, F2 healthy adults, were isolated and transferred to separate plates and screened for GFP in the ASE right (ASER) neuron using a SZX7 Olympus microscope equipped with a digital camera and dedicated software. Twenty-five mutants were isolated and 22 were discarded for several reasons. The remaining 3 mutants were saved for further analysis. Mutant ses2509 was used in this study.
Forward genetic screen
We mapped the ses2509 mutant allele using the single-nucleotide polymorphism method (Davis et al. 2005). Thus, FDX2509 worms were crossed with CB4856 Hawaiian strain. F2 worms were singled out and screened for the lost GFP phenotype (control) or the suppression of it (test); thus, two groups of 40 animals each were collected. The DNA from the control and the test group were extracted in DNA extraction buffer and screened using 48 pairs of primers listed in Davis et al. (2005) by PCR. This screening led us to locating the mutation in a region of ∼4 Mb in chromosome III [physical location from 3,359,033 (F45H17) to 7,320,107 (F56C9), Supporting Information, Figure S1]. We screened this region using, first, an additional set of SNP primers that we designed and that are listed in Table 1 and then bacterial RNAi (Source Bioscience Lifesciences, CA) of 52 genes located in a 0.2 Mb region between S5 and S6 SNPs (Table 1).
Table 1. Additional chromosome III SNP primers.
| Primers set | Sequence (5′ to 3′) | ||
|---|---|---|---|
| S1 | CCAACTGTCGACGATAGAGATG | ||
| AATTCCGTGTAGCGAATCG | |||
| S2 | TTTCAAGACAAGGCAAGTATCC | ||
| TTTGATCAATCAGTGGGTCTTG | |||
| S3 | AATTGCCGAACAGGAAAATG | ||
| CACGCGTGTAACTCTTCCAA | |||
| S4 | CATCGGATACATGGTGCATCG | ||
| GGCAAGTTTTCCTTCTCGTCC | |||
| S5 | TGGTCCCCAGAGATTTCAGA | ||
| GAGTACGCGGGTCATTTTTG | |||
| S6 | GACCGAATCGAATCGGAAG | ||
| AAACAAGGATCAGTGGAATGTG | |||
| S7 | ATGGCCGTGGAATTGATTGC | ||
| ACCCGGAACCCATTTGTAGA | |||
| S8 | GCGGATTCTCGATAAAAACTAC | ||
| AAGCAGAAAAGTGGCATTCA | |||
| S9 | CATCTGAAACTCACGGGAAGTC | ||
| AGTTACAACCTCGGGTCGTG | |||
| S10 | GAATTTTGCTGTGCCAAGCC | ||
| CGCTCACTGTGTCAAAGTGT | |||
| S11 | CATTAGGAAGTGATGCAAGTGG | ||
| TGGATTTGAGAGGTGTCCATAG | |||
| S12 | TCACATCATAAGTCAAGCGGA | ||
| TCGATCTCTAGGCGGATAATCA | |||
| RFLP | N2 digest (bp) | CB4856 digest (bp) | |
| S1 | EcoRI | 262, 205 | 467 |
| S2 | RsaI | 31, 299 | 334 |
| S3 | PsiL | 60, 407 | 467 |
| S4 | XmnI | 442 | 138, 304 |
| S5 | MseI | 419 | 255, 164 |
| S6 | HpyCH4 V | 434 | 244, 190 |
| S7 | RsaI | 423 | 165, 258 |
| S8 | AciI | 463 | 236, 227 |
| S9 | RsaI | 243, 278 | 521 |
| S10 | AclI | 276, 193 | 469 |
| S11 | AvaII | 196, 294 | 490 |
| S12 | HpyCH4 III | 344 | 237, 107 |
SNP primers sequences. The forward primer is listed first; restriction enzymes and band sizes are indicated.
Age synchronization
Age-synchronization was performed as previously described (Duan and Sesti 2013). Briefly, nematodes were grown in standard 10-cm NGM plates + OP50 Escherichea coli until a large population of gravid adults was reached (3–5 days). The animals were collected in 50-ml Falcon tubes, washed in M9 buffer, and treated with 10 vol of basic hypochlorite solution (0.25 M NaOH, 1% hypochlorite freshly mixed; no significant differences were observed in bleach-free preparations, obtained by isolating laid eggs with SDS/NaOH). Worms were incubated at room temperature for 10 min and then the eggs (and carcasses) were collected by centrifugation at 400 g for 5 min at 4°, incubated overnight in M9 buffer, and seeded on standard NMG plates. The age of the worms was counted starting from seeding day (day 0).
Behavioral assays
Behavioral tests were carried out blind, using age-synchronized worms.
In lost GFP phenotype assays, worms were analyzed with a SZX7 Olympus microscope equipped with a digital camera in a dark room (to minimize and maintain invariable background light). A worm had lost its GFP signal when not even the faintest GFP fluorescence could be detected by eye. Representative images are shown in the inset of Figure 1A.
Figure 1.
The lost GFP phenotype is worms expressing Aβ42 in ASE neurons. (A) Percent of Aβ42-expressing (FDX25) and control (FDX2512) animals with detectable GFP fluorescence in ASER as a function of age (lost GFP phenotype). n = 4 experiments/genotype. Inset: Representative pictures of ASER neurons of 5- and 12-day-old FDX25 and FDX2512 control worms. (B) Lost GFP phenotype in FDX25 worms in control or subjected to CED-4 RNAi. n = 3 experiments. (C) Lost GFP phenotype in FDX25 worms grown in absence or presence of 1 μg/ml Q-VD-Oph dissolved in the NMG agar media. n = 3 experiments. In a single experiment, 30–50 age-synchronized worms were grown under standard conditions and scored every day for GFP in the ASER neuron. In all figures, mean ±SEM. Error bars smaller than the size of the symbols are not visible in the figure.
In loss of sensory dendrite assays, and in the experiments shown in Figure 6, worms were photographed with a BX61 Olympus microscope and the intensity of the GFP signal was measured using ImageJ1.46 software.
Figure 6.
