Abstract
Copper is a micronutrient essential for growth due to its role as a cofactor in enzymes involved in respiration, defense against oxidative damage, and iron uptake. Yet too much of a good thing can be lethal, and yeast cells typically do not have tolerance to copper levels much beyond the concentration in their ancestral environment. Here, we report a short-term evolutionary study of Saccharomyces cerevisiae exposed to levels of copper sulfate that are inhibitory to the initial strain. We isolated and identified adaptive mutations soon after they arose, reducing the number of neutral mutations, to determine the first genetic steps that yeast take when adapting to copper. We analyzed 34 such strains through whole-genome sequencing and by assaying fitness within different environments; we also isolated a subset of mutations through tetrad analysis of four lines. We identified a multilayered evolutionary response. In total, 57 single base-pair mutations were identified across the 34 lines. In addition, gene amplification of the copper metallothionein protein, CUP1-1, was rampant, as was chromosomal aneuploidy. Four other genes received multiple, independent mutations in different lines (the vacuolar transporter genes VTC1 and VTC4; the plasma membrane H+-ATPase PMA1; and MAM3, a protein required for normal mitochondrial morphology). Analyses indicated that mutations in all four genes, as well as CUP1-1 copy number, contributed significantly to explaining variation in copper tolerance. Our study thus finds that evolution takes both common and less trodden pathways toward evolving tolerance to an essential, but highly toxic, micronutrient.
Keywords: Saccharomyces cerevisiae, genetic basis of adaptation, copper tolerance, aneuploidy, CUP1, fitness, parallel adaptation
IN his book, Wonderful Life (Gould 1989, p. 51), Stephen J. Gould famously opined that evolution is a historical and contingent process, so much so that “any replay of the tape would lead evolution down a pathway radically different from the road actually taken.” While this is undoubtedly true when one considers the full complexity of an organism, refrains are often observed in evolution at the trait level. Repeated evolution, defined as “the independent appearance of similar phenotypic traits in distinct evolutionary lineages” (Gompel and Prud’homme 2009) has been documented in both ecological and clinical environments at all taxonomic levels, e.g., repeated loss of stickleback lateral plates in freshwater (Schluter et al. 2004), ecomorphs of Anolis lizards (Losos 1992), the acquisition of “cystic fibrosis lung” phenotypes in Pseudomonas aeruginosa in patients with cystic fibrosis (Huse et al. 2010), to name but a few. The development of sequencing technologies has recently allowed biologists to ask whether parallel genetic changes underlie observations of parallel phenotypic change. In some cases, parallel phenotypic evolution has been attributed to parallel genotypic evolution, for example, repeated changes to cis-regulatory regions of the same gene—the pigmentation gene yellow—underlie changes in wing pigmentation in male Drosophila (Prud’homme et al. 2006). At the other extreme are cases where different genetic targets underlie similar phenotypic shifts; for example, yeast adapting to rich media converged in fitness via a variety of genetic mechanisms (Kryazhimskiy et al. 2014), and beach mice adapting to sandy coastal dunes from the Gulf and Atlantic coasts of Florida converged in coat coloration via different mutations (Manceau et al. 2010). In such cases, unique evolutionary trajectories at the genetic level appear repeatable at the phenotypic level.
The degree of phenotypic repeatability is inherently linked with the genomic target size of appropriate mutations, with single-locus Mendelian traits with fewer target sites (and hence higher repeatability) at one extreme and quantitative traits at the other extreme. Even when multiple genes underlie a selected trait, however, there may be relatively few sites that, when mutated, have the magnitude of effect and sufficiently minor deleterious side effects to improve fitness overall (Stern 2013). Such pleiotropic constraints are thought to explain why cis-regulatory sites more often contribute to adaptation than trans-regulatory changes (Stern 2000; Gompel et al. 2005). The size of the population and the manner in which it reproduces are also critical. Large populations have access to rarer mutations, particularly those of large effect (Burch and Chao 1999), increasing the chance that the best of these mutations will fix in independent evolutionary trials (Bell and Collins 2008). Mutations with particularly high fitness are also more likely to fix in asexual populations, because clonal interference reduces the chance that minor-effect mutations establish (Rozen et al. 2002), unless adaptive mutations are so common that coalitions of mutations establish together (Fogle et al. 2008; Lang et al. 2013).
The nature and severity of environmental challenge will also affect the degree of repeatability at both the genotypic and phenotypic levels. If the environmental change is so severe that the population cannot replace itself and there are only a small fraction of mutations whose benefits are large enough to bring absolute fitness above one (Bell and Collins 2008), then adaptation would be more repeatable. On the other hand, if an organism is adapting via mutations whose effects are small relative to the distance to the fitness optimum, nearly half of mutations are predicted to be beneficial (Fisher 1930), and adaptation would be less repeatable. The genomic target size must also depend on the nature of mutations required: when adaptation can be accomplished by the loss of a function, adaptive mutations can potentially arise in any step along the pathway leading to that function via a variety of mechanisms [e.g., single base-pair changes leading to premature stop codons early within a gene, movement of transposable elements within a gene, mutations in the promoter that alter transcription factor (TF) binding sites, etc.]. In contrast, if the environmental challenge requires the appearance of a novel trait, or an alteration of an existing trait, the number of genomic targets is likely diminished. Despite these long-standing theoretical predictions, empirical data have only recently been catching up, largely due to breakthroughs in sequencing technology (e.g., Schneeberger 2014).
In this study, we set out to determine the repeatability of adaptive evolution at the genotypic and phenotypic levels using short-term experiments with the yeast, Saccharomyces cerevisiae. We purposefully employed a short-term experimental design in an attempt to avoid the potential influence of epistasis limiting the mutations that are sampled (Chou et al. 2011; Kvitek and Sherlock 2011). The design of the experiment was similar to a previous study in our group where we examined adaptation to the fungicide nystatin (Gerstein et al. 2012). In both cases, multiple isogenic lines of yeast were exposed to inhibitory levels of either nystatin or copper, with levels chosen to be slightly higher than those in which growth occurred reliably. Lines that showed growth were isolated and analyzed. Through whole-genome sequencing of 35 lines that evolved tolerance in the nystatin experiment, we found that adaptation repeatedly involved the same four genes in a single pathway leading to the production of ergosterol (Gerstein et al. 2012), the membrane-bound target of nystatin (Woods 1971). Indeed, of the 20 unique mutations identified, 18 involved the same two genes (11 different sites in ERG3 and 7 in ERG6). In hindsight, the highly repeated nature of this adaptation may well be explained by the narrowness of the environmental challenge: the cells can survive by blocking the production of ergosterol, and this can be accomplished through loss-of-function mutations in the ergosterol biosynthesis pathway (particularly in ERG3 or ERG6). We thus set out to assess the degree of repeatability in the face of an entirely different environmental challenge: high copper concentrations, where loss-of-function mutations are less expected.
