Abstract
As a common cause of reproductive isolation in diverse taxa, hybrid incompatibilities are fundamentally important to speciation. A key question is which evolutionary forces drive the initial substitutions within species that lead to hybrid dysfunction. Previously, we discovered a simple genetic incompatibility that causes nearly complete male sterility and partial female sterility in hybrids between the two closely related yellow monkeyflower species Mimulus guttatus and M. nasutus. In this report, we fine map the two major incompatibility loci—hybrid male sterility 1 (hms1) and hybrid male sterility 2 (hms2)—to small nuclear genomic regions (each <70 kb) that include strong candidate genes. With this improved genetic resolution, we also investigate the evolutionary dynamics of hms1 in a natural population of M. guttatus known to be polymorphic at this locus. Using classical genetic crosses and population genomics, we show that a 320-kb region containing the hms1 incompatibility allele has risen to intermediate frequency in this population by strong natural selection. This finding provides direct evidence that natural selection within plant species can lead to hybrid dysfunction between species.
Keywords: speciation, hybrid incompatibilities, postzygotic reproductive isolation, Mimulus, monkeyflower
SPECIATION occurs when diverging populations accumulate genetic differences that cause reproductive isolation. Many forms of prezygotic reproductive isolation likely evolve as byproducts of adaptation to different ecological conditions (e.g., habitat or behavioral differences), but the evolutionary dynamics of intrinsic postzygotic isolation are less clear. This is because the production of dead or sterile hybrids cannot be favored by natural selection. Dobzhansky (1937) and Muller (1942) proposed a solution to this long-standing mystery (Darwin 1859), explaining that a new mutation might function perfectly well with alleles present in its native species, and only cause sterility or inviability when found in a hybrid genetic background. The so-called Dobzhansky–Muller model shows that natural selection need not oppose the evolution of hybrid dysfunction, but it makes no predictions about the nature of the genetic changes or the evolutionary forces that give rise to hybrid incompatibilities.
The recent identification of several hybrid incompatibility genes in diverse taxa has begun to reveal some insights into the evolution of hybrid dysfunction (reviewed in Presgraves 2010; Maheshwari and Barbash 2011; Sweigart and Willis 2012). In Arabidopsis and rice, genetic incompatibilities have been mapped to duplicate genes that carry loss-of-function alleles in alternate copies (Bikard et al. 2009; Mizuta et al. 2010; Yamagata et al. 2010), suggesting that divergence among paralogs via mutation and genetic drift might cause postzygotic reproductive isolation (Lynch and Force 2000). There are also hints that natural selection can contribute to the spread of incompatible alleles within populations and species. For example, plant hybrid necrosis has been mapped repeatedly to disease resistance genes (Krüger et al. 2002; Bomblies et al. 2007; Alcazar et al. 2009; Jeuken et al. 2009; Yamamoto et al. 2010; Chen et al. 2014), which are likely targets of adaptive divergence to unique pathogen communities (Bomblies and Weigel 2007). Additionally, many hybrid incompatibility genes show molecular signatures of positive selection (Presgraves et al. 2003; Brideau et al. 2006; Maheshwari et al. 2008; Oliver et al. 2009; Phadnis and Orr 2009; Tang and Presgraves 2009), but, interestingly, few of these genes seem to be involved in classical ecological adaptation. Instead, it has been proposed that rapid divergence at hybrid incompatibility loci might be driven by recurrent bouts of intragenomic conflict involving segregation distorters (Frank 1990; Hurst and Pomiankowski 1991). Consistent with this idea, studies in Drosophila and rice find that hybrid segregation distortion maps to the same genomic locations as hybrid sterility (Tao et al. 2001; Long et al. 2008; Phadnis and Orr 2009; Zhao et al. 2010; Yang et al. 2012). Ultimately, an important challenge for speciation geneticists is to determine which evolutionary forces cause the initial spread of incompatibility alleles within species.
A promising way forward is to focus on young species pairs that are not yet fixed for hybrid incompatibilities. There is now abundant evidence from diverse systems that polymorphic loci contribute to variation in hybrid dysfunction (Cutter 2012); in some cases, it is feasible to both genetically map hybrid incompatibilities and investigate their evolutionary dynamics in natural populations. This combination of approaches was used in a recent study of Mimulus guttatus, which showed that a hybrid lethality allele at the NecI locus hitchhiked to high frequency during the adaptive fixation of a copper tolerance allele at a tightly linked gene (Wright et al. 2013). Additional studies of this sort are needed to determine which population genetic forces and selective agents are most important for the evolution of postzygotic reproductive isolation.
Here we investigate the genetics and evolution of a well-characterized hybrid incompatibility between two closely related species of monkeyflower, M. guttatus and M. nasutus. In this system, two major incompatibility loci—hybrid male sterility 1 (hms1) and hybrid male sterility 2 (hms2)—cause nearly complete male sterility and partial female sterility in Mimulus hybrids (Sweigart et al. 2006). Additionally, we know how these loci vary in nature: the hms1 incompatibility allele has a limited distribution in M. guttatus and is even polymorphic within populations, whereas the hms2 incompatibility allele is geographically widespread, and potentially fixed, in M. nasutus (Sweigart et al. 2007). This genetically simple, polymorphic hybrid incompatibility system in a young species pair sets the stage to directly link the intraspecific causes of divergence to hybrid dysfunction.
In the present study, we perform fine-scale genetic mapping to narrow the hms1 and hms2 intervals. In both genomic regions, we identify strong candidate genes for Mimulus hybrid sterility. We also take advantage of natural variation within M. guttatus to investigate the evolutionary dynamics of hms1. Using a population genomics approach, we discover that strong natural selection has driven the hms1 incompatibility allele to intermediate frequency within a population of M. guttatus. This study provides an especially detailed look at the evolution of a hybrid incompatibility locus that is still in the early stages of divergence.
Materials and Methods
Study system
The M. guttatus species complex is a group of closely related, phenotypically diverse wildflowers that exhibits tremendous variation in reproductive isolation between populations and species. Our study focuses on the most geographically widespread and morphologically extreme members of the complex: M. guttatus, an outcrosser with large, bee-pollinated flowers, and M. nasutus, a selfer with reduced, mostly closed flowers. Despite their phenotypic differences, these species are closely related, having diverged ∼200,000 years ago (Brandvain et al. 2014). Natural populations of both species are abundant throughout much of western North America but the range of M. nasutus is more restricted. In areas of overlap, the two Mimulus species are partially reproductively isolated by differences in floral morphology, flowering phenology, and pollen–pistil interactions (Diaz and MacNair 1999; Martin and Willis 2007; Fishman et al. 2014). Nevertheless, hybrids are frequently observed at sympatric sites (Vickery 1964; Martin and Willis 2007; A. M. Kenney and A. L. Sweigart, unpublished results), and we find evidence of genome-wide introgression (Sweigart and Willis 2003; Brandvain et al. 2014). Hybrid incompatibilities are also common, but variable (Vickery 1978; Christie and MacNair 1987; Sweigart et al. 2007; Case and Willis 2008; Martin and Willis 2010).
Specific lines and previous genetic mapping
Previously, we identified a pair of nuclear incompatibility loci that causes nearly complete male sterility and partial female sterility in a fraction of F2 hybrids between an inbred line of M. guttatus from Iron Mountain, Oregon (IM62), and an M. nasutus line from Sherar’s Falls, Oregon (SF5) (Sweigart et al. 2006). The incompatibility is between a semidominant IM62 allele at hms1 and recessive SF5 alleles at hms2. Initially, we mapped hms1 to a region of 12 cM on linkage group 6 and hms2 to 8 cM on linkage group 13 (Sweigart et al. 2006). Following the release of the M. guttatus reference genome (generated from the IM62 line, www.phytozome.net), we identified previously unmapped, gene-based MgSTS markers (http://www.mimulusevolution.org) in these intervals. We also designed new, intron-spanning, length-polymorphic markers targeted to each locus. Using F2 recombinants generated in our previous study and these new markers, we refined each hybrid sterility locus: hms1 was mapped between flanking markers M8 and M24, and hms2 between MgSTS193 and M51 (primer sequences for these markers are in Supporting Information, Table S1).