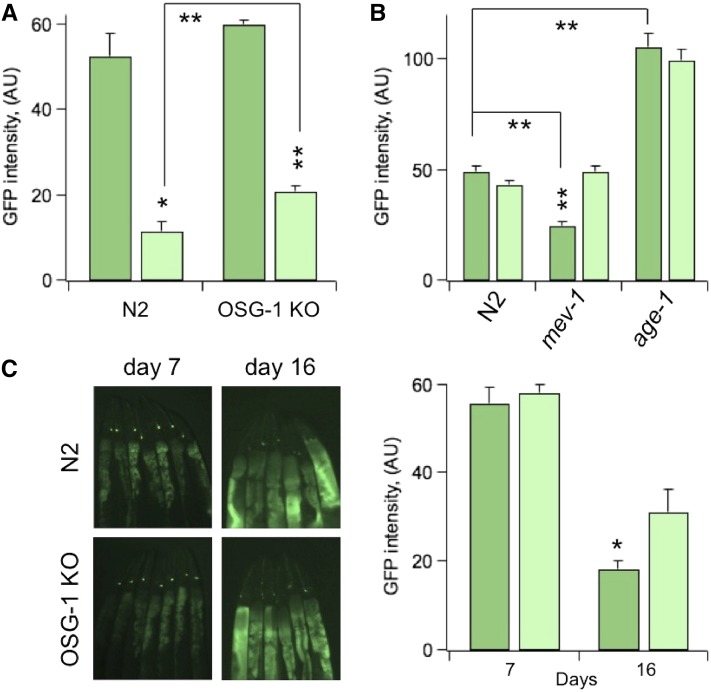
OSG-1 is recruited under conditions of oxidative stress. (A) GFP intensity (arbitrary units) in ASER, in 10-day-old N2 (sesIs2512) or OSG-1 KO (sesIs2528) genotypes in control or treated with 1.0 mM H2O2 3 days earlier. n = 3 experiments/genotype. (B) GFP intensity in ASER in 10-day-old N2 (sesIs2512) or high oxidative stress expressing mev-1 (sesIs2530) or low oxidative stress expressing age-1 (sesIs2529) genotypes, in control or subjected to OSG-1 RNAi from day 7 to day 10. n = 3–4 experiments/genotype. (C) GFP intensity in ASER in N2 (sesIs2512) or OSG-1 KO (sesIs2528) genotypes as a function of their age. n = 3 experiments/genotype. Inset: Representative images of GFP-fluorescent ASER neurons in N2 (sesIs2512) and OSG-1 KO (sesIs2528) genotypes at the indicated ages. In a single experiment, between 30 and 60 worms/genotype were analyzed. Animals were anesthetized with 20 μl of 20 mM NaN3 and photographed with a SZX7 Olympus microscope and a digital camera. Pictures were analyzed with ImageJ1.46 software (National Institutes of Health).
Chemotaxis experiments were carried out as previously described (Bianchi et al. 2003). Briefly, a chunk of agar 0.5 cm in diameter was removed from 10-cm plates and soaked in 0.5 M NH4Cl (pH 7.0 with NH4OH) for 2 hr. Chunks were put back in the plate overnight to allow equilibration and formation of a gradient. Roughly 20 age-synchronized worms were placed between the test spot and a control spot on the opposite side of the plate. A total of 10 µl of 20 mM NaN3 was placed on both spots. After 1 hr, animals on the test/control spot were counted, and a chemotaxis index, C.I., calculated as the number of animals at the test spot (Ntest) minus the number of animals at the control spot (NCnt.), divided by the total number of animals (N). An experiment was carried out with ∼60 worms/genotype distributed in three test plates. To study chemotaxis during aging, experiments were started with ∼600–1000 worms per genotype (mean life span ∼20 days). Worms were examined every day until death and were scored as dead when they were no longer able to move even in response to prodding with a platinum pick. Each day, worms were transferred to a fresh plate containing bacteria.
Thrashing experiments were carried out as previously described (Bianchi et al. 2003). Briefly, worms were picked in a drop of M9 buffer on an agar plate. After 2 min of recovery, thrashes were counted for 1 min. A thrash is defined as a change in the body bend at the midbody point.
DiD staining
Dye-filling experiments were performed as previously described (Bianchi et al. 2003). Briefly, worms were transferred to a plate containing 0.01 mg/ml DiD (Molecular Probes) in M9 buffer and allowed to stain for 2–3 hr at room temperature. Then worms were transferred to a glass cover slip and photographed with an Olympus BX61 microscope equipped with a digital camera.
Bacterial RNAi
RNAi screening was performed as described in Kamath et al. (2001). Briefly, age-synchronized worms were grown in NGM plates containing 25 µg/ml carbenicillin and 1.0 mM IPTG seeded with HT115 double-strand RNAi synthetizing E. coli.
Statistical analysis
Quantitative data are presented as mean ±SEM. The level of significance was calculated using two-tailed Student’s t-test (http://studentsttest.com). Statistical significance was assumed at the 95% confidence threshold. * = P<0.05; ** = P<0.01
Results and Discussion
The lost GFP phenotype in transgenic worms expressing Aβ42
To identify genes that promote loss of neuronal function during aging we followed a discovery driven approach using C. elegans. For these studies, we employed the FDX25 worm that expresses human Aβ42—a potent neurodegenerative factor (Mattson 2004)—in ASE sensory neurons (Cotella et al. 2012). In addition, we labeled the right ASE neuron (ASER) with a specific GFP reporter (Pgcy-5::GFP) (Yu et al. 1997). Figure 1A shows the percentage of worms with detectable fluorescence in the ASER neuron as a function of age. Control worms exhibited normally viable and fluorescent ASER neuron. In contrast, ASER fluorescence quickly faded in worms expressing Aβ42. We often refer to this phenotype as the “lost GFP” phenotype.
We previously proposed that the lost GFP phenotype reflects neuronal apoptosis (Cotella et al. 2012; Duan and Sesti 2013). Here, we sought to corroborate this notion by genetic and pharmacological evidence. Thus, knockdown of the CED-4 gene, which acts upstream of many apoptotic pathways in C. elegans (Ellis and Horvitz 1986; Yuan and Horvitz 1992), ameliorated the lost GFP phenotype (Figure 1B). Activation of caspases and cleavage of critical cellular proteins is crucially associated with apoptotic death (Rohn and Head 2008, 2009). In consequence, treatment with generic caspase inhibitor N-(2-quinolyl)valyl-aspartyl-(2,6-difluorophenoxy)methyl ketone (Q-VD-Oph) prolonged the life of GFP signals (Figure 1C). Q-VD-Oph was more efficient in cultured ASER neurons obtained from embryos (Duan and Sesti 2013) than in live worms. However, these results are largely expected, because many factors contribute to hamper drug uptake in C. elegans. Furthermore, compelling evidence shows that the toxic effects of Aβ42 in C. elegans cells are protein specific and not due to overexpression (reviewed in Link 2006). Our data further support Aβ42-specific toxicity because the ASER neurons of FDX2512 worms that overexpressed GFP were perfectly viable. In conclusion, like in mammalian neurons, in C. elegans neurons Aβ42 can induce apoptosis.