Copper is a micronutrient that is essential for several different enzymatic processes in yeast (cytochrome oxidase involved in respiration, superoxide dismutase involved in defense against oxidative damage, and the Fet3p ferro-oxidase involved in iron uptake, Graden and Winge 1997). Thus, unlike nystatin, cells cannot entirely block copper uptake. On the other hand, copper is extremely toxic at high concentrations, both because it displaces other metal cofactors from proteins and because it produces highly reactive oxygen species, including damaging hydroxyl radicals (Peña et al. 1999). The fact that multiple cellular processes require copper, that multiple cellular compartments are involved in copper sequestration (especially vacuoles and mitochondria), and that multiple processes are impacted negatively by copper (Peña et al. 1999) suggests that adaptation to high copper concentrations may occur through a variety of mechanisms. Here we report the results of a short-term adaptation experiment to this toxic but essential metal. Through whole-genome sequencing, we identify the nature of the genetic changes that underlay the evolutionary rescue of 34 lines of S. cerevisiae exposed to inhibitory copper concentrations.
Materials and Methods
Evolution of haploid mutation lines
Mutations were acquired in haploid lines of the common lab strain, BY4741 (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0), obtained from Open Biosystems in 2009. Preliminary experiments determined that BY4741 grown in liquid YPD + 12.5 mM CuSO4 does not show consistent growth. Instead, some populations begin growing at different times, a stochastic pattern of growth we have previously shown to be consistent with beneficial mutations in other environments (Gerstein et al. 2012). To initiate mutation acquisition, we streaked BY4741 from frozen onto a YPD plate and randomly chose a single colony to grow overnight in 10 ml YPD, shaking at 30°. We added 4.5 ml of this common wild-type stock to 185.5 ml YPD + 12.5 mM CuSO4 (hereafter referred to as “copper12” medium). We placed 1- ml aliquots into 180 inner wells of three 96 deep-well boxes, with 1 ml of dH2O in the outer wells. Boxes were maintained shaking on a platform shaker at 30°. All boxes were checked daily by visual examination of the bottom of the wells. Growth was recorded when we saw precipitate on the bottom of a well and was first observed after 7 days of incubation (Supporting Information, Table S1). Twenty-four hours after growth was first seen, we manually mixed the well and froze 500 μl of culture in 15% glycerol. In this way we isolated 56 “putative mutation lines” within 14 days, postinoculation.
At the end of the mutation-accumulation phase, we struck each putative mutation line from the freezer stock onto a single YPD plate and grew them at 30°. Two of the putative mutation lines did not grow within 72 hr and were excluded from this point forward. Fourteen lines exhibited very small colonies, typical of petite colonies that have lost mitochondrial function. One of our initial goals in acquiring these mutation lines was to measure their fitness in heterozygous form; to avoid assaying nonnuclear mutations, these lines were also excluded from the set of lines we genotyped and phenotyped. From each remaining line we haphazardly chose eight colonies and inoculated each colony into 1 ml copper12 medium and 1 ml YPD (by using the same pipette tip) and incubated them at 30° with shaking. In six cases, none of the eight colonies grew in copper12 medium within 72 hr, leaving us with 34 copper-adapted mutation lines [copper beneficial mutation (CBM) lines, Table 1]. From the paired YPD culture descended from the same colony (limiting exposure of our stocks to copper), 500 μl was added to 500 μl 30% glycerol and frozen.
Table 1. Mutations identified in the CBM lines.
| CUP1 | Genome position | Mutation | Position | Amino acid | |||
|---|---|---|---|---|---|---|---|
| CBM line | coverage | (chr.bp) | Gene | (Watson strand) | (from 5′ end) | change | Exchangeability |
| CBM1 | 1.61 | X.412600 | VTC4 | C > T | 800 | Trp > Stop | |
| XI.105507 | FAS1 | G > T | 4837 | Val > Phe | 0.207 | ||
| XVI.420661 | Intergenic | A > T | |||||
| CBM2 | 2.00 | ChrII aneuploidy | |||||
| CBM3 | 2.48 | VII.150650 | Intergenic | G > T | |||
| ChrII aneuploidy | |||||||
| CBM4 | 3.26 | mito.24277a | COX1b | 1D indel (GG C/- CC) | 10460 | Intron | |
| CBM5 | 3.78 | X.413174 | VTC4 | C > A | 226 | Glu > Stop | |
| X.654261 | Intergenicc | T > C | |||||
| XIV.284255 | Intergenic | T > G | |||||
| CBM6 | 3.69 | III.100061 | BUD3 | G > A | 3781 | Gly > Arg | 0.178 |
| IV.319466 | VAM6 | T > A | 655 | Lys > Stop | |||
| mito.59168 | 21S_RRNA | A > G | 1160 | Lys > Arg | 0.440 | ||
| mito.69322 | tRNA-Arg | C > G | 34 | Arg > Gly | 0.251 | ||
| CBM7 | 0.91 | II.365359 | TRM7 | C > T | 361 | Val > Ile | 0.537 |
| III.306327 | Intergenic | G > T | |||||
| IV.143017 | YDL176W | G > T | 921 | Ser > Ser | |||
| IV.177435 | CLB3 | T > G | 663 | Thr > Thr | |||
| V.392908 | BOI2 | C > T | 805 | Glu > Lys | 0.323 | ||
| VII.949946 | SMI1 | C > A | 954 | Lys > Asn | 0.457 | ||
| IX.370383 | Intergenic | C > G | |||||
| XV.215888 | MAM3 | C > G | 250 | Gly > Arg | 0.178 | ||
| ChrVIII aneuploidy | |||||||
| ChrXVI aneuploidy | |||||||
| CBM11 | 2.46 | X.413020 | VTC4 | 1D indel (GG A/- AA) | 380 | Phe > Ser+frameshift | |
| XI.566200 | CCP1 | A > G | 999 | Phe > Phe | |||
| XII.605283 | Intergenic | 1D indel (TT A/- AA) | |||||