Fine mapping
To fine map hms1 and hms2, we generated two large SF5 × IM62 F2 mapping populations in consecutive years (in 2011, N = 4220 and in 2012, N = 4894). The first F2 population was grown in the greenhouse at the University of Montana (in 2011) and the second at the University of Georgia (in 2012). Seeds were planted into 96-well flats containing Fafard 3b potting mix, chilled for 7 days at 4° to promote germination, and then placed in a greenhouse with supplemental lights set to 16-hr days. Plants were bottom watered daily and temperatures were maintained at 24° during the day and 16° at night.
We collected leaf tissue from all F2 hybrids and isolated genomic DNA using a rapid extraction protocol (Cheung et al. 1993) modified for 96-well format. We then genotyped these F2 hybrids for a multiplexed set of fluorescently labeled markers that flank hms1 (M8 and M24) and hms2 (MgSTS193 and M51) following amplification protocols used previously (Sweigart et al. 2006, 2007). All individuals with informative recombination events in either interval were retained and genotyped at additional markers designed by walking through the hms1 and hms2 regions of the IM62 M. guttatus genome sequence assembly. Publically available resequence data from the SF5 M. nasutus parent (Table S2) allowed us to develop new markers that span known SNPs and/or indel polymorphisms (Table S1). All genotypes were scored automatically using GeneMarker (SoftGenetics), with additional hand scoring where necessary. Male fertility (i.e., pollen viability) for informative recombinants was assessed as described previously (Sweigart et al. 2006, 2007).
To map hms1, we retained individuals that were homozygous for SF5 alleles at both hms2 flanking markers and recombinant for hms1 markers. Because these individuals are homozygous for the SF5 incompatibility allele at hms2, they will be male sterile if they also carry at least one IM62 allele at the causal hms1 locus. For one class of hms1 recombinants—those that are homozygous for SF5 alleles at one flanking marker and heterozygous at the other—it is straightforward to assess the effect of genotype on male fertility phenotype. However, for the other class of hms1 recombinants—those that are homozygous for IM62 alleles at one marker and heterozygous at the other—progeny testing is required (this is because there is no phenotypic variation among this class of recombinants; all are highly male sterile, having inherited at least one copy of the IM62 allele). We used these male sterile F2 recombinants as maternal parents in crosses to SF5: individuals that are homozygous for IM62 alleles at hms1 produce only male sterile progeny (all are heterozygous at hms1 and homozygous for SF5 alleles at hms2), and those that are heterozygous at hms1 produce progeny that segregate 1:1 for male fertility.
To map hms2, we identified individuals from the 2011 F2 mapping population that carried at least one IM62 allele at hms1 (heterozygous or homozygous for IM62 alleles at hms1 flanking markers) and were recombinant for hms2 markers. For hms2, we retained only one class of recombinants—those homozygous for SF5 alleles at one hms2 marker and heterozygous at the other. Because these F2 recombinants all carry at least one IM62 allele at hms1, they are expected to be male sterile if they are homozygous for SF5 alleles at hms2 and male fertile if they are heterozygous. However, male fertility among these hms2 recombinants is not entirely discrete, due to both incomplete dominance at hms1 and variation at additional, small-effect hybrid sterility loci (see figure 5 in Sweigart et al. 2006). To address this issue, we used a QTL mapping approach to localize hybrid sterility effects in the hms2 region. Linkage analysis was performed using JoinMap 4.1 (Van Ooijen 2006) by genotyping a subset of informative recombinants for hms2. To preserve realistic genetic distances, we also included nonrecombinant F2 genotypes (i.e., individuals with the same genotype at both hms2-flanking markers) in the linkage analysis and assumed no recombination among intervening markers. A genetic map was constructed using the maximum likelihood mapping algorithm and Haldane mapping function. Markers were given a fixed order (based on physical locations) and grouped with a LOD score threshold of 10.0. To detect QTL, we used the R/qtl package (Broman et al. 2003) to run a “scanone” analysis in 1-cM steps using the Haley–Knott regression.
For a subset of hms1 and hms2 recombinants, we performed additional progeny testing to improve our confidence in phenotypic assignments (Table S3 and Table S4). The idea was to minimize variation in hybrid male fertility due to the environment (e.g., common greenhouse pests like thrips can lower pollen viability). To generate progeny, fertile recombinants were self-fertilized and male sterile recombinants were crossed to SF5 (using the F2 hybrid as the maternal parent). For 10 hms1 recombinants and 12 hms2 recombinants, we scored between 10 and 106 progeny for hms1-2 genotypes and male fertility phenotypes (an average of 43 progeny were measured).
Crosses to detect carriers of the hms1 incompatibility allele
Previously, we showed that hms1 is polymorphic within the Iron Mountain population and that the hybrid sterility allele occurs at intermediate frequency (Sweigart et al. 2007). To test for the presence of hms1 incompatibility alleles in additional M. guttatus inbred lines derived from the IM population, we performed backcrosses between them and SF5. If a particular IM inbred line carries an IM62-like hybrid sterility allele at hms1, roughly one-fourth of its progeny in an F1 backcross to SF5 is expected to have the incompatible genotype. If instead, the IM inbred line carries a compatible allele at hms1, genotypes at hms1 and hms2 should not affect male fertility. For each IM inbred line-SF5 backcross, we measured pollen viability and determined hms1 and hms2 genotypes for at least 24 progeny (an average of 42 were measured). We then performed an ANOVA for each cross to assess the contribution of hms1 and hms2 to variation in hybrid male fertility.
Sequencing and population genomic analyses
To examine population genomic variation for hms1 and hms2, we used whole genome resequence data from 10 IM inbred lines (Flagel et al. 2014) and 10 M. guttatus complex accessions sampled from throughout the species range (Table S2; for accession information see Brandvain et al. 2014). For the IM inbred lines, full protocols for genomic DNA isolation, Illumina sequencing, and sequence alignment can be found in Flagel et al. 2014. Briefly, each line was sequenced to ∼7–15 times the genomic coverage using Illumina paired-end sequences. These sequences were aligned to the IM62 v2.0 reference genome assembly (ftp://ftp.jgi-psf.org/pub/compgen/phytozome/v9.0/early_release/Mguttatus_v2.0/assembly/) using the Burrows-Wheeler Aligner (Li and Durbin 2010). We then performed base calls using the UnifiedGenotyper method in GATK v3.2 (Depristo et al. 2011). We retained sites with a genotype quality score of ≥30. Raw sequence data can be found at the National Center for Biotechnology Information Sequence Read Archive accessions listed in Table S2. For population genomic analyses, we fit an integrated Mimulus genetic map (described in Brandvain et al. 2014) to physical positions from the IM62 sequence assembly.
Because we resequenced the inbred line used to create the reference genome (IM62) we have a direct estimate of the sequencing/genotyping error rate for our samples. Genome-wide, the difference between genotype calls for the resequenced IM62 and the IM62 reference genome was 2.6 SNPs every 10 kb (i.e., total πerror = 2.6 × 10−4). Assuming half the errors belong to the reference and half belong to our method, we can estimate our lower detection limit for diversity to be approximately π = 1.3 × 10−4.
To design genetic markers that distinguish hms1 haplotype groups (see Results), we screened for SNP differences that create polymorphic restriction sites. These cleaved amplified polymorphic sequence (CAPS) markers (Table S1) were used to examine hms1 genomic variation in a large collection of inbred lines derived from the IM population.
Results
Fine mapping localizes hybrid sterility to strong candidate genes
To fine map Mimulus hybrid sterility loci, we genotyped all individuals from two large F2 mapping populations at gene-based markers (see www.mimulusevolution.org and Table S1) known to flank hms1 (M8 and M24) and hms2 (M51 and MgSTS193). Consistent with previous findings (Sweigart et al. 2006), we observed significant transmission ratio distortion at hms2 with a deficit of M. nasutus alleles (frequency of M. guttatus to M. nasutus alleles: expected = 0.5:0.5, observed = 0.62:0.38, χ2 = 177.14, d.f. = 1, P < 0.0001, N = 6137). At hms1, allelic transmission did not differ from the Mendelian expectation (M. guttatus to M. nasutus alleles: expected = 0.5:0.5, observed = 0.49:0.51, χ2 = 0.44, d.f. = 1, P = 0.51, N = 7028).