Genetic screen maps locus of resistance to Aβ42 to a guanine exchange factor gene
We speculated that if we randomly mutated FDX25 worms, we could select Aβ42-resistant mutants by singling out the worms with longer lasting GFP signals. Then, by exploiting the genetic tools of the worm, we could identify genes that confer resistance to Aβ42-induced toxicity. This approach led us to isolate several mutant lines, each exhibiting partial suppression of the lost GFP phenotype. In what follows, we describe studies with mutant line FDX2509 (Figure 2A). A forward genetic screen of this mutant by the single-nucleotide polymorphisms method (Figure S1) (Davis et al. 2005) mapped to an adenine deletion at position 10187 in the R02F2.2 gene, leading to a premature stop codon in exon 14 (ses2509 allele). R02F2.2 encodes a putative Dbl GEF (Figure 2B), which we dubbed OSG-1. To confirm that OSG-1 is responsible for the partial suppression of the lost GFP phenotype in worms expressing Aβ42 peptide, we knocked down this gene by RNAi-producing bacteria (Kamath et al. 2001). As expected, GFP signals lasted longer in worms subjected to OSG-1 RNAi (Figure 2C). We obtained a OSG-1 knockout (KO) worm from the Caenorhabditis Genetic Center. We expressed Aβ42 and GFP in OSG-1 KO worms (FDX2531 strain) by crossing them with FDX25 worms. Consistent with results obtained using RNAi, genetic ablation of OSG-1 led to a significant suppression of the lost GFP phenotype (Figure 2C). In fact, the time course of GFP fluorescence in worms subjected to OSG-1 knockout or knockdown recapitulated the phenotype of the FDX2509 mutant worm, that is, the one that harbors the ses2509 allele (compare Figure 2A with Figure 2C). Further, OSG-1 RNAi in control worms, which express GFP but not Aβ42 (FDX2512), had no effect on the duration of the GFP signal (data not shown). Results pertaining to FDX2532 worms in Figure 2C are discussed below.
Figure 2.
R02F2.2 (OSG-1) partially controls the lost GFP phenotype. (A) Lost GFP phenotype in FDX25 and FDX2509 mutant worms as a function of age. n = 5 experiments/genotype. Error bars smaller than the size of the symbols are not visible in the figure. (B) OSG-1 is a predicted 1182-amino-acid polypeptide with an estimated molecular weight of 133 kDa. The predicted primary structure contains a DH homology domain, a PH domain, and a WD-40 repeats domain. The adenine deletion in mutant allele ses2509 introduces a premature stop at position 917 in the amino acids sequence, which is located in the putative WD-40 domain. The structure was calculated using bioinformatic tools available at the SIB bioinformatics Resources Portal (http://www.expasy.org/tools/). (C) Lost GFP phenotype in FDX25 worms, FDX25 worms subjected to OSG-1 RNAi, OSG-1 KO worms expressing Aβ42 in ASE neurons (FDX2531) and in worms that express Aβ42 in the ASE neurons in the age-1 background (FDX2532). n = 3–5 experiments/genotype.
OSG-1 is expressed in neurons and muscle
We determined the expression pattern of OSG-1 by gene reporter analysis. A transcriptional reporter expressing GFP driven by the OSG-1 promoter (Posg-1::GFP, FDX2533 strain) yielded signals in body-wall muscle, in head muscle, and in several neurons in the head including FLP neurons (Figure 3, A and B). Posg-1::GFP expression was also detected in the ASE neurons whose identification was confirmed by DiD dye staining (Figure 3C. Note that the fluorescence in head muscle partially masked that in the ASE neurons. ASE neurons are extensively characterized (Bargmann and Mori 1997) and therefore provided an excellent system to study OSG-1.
Figure 3.
OSG-1 is expressed in muscle and nervous system. (A) Fluorescence microscopy image taken from a Posg-1::GFP nematode (FDX2533), showing Posg-1 expression in head and body wall muscle. (B) Image of the head of a Posg-1::GFP nematode demonstrating Posg-1::GFP expression in head muscle and FLP neurons. (C) DiD staining of amphid neurons of a Posg-1::GFP nematode, demonstrating OSG-1 expression in the ASE neuron. (D) Number of thrashes in 4-day-old N2 or OSG-1 KO worms. n ≥ 20 animals/genotype. (E) Representative images of ASER neurons marked with GFP in 4-day-old N2 or OSG-1 KO genotypes. Images were taken with a BX61 Olympus microscope equipped with Nomarsky optics and a digital camera.
OSG-1 KO worms are normally viable
OSG-1 KO worms underwent normal development and were normal in size (Figure S2A) and brood (not shown). Mutations in body-wall muscle genes can give rise to Pat (paralyzed arrested at twofold) or Unc (uncoordinated) phenotypes, but we did not detect these defects in OSG-1 KO worms. Indeed they exhibited normal locomotion behavior (File S1 and File S2) and this impression was corroborated by counting thrashes (Figure 3D). Most importantly, we could not detect any morphological abnormality in the ASER neuron of OSG-1 KO animals (Figure 3E). These observations seem to indicate that OSG-1 is not obviously required for development.