| ChrII aneuploidy | |||||||
| CBM13 | 4.02 | X.412247 | VTC4 | C > A | 1153 | Glu > Stop | |
| X.654261 | Intergenicc | T > C | |||||
| CBM14 | 2.15 | XV.215018 | MAM3 | C > T | 1120 | Val > Ile | 0.537 |
| CBM16 | 0.28 | VII.480836 | PMA1 | A > T | 1831 | Phe > Ile | 0.181 |
| ChrII aneuploidy | |||||||
| CBM17 | 0.98 | X.412325 | VTC4 | A > G | 1075 | Tyr > His | 0.197 |
| XIII.711207 | ESC1 | C > T | 4075 | Leu > Phe | 0.336 | ||
| XIII.821262 | FCP1 | T > C | 1007 | Leu > Ser | 0.212 | ||
| ChrVIII aneuploidy | |||||||
| ChrXVI aneuploidy | |||||||
| CBM18 | 2.80 | V.303094 | VTC1 | G > T | 289 | Asp > Tyr | 0.227 |
| VII.548326 | GSC2 | C > T | 63 | Asp > Asp | |||
| XI.646464-onwards | FLO10d | ||||||
| CBM20 | 1.82 | VII.480463 | PMA1e | G > T | 2204 | Ala > Asp | 0.193 |
| XV.215332 | MAM3 | C > T | 806 | Ser > Asn | 0.390 | ||
| XVI.84024 | YPL247C | C > T | 173 | Gly > Asp | 0.188 | ||
| ChrII aneuploidy | |||||||
| CBM21 | 1.12 | VII.971165 | PFK1 | G > C | 2570 | Pro > Arg | 0.254 |
| X.654261 | Intergenicc | T > C | |||||
| ChrII aneuploidy | |||||||
| ChrIII aneuploidy | |||||||
| ChrVIII aneuploidy | |||||||
| CBM22 | 0.78 | V.302818 | VTC1 | 1D indel (CA C/- CA) | 13 | Pro > His+frameshift | |
| ChrVIII aneuploidy | |||||||
| ChrXVI aneuploidy | |||||||
| CBM24 | 0.77 | IV.805485 | Intergenic | A > G | |||
| IV.805517 | Intergenic | G > A | |||||
| CBM25 | 2.28 | IV.527743-onwards | ENA5f | ||||
| XI.621992 | MLP1 | G > T | 2188 | Glu > Stop | |||
| CBM26 | 0.66 | VII.480470 | PMA1 | T > G | 2197 | Thr > Pro | 0.164 |
| ChrI aneuploidy | |||||||
| ChrV aneuploidy | |||||||
| ChrVIII aneuploidy | |||||||
| CBM29 | 1.08 | VII.1376 | Intergenic | A > C | |||
| VII.480463 | PMA1e | G > T | 2204 | Ala > Asp | 0.193 | ||
| XV.566240 | Intergenic | G > C | |||||
| ChrII aneuploidy | |||||||
| CBM30 | 3.30 | ChrII aneuploidy | |||||
| CBM33 | 2.40 | VII.618173 | VHT1 | G > C | 1686 | Ile > Met | 0.279 |
| VIII.321332 | SBE22 | A > T | 919 | Met > Leu | 0.513 | ||
| X.412080 | VTC4g | C > T | 1320 | Trp > Stop | |||
| mito.24277a | COX1b | 1D indel (GG C/- CC) | 10460 | Intron | |||
| CBM34 | 2.92 | X.412080 | VTC4g | C > T | 1320 | Trp > Stop | |
| XI.364518 | Intergenic | Complex 1I indel (GA > AAT) | |||||
| mito.24277a | COX1b | 1D indel (GG C/- CC) | 10460 | Intron | |||
| CBM36 | 2.15 | X.412080 | VTC4g | C > T | 1320 | Trp > Stop | |
| mito.24277a | COX1b | 1D indel (GG C/- CC) | 10460 | Intron | |||
| CBM37 | 2.04 | X.412080 | VTC4g | C > T | 1320 | Trp > Stop | |
| mito.24277a | COX1b | 1D indel (GG C/- CC) | 10460 | Intron | |||
| CBM44h | 1.39 | X.412080 | VTC4g | C > T | 1320 | Trp > Stop | |
| mito.24277a | COX1b | 1D indel (GG C/- CC) | 10460 | Intron | |||
| CBM45 | 2.68 | X.412080 | VTC4g | C > T | 1320 | Trp > Stop | |
| mito.24277a | COX1b | 1D indel (GG C/- CC) | 10460 | intron | |||
| CBM46 | 2.75 | X.412643 | VTC4 | T > A | 757 | Arg > Stop | |
| XI.438478 | DID4 | A > G | 701 | Gln > Arg | 0.366 | ||
| CBM47 | 2.34 | V.302909 | VTC1 | C > A | 104 | Ser > Stop | |
| CBM49 | 3.14 | V.438349 | Intergenic | G > C | |||
| XII.1034221 | HMG2 | C > T | 1595 | Pro > Leu | 0.258 | ||
| XIII.420239 | Intergenic | A > C | |||||
| XIV.265933 | GCR2 | T > A | 598 | Lys > Stop | |||
| CBM51 | 2.50 | II.444465 | FES1 | C > T | 229 | Asp > Asn | 0.201 |
| IV.310552 | Intergenic | A > G | |||||
| CBM53 | 2.88 | V.180433 | PRP22 | C > T | 1593 | Ile > Ile | |
| CBM54 | 1.98 | VII.1077964 | MAL12 | G > T | 1366 | Gly > Stop | |
| CBM55 | 2.25 | (no mutations except to CUP1) |
CUP1 coverage for each line is provided in the second column and does not account for additional copies via chrVIII aneuploidy.
This mutation falls in an intron of COX1 but causes a frameshift in an overlapping predicted gene, A15_Beta.
Identical COX1 mutation observed in seven different lines.
Identical intergenic mutation observed in three different lines.
The alignment formed a 100% match to the beginning of FLO10 until XI.647464, at which point the alignment switched to a perfect match to a similar region downstream, starting at XI.648031. In silico qPCR confirmed the absence of unique intervening sites (CACCAGCTCTTCCTGGTCGT and CACCAGCTCTTCCTGGTCGT) within the FASTQ files for CBM18 (but present in other CBM lines), indicating a deletion in this region.
Identical PMA1 mutation observed in two different lines.
The alignment within this region exhibited a 100% match to the beginning of ENA5 but switched to a 100% match to ENA1 from approximately site IV.527743, suggesting a deletion. Because of the highly repetitive nature of this array, in silico qPCR was unable to uniquely identify the missing positions.
Identical VTC4 mutation observed in six different lines.
CBM44 was sequenced from the original population, not the representative colony.
Sequencing of haploid mutation lines
Freezer culture from each CBM line was streaked onto YPD plates and grown for 48 hr at 30°. We haphazardly picked a single colony for each line and grew it for 24 hr in 50 ml of YPD at 30° with shaking. Genomic DNA was extracted using standard protocols (Sambrook and Russell 2001). Protocols supplied by Illumina were followed to create barcoded libraries for each line (2011 Illumina, all rights reserved). We sequenced 100-bp single-end fragments for each line, pooling 12 uniquely barcoded strains in each lane on an Illumina HiSeq 2000. Twelve samples were rerun to obtain sufficient depth of coverage using 100-bp paired-end fragments: CBM18, 20, 21, 22, 24, 25, 26, 29, 30, 34, 36, and 44.