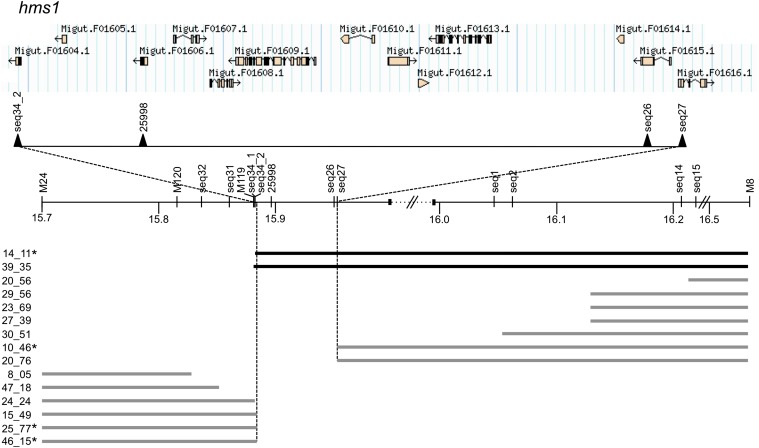
To dissect the hms1 locus, we focused on a small subset of F2 hybrids that were both recombinant at hms1 and homozygous for M. nasutus alleles at hms2 (note that this hms2 genotypic class is small due to the transmission ratio distortion reported above). We then genotyped these recombinants at additional markers targeted to the hms1 region. As shown in Figure 1, F2 recombination within this region localizes the hms1 sterility effects to an interval of only 67,893 bp with 11 predicted genes (Table 1).
Figure 1.
Genetic dissection and physical map of the hms1 locus in Mimulus. Informative recombinants from an F2 mapping population are shown with horizontal bars representing heterozygous genotypes, or, for recombinants that required progeny testing (indicated by asterisks), regions homozygous for M. guttatus alleles (see Materials and Methods for details). Solid bars indicate male-sterile individuals and shaded bars indicate male-fertile individuals. Physical locations of genetic markers (1 Mb shown) map hybrid male sterility to a region of only 68 kb (between markers seq34_2 and seq27) that includes 11 predicted genes. Annotation is based on M. guttatus Annotation v2.0, phytozome.net (M. guttatus v1.1 assembly scaffolds are separated by dotted lines with double hash marks).
Table 1. Predicted genes found in fine mapped regions of hms1 and hms2.
| Locus | Gene namea | Gene annotationa | Predicted functional roleb | Sterility phenotype in Arabidopsis? |
|---|---|---|---|---|
| hms1 | Migut.F01605 and Migut.F01606 | Skp1 family | Cell cycle regulation, component of SCF ubiquitin–ligase complexc | Arabidopsis Skp1 homolog (ASK1) mutant causes male sterility via defects in male meiosisd |
| Migut.F01607 | Iron–sulfur cluster biosynthesis | Iron–sulfur cluster binding, structural molecule activity | ||
| Migut.F01608 | 4′-phosphopantetheinyl transferase superfamily | Involved in metabolic processes, macromolecule biosynthesis | ||
| Migut.F01609 | None | |||
| Migut.F01610 | Leucine rich repeat (LRR) | Kinase activity, signal transduction; diverse roles in plant development and disease resistancee | Mutations in EMS1/EXS (1 of >200 LRR genes in Arabidopsis) causes male sterility when microsporocytes do not undergo proper cytokinesisf | |
| Migut.F01611 | PPR repeat | Organellar RNA processing, embryonic developmentg | Some members of this large gene family (450 in Arabidopsis) are involved in fertility restoration of CMS plantsh | |
| Migut.F01612 | F-box | Component of SCF ubiquitin–ligase complex, targets specific proteins for degradation | Among this large gene family (>600 in Arabidopsis), COI1 mutants are male sterilei, FBL17 mutants inhibit male germline proliferationj, the rice hybrid sterility gene SaF encodes an F-box proteink | |
| Migut.F01613 | None | |||
| Migut.F01614 | Yip1 domain; Golgi membrane protein | Involved in the trans-Golgi networkl | ||
| Migut.F01615 | Amino acid kinase family; gamma-glutamyl kinase | Amino acid biosynthesis | ||
| hms2 | Migut.M00294 | Protein kinase domain; cdc2-related protein kinase | Cyclin-dependent kinase (CDK)-like proteinm, cell cycle regulation | Arabidopsis cdc2 mutant (CDKA;1) is male sterile because it fails to progress through the second mitosis in male gametogenesisn |
| Migut.M00295 | Cellulase (glycosyl hydrolase family 5) | Involved in carbohydrate metabolism, hydrolase activity | ||
| Migut.M00296 | None | |||
| Migut.M00297 | RNA polymerase Rpb2 | Encodes a subunit of DNA-dependent RNA polymerase II | ||
| Migut.M00298 | GH3 auxin-responsive promoter | Involved in red light-specific hypocotyl elongation |
Unless a citation is provided, functional descriptions are taken from TAIR (arabidopsis.org) for the gene’s closest Arabidopsis thaliana BLAST hit.
Xie et al. 1999.
The genes that map to the hms1 locus include three strong candidates for involvement in hybrid sterility. Migut.F01605 and Migut.F01606 are tandem duplicates of SKP1-like genes. In Arabidopsis, SKP1 (ASK1) mutants cause male sterility and the SKP1 gene functions during meiosis (Yang et al. 1999). Migut.F01612 is annotated as a member of the F-box gene family, which mediates protein–protein interactions and influences diverse developmental traits including fertility (e.g., Devoto et al. 2002; Kim et al. 2010). Intriguingly, of the few hybrid sterility genes cloned in rice, one encodes an F-box gene (Sa, Long et al. 2008). Although none of the other hms1-interval genes have such clear connections to gametogenesis, we cannot yet rule out their involvement in Mimulus hybrid sterility. Indeed, two of the predicted genes in the hms1 interval (Migut.F01610 and Migut.F01611) are members of large gene families that play a variety of roles during plant development. Two others lack functional annotations altogether, so it is unclear how they might affect fertility. Going forward, additional rounds of fine mapping and/or functional experiments will be needed to identify which of these hms1 genes causes Mimulus hybrid sterility.
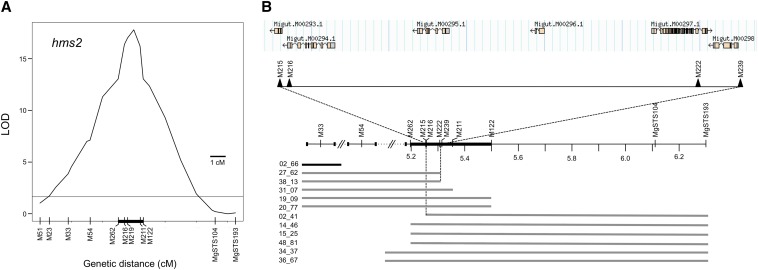
To fine map hms2, we identified F2 hybrids that were recombinant at hms2 and also carried at least one M. guttatus allele at hms1. Against this hms1 background, which is carried by roughly three-quarters of the F2 hybrids, we identified a large number of informative recombinants in the 2011 F2 mapping population (N = 130 recombinants homozygous for M. nasutus alleles at one hms2 flanking marker and heterozygous at the other). To narrow the genomic region associated with hybrid male sterility, we took a QTL mapping approach, genotyping the hms2 recombinants for newly designed markers spanning the region between the original flanking markers MgSTS193 and M51. The QTL for hybrid male sterility is highly significant [likelihood ratio (LR) = 17.8 vs. an LR threshold of 1.65; Figure 2] and maps to a 98,503-bp interval between markers M216 and M211 (1 LOD-drop boundaries). Further progeny testing for 12 of the hms2 recombinants suggests that the QTL resides in a region of only 60,052 bp containing five predicted genes (Table 1). One of these, Migut.M00294, is annotated as a cdc2-related protein kinase and is a strong candidate for hms2. Highly conserved among all eukaryotes, cdc2 is a key regulator of the cell cycle. In Arabidopsis, a loss-of-function mutation in a homolog of cdc2 (CDKA; 1) results in pollen lethality due to a failure of generative cell division in male gametogenesis (Nowack et al. 2005; Iwakawa et al. 2006). Although the other four candidate genes in the hms2 region have no known roles in pollen development, as with hms1, additional experiments are needed to determine with certainty which gene underlies hms2.