OSG-1 activates a RHO-1 GTPase signaling network
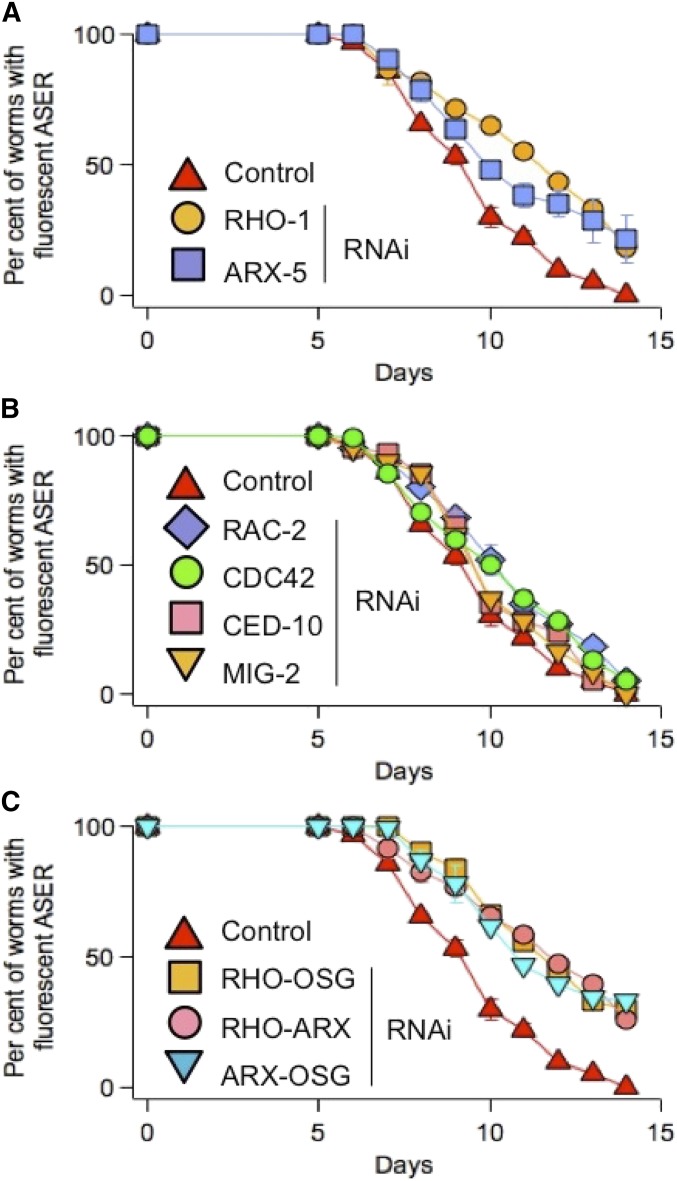
The predicted primary structure of OSG-1 (Figure 2A) places this gene in the Dbl family of GEFs that endow Rho GTPases. The Rho family is conserved in C. elegans (Table 2) and these GTPases are expressed in ASE neurons (Chen and Lim 1994; Lundquist 2006). Therefore, to determine whether OSG-1 can interact with Rho GTPases, as predicted by sequence analysis, we individually knocked down these genes by bacterial RNAi. The ability to turn ON/OFF RNAi at any desired age by feeding C. elegans with RNAi-producing bacteria was a crucial asset to these experiments, because eliminating GTPase activities too early disrupts embryogenesis (Spencer et al. 2001). Indeed, RHO-1 or CED42 RNAi treatment from day 0 was lethal (data not shown). Worms survived triple RAC-2/CED-10/MIG-2 RNAi (these GTPases function redundantly Lundquist et al. 2001) but they were Unc and egg-laying defective (Egl) and had a dumpy (Dpy) body shape (data not shown). Accordingly, the data shown in Figure 4 were obtained by subjecting the worms to a bacterial RNAi diet after day 4. Thus, the RHO-1 knockdown, individually (Figure 4A) or together with OSG-1 (Figure 4C), led to a significant suppression of the lost GFP phenotype. In contrast, RNAi of the other Rho GTPases listed in Table 2, including triple RAC-2/CED-10/MIG-2 RNAi, had little effect on the duration of GFP signals (Figure 4B; Triple RAC-2/CED-10/MIG-2 not shown). Most importantly, the double OSG-1/RHO-1 RNAi was not additive, consistent with the notion that OSG-1 activates RHO-1. Dysregulated RHO-1 activity may affect actin-binding proteins (ABS), such as the Arp2/3 complex, and consequently induce dendritic loss and apoptosis by shifting the relative fractions of G-actin and F-actin (reviewed in Desouza et al. 2012). Accordingly, when subunits ARX-3 and ARX-5, orthologs of human Arp2/3 subunit 1A and subunit 3, respectively (Table 2) (Sawa et al. 2003; Roh-Johnson and Goldstein 2009; Shaye and Greenwald 2011), were knocked down, the lost GFP phenotype was ameliorated (Figure 4A, ARX-3 not shown). Double knockdown of ARX-5 and OSG-1 and of ARX-5 and RHO-1 gave similar results and were not additive (Figure 4C), suggesting that OSG-1 promotes loss of neuronal function by altering actin dynamics via its interaction with RHO-1.
Table 2. Homology between human and C. elegans cytoskeletal genes.
| C. elegans | H. sapiens | Blast E-value |
|---|---|---|
| R02F2.2 | ARHGEF10 | 8.4e-85 |
| RHO-1 | RhoA | 1.1e-90 |
| RAC-2 | Rac1 | 9.2e-97 |
| MIG-2 | Rac1 | 2.2e-69 |
| CED-10 | Rac1 | 8.2e-79 |
| CDC-42 | Cdc42 | 1.1e-90 |
| ARX-1/-7 | Arp 2/3 complex | 2.9e-177 |
Homologies and blast E-values were taken from http://www.wormbase.org/. E-blast value ≤1e-4 is commonly accepted as a good match.
Figure 4.
OSG-1 controls the RHO-1/ARX2/3 complex pathway. (A) Lost GFP phenotype in FDX25 worms in control or subjected to RHO-1 RNAi, or ARX-5 RNAi. n = 3–4 experiments. (B) Lost GFP phenotype in FDX25 worms in control or subjected to RAC-2, RNAi, or MIG-2 RNAi or CED-10 RNAi or CDC-42 RNAi. n = 3 experiments each. (C) Lost GFP phenotype in FDX25 worms in control or subjected to RHO-1/OSG-1 RNAi or RHO-1/ARX-5 RNAi or ARX-5/OSG-1 RNAi. n = 3 experiments each. Worms were fed with RNAi expressing bacteria at 4 days of age to avoid Rho GTPase-mediated disruption of embryogenesis. Error bars smaller than the size of the symbols are not visible in the figure
OSG-1 mediates loss of GFP signal in the sensory dendrite via RHO-1 and ARX2/3 complex activation
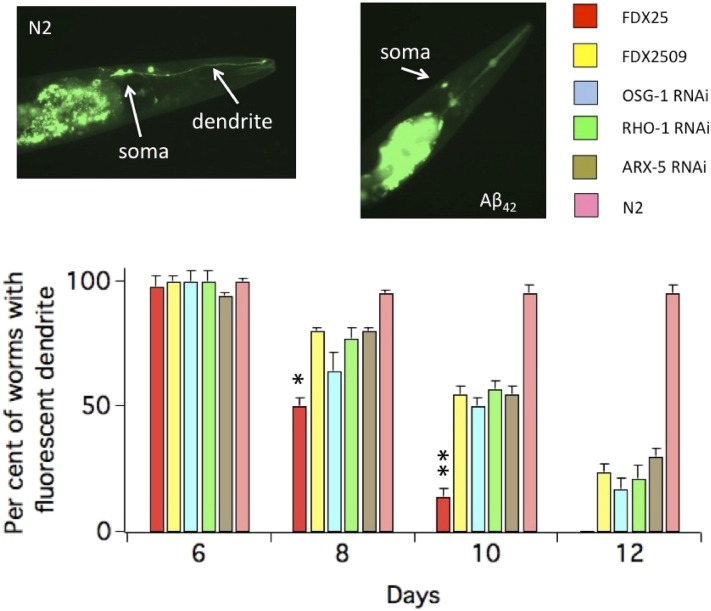
In mammalian neurons, Rho-mediated alterations in actin dynamics begin with loss of spines and as this process continues, can end with apoptosis (reviewed in Tolias et al. 2011). Also C. elegans Rho GTPases control actin dynamics (Lundquist 2006). Therefore we asked whether the GFP signal would begin to fade in the sensory dendrite of the ASER neuron and whether OSG-1 would affect this process via its interaction with RHO-1. Thus the loss of GFP signal in the sensory dendrite was markedly enhanced in worms expressing Aβ42 (FDX25) compared to control worms (FDX2512, Figure 5). This loss was delayed in FDX25 worms subjected to either OSG-1 or RHO-1 RNAi as well as in the FDX2509 mutant that bears a premature stop codon in the OSG-1 gene. Moreover, when subunits ARX-3 and ARX-5 were knocked down, the loss of GFP in the sensory dendrite was markedly reduced (Figure 5, ARX-3; not shown).