The resulting genomic sequence data were processed using Illumina’s CASAVA-1.8.0 as in Gerstein et al. (2012). We called SNPs and small insertions and deletions using configure-Build.pl and parsed the output files with custom UNIX and perl scripts. We took advantage of Illumina data from the previous set of experiments with nystatin (Gerstein et al. 2012), which were initiated from the same BY4741 culture, to determine the mutations that are common to our strain background yet different from the S288C reference genome (scergenome.fasta release R64-1-1 downloaded from the Saccharomyces Genome Database (SGD), http://downloads.yeastgenome.org/sequence/S288C_reference/genome_releases/); all such common mutations were removed from the dataset. Given that our lines were haploid, mutations called as heterozygous were discarded (likely alignment errors), as were SNP and indel calls of low quality (Q < 20). Remaining variants were checked in the alignments, using tview in samtools 0.1.7a (Li et al. 2009). SNPs were independently called using the bwa software package to perform the alignment along with samtools 0.1.7a to identify SNPs, using the -bq 1 option to limit data to reliable alignments (Li et al. 2009), confirming the set of SNPs found by CASAVA (Table 1).
To assess chromosomal aneuploidy events, the total depth of coverage for each chromosome was calculated as the proportion of sequenced sites mapping to a particular chromosome, relative to the proportion of known mapped sites located on that chromosome within the yeast reference genome (as reported by configureBuild.pl in Illumina’s CASAVA 1.8.0 package).
Intergenic mutations were analyzed for gains and losses of predicted TF binding sites using Cis-BP, a new tool offered by the online Catalog of Direct and Inferred Sequence Binding Preferences (available at http://cisbp.ccbr.utoronto.ca/TFTools.php). Cis-BP compares two sequences (i.e., one wild type and one mutant allele) for differential transcription factor binding inferred based on the relationship between similarity in DNA binding domain amino acid sequence and DNA sequence preferences (Weirauch et al. 2014).
Expected frequency of mutations causing nonsynonymous and stop codons
The expected frequency of mutations that would generate a particular type of amino acid change (synonymous, nonsynonymous, or stop) was calculated from the observed codon frequency in S. cerevisiae [http://downloads.yeastgenome.org/unpublished_data/codon/ysc.orf.cod; produced by J. Michael Cherry based on the 6216 ORFs within SGD as of January 1999]. For each position in each codon, the frequency of all possible mutations was calculated according to the observed spectrum of mutations reported by Lynch et al. (2008) based on previous studies in yeast. (Similar results were obtained using other mutation spectra, including a uniform distribution, the spectrum observed by Lynch et al. (2008) in their mutation-accumulation study, and the observed mutation spectrum in this study.)
Summing over the whole genome, the expected frequency of mutations leading to stop codons is 5.78%. The expected frequency of nonsynonymous mutations is 73.0% among all possible codon changes or 77.6% among only the synonymous and nonsynonymous changes (excluding those going to or from a stop codon). The expected frequency of mutations causing any change to the amino acid sequence is 78.9%, which is similar to the expectation used previously (78.7%) based on a uniform frequency of mutations (Wenger et al. 2011). Because mutations are biased toward transitions and away from G/C, we recommend using the estimates reported here, which are based on the greatest amount of data regarding the mutational spectrum (Lynch et al. 2008).
Determination of CUP1 copy number
Using samtools 0.1.7a, the alignments of all CBM lines were manually checked at genes that are known or suspected to be important for acclimation to high levels of copper in S. cerevisiae, with a particular focus on genes that were previously identified to be up-regulated under high levels of copper: BSD2, CCC2, COX23, CTR2, CUP1-1, CUP1-2, CUP2, FET3, FMP23, GEF1, HAA1, PCA1, SCO1, SCO2, SLF1, and VMA3. The alignments were normal for all of these genes (including 500 bp up- and downstream), except CUP1-1 and CUP1-2 on chromosome VIII. In this region, large gaps were consistently found spanning the duplicated copies of these genes, caused by alignment ambiguities in this tandem repeat region.
To measure CUP1 copy number without having to rely on alignments, we carried out the bioinformatics equivalent of a qPCR analysis (in silico qPCR; Gerstein et al. 2014) by using the unix command “grep” to directly count the number of fastq fragments containing “primers” in the CUP1 region. Specifically, we summed the number of fragments containing the 16-bp fragment from the very beginning and from the very end of CUP1, plus two 16-bp fragments between CUP1-1 and CUP1-2 (TTTCAAGAGAACATTT and GGGTGGTGAAGTAATA), searching for all four in the forward and reverse directions (e.g., using “zgrep TTTCAAGAGAACATTT *fastq*”). We then repeated this in silico qPCR procedure for three unique genes on chromosome VIII as controls (using the first 16 bp of DED81, DUR3, and RIX1 in both the forward and reverse directions). A BLAST search was used to confirm that these fragments aligned only to the appropriate genes (http://www.yeastgenome.org/cgi-bin/blast-sgd.pl). We also conducted this procedure with the 35 BMN lines isolated in nystatin [“beneficial mutation nystatin”; Gerstein et al. (2012)], which we initiated from the same ancestral genetic background, providing a baseline for comparison. Relative to the three control genes, the BMN lines had an average of 18.13 copies of CUP1 (range: 12.40–30.45). Note that although the S288C reference genome on the SGD reports only two CUP1 copies, an isolate of S288C was recently found to contain ∼14 copies by Southern analysis (Zhao et al. 2014). Our data are thus consistent with our ancestral BY4741 strain having undergone amplification in this region, and we report the number of CUP1 copies in our copper adaptation strains relative to the average across the BMN lines.
To test whether levels of CUP1 inferred from in silico qPCR were consistent with levels of CUP1 transcription, we assayed RNA levels using quantitative real-time PCR (qPCR). Detailed methods are provided in File S1. Briefly, we chose 10 CBM lines that spanned the range of CUP1 copy number. A single colony of each CBM line and two colonies of BY4741 were inoculated into 1 ml YPD + 5.5 mM CuSO4 (a lower concentration was used to allow growth of all lines, including BY4741) and grown for 12 hr at 30° with shaking, at which point RNA was isolated using the RNEasy Mini kit from Qiagen, following the yeast protocol. Oligonucleotides for qPCR (Table 2) were designed using Primer Express (ABI). mRNA levels of TAF10 were used for normalization, because TAF10 exhibits stable expression across strains and conditions (Teste et al. 2009).