Figure 2.
Genetic dissection and physical map of the hms2 locus in Mimulus. (A) QTL profile generated from informative hms2 F2 recombinants that also carry at least one M. guttatus allele at hms1. The positions of molecular markers are indicated along the bottom. Hybrid male sterility effects were mapped to an interval of roughly 1 cM between M216 and M211. The thick black bar along the x-axis indicates a 2-LOD drop on either side of the peak, corresponding to 1.6 cM. The horizontal line at 1.65 shows the 5% significance LOD threshold generated using 1000 permutations. (B) Progeny testing for a subset of the informative hms2 recombinants further refined the sterility locus. The hms2 F2 recombinants are shown with horizontal bars representing heterozygous genotypes (regions left blank indicate markers homozygous for M. nasutus alleles). Because all of these individuals carry at least one M. guttatus allele at hms1, they are sterile if they are also homozygous for M. nasutus alleles at hms2. Black bar indicates a male-sterile individual and shaded bars indicate male-fertile individuals. (Note that the larger number of male fertiles reflects a bias in seed production between these phenotypic classes; we were more likely to collect seeds from male-fertile recombinants.) Physical locations of genetic markers (∼1 Mb shown) map hms2 to a region of only 60,052 bp (between markers M215 and M239) that includes five predicted genes. Annotation is based on M. guttatus Annotation v2.0, phytozome.net (M. guttatus v1.1 assembly scaffolds are separated by dotted lines with double hash marks).
The hms1 sterility allele is at intermediate frequency within the Iron Mountain population
To examine natural variation for hms1 within the Iron Mountain population of M. guttatus, we tested for the presence of the incompatibility allele in 18 inbred lines derived from this locale. Consistent with our previous results (Sweigart et al. 2007), we find that the hms1 incompatibility allele is at intermediate frequency in the Iron Mountain population: 9 of the 18 IM inbred lines segregate male sterile progeny when backcrossed to SF5 and variation in male fertility is strongly associated with hms1 inheritance (Table 2). The other 9 IM lines produce mostly fertile progeny when backcrossed to SF5 and show no effect of genotype at the hms1 locus. Although our sample sizes were often too small to detect a significant interaction between hms1 and hms2, the backcross progeny that carried the hms1-hms2 incompatibility (i.e., heterozygous at hms1 and homozygous for M. nasutus alleles at hms2) were almost always the most sterile genotype.
Table 2. Variation at the hms1 locus within the Iron Mountain population of M. guttatus.
| PV hms1: GN | PV hms1: NN | |||||
|---|---|---|---|---|---|---|
| IM line | hms2: GN | hms2: NN | hms2: GN | hms2: NN | Fhms1 | Fhms1-2 |
| 624 | 0.410 (0.064, 17) | 0.535 (0.088, 9) | 0.400 (0.088, 9) | 0.594 (0.099, 7) | 0.081 | 0.165 |
| 617 | 0.717 (0.055, 6) | 0.732 (0.030, 21) | 0.819 (0.096, 2) | 0.677 (0.034, 16) | 0.153 | 1.724 |
| 1189 | 0.627 (0.078, 10) | 0.477 (0.068, 13) | 0.528 (0.087, 8) | 0.453 (0.078, 10) | 0.625 | 0.235 |
| 155 | 0.751 (0.062, 9) | 0.767 (0.066, 8) | 0.696 (0.071, 7) | 0.708 (0.048, 15) | 0.830 | 0.002 |
| 767 | 0.707 (0.058, 14) | 0.615 (0.060, 13) | 0.717 (0.062, 12) | 0.737 (0.065, 11) | 1.188 | 0.845 |
| 323 | 0.740 (0.076, 7) | 0.783 (0.061, 11) | 0.600 (0.064, 10) | 0.753 (0.082, 6) | 1.428 | 0.602 |
| 591 | 0.486 (0.074, 16) | 0.479 (0.085, 12) | 0.538 (0.072, 17) | 0.669 (0.099, 9) | 2.115 | 0.692 |
| 1152 | 0.722 (0.070, 12) | 0.644 (0.085, 8) | 0.492 (0.067, 13) | 0.638 (0.097, 13) | 2.623 | 2.326 |
| 1145 | 0.287 (0.180, 1) | 0.733 (0.052, 12) | 0.691 (0.074, 6) | 0.677 (0.060, 9) | 2.734 | 4.776 |
| 1724 | 0.502 (0.176, 1) | 0.412 (0.049, 13) | 0.810 (0.072, 6) | 0.761 (0.059, 9) | 10.309** | 0.040 |
| 693 | 0.556 (0.084, 7) | 0.263 (0.111, 4) | 0.833 (0.100, 5) | 0.703 (0.079, 8) | 14.390** | 0.740 |
| 160 | 0.537 (0.064, 11) | 0.384 (0.061, 12) | 0.790 (0.095, 5) | 0.780 (0.075, 8) | 18.858**** | 0.912 |
| 245 | 0.605 (0.056, 14) | 0.188 (0.075, 8) | 0.899 (0.094, 5) | 0.720 (0.094, 5) | 25.751**** | 2.144 |
| 412 | 0.661 (0.049, 12) | 0.369 (0.070, 6) | 0.767 (0.070, 6) | 0.886 (0.051, 11) | 26.277**** | 11.546** |
| 62 | 0.656 (0.062, 10) | 0.166 (0.053, 14) | 0.685 (0.053, 14) | 0.757 (0.053, 14) | 31.440**** | 25.936**** |
| 232 | 0.494 (0.040, 10) | 0.528 (0.038, 11) | 0.792 (0.048, 7) | 0.725 (0.036, 12) | 37.182**** | 1.538 |
| 320 | 0.581 (0.036, 23) | 0.252 (0.036, 23) | 0.689 (0.052, 11) | 0.671 (0.033, 27) | 43.818**** | 15.338** |
| 294 | 0.704 (0.055, 7) | 0.169 (0.042, 12) | 0.887 (0.051, 8) | 0.784 (0.042, 12) | 69.145**** | 20.179**** |
Least squared means of pollen viability (PV) for each of four hms1–hms2 genotypic classes in SF5-backcross progeny using various Iron Mountain (IM) inbred lines (GN = heterozygote and NN = SF5 homozygote). Values in parentheses are standard errors and number of progeny for each genotypic class. The last two columns show results of ANOVAs to test the effect of genotype at hms1 (Fhms1) and of both hms1 and hms2 (Fhms1-2) on pollen viability. IM lines in boldface type indicate those with whole genome resequence data. **P < 0.005, ****P < 0.0001.
Evidence for a selective sweep at hms1
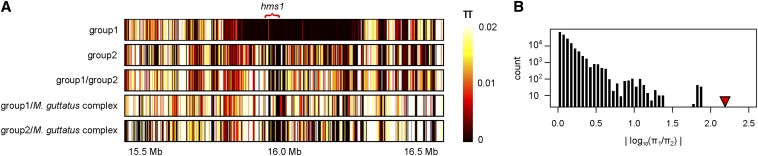
Using whole genome resequence data from 10 IM inbred lines (including IM62), we discovered an intriguing pattern of population genomic variation within and around hms1. Among this sample of lines, five carry a nearly invariant haplotype that extends for 320 kb and includes the hms1 locus (group 1, Figure 3A; π in the 320-kb region = 9.7 × 10−5). In this same genomic region, the other five IM lines show typical levels of nucleotide variation (group 2, Figure 3A; π in the 320-kb region = 0.016, among all IM lines π genome-wide = 0.014). Moreover, the group 1 IM haplotype does not appear to be a common variant throughout the species’ range: in this 320-kb region, group 1 and group 2 IM lines are similarly divergent from a sample of several M. guttatus complex lines (including SF5) collected from diverse locales (see Table S2; π in the 320-kb region: group 1 vs. M. guttatus complex = 0.026, group 2 vs. M. guttatus complex = 0.024). Note that although the sharp “borders” of the 320-kb haplotype might seem to suggest an inversion, this possibility is inconsistent with our earlier finding of several SF5 × IM62 F2 hybrid hms1 recombinants within this same interval, which indicate this region is collinear (see Figure 1).