Figure 5.
OSG-1 induces loss of GFP signal in the ASER sensory dendrite. The bar graph shows the percentage of animals expressing GFP in the ASER sensory dendrite as a function of age. Indicated RNAi was performed on Aβ42-expressing (FDX25) worms. In control conditions, worms were fed with HT115 bacteria containing empty vector. n = 3 experiments/genotype. In a single experiments 20–30 worms were analyzed for each time point. Inset: Representative images of GFP fluorescence in the ASER neuron of 8-day-old worms in the absence (FDX2512) or presence of Aβ42 (FDX25. The fluorescent line in the middle is the pharyngeal duct. In all figures, * = P, 0.05; ** = P, 0.01
OSG-1 and oxidative stress are tightly linked
Aβ oligomers cause oxidative damage through multiple mechanisms and pathways that are conserved to a considerable extent between mammalian and C. elegans neurons (Butterfield and Lauderback 2002; Wu et al. 2006). Indeed, the lost GFP phenotype was mitigated in the age-1 background (Figure 2C, FDX2532), which is rich in antioxidant defenses (Larsen 1993), suggesting that oxidative stress is one of the routes through which Aβ42 induces toxicity in ASE neurons (Wu et al. 2006). Based on these results, we speculated that the driving force behind the activation of OSG-1 was oxidative stress. In a first approach, young 7-day-old worms were soaked in a solution containing 1.0 mM hydrogen peroxide (H2O2) for 20 min and scored for GFP fluorescence in the ASER neuron 3 days later. This treatment induced a robust decrease in the intensity of the GFP signal in control (Figure 6A and Figure S2B), whereas in OSG-1 KO background, decrease was significantly attenuated (Figure 6A). To maintain equal levels of GFP expression, OSG-1 KO and the other strains in Figure 6 were crossed with FDX2512. Abnormal amounts of reactive oxygen species (ROS) can be induced in C. elegans by genetic manipulation. For example, the TK22 mutant worm is subjected to oxidative stress and has a shorter life span, due to a mutation deletion in cytochrome b (mev-1) in mitochondrial respiratory chain complex II (Ishii et al. 1990, 1998, 2004). Accordingly, GFP intensity at baseline was diminished in 10-day-old TK22 background compared to control (Figure 6B and Figure S2C). However, 3-day-OSG-1 RNAi (from day 7 to day 10), which was too mild to exert appreciable effects on control worms (FDX2512), was sufficient to partially rescue the loss of GFP signal in TK22 worms. Moreover, GFP intensity was stronger in the age-1 genotype compared to control and was not affected by OSG-1 RNAi (Figure 6B and Figure S2C). We further speculated that since oxidative stress naturally increases in neurons during aging, the intensity of GFP signals should decrease in old worms and suppression of OSG-1 should mitigate this decline. Indeed, whereas GFP intensity gradually decreased in aging worms, this decline was significantly attenuated in OSG-1 KO worms (Figure 6C). It must be noted that there is no contradiction between these results and those in Figure 1A, where all FDX2512 control worms had fluorescent ASER neurons, because different experimental methodologies were used. In lost GFP phenotype assays, we counted the number of worms that had lost the GFP signal. In contrast, in aging assays we measured changes in the intensity of the GFP signal. Thus in lost GFP phenotype assays, changes in GFP intensity were not taken into account as long as the signal was detectable.
OSG-1 promotes loss of ASE function during aging
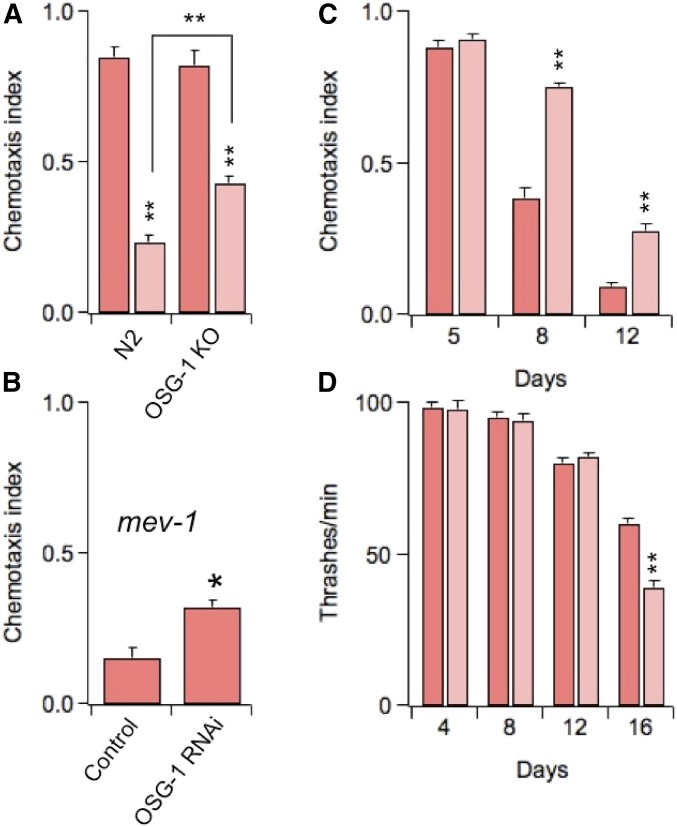
C. elegans uses chemotaxis to perform a number of vital functions, including searching for food, avoiding toxic substances, and mating (Bargmann and Mori 1997). The ASE neurons play an important role in the chemosensory activities of the worm by mediating the animal’s attraction for salts, vitamins, amino acids, etc. (chemotaxis to water-soluble attractants), by-products of bacterial metabolism. ASE neurons function is commonly assessed in chemotaxis assays, which give a measure of the ability of the worms to track to the point source of a test attractant (Bargmann and Horvitz 1991; Bargmann and Mori 1997). Thus, Figure 7A shows chemotaxis to chloride (ASER is primarily sensitive to this anion; Pierce-Shimomura et al. 2001) in young worms that had been subjected to a 1.0 mM H2O2 oxidative insult the day before. Similar to its effects on GFP signal (Figure 6B), H2O2 treatment induced a significant decline in the chemosensory response of the worms and this decline was ameliorated in OSG-1 OK worms. Moreover, chemotaxis was impaired in the TK22 genotype, and this impairment could be partially suppressed by knocking down OSG-1 (Figure 7B). We and others previously showed that oxidative stress contributes to the decline of chemotaxis in aging worms (Wu et al. 2006; Cai and Sesti 2009; Minniti et al. 2009; Maglioni et al. 2014). Therefore we speculated that if OSG-1 is activated under conditions of oxidative stress, chemotaxis should improve in aging worms lacking this gene. As expected, whereas aging worms gradually lost the ability to track chloride, OSG-1 KO worms performed significantly better (Figure 7C). Defects in locomotion behavior may affect chemotaxis in aging worms. However, worms appeared to move normally and no differences in the speeds of control and OSG-1 KO worms could be detected (File S3 and File S4). To