Table 2. Oligonucleotides employed for RT-PCR, Southern blot analysis, and genotyping in the forward and reverse directions.
| Primer name | Sequence | Experiment |
|---|---|---|
| CUP1-F | AGCTGCAAAAATAATGAACAATGC | RT |
| CUP1-R | GCATTTGTCGTCGCTGTTACA | RT |
| TAF10-F | AAGTTGTTCTGACGGTGAACGA | RT |
| TAF10-R | GCGACCTATATTGAGCCCGTATT | RT |
| CUP1-F | 5Biosg/TTAATTAACTTCCAAAATGAAGGTCA | SB |
| CUP1-R | 5Biosg/AGACTATTCGTTTCATTTCCCAGAG | SB |
| MAM3-F | AATGAGTGCCGATACCATCC | GT |
| MAM3-R | GATTCGTCCCAATCTTTTGC | GT |
| VTC4-F | GTTCATGATCTAGCAAAGTTTTCG | GT |
| VTC4-R | GGTAACCAAAATGGGATTGAA | GT |
| LYS2-F | TCAAGGGCTGAAAAGACAATCAA | GT |
| LYS2-R | CGACGCAAAGAGATGAAACCA | GT |
RT, real-time PCR; SB, Southern blot; GT, genotyping; F, forward; R, reverse.
Phenotypic assays of CBM lines
To determine the extent of copper tolerance acquired, we conducted dose–response experiments in deep-well boxes. Each CBM line was struck from frozen onto YPD and grown for 48 hr at 30°. A single colony was then haphazardly chosen from each line and inoculated into 10 ml YPD, shaking overnight at 30°. The optical density of all lines was standardized to the least dense line and 200 μl of standardized culture was added to 400 μl YPD; 15μl was then inoculated into 1 ml of eight different levels of YPD + CuSO4 (0 mM, 4 mM, 8 mM, 9 mM, 10 mM, 11 mM, 12 mM, and 14 mM). Four replicates were grown for each line in each level of copper. Boxes were maintained shaking on a bench top shaker at 30°. After 72 hr, we manually mixed each well and the optical density (OD) of 200 μl of culture was measured on a BioTek plate reader. With these data, we determined the IC50 (half-maximal inhibitory concentration) of copper using a maximum likelihood fitting procedure, as previously described (Gerstein et al. 2012).
To assess whether there was a correlation between the ability to grow in elevated levels of copper and fitness in other environments, we conducted a series of growth rate experiments using the Bioscreen C Microbiological Workstation (Thermo Labsystems) to automate OD readings. From the rise in OD, growth rates were estimated under multiple environmental conditions: YPD + 8 mM CuSO4 (“copper8”); YPD, a standard laboratory rich medium; YPG, a medium that requires yeast cells to respire; and YPD + iron (ferric citrate). The latter environment was of particular interest because of copper’s role in iron uptake via the Fet3p ferro-oxidase, so growth was assayed at three levels of ferric citrate: 10 mM, 40 mM, and 60 mM; we only present the 40 mM results in the main text because results were highly correlated across the iron concentrations (Table S2). Copper (0.2 M Cu(II)SO4⋅5H2O) and iron (1M C6H5FeO7) stocks were made in distilled water. Iron stock was made at least 3 days prior to use with occasional vortexing and mild heating to keep the ferric citrate in solution. In both cases, copper or iron stock was added after YPD was autoclaved, roughly 1 hr before the addition of yeast culture.
Each growth rate assay was initiated in a similar manner to the IC50 assays. Cultures from BY4741 and all CBM lines were struck from frozen and grown on YPD plates incubated at 30° for 2–3 days. Four or five colonies from BY4741 and a single colony from each CBM line was then inoculated into 10 ml YPD, shaking overnight at 30°. Optical density from overnight culture was standardized, and a 1:101 dilution was conducted into the appropriate medium. For each line, five random wells spanning two 100-well honeycomb plates were filled with 150 μl of diluted culture. Plates were incubated at 30° with maximum shaking for 24 hr in a Bioscreen C, with automated OD readings every 30 min. From the raw data, we extracted the maximum growth rate using a nonparametric spline fit performed by a custom R script (Gerstein et al. 2012). The maximum growth rates from the replicates of each CBM line were statistically compared against all replicates initiated from BY4741 using a t-test (replicates involving separate wells from a single Bioscreen C plate).
Copper tolerance of deletion lines
To assess whether intragenic mutations that arose within our CBM lines are phenotypically similar to knockout mutations, we measured copper tolerance (IC50) of 21 gene deletion lines (Giaever et al. 2002), representing all of the available knockouts for the characterized genes that had mutated in our study (excluding the uncharacterized YPL247C and YDL176W). BY4741 is the progenitor of both the deletion collection and our ancestral strain background, allowing a direct comparison of the impact of deleting these genes. Tolerance (IC50) was determined as above from OD measurements taken across an array of copper concentrations at 24 hr. Tolerance assays were conducted in the Bioscreen C and replicated twice, running simultaneously on two different machines.
Tetrad dissections to isolate single mutations
To separate the effects of single mutations from other mutations present in the evolved lines (including extra copies of CUP1), we crossed all of the CBM lines with BY4739, which has a common genotype yet opposite mating type and different auxotrophies than BY4741, the progenitor of our lines. We then attempted to sporulate the resulting diploid lines, focusing on a subset that contained each common mutation or aneuploidy and the fewest number of additional mutations (∼1/3 of the lines). Detailed methods are provided in File S1.
We encountered substantial difficulties in obtaining tetrads from our strains; BY4741, a derivative of S288C, is known to be a poor sporulator (Deutschbauer and Davis 2005; Ben-Ari et al. 2006). In particular, despite many attempts, no tetrads were obtained for CBM16 (PMA1 mutation plus chrII aneuploidy), CBM26 (PMA1 mutation plus chrI, chrV, and chrVIII aneuploidy), CBM29 (PMA1 mutation plus chrII aneuploidy), CBM47 (VTC1 mutation), or CBM55 (no mutation identified other than extra copies of CUP1).
We were able to sporulare CBM2 (chrII aneuploidy), CBM14 (MAM3 mutation), CBM25 (MLP1 and ENA5 mutations), and CBM34 (VTC4 mutation). CBM25 was not initially chosen for tetrad dissection but was dissected as a contaminate of CBM22 (VTC1 plus chrVIII and chrXVI aneuploidy), as detected by subsequent sequencing. CBM25 contaminating cells were likely positively selected during the sporulation procedure given that the aneuploid lines in our experiment, like CBM22, had very low sporulation rates.