Figure 3.
Signature of a selective sweep at the hms1 locus. (A) Each of the five panels shows nucleotide diversity (π) on linkage group 6 near the hms1 locus at 1-kb resolution (heat map color scheme is to the right). The hms1 sterility allele is embedded in a low-variation, 320-kb haplotype (top panel) that appears to have evolved recently in the IM population. Panels are as follows: group 1 = 5 IM lines that carry the 320-kb haplotype associated with hybrid sterility, group 2 = 5 IM lines with typical levels of nucleotide diversity, and M. guttatus complex = 10 M. guttatus complex lines collected from diverse locales (Table S2). (B) A histogram of the genome-wide distribution of the abs [ log10 (π 1/π2) ] test statistic, excluding sites in the 320-kb region. The maximum value of the test statistic from within the 320-kb haplotype (red triangle) is 2.2. Note that this value comes from a site near the center of the low-diversity region.
Using hms1-linked genomic sequence from these 10 IM lines, we designed two CAPS markers (Table S1) targeted to the outside edges of the 320-kb region that were diagnostic for haplotype group in this sample. We then used these CAPS markers to genotype a large panel of inbred lines from Iron Mountain. In this larger sample, the group 1 haplotype segregates at intermediate frequency (44%, N = 126 individuals).
To determine whether or not polymorphism for male sterility at hms1 is associated with variation at this 320-kb haplotype, we focused on the 18 IM inbred lines with known hms1 phenotypes (Table 2). Among this sample, allelic status at hms1 is highly predictive of haplotype group. Of the 9 IM inbred lines that carry the hybrid sterility allele at hms1, 8 carry the group 1 haplotype and only 1 line carries a group 2 sequence. The opposite pattern is seen for the 9 IM inbred lines that carry a compatible allele at hms1: 8 of these lines carry group 2 haplotypes and only 1 line carries a group 1 haplotype. This association between hms1 haplotype and hybrid sterility phenotype in these 18 IM inbred lines is highly significant (Fisher’s exact test, P = 0.0034). Note that the two exceptions to the general pattern (IM232 and IM591) are IM lines that show somewhat atypical hybrid sterility, which might be caused by environmental and/or genetic background effects. Of all the IM inbred lines that were identified as carriers of the hybrid sterility allele at hms1 (Table 2), IM232 is the only one for which the hms1-hms2 incompatibility genotype (i.e., heterozygous at hms1 and homozygous for M. nasutus alleles at hms2) is not the most sterile genotypic class. Additionally, among the lines identified as carrying compatible alleles at hms1, IM591 shows fairly high backcross sterility and a modest (nonsignificant) effect of hms1 genotype. Taken together, these results provide strong evidence that the hms1 sterility allele resides exclusively within the nearly invariant group 1 haplotype.
But is this pattern of nucleotide diversity at hms1 unique, or might we find other such low-variation haplotypes throughout the genome? To address this question, we examined genomic variation among the 10 resequenced IM inbred lines (5 with the group 1 haplotype, 5 with group 2), conditioning on two key attributes of the hms1 haplotype: its genetic size (1.6 cM, estimated from an integrated genetic map fit to physical positions) and allele frequency (50% among the 10 resequenced lines). Centered on SNPs with alleles at 50% frequency (N = 181,659 SNPs), we compared nucleotide diversity in 1.6-cM windows among haplotype groups (defined by alternate alleles at the central SNP). Using this approach, we generated a null expectation for hms1 diversity:
where π1 and π2 is the pairwise diversity in a 1.6-cM block for the five haplotypes associated with allele 1 and the five haplotypes associated with allele 2, respectively. Comparison with this null distribution reveals that the pattern of variation at the hms1-associated haplotype is indeed exceptional (Figure 3B).
This pattern is consistent with a strong, partial selective sweep of the group 1 haplotype in the Iron Mountain population (e.g., Voight et al. 2006). Assuming typical rates of recombination (one crossover/chromosome), the group 1 haplotype is likely to be young (∼63 generations old, for a 1.6-cM block). Its recent origin explains the extreme paucity of variation we observe (<1 mutation expected for 63 generations with mutation rate µ = 1 × 10−8; observed SNPs ∼0, after accounting for sequencing error) (see Materials and Methods). How strong would natural selection have to be for the group 1 haplotype to increase in frequency so dramatically over such a short time scale? Population genetic theory predicts that a newly derived allele with a selection coefficient s will take time t ≈ 2log(2Ne)/s generations to reach fixation (note that because the group 1 haplotype is at 44% frequency, the equation provides only a crude approximation). Assuming t ∼ 63 generations and Ne ∼ 10,000 (a reasonable estimate for the large IM population), the strength of selection should be on the order of 0.15.
Discussion
In recent years, speciation geneticists have made a great deal of progress toward understanding the molecular functions and evolutionary histories of genes involved in hybrid dysfunction. Nevertheless, there is still much to learn about the population genetic forces and selective agents that initially drive substitutions at these genes within species. In this report, we fine map hybrid sterility between two evolutionarily young species of Mimulus, narrowing the hms1 and hms2 incompatibility loci to small genomic regions with strong candidate genes. We take advantage of polymorphism at hms1 to investigate the evolutionary dynamics of this locus within a natural population of M. guttatus. Our population genomics analyses provide strong evidence that the hms1 incompatibility allele is involved in a partial selective sweep. Indeed, to our knowledge, we provide the first estimate of the strength of selection acting on a polymorphic hybrid incompatibility allele. Further work will be needed to identify both the underlying cause of selection at hms1 and the mechanism of interaction between hybrid sterility loci, but this study provides a strong framework for direct investigations of the evolution of hybrid dysfunction.
We have now fine mapped both partners of the hms1-hms2 incompatibility to small genomic intervals, each with only a handful of genes. Because the incompatibility affects the fertility of both sexes in Mimulus hybrids (Sweigart et al. 2006), we expect the causal genes to be involved in a process common to both male and female gametogenesis. Mimulus species are hermaphroditic and have flowers that contain male and female reproductive structures. In higher plants, the development of both male and female gametes initiates with the differentiation of archisporial cells and meiosis. The meiotic products then undergo two or three rounds of mitosis to produce multicellular gametophytes (the pollen grain and embryo sac). In Arabidopsis, a number of meiotic and mitotic cell cycle mutants have been isolated that cause sterility in one or both sexes (Liu and Qu 2008). Intriguingly, both hms1 and hms2 contain genes predicted to play key roles in cell cycle regulation.
The hms1 locus has three strong candidate genes: Migut.F01605, Migut.F01606, and Migut.F01612. The first two are tandem duplicates of SKP1-like genes (M. guttatus has 13 predicted SKP1-like genes genome-wide). Conserved throughout eukaryotes, SKP1 is a subunit of the SKP1–Cullin–F-box protein (SCF) E3 ubiquitin ligase, a complex that regulates diverse developmental processes including the cell cycle (Hellmann and Estelle 2002). In Arabidopsis and Oryza, this gene family has undergone independent expansions and tandem duplicates are common in both species (Kong et al. 2007). Although most of the 21 Arabidopsis-SKP1-like (ASK) genes have not yet been functionally characterized, mutants in ASK1 show defects in meiotic chromosome segregation and are male sterile (Yang et al. 1999). Interestingly, the other strong candidate for the hms1 sterility phenotype, an F-box gene, is also a component of the SCF regulatory complex. A key question is whether hms1 is caused by a single gene or instead by two or more tightly linked genes. In rice, adjacent genes are involved in two different cases of hybrid sterility between the indica and japonica subspecies (Long et al. 2008; Yang et al. 2012).