corroborate these qualitative impressions we assessed muscle contractility by counting thrashes. In agreement with previous reports (Cai and Sesti 2009; Maglioni et al. 2014) the number of thrashes moderately decreased during the first 12 days of life, a period of time in which loss of chemosensory function was almost complete (compare Figure 7C with Figure 7D). The decrease was similar in both genotypes suggesting that deficits in locomotion behavior could only marginally affect chemotaxis. However, contractility began to sharply decline in late life. At day 16, loss was significantly more marked in OSG-1 KO worms than in N2 worms. Together, these results corroborate the notion that in C. elegans, like in mammals, sensory function declines faster than muscle function (Liu et al. 2013; Maglioni et al. 2014) and may further imply that OSG-1 plays a role in the aging muscle. However, there is no evidence that the conditions of oxidative stress that lead to OSG-1 recruitment in neurons are also present in muscle cells. In this study we did not attempt to elucidate the mechanisms through which oxidative stress leads to activation of OSG-1. This will require a significant effort and will be the matter of a separate study.
Figure 7.
OSG-1 accelerates loss of ASE function during aging. (A) Chemotaxis to Cl− in 5-day-old N2 or OSG-1 KO worms and in 5-day-old N2 or OSG-1 KO worms that were exposed to 1.0 mM H2O2 the day before. n = 4 experiments/genotype. (B) Chemotaxis to Cl− in 11-day old TK22 (mev-1) worms in control or subjected to OSG-1 RNAi. n = 3 experiments. (C) Chemotaxis to Cl− in N2 or OSG-1 KO worms as a function of their age. n = 3 experiments/genotype. (D) Number of thrashes/minute in N2 or OSG-1 KO worms as a function of their age. n ≥ 20 animals/genotype/time point.
OSG-1 has no effect on life span
The amphid sensory neurons contribute to the regulation of the life span of C. elegans through complex mechanisms (Apfeld and Kenyon 1999; Alcedo and Kenyon 2004). The fact OSG-1 is expressed in ASE neurons and is activated during aging argues that OSG-1 might affect life span (we assume that low-level expression elsewhere, below the limit of detection, is absent). However, OSG-1 KO worms exhibited both normal mean life span at 20° (20.7 ± 0.5 days and 20.2 ± 0.4 days for N2 and OSG-1 KO worms, respectively; Figure S3A) and maximal life span (27.0 ± 0.5 days and 27.3 ± 0.7 days for N2 and OSG-1, respectively; Figure S3C). Moreover, egg laying, a phenotype that has been linked to increases in life span in C. elegans (Kenyon 1997) was normal in the OSG-1 KO worm (not shown). We conclude that OSG-1 does not contribute to the regulation of life span in C. elegans.
The nervous system of C. elegans undergoes several age-dependent morphological and functional changes including cytoskeletal disorganization, axon beading, defasciculation, deteriorations in synaptic transmission, and neurite branching (Pan et al. 2011; Tank et al. 2011; Toth et al. 2012; Liu et al. 2013), and the results of this study add new insight by implicating Rho signaling in neuronal aging. In particular, the actin cytoskeleton appears to be one of the targets of OSG-1/RHO-1 as evidenced by the fact that knockdown of ARX-3 and ARX-5 alone or with RHO-1 or with OSG-1 ameliorated the loss of GFP signal in the ASER neuron in a nonadditive fashion. Furthermore, the premature stop codon in the ses2509 mutant allele is located in the WD-40 domain, which can promote GEF binding to actin filaments (Li and Roberts 2001). A number of studies, primarily in yeast, have shown that the formation of aggregates of F-actin—as a result of actin dynamics slowdown—correlates with apoptosis (reviewed in Gourlay and Ayscough 2005a). For example, pharmacological agents that stabilize actin turnover enhance apoptotic death. In yeast, genetic manipulation of genes that control the formation of actin aggregates, such as act1-157 and Δscp1, results in decreased levels of ROS and longer life span. Shifts in apoptosis rates can similarly be obtained by interfering with actin regulatory proteins such as Gelsolin and Cofilin. The mechanisms by which F-actin aggregates promote apoptosis are subjected to intense investigation. The current model predicts that they indirectly induce oxidative stress and apoptosis by targeting and destabilizing mitochondria (Gourlay and Ayscough 2005a). This body of evidence led us to propose that OSG-1 induces apoptosis by leading to stabilization of actin turnover, via dysregulated RHO-1 activity. Further studies will rigorously test this model.
OSG-1 has a human homolog, ARHGEF10, which is widely expressed in the brain and which has been linked to a number of neuronal pathologies (Tahvanainen et al. 1994; Ranta et al. 1997; Verhoeven et al. 2003; Matsushita et al. 2010). Our findings may imply that ARHGEF10 as well as other GEFs might be implicated in aging and in neurodegenerative conditions characterized by extensive oxidative stress such as Alzheimer’s disease. In fact, Mendoza-Naranjo (et al. 2007) have shown that in murine hippocampal neurons, Aβ42 induces apoptosis via Rac1 and Cdc42 signaling. Both the GTPases are recruited by Tiam1, a member of the Dbl family. Moreover, Rho and one of its effectors, Rho-associated kinase, ROCK, have been proposed to reduce Aβ synthesis via inhibition of γ-secretase activity (reviewed in Tang 2005).