The genotype of resulting spores was then determined (see File S1; PCR primer information in Table 2). In brief, for CBM14 and CBM34 tetrad lines, MAM3 and VTC4, respectively, were amplified by PCR. All SNPs showed the expected 2:2 segregation pattern in the four spores of each dissected tetrad. CBM25 spores were sequenced on Illumina HiSeq 2000, which is when the strain was discovered to be CBM25 (bearing a mutation in MLP1 and ENA5), not CBM22. The segregation pattern for the additional copy of chrII in CBM2 spores was determined by the segregation patterns of LYS2 alleles. To quantify the segregation patterns of CUP1 among the spores, Southern blots with CUP1-specific probes were performed. We isolated genomic DNA and ran a Southern blot on three separate occasions for each spore. Band intensity was quantified in ImageJ (Abramoff et al. 2004) using the “background corrected density” macro to estimate CUP1 copy number.
Fitness effect of single mutations on growth rate and copper tolerance
To measure the fitness effects of the mutations isolated by tetrad dissection, growth rate assays were conducted within the Bioscreen C using either YPD + 9 mM CuSO4 (“copper9”) or YPD, as described above with the following exceptions. Yeast was occasionally taken from a lawn plated from frozen cells rather than from single colonies (the sporulated lines had been bottlenecked to a single colony just prior to freezing, and so a second bottleneck at this stage was less essential). Optical density was not standardized for the copper9 experiments as this was deemed to have little effect on inferred growth rates. For each line, two (copper9) or four (YPD) nonadjacent wells were filled with 150 μl of diluted culture and allowed to grow for 24 hr. This procedure was performed three times in the copper9 environment, and once in YPD to determine whether these lines were affected in their ability to grow in the nutrient-rich environment. The mean maximum growth rate was determined for each Bioscreen C assay in the copper9 environment, and statistics were performed using these means as data points.
Copper tolerance was determined for a subset of spores through dose–response experiments as described for the knockout lines except that two separate Bioscreen C runs were performed, with two replicate wells per run (Figure S1). Specifically, we assayed IC50 for two spores (among all of the tetrads for each line) that carried mutations of interest but had low CUP1 copy number.
Results
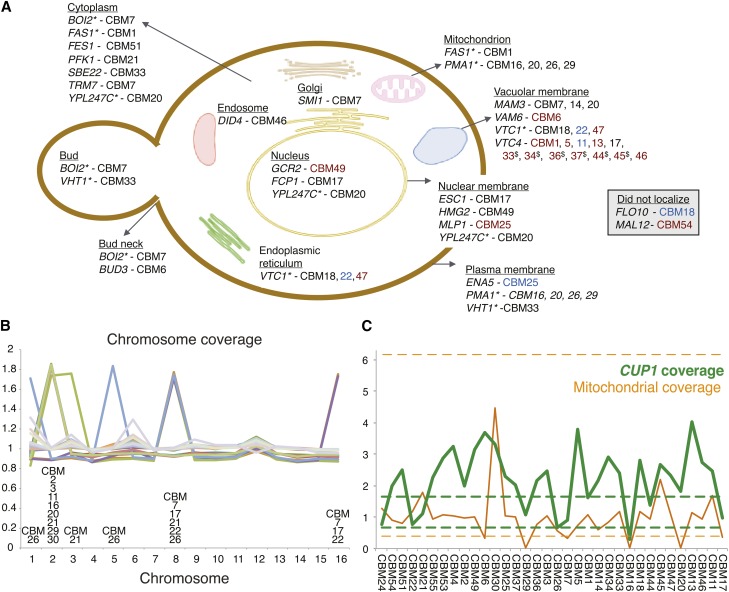
We recovered a broad spectrum of genetic changes across 34 lines exposed to initially inhibitory levels of copper (Figure 1, Table 1). Most lines contained multiple mutations, in contrast to our previous results in nystatin (Gerstein et al. 2012), which is consistent with the longer waiting period before growth observed (4–7 days with nystatin, 7–14 days with copper). All lines except for two (CBM2, one of the five lines isolated on the first day, and CBM55, one of the 11 lines isolated on the last day) contained one or more single base-pair mutations. In total, there were 57 unique base-pair changes, including four single base-pair deletions and one single base-pair insertion (the latter also resulted in an adjacent base-pair change). Beyond changes to single sites, there were several large-scale mutations. Twelve lines exhibited chromosomal aneuploidy (Figure 1B), and three lines (CBM6, CBM7, and CBM17) appeared to have low mtDNA coverage, outside of the range of lines from our previous study with nystatin (Figure 1C). In addition, two changes involved deletions within repetitive regions, one in CBM25 involving the tandem array of P-type ATPase sodium pumps (ENA5, ENA2, and ENA1) and the second in CBM18 involving the flocculation gene FLO10 (see details in Table 1).
Figure 1.
Observed mutations in copper-adaptation lines. (A) Genes mutated within the CBM lines (see Table 1 for specific mutations) illustrated based on previous localization studies of the genes involved (based on cellular component information curated at the Yeast Resource Center, http://www.yeastrc.org). The color of line names reflects the type of mutation: black for a nonsynonymous amino acid change, red for a premature stop codon, and blue for an indel or rearrangement (synonymous changes and RNA genes not shown). Genes that localized to more than one location are listed multiply and identified by “*.” CBM lines 33, 34, 36, 37, and 45 contained the same VTC4 mutation, indicated by “$.” (B) Chromosomal coverage. Aneuploidy was prevalent, appearing in 12 of 34 lines. Coverage is determined by the average number of reads across the chromosome, compared to the reference strain. (C) CUP1 copy number (green line) and mitochondrial coverage (orange line) for each CBM line. CUP1 copy number is measured relative to the average level observed in our parallel nystatin study, which showed negligible variation in copy number (range of the 35 BMN lines shown as dashed green lines). Mitochondrial depth of coverage in the CBM lines was divided by 10 and is presented relative to the average depth of coverage from mapped nuclear DNA (the equivalent range for the 35 BMN lines shown as dashed orange lines). Lines are ordered according to increasing copper tolerance (Figure 2).
The most frequent mutation across all lines was copy number alteration of the CUP1 locus. Based on in silico qPCR, CUP1 estimates were, on average, 2.17 times higher than estimates from the 35 lines obtained in nystatin. CUP1 copy number was estimated to be above the entire range of nystatin lines for 24 of the 34 CBM lines, while two lines (CBM16 and 26) both exhibited CUP1 levels lower than the range of nystatin lines (Figure 1C). CUP1 is a metallothionein protein that binds copper in S. cerevisiae. It is present as a tandem duplication on chrVIII in the S288C reference strain, and amplification of this locus is known to be a common mutation that confers increased resistance to copper (Fogel and Welch 1982; Fogel et al. 1983; Adamo et al. 2012). Disomy for chrVIII has also been shown to increase copper tolerance (Fogel and Welch 1982). Indeed, including cases with chrVIII aneuploidy, 27 of the 34 CBM lines have increased CUP1 copy number above the range of BMN lines (one of the exceptions, CBM24, is the least copper tolerant of our CBM lines).