Based on predicted molecular function, only one of the five hms2 genes is a strong candidate for involvement in Mimulus hybrid sterility. The annotation for this gene (Migut.M00294) identifies it as related to a cyclin-dependent kinase (CDK) and its top BLAST hit in Arabidopsis thaliana encodes a CDK-like (CKL) protein. In Arabidopsis, 15 evolutionarily related CKLs were identified as core regulators of the cell cycle by their gene expression profiles (Menges et al. 2005). Although there is not yet information on the precise molecular functions of CKLs, they share high sequence similarity with CDKs, which control cell cycle progression in all eukaryotes (Dewitte and Murray 2003). Moreover, Arabidopsis mutants for an A-type CDK (CDKA; 1) are male sterile due to defects in pollen meiosis II (Nowack et al. 2005; Iwakawa et al. 2006. Beyond its molecular function, we have discovered a potentially important, M. nasutus-specific substitution in Migut.M00294: in the third codon of this gene, M. nasutus carries a SNP that changes a cysteine to a glycine (N = 5 M. nasutus lines). Although the function of this cysteine is unknown, it is generally conserved among homologous sequences from higher plants (Figure S1) and a change to glycine is predicted to be functionally deleterious (protein variation effect analyzer, PROVEAN score = −7.7; Choi et al. 2012). In selfing populations, the efficacy of purifying selection is expected to be reduced due to a drop in effective population size and recombination rates (Nordborg 2000; Charlesworth and Wright 2001). One intriguing possibility is that this Cys-to-Gly mutation has reached high frequency in M. nasutus as a consequence of the transition to self-fertilization, as appears to be the case for numerous other putatively deleterious mutations (Brandvain et al. 2014). Further experiments will be needed to determine the functional significance of this substitution, but like this Migut.M00294 glycine variant, previous genetic analyses (Sweigart et al. 2007) suggest that the hms2 incompatibility allele is specific to M. nasutus.
Even before getting to the gene level, fine mapping has provided sufficient resolution to begin investigating the evolutionary dynamics of the polymorphic hms1 locus. Together, the results from our genetic crosses and population genomic analyses suggest that the hms1 incompatibility allele is associated with a partial selective sweep. The selective advantage of this allele (or one at a tightly linked locus) is apparently substantial (s ∼ 0.15). In Drosophila, most of the hybrid incompatibility genes that have been cloned have also been shown to be rapidly evolving (Ting et al. 1998; Presgraves et al. 2003; Barbash et al. 2006; Brideau et al. 2006; Phadnis and Orr 2009; Tang and Presgraves 2009). Similarly, a gene that causes hybrid sterility between mouse subspecies (Prdm9) has undergone recurrent bouts of positive selection (Oliver et al. 2009). In plants, it is not yet clear how often natural selection is a primary driver of hybrid incompatibilities. In several cases, hybrid dysfunction is caused by divergence at duplicate genes in a manner consistent with the action of random genetic drift (Bikard et al. 2009; Mizuta et al. 2010; Yamagata et al. 2010). On the other hand, it was recently shown that a hybrid lethality allele has risen to high frequency in a population of M. guttatus due to linked selection for an allele that confers copper tolerance (Wright et al. 2013). Natural selection is also presumed to play a role in divergence among disease resistance genes that cause hybrid necrosis in Arabidopsis and rice (Kruger et al. 2002; Bomblies et al. 2007; Alcazar et al. 2009; Jeuken et al. 2009; Yamamoto et al. 2010; Chen et al. 2014), although direct population genetic evidence is lacking. Likewise, the genes that cause cytoplasmic male sterility between Mimulus species (involving the same IM62 and SF lines used in this study) seem more likely to have evolved by selfish mitochondrial evolution and compensatory nuclear coevolution than by genetic drift (Barr and Fishman 2010). Adding to these previous studies, our finding that hms1 is involved in a selective sweep provides some of the strongest evidence to date that natural selection within plant species (in this case, M. guttatus) can lead to postzygotic reproductive isolation between species.
The selective sweep at hms1 raises several additional questions. For one, it is not clear whether hms1 or a linked gene is the target of selection. Might the hms1 sterility allele be spreading by genetic hitchhiking as has been seen for M. guttatus hybrid lethality and copper tolerance alleles (Wright et al. 2013)? Additionally, we do not yet know the underlying cause of natural selection. Is there an ecological advantage of the sweeping hms1-associated haplotype? Alternatively, might the hms1 incompatibility allele provide no direct benefit, but instead evolve selfishly to bias its own transmission? Consistent with this second possibility, we observe non-Mendelian inheritance at hms1 in certain M. guttatus–M. nasutus hybrids (A. L. Sweigart, unpublished data). However, we do not yet know whether transmission ratio distortion at hms1 also occurs within the Iron Mountain population of M. guttatus, as has been found for a female meiotic drive locus associated with the LG11 centromere (Fishman and Saunders 2008). Another issue is whether the hms1 incompatibility allele is on its way to fixation or maintained at an intermediate frequency by short-term balancing selection. With such strong selection and no counteracting fitness effects, the hms1 incompatibility allele is expected to spread quickly. Fortunately, we can test this prediction: future field studies at Iron Mountain will measure the fitness effects of alternative hms1 alleles and track allele frequency changes across years. Finally, is there any evidence that natural selection has also contributed to the spread of the hms2 incompatibility allele? Demographic effects of selfing (e.g., population bottlenecks) make it challenging to detect selection in M. nasutus, but functional experiments to determine allelic effects on fitness may help resolve this issue.
Another question concerns the evolutionary origins of the hms1-hms2 incompatibility. Did the hms1 and hms2 incompatibility alleles arise as new mutations or instead from standing variation? There is evidence that the hms1 hybrid incompatibility allele is present in other populations near Iron Mountain (Sweigart et al. 2007; A. L. Sweigart, unpublished data). One possibility is that this allele was introduced to Iron Mountain through a recent admixture event. As for hms2, the incompatibility allele is specific to M. nasutus among the samples examined thus far (Sweigart et al. 2007). However, it is certainly possible that more intensive sampling will show this allele is also present at low frequency in M. guttatus, particularly in the region of California where M. nasutus likely originated (Brandvain et al. 2014). Given the recent divergence of these Mimulus species, this system holds great promise for understanding the evolution of postzygotic reproductive isolation as a continuum, from the initial dynamics within species to the mechanisms of hybrid breakdown between species.
Acknowledgments
We thank John Willis and John Kelly for sharing their Iron Mountain inbred lines. We are grateful to Amanda Kenney for assistance with rQTL. We thank Lila Fishman and Graham Coop for valuable discussions. Lila Fishman, Dave Hall, John Willis, Matt Zuellig, and two anonymous reviewers made thoughtful comments on an earlier draft, which greatly improved the manuscript. We are especially indebted to Rachel Hughes for expert greenhouse care and genotyping assistance. We also thank the Duke Genome Sequencing and Analysis Core, the University of North Carolina Chapel Hill High-Throughput Sequencing Facility, and the Joint Genome Institute for generating the sequences used in this study. This work was supported by National Science Foundation (NSF) grant DEB-1350935 and funds from the University of Georgia Research Foundation to A.L.S., a National Institutes of Health Ruth L. Kirschstein NRSA award (F32GM090763) to L.E.F., and NSF grant DEB-0918902 to Lila Fishman.
Footnotes
Supporting information is available online at http://www.genetics.org/lookup/suppl/doi:10.1534/genetics.114.171819/-/DC1.