In summary, our studies underscore a novel mechanism of functional aging. This mechanism involves Rho signaling and is unique in that it is initiated by a GEF that appears to be specifically activated in response to conditions of oxidative stress. While our results support a leading role for Rho GTPases in the aging process they also raise many questions. For example, through which molecular mechanisms does oxidative stress lead to OSG-1 activation? How does OSG-1 activity affect the actin cytoskeleton? Does OSG-1 have a role in muscle? Future studies will tackle these fascinating questions.
Acknowledgments
We thank Maria Riego and Joseph Ifrach and Dr. Shuang Liu for critical reading of the manuscript and the Caenorhabditis Genetic Center for the DA1262, RB1560, TK22, and TJ1052 strains. The Pgcy-5::GFP construct was a kind gift of Dr. Oliver Hobert. This work was supported by a National Science Foundation grant (1026958) to F.S.
Footnotes
Supporting information is available online at http://www.genetics.org/lookup/suppl/doi:10.1534/genetics.114.173500/-/DC1.
Communicating editor: B. Goldstein
Literature Cited
- Alcedo J., Kenyon C., 2004. Regulation of C. elegans longevity by specific gustatory and olfactory neurons. Neuron 41: 45–55. [DOI] [PubMed] [Google Scholar]
- Apfeld J., Kenyon C., 1999. Regulation of lifespan by sensory perception in Caenorhabditis elegans. Nature 402: 804–809. [DOI] [PubMed] [Google Scholar]
- Bargmann C. I., Horvitz H. R., 1991. Chemosensory neurons with overlapping functions direct chemotaxis to multiple chemicals in C. elegans. Neuron 7: 729–742. [DOI] [PubMed] [Google Scholar]
- Bargmann C. I., Mori I., 1997. Chemotaxis and thermotaxis, pp. 645–677 in C. elegans II, edited by Riddle D. L., Blumenthal T., Meyer B. J., Priess J. R. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [PubMed] [Google Scholar]
- Bianchi L., Kwok S. M., Driscoll M., Sesti F., 2003. A potassium channel-MiRP complex controls neurosensory function in Caenorhabditis elegans. J. Biol. Chem. 278: 12415–12424. [DOI] [PubMed] [Google Scholar]
- Bustelo, X. R., V. Sauzeau and I. M. Berenjeno, 2007 GTP-binding proteins of the Rho/Rac family: regulation, effectors and functions in vivo. BioEssays 29: 356–370. [DOI] [PMC free article] [PubMed]
- Butterfield D. A., Lauderback C. M., 2002. Lipid peroxidation and protein oxidation in Alzheimer’s disease brain: potential causes and consequences involving amyloid beta-peptide-associated free radical oxidative stress. Free Radic. Biol. Med. 32: 1050–1060. [DOI] [PubMed] [Google Scholar]
- Cai S. Q., Sesti F., 2009. Oxidation of a potassium channel causes progressive sensory function loss during aging. Nat. Neurosci. 12: 611–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W., Lim L., 1994. The Caenorhabditis elegans small GTP-binding protein RhoA is enriched in the nerve ring and sensory neurons during larval development. J. Biol. Chem. 269: 32394–32404. [PubMed] [Google Scholar]
- Cotella, D., B. Hernandez-Enriquez, X. Wu, R. Li, Z. Pan et al., 2012 Toxic role of k+ channel oxidation in mammalian brain. J. Neurosci. 32: 4133–4144. [DOI] [PMC free article] [PubMed]
- Davis M. W., Hammarlund M., Harrach T., Hullett P., Olsen S., et al. , 2005. Rapid single nucleotide polymorphism mapping in C. elegans. BMC Genomics 6: 118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desouza M., Gunning P. W., Stehn J. R., 2012. The actin cytoskeleton as a sensor and mediator of apoptosis. BioArchitecture 2: 75–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan Z., Sesti F., 2013. A Caenorhabditis elegans model system for amylopathy study. J. Vis. Exp. 75: 10.3791/50435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis H. M., Horvitz H. R., 1986. Genetic control of programmed cell death in the nematode C. elegans. Cell 44: 817–829. [DOI] [PubMed] [Google Scholar]
- Gourlay C. W., Ayscough K. R., 2005a The actin cytoskeleton in ageing and apoptosis. FEMS Yeast Res. 5: 1193–1198. [DOI] [PubMed] [Google Scholar]
- Gourlay C. W., Ayscough K. R., 2005b The actin cytoskeleton: a key regulator of apoptosis and ageing? Nat. Rev. Mol. Cell Biol. 6: 583–589. [DOI] [PubMed] [Google Scholar]
- Ishii N., Takahashi K., Tomita S., Keino T., Honda S., et al. , 1990. A methyl viologen-sensitive mutant of the nematode Caenorhabditis elegans. Mutat. Res. 237: 165–171. [DOI] [PubMed] [Google Scholar]
- Ishii N., Fujii M., Hartman P. S., Tsuda M., Yasuda K., et al. , 1998. A mutation in succinate dehydrogenase cytochrome b causes oxidative stress and ageing in nematodes. Nature 394: 694–697. [DOI] [PubMed] [Google Scholar]
- Ishii N., Senoo-Matsuda N., Miyake K., Yasuda K., Ishii T., et al. , 2004. Coenzyme Q10 can prolong C. elegans lifespan by lowering oxidative stress. Mech. Ageing Dev. 125: 41–46. [DOI] [PubMed] [Google Scholar]
- Jaffe A. B., Hall A., 2005. Rho GTPases: biochemistry and biology. Annu. Rev. Cell Dev. Biol. 21: 247–269. [DOI] [PubMed] [Google Scholar]
- Kamath, R. S., M. Martinez-Campos, P. Zipperlen, A. G. Fraser, and J. Ahringer, 2001 Effectiveness of specific RNA-mediated interference through ingested double-stranded RNA in Caenorhabditis elegans. Genome Biol. 2: RESEARCH0002. [DOI] [PMC free article] [PubMed]
- Kenyon C. (Editor), 1997. Environmental Factors and Gene Activities That Influence Life Span. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [PubMed] [Google Scholar]
- Larsen P. L., 1993. Aging and resistance to oxidative damage in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 90: 8905–8909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D., Roberts R., 2001. WD-repeat proteins: structure characteristics, biological function, and their involvement in human diseases. Cellular and molecular life sciences. Cell. Mol. Life Sci. 58: 2085–2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Link C. D., 2006. C. elegans models of age-associated neurodegenerative diseases: lessons from transgenic worm models of Alzheimer’s disease. Exp. Gerontol. 41: 1007–1013. [DOI] [PubMed] [Google Scholar]
- Liu J., Zhang B., Lei H., Feng Z., Hsu A. L., et al. , 2013. Functional aging in the nervous system contributes to age-dependent motor activity decline in C. elegans. Cell Metab. 18: 392–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundquist, E. A., 2006 Small GTPases. WormBook, ed. The C. elegans Research Community WormBook, /10.1895/wormbook.1.6,7.1, http://www.wormbook.org.