When expression level of CUP1 was investigated by qPCR in a subset of lines and compared with the in silico qPCR estimates, it was found that the slope was positive and significant when forced through the point (1,1), which assumes that both axes are scaled to the ancestor (even though, technically, the derived nystatin lines and not BY4741 were used as the control in the in silico qPCR assays; P = 0.02, Figure S2A). The slope was still positive, but not significant otherwise (P = 0.27).
Single base-pair changes
Of the 57 unique single base-pair changes (Table 1), 15 were present in intergenic regions (2 of which were indels), and one single base-pair deletion was present in the intron of COX1. The remaining 41 unique single base-pair mutations were found within 29 different genes, whose products localize to many different cellular structures (Figure 1A). Five of these mutations were synonymous changes, 24 were nonsynonymous changes, 10 were premature stop codons, and 2 were frameshift mutations caused by single base-pair deletions.
Four sites were altered in the exact same way in multiple lines (a mutation at the intergenic site X.654261 in three lines, mito.24277 within an intron of COX1 in seven lines, VII.480463 causing an amino acid change in PMA1 in two lines, and X.412080 causing a stop codon in VTC4 in six lines). As discussed previously (Gerstein et al. 2012), we cannot distinguish between repeated mutational hits and either a single ancestral mutation that amplified during the growth of the strain prior to being separated into lines (i.e., during growth of the ancestral colony, followed by overnight growth in 10 ml YPD) or contamination during the sampling of lines on previous days. To be conservative, we consider the same mutation in multiple lines to be nonindependent and count them as having arisen only once in the statistical analyses below.
Four genes acquired multiple independent mutations, involving different positions in different strains. Fourteen lines acquired mutations in one of two subunits of the vacuolar transporter chaperone complex, VTC1 (3 unique mutations in 3 lines) or VTC4 (7 unique mutations in 12 lines); 4 lines acquired mutations in the plasma membrane H+-ATPase PMA1 (3 unique mutations); and 3 lines acquired unique mutations in MAM3, a protein required for normal mitochondrial morphology (Entian et al. 1999).
Given the 6607 ORFs within the S. cerevisiae genome (http://www.yeastgenome.org/genomesnapshot), the data are enriched for multiply hit genes. Specifically, there is a 99% chance that the 41 genic mutations would either hit different genes (first line in equation 1) or would hit one gene twice but no more (second line in equation 1):
| (1) |
assuming that ORFs are roughly equal in length. Thus, seeing even one gene bearing mutations at three or more independent sites is highly unlikely, and we conclude that positive selection acted upon the mutations in VTC1, VTC4, PMA1, and MAM3.
Excluding the indels, the single base-pair mutations that occurred within exons generated a stop codon much more often than predicted by chance (10/39 = 25.6%, P = 0.00006, exact one-tailed binomial test with expectation of 5.78% based on the mutational spectrum in yeast, see Materials and Methods). This result remains marginally significant when we focus only on genes hit once and exclude the four multiply hit genes (4/25 = 16%, P = 0.053, expectation of 5.78%).
On the other hand, the fraction of unique mutations that fall within an exon rather than a noncoding region is not significantly greater than the expected fraction in S. cerevisiae (41/57 = 72.0%, P = 0.63, expectation of 72.9% from Alexander et al. 2010). Similarly, among the synonymous and nonsynonymous mutations, nonsynonymous changes did not occur more often than expected (including all changes: 24/29 = 82.8%, P = 0.34; excluding multiply hit genes: 16/21 = 76.2%, P = 0.67; both exact one-tailed binomial tests with an expected fraction of 77.6%). Furthermore, the mean exchangeability score (an empirically based measure of the change in protein function following a particular amino acid change; Yampolsky and Stoltzfus 2005) of our observed amino acid changes (0.294) was within one standard error of the grand mean for mutations in yeast (0.31, calculated based on the mutational spectrum reported in Lynch et al. 2008). These tests are likely conservative, however, because selection against deleterious amino acid changes would have eliminated nonsynonymous mutations from our dataset, making it difficult to detect an enrichment of amino acid changes due to positive selection.
The set of genes whose protein products were altered is not enriched for either a specific GO term or a particular pathway (based on YeastMine analysis; Balakrishnan et al. 2012), although a significant number of mutated genes localize to the plasma membrane (PMA1, ENA5, and VHT1), the nuclear membrane (ESC1, HMG2, MLP1, and YPL247C), and the vacuolar membrane (MAM3, VAM6, VTC1, and VTC4). The set of genes is also enriched for three of the MIPS functional classification groups: vacuole or lysosome (VAM6, VTC1, and VTC4), cation transport (ENA5, PMA1, and VTC1), and protein synthesis (TRM7 and FES1) (identified using Funspec; Robinson et al. 2002).
Of the characterized genes that bore mutations, 21 were available from the yeast knockout collection (Giaever et al. 2002). Relative to BY4741, 13 lines showed a significant increase in copper tolerance, and two showed a significant decrease in copper tolerance (Figure S3). This assay supports the idea that a number of the singly hit genes might contain mutations that influenced copper tolerance.
To identify potential regulatory changes caused by the 15 intergenic mutations we found, we assessed whether predicted TF binding sites were gained or lost using Cis-BP (Weirauch et al. 2014) (Table S3). One of the positions (in CBM1) is not predicted to be at a TF binding site, while the remaining 14 were split among changes that caused both gains and losses (five mutations), only gains (four mutations), and only losses (five mutations). We identified a number of commonalities among the mutations, including two sets of TF binding sites that were each lost together three times (one set involved members of the Forkhead family, FKH2 and HCM1; and a second set involved NHP6A, NHP6B, and PHO2), two that were gained together three times (GAT1 and GLN3, members of the GATA family), and some that were both gained and lost (particularly ORC2 and SUM1). Among TF binding site mutations that were within 500 bp and 5′ of the start site of a gene, only one gene (RPP1A) was listed in the SGD as having an effect on metal tolerance, although two others affected vacuolar functioning (MUK1 and YIR007W) and one affected mitochondrial functioning (COX4). We did not, however, directly measure the effects of the intergenic mutations.
Aneuploidies
Chromosomal aneuploidy was common, appearing in one-third of all CBM lines (Figure 1B). All aneuploid lines had an extra copy of either chrII or chrVIII (one line had both). chrII aneuploidy was generally found by itself, only one of the eight lines with chrII aneuploidy carried additional aneuploid chromosomes (chrIII and chrVIII). In contrast, chrVIII aneuploidy never appeared in isolation. Three of the five cases of chrVIII aneuploidy also contained an extra copy of chrXVI and one line contained additional copies of chrI and chrV (in addition to the aforementioned line containing chrII and chrIII).