Communicating editor: L. C. Moyle
Literature Cited
- Alcázar R., García A. V., Parker J. E., Reymond M., 2009. Incremental steps toward incompatibility revealed by Arabidopsis epistatic interactions modulating salicylic acid pathway activation. Proc. Natl. Acad. Sci. USA 106: 334–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai C., Sen P., Hofmann K., Ma L., Goebl M., et al. , 1996. SKP1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-box. Cell 86: 263–274. [DOI] [PubMed] [Google Scholar]
- Barbash D. A., Awadalla P., Tarone A. M., 2006. Functional divergence caused by ancient positive selection of a Drosophila hybrid incompatibility locus. PLoS Biol. 2: e142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkan A., Small I., 2014. Pentatricopeptide repeat proteins in plants. Annu. Rev. Plant Biol. 65: 415–442. [DOI] [PubMed] [Google Scholar]
- Barr C. M., Fishman L., 2010. The nuclear component of a cytonuclear hybrid incompatibility in Mimulus maps to a cluster of pentatricopeptide repeat genes. Genetics 184: 455–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bikard D., Patel D., Metté C. L., Giorgi V., Camilleri C., et al. , 2009. Divergent evolution of duplicate genes leads to genetic incompatibilities within A. thaliana. Science 323: 623–626. [DOI] [PubMed] [Google Scholar]
- Bomblies K., Weigel D., 2007. Hybrid necrosis: autoimmunity as a potential gene-flow barrier in plant species. Nat. Rev. Genet. 8: 382–393. [DOI] [PubMed] [Google Scholar]
- Bomblies K., Lempe J., Epple P., Warthmann N., Lanz C., et al. , 2007. Autoimmune response as a mechanism for a Dobzhansky-Muller-type incompatibility syndrome in plants. PLoS Biol. 5: e236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandvain Y., Kenney A. M., Flagel L., Coop G., Sweigart A. L., 2014. Speciation and introgression between Mimulus nasutus and Mimulus guttatus. PLoS Genet. 10: e1004410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brideau N. J., Flores H. A., Wang J., Maheshwari S., Wang X., et al. , 2006. Two Dobzhansky-Muller genes interact to cause hybrid lethality in Drosophila. Science 314: 1292–1295. [DOI] [PubMed] [Google Scholar]
- Broman K. W., Wu H., Sen Ś., Churchill G. A., 2003. R/qtl: QTL mapping in experimental crosses. Bioinformatics 19: 889–890. [DOI] [PubMed] [Google Scholar]
- Canales C., Bhatt A. M., Scott R., Dickinson H., 2002. EXS, a putative LRR receptor kinase, regulates male germline cell number and tapetal identity and promotes seed development in Arabidopsis. Curr. Biol. 12: 1718–1727. [DOI] [PubMed] [Google Scholar]
- Case A. L., Willis J. H., 2008. Hybrid male sterility in Mimulus (Phrymaceae) is associated with a geographically restricted mitochondrial rearrangement. Evolution 62: 1026–1039. [DOI] [PubMed] [Google Scholar]
- Charlesworth D., Wright S. I., 2001. Breeding systems and genome evolution. Curr. Opin. Genet. Dev. 11: 685–690. [DOI] [PubMed] [Google Scholar]
- Chen C., Chen H., Lin Y. S., Shen J. B., Shan J. X., et al. , 2014. A two-locus interaction causes interspecific hybrid weakness in rice. Nat. Commun. 5: 3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung W. Y., Hubert N., Landry B. S., 1993. A simple and rapid DNA microextraction method for plant, animal, and insect suitable for RAPD and other PCR analyses. Genome Res. 3: 69–70. [DOI] [PubMed] [Google Scholar]
- Choi Y., Sims G. E., Murphy S., Miller J. R., Chan A. P., 2012. Predicting the functional effect of amino acid substitutions and indels. PLoS ONE 7: e46688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie P., MacNair M. R., 1987. The distribution of postmating reproductive isolating genes in populations of the yellow monkey flower, Mimulus guttatus. Evolution 41: 571–578. [DOI] [PubMed] [Google Scholar]
- Cutter A. D., 2012. The polymorphic prelude to Bateson-Dobzhansky-Muller incompatibilities. Trends Ecol. Evol. 27: 209–218. [DOI] [PubMed] [Google Scholar]
- Darwin C., 1859. On the Origin of Species by Means of Natural Selection. J. Murray, London. [Google Scholar]
- DePristo M. A., Banks E., Poplin R., Garimella K. V., Maguire J. R., et al. , 2011. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 5: 491–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devoto A., Nieto-Rostro M., Xie D., Ellis C., Harmston R., et al. , 2002. COI1 links jasmonate signaling and fertility to the SCF ubiquitin-ligase complex in Arabidopsis. Plant J. 32: 457–466. [DOI] [PubMed] [Google Scholar]
- Dewitte W., Murray J. A., 2003. The plant cell cycle. Annu. Rev. Plant Biol. 54: 235–264. [DOI] [PubMed] [Google Scholar]
- Diaz A., MacNair M. R., 1999. Pollen tube competition as a mechanism of prezygotic reproductive isolation between Mimulus nasutus and its presumed progenitor M. guttatus. New Phytol. 144: 471–478. [DOI] [PubMed] [Google Scholar]
- Diévart A., Clark S. E., 2004. LRR-containing receptors regulating plant development and defense. Development 131: 251–261. [DOI] [PubMed] [Google Scholar]
- Dobzhansky T. H., 1937. Genetics and the Origin of Species, Columbia University Press, New York. [Google Scholar]
- Drakakaki G., van de Ven W., Pan S., Miao Y., Wang J., et al. , 2012. Isolation and proteomic analysis of the SYP61 compartment reveal its role in exocytic trafficking in Arabidopsis. Cell Res. 22: 413–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishman L., Saunders A., 2008. Centromere-associated female meiotic drive entails male fitness costs in monkeyflowers. Science 322: 1559–1562. [DOI] [PubMed] [Google Scholar]
- Fishman L., Sweigart A. L., Kenney A. M., Campbell S., 2014. Major quantitative trait loci control divergence in critical photoperiod for flowering between selfing and outcrossing species of monkeyflower (Mimulus). New Phytol. 201: 1498–1507. [DOI] [PubMed] [Google Scholar]
- Flagel L. E., Willis J. H., Vision T. J., 2014. The standing pool of genomic structural variation in a natural population of Mimulus guttatus. Genome Biol. Evol. 6: 53–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank S. A., 1990. Divergence of meiotic drive-suppression systems as an explanation for sex-biased hybrid sterility and inviability. Evolution 45: 262–267. [DOI] [PubMed] [Google Scholar]
- Gusti A., Baumberger N., Nowack M., Pusch S., Eisler H., et al. , 2009. The Arabidopsis thaliana F-box protein FBL17 is essential for progression through the second mitosis during pollen development. PLoS ONE 4: e4780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellmann H., Estelle M., 2002. Plant development: regulation by protein degradation. Science 297: 793–797. [DOI] [PubMed] [Google Scholar]
- Hurst L. D., Pomiankowski A., 1991. Causes of sex ratio bias may account for unisexual sterility in hybrids: a new explanation of Haldane’s rule and related phenomena. Genetics 128: 841–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwakawa H., Shinmyo A., Sekine M., 2006. Arabidopsis CDKA; 1, a cdc2 homologue, controls proliferation of generative cells in male gametogenesis. Plant J. 45: 819–831. [DOI] [PubMed] [Google Scholar]
- Jeuken M. J. W., Zhang N. W., McHale L. K., Pelgrom K., Den Boer E., et al. , 2009. Rin4 causes hybrid necrosis and race-specific resistance in an interspecific lettuce hybrid. Plant Cell 21: 3368–3378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H. J., Oh S. A., Brownfield L., Hong S. H., Ryu H., et al. , 2008. Control of plant germline proliferation by SCFFBL17 degradation of cell cycle inhibitors. Nature 455: 1134–1137. [DOI] [PubMed] [Google Scholar]
- Kim O. K., Jung J. H., Park C. M., 2010. An Arabidopsis F-box protein regulates tapetum degeneration and pollen maturation during anther development. Planta 232: 353–366. [DOI] [PubMed] [Google Scholar]
- Kong H., Landherr L. L., Frohlich M. W., Leebens-Mack J., Ma H., et al. , 2007. Patterns of gene duplication in the plant SKP1 gene family in angiosperms: evidence for multiple mechanisms of rapid gene birth. Plant J. 50: 873–885. [DOI] [PubMed] [Google Scholar]