- Lundquist E. A., Reddien P. W., Hartwieg E., Horvitz H. R., Bargmann C. I., 2001. Three C. elegans Rac proteins and several alternative Rac regulators control axon guidance, cell migration and apoptotic cell phagocytosis. Development 128: 4475–4488. [DOI] [PubMed] [Google Scholar]
- Maglioni S., Schiavi A., Runci A., Shaik A., Ventura N., 2014. Mitochondrial stress extends lifespan in C. elegans through neuronal hormesis. Exp. Gerontol. 56: 89–98. [DOI] [PubMed] [Google Scholar]
- Matsushita T., Ashikawa K., Yonemoto K., Hirakawa Y., Hata J., et al. , 2010. Functional SNP of ARHGEF10 confers risk of atherothrombotic stroke. Hum. Mol. Genet. 19: 1137–1146. [DOI] [PubMed] [Google Scholar]
- Mattson M. P., 2004. Pathways towards and away from Alzheimer’s disease. Nature 430: 631–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendoza-Naranjo A., Gonzalez-Billault C., Maccioni R. B., 2007. Abeta1–42 stimulates actin polymerization in hippocampal neurons through Rac1 and Cdc42 Rho GTPases. J. Cell Sci. 120: 279–288. [DOI] [PubMed] [Google Scholar]
- Minniti A. N., Cataldo R., Trigo C., Vasquez L., Mujica P., et al. , 2009. Methionine sulfoxide reductase A expression is regulated by the DAF-16/FOXO pathway in Caenorhabditis elegans. Aging Cell 8: 690–705. [DOI] [PubMed] [Google Scholar]
- Pan C. L., Peng C. Y., Chen C. H., McIntire S., 2011. Genetic analysis of age-dependent defects of the Caenorhabditis elegans touch receptor neurons. Proc. Natl. Acad. Sci. USA 108: 9274–9279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce-Shimomura J., Faumont S., Gaston M., Pearson B., Lockery S., 2001. The homeobox gene lim-6 is required for distinct chemosensory representations in C. elegans. Nature 410: 694–698. [DOI] [PubMed] [Google Scholar]
- Ranta S., Lehesjoki A. E., de Fatima Bonaldo M., Knowles J. A., Hirvasniemi A., et al. , 1997. High-resolution mapping and transcript identification at the progressive epilepsy with mental retardation locus on chromosome 8p. Genome Res. 7: 887–896. [DOI] [PubMed] [Google Scholar]
- Roh-Johnson M., Goldstein B., 2009. In vivo roles for Arp2/3 in cortical actin organization during C. elegans gastrulation. J. Cell Sci. 122: 3983–3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohn T. T., Head E., 2008. Caspase activation in Alzheimer’s disease: early to rise and late to bed. Rev. Neurosci. 19: 383–393. [DOI] [PubMed] [Google Scholar]
- Rohn T. T., Head E., 2009. Caspases as therapeutic targets in Alzheimer’s disease: Is it time to “cut” to the chase? Int. J. Clin. Exp. Pathol. 2: 108–118. [PMC free article] [PubMed] [Google Scholar]
- Sawa M., Suetsugu S., Sugimoto A., Miki H., Yamamoto M., et al. , 2003. Essential role of the C. elegans Arp2/3 complex in cell migration during ventral enclosure. J. Cell Sci. 116: 1505–1518. [DOI] [PubMed] [Google Scholar]
- Schmidt A., Hall A., 2002. Guanine nucleotide exchange factors for Rho GTPases: turning on the switch. Genes Dev. 16: 1587–1609. [DOI] [PubMed] [Google Scholar]
- Shaye D. D., Greenwald I., 2011. OrthoList: a compendium of C. elegans genes with human orthologs. PLoS ONE 6: e20085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer A. G., Orita S., Malone C. J., Han M., 2001. A RHO GTPase-mediated pathway is required during P cell migration in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 98: 13132–13137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahvanainen E., Ranta S., Hirvasniemi A., Karila E., Leisti J., et al. , 1994. The gene for a recessively inherited human childhood progressive epilepsy with mental retardation maps to the distal short arm of chromosome 8. Proc. Natl. Acad. Sci. USA 91: 7267–7270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang B. L., 2005. Alzheimer’s disease: channeling APP to non-amyloidogenic processing. Biochem. Biophys. Res. Commun. 331: 375–378. [DOI] [PubMed] [Google Scholar]
- Tank, E. M., K. E. Rodgers and C. Kenyon, 2011 Spontaneous age-related neurite branching in Caenorhabditis elegans. J. Neurosci. 31: 9279–9288. [DOI] [PMC free article] [PubMed]
- Tolias K. F., Duman J. G., Um K., 2011. Control of synapse development and plasticity by Rho GTPase regulatory proteins. Prog. Neurobiol. 94: 133–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toth, M. L., I. Melentijevic, L. Shah, A. Bhatia, K. Lu et al., 2012 Neurite sprouting and synapse deterioration in the aging Caenorhabditis elegans nervous system. J. Neurosci. 32: 8778–8790. [DOI] [PMC free article] [PubMed]
- Verhoeven K., De Jonghe P., Van de Putte T., Nelis E., Zwijsen A., et al. , 2003. Slowed conduction and thin myelination of peripheral nerves associated with mutant rho Guanine-nucleotide exchange factor 10. Am. J. Hum. Genet. 73: 926–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, Y., Z. Wu, P. Butko, Y. Christen, M. P. Lambert et al., 2006 Amyloid-beta-induced pathological behaviors are suppressed by Ginkgo biloba extract EGb 761 and ginkgolides in transgenic Caenorhabditis elegans. J. Neurosci. 26: 13102–13113. [DOI] [PMC free article] [PubMed]
- Yu S., Avery L., Baude E., Garbers D., 1997. Guanylyl cyclase expression in specific sensory neurons: a new family of chemosensory receptors. Proc. Natl. Acad. Sci. USA 94: 3384–3387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J., Horvitz H. R., 1992. The Caenorhabditis elegans cell death gene ced-4 encodes a novel protein and is expressed during the period of extensive programmed cell death. Development 116: 309–320. [DOI] [PubMed] [Google Scholar]