- Krüger J., Thomas C. M., Golstein C., Dixon M. S., Smoker M., et al. , 2002. A tomato cysteine protease required for cf-2-dependent disease resistance and suppression of autonecrosis. Science 296: 744–747. [DOI] [PubMed] [Google Scholar]
- Li H., Durbin R., 2010. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26: 589–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Qu L. J., 2008. Meiotic and mitotic cell cycle mutants involved in gametophyte development in Arabidopsis. Mol. Plant 1: 564–574. [DOI] [PubMed] [Google Scholar]
- Long Y., Zhao L., Niu B., Su J., Wu H., et al. , 2008. Hybrid male sterility in rice controlled by interaction between divergent alleles of two adjacent genes. Proc. Natl. Acad. Sci. USA 105: 18871–18876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M., Force A., 2000. The probability of duplicate gene preservation by subfunctionalization. Genetics 154: 459–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maheshwari S., Wang J., Barbash D. A., 2008. Recurrent positive selection of the Drosophila hybrid incompatibility gene Hmr. Mol. Biol. Evol. 25: 2421–2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maheshwari S., Barbash D., 2011. The genetics of hybrid incompatibilities. Annu. Rev. Genet. 45: 331–355. [DOI] [PubMed] [Google Scholar]
- Martin N. H., Willis J. H., 2007. Ecological divergence associated with mating system causes nearly complete reproductive isolation between sympatric Mimulus species. Evolution 61: 68–82. [DOI] [PubMed] [Google Scholar]
- Martin N. H., Willis J. H., 2010. Geographical variation in postzygotic isolation and its genetic basis within and between two Mimulus species. Philos. Trans. R. Soc. Lond. B Biol. Sci. 365: 2469–2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menges M., De Jager S. M., Gruissem W., Murray J. A., 2005. Global analysis of the core cell cycle regulators of Arabidopsis identifies novel genes, reveals multiple and highly specific profiles of expression and provides a coherent model for plant cell cycle control. Plant J. 41: 546–566. [DOI] [PubMed] [Google Scholar]
- Mizuta Y., Harushima Y., Kurata N., 2010. Rice pollen hybrid incompatibility caused by reciprocal gene loss of duplicated genes. Proc. Natl. Acad. Sci. USA 107: 20417–20422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller H. J., 1942. Isolating mechanisms, evolution, and temperature. Biol. Symp. 6: 71–125. [Google Scholar]
- Nordborg M., 2000. Linkage disequilibrium, gene trees and selfing: an ancestral recombination graph with partial self-fertilization. Genetics 154: 923–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowack M. K., Grini P. E., Jakoby M. J., Lafos M., Koncz C., et al. , 2005. A positive signal from the fertilization of the egg cell sets off endosperm proliferation in angiosperm embryogenesis. Nat. Genet. 38: 63–67. [DOI] [PubMed] [Google Scholar]
- Oliver P. L., Goodstadt L., Bayes J. J., Birtle Z., Roach K. C., et al. , 2009. Accelerated evolution of the Prdm9 speciation gene across diverse metazoan taxa. PLoS Genet. 5: e1000753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phadnis N., Orr H. A., 2009. A single gene causes both male sterility and segregation distortion in Drosophila hybrids. Science 323: 376–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presgraves D. C., 2010. The molecular evolutionary basis of species formation. Nat. Rev. Genet. 11: 175–180. [DOI] [PubMed] [Google Scholar]
- Presgraves D. C., Balagopalan L., Abmayr S. M., Orr H. A., 2003. Adaptive evolution drives divergence of a hybrid inviability gene between two species of Drosophila. Nature 423: 715–719. [DOI] [PubMed] [Google Scholar]
- Saha D., Prasad A. M., Srinivasan R., 2007. Pentatricopeptide repeat proteins and their emerging roles in plants. Plant Physiol. Biochem. 45: 521–534. [DOI] [PubMed] [Google Scholar]
- Sweigart A. L., Willis J. H., 2003. Patterns of nucleotide diversity in two species of Mimulus are affected by mating system and asymmetric introgression. Evolution 57: 2490–2506. [DOI] [PubMed] [Google Scholar]
- Sweigart A. L., Willis J. H., 2012. Molecular evolution and genetics of postzygotic reproductive isolation in plants. F1000 Biol. Rep. 4: 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweigart A. L., Fishman L., Willis J. H., 2006. A simple genetic incompatibility causes hybrid male sterility in Mimulus. Genetics 172: 2465–2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweigart A. L., Mason A. R., Willis J. H., 2007. Natural variation for a hybrid incompatibility between two species of Mimulus. Evolution 61: 141–151. [DOI] [PubMed] [Google Scholar]
- Tang S., Presgraves D. C., 2009. Evolution of the Drosophila nuclear pore complex results in multiple hybrid incompatibilities. Science 323: 779–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Y., Hartl D. L., Laurie C. C., 2001. Sex-ratio segregation distortion associated with reproductive isolation in Drosophila. Proc. Natl. Acad. Sci. USA 98: 13183–13188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ting C. T., Tsaur S. C., Wu M. L., Wu C. I., 1998. A rapidly evolving homeobox at the site of a hybrid sterility gene. Science 282: 1501–1504. [DOI] [PubMed] [Google Scholar]
- Van Ooijen, J. W., 2006 JoinMap 4, software for the calculation of genetic linkage maps in experimental populations. Kyazma B. V., Wageningen, The Netherlands. [Google Scholar]
- Vickery R. K., Jr, 1964. Barriers to gene exchange between members of the Mimulus guttatus complex Scrophulariaceae. Evolution 18: 52–69. [Google Scholar]
- Vickery R. K., Jr, 1978. Case studies in the evolution of species complexes in Mimulus. Evol. Biol. 11: 405–507. [Google Scholar]
- Voight B. F., Kudaravalli S., Wen X., Pritchard J. K., 2006. A map of recent positive selection in the human genome. PLoS Biol. 4: e72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright K. M., Lloyd D., Lowry D. B., MacNair M. R., Willis J. H., 2013. Indirect evolution of hybrid lethality due to linkage with selected locus in Mimulus guttatus. PLoS Biol. 11: 1001497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie D. X., Feys B. F., James S., Nieto-Rostro M., Turner J. G., 1998. COI1: an Arabidopsis gene required for jasmonate-regulated defense and fertility. Science 280: 1091–1094. [DOI] [PubMed] [Google Scholar]
- Yamagata, Y., E. Yamamoto, K. Aya, K. T. Win, and K. Doi et al 2010 Mitochondrial gene in the nuclear genome induces reproductive barrier in rice. Proc. Natl. Acad. Sci. USA 107: 1494–1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto E., Takashi T., Morinaka Y., Lin S., Wu J., et al. , 2010. Gain of deleterious function causes an autoimmune response and Bateson-Dobzhansky-Muller incompatibility in rice. Mol. Genet. Genomics 283: 305–315. [DOI] [PubMed] [Google Scholar]
- Yang M., Hu Y., Lodhi M., McCombie W. R., Ma H., 1999. The Arabidopsis SKP1-LIKE1 gene is essential for male meiosis and may control homologue separation. Proc. Natl. Acad. Sci. USA 96: 11416–11421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Zhao X., Cheng K., Du H., Ouyang Y., et al. , 2012. A killer-protector system regulates both hybrid sterility and segregation distortion in rice. Science 337: 1336–1340. [DOI] [PubMed] [Google Scholar]
- Zhao D. Z., Wang G. F., Speal B., Ma H., 2002. The EXCESS MICROSPOROCYTES1 gene encodes a putative leucine-rich repeat receptor protein kinase that controls somatic and reproductive cell fates in the Arabidopsis anther. Genes Dev. 16: 2021–2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z. G., Zhu S. S., Zhang Y. H., Bian X. F., Wang Y., et al. , 2010. Molecular analysis of an additional case of hybrid sterility in rice (Oryza sativa L.). Planta 233: 485–494. [DOI] [PubMed] [Google Scholar]