Mutagenic effects of copper
While selection must underlie the repeated spread of mutations affecting the genes that were multiply hit, it is possible that copper exposure directly altered the rate and nature of mutations that arose during the experiment. Indeed, exposure to high concentrations of copper is known to be mutagenic in experiments that directly expose DNA to copper (Tkeshelashvili et al. 1991). There is no evidence, however, for an elevated base-pair mutation rate in our experiment. Focusing only on nucleotide changes (not indels), we observed 52 unique single base-pair changes across the 34 lines isolated over the course of 7–14 days (average 11.0 days until isolation; Table S1). By comparison, in our previous study where the same ancestral strain was exposed to nystatin, we observed 35 mutations among 35 lines isolated over the course of 4–7 days (average 4.7 days until isolation). Thus, if anything, slightly more mutations accumulated per line per day in nystatin (0.21) than in copper (0.14), although the difference is not significant (P = 0.076, two-tailed exact binomial test with n = 52 + 35 mutation events and a proportion expected in copper given by 0.693 given that there were 34 lines × 11.0 days in copper and 35 × 4.7 in nystatin). Furthermore, while previous in vitro work indicates that copper should induce an excess of G:C → A:T mutations (Tkeshelashvili et al. 1991), the spectrum of single base-pair mutations observed within this study (8 A:T → G:C, 14 G:C → A:T, 6 A:T → T:A, 12 G:C → T:A, 5 A:T → C:G, 7 G:C → C:G) is not significantly different from the mutational spectra for yeast reported by Lynch et al. (2008) (see their Table 1), either based on prior studies (χ2 = 6.76, d.f. = 5, P = 0.239) or based on their mutation-accumulation experiment (χ2 = 1.48, d.f. = 5, P = 0.915).
We did, however, observe many more aneuploid events with copper (affecting 12/34 lines) than with nystatin (affecting 1/35 lines; Gerstein et al. 2012), but this is only marginally significant if we account for the greater number of days until isolation (P = 0.11, two-tailed exact binomial test with n = 12 + 1 aneuploid lines, where the proportion expected in copper is 0.693). Here, we have treated multiple aneuploid chromosomes within a line as a single event, in the absence of information about their independence; if they were independent, the excess of aneuploid events in the presence of copper would be very significant (P = 0.013, two-tailed exact binomial test with n = 19 + 1 aneuploid chromosomes). An enrichment of aneuploid events in copper may well be due to selection for aneuploidy rather than an increased mutation rate, consistent with the frequent occurrence of additional copies of chrVIII, bearing CUP1. Nevertheless, previous studies with mice have found copper to be mutagenic, using a micronuclei assay that is sensitive to errors in chromosome segregation during mitosis (Prá et al. 2008). We thus consider it plausible that the high frequency of aneuploidy observed in this study may have been directly due to copper exposure.
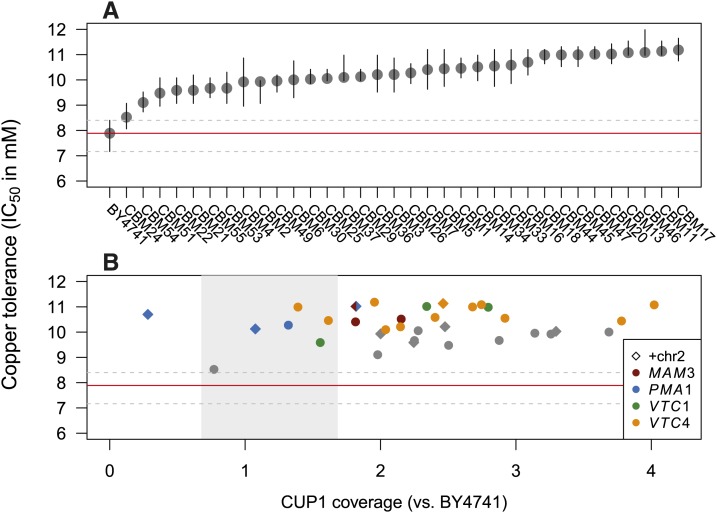
Phenotypic assays of CBM lines
Copper tolerance (measured as IC50 in deep-well boxes grown for 72 hr) was fairly similar across the 34 copper-adapted CBM lines, ranging from 8.5 mM to 11.2 mM (Figure 2A). The date that mutations were isolated does not correlate with copper tolerance (mutations are numbered based on the date of isolation; Table S1). The number of copies of CUP1 inferred from in silico qPCR (Figure 1C, corrected to include chrVIII aneuploidy) does not directly correlate with copper tolerance (Figure 2B; r = 0.16, t32 = 0.92, P = 0.36), yet this is likely due to the confounding effects of the other genetic changes. For example, three of the lines with the lowest CUP1 copy number carried mutations in PMA1 (excluding CBM24, which had low tolerance; Figure 2B). To tease apart the effects of these mutations, we both statistically analyzed the tolerance data collected for all lines and physically dissected the mutations via tetrad analysis in a subsample of four CBM lines (see below). A linear model with the four multiply hit genes as well as CUP1 copy number and chrII aneuploidy as factors indicated that CUP1 copy number (adjusted to include chrVIII aneuploidy) as well as the presence of a mutation in MAM3, PMA1, VTC1, or VTC4 were all significant predictors of copper tolerance, while chrII aneuploidy was not (CUP1 coverage: t27 = 2.58, P = 0.016; VTC1: t27 = 3.73, P = 0.0009; PMA1: t27 = 2.99, P = 0.0060; MAM3: t27 = 3.11, P = 0.0044; VTC4: t27 = 6.66, P < 0.001; and chrII: t27 = 1.67, P = 0.11).
Figure 2.
Copper tolerance across 34 copper-adapted lines (“CBM lines”). (A) Lines are numbered based on the date the mutant line was isolated following exposure to copper. The order of CBM lines in other graphs is based on the order of copper tolerance depicted here, which gives the IC50 after 72 hr of growth in deep-well boxes (bars represent 95% confidence intervals). Tolerance of BY4741 (the ancestor) is indicated by the horizontal red line with its confidence interval indicated by dashed gray lines. (B) Copper tolerance of each CBM line is generally high, regardless of the CUP1 copy number (x-axis, accounting for duplication of chrVIII). The absence of a correlation between CUP1 level and copper tolerance is due to the existence of additional mutations in the CBM lines, particularly in the four genes that were mutated independently (colors). Gray shading shows the range of CUP1 observed among the BMN lines (see Materials and Methods and Figure 1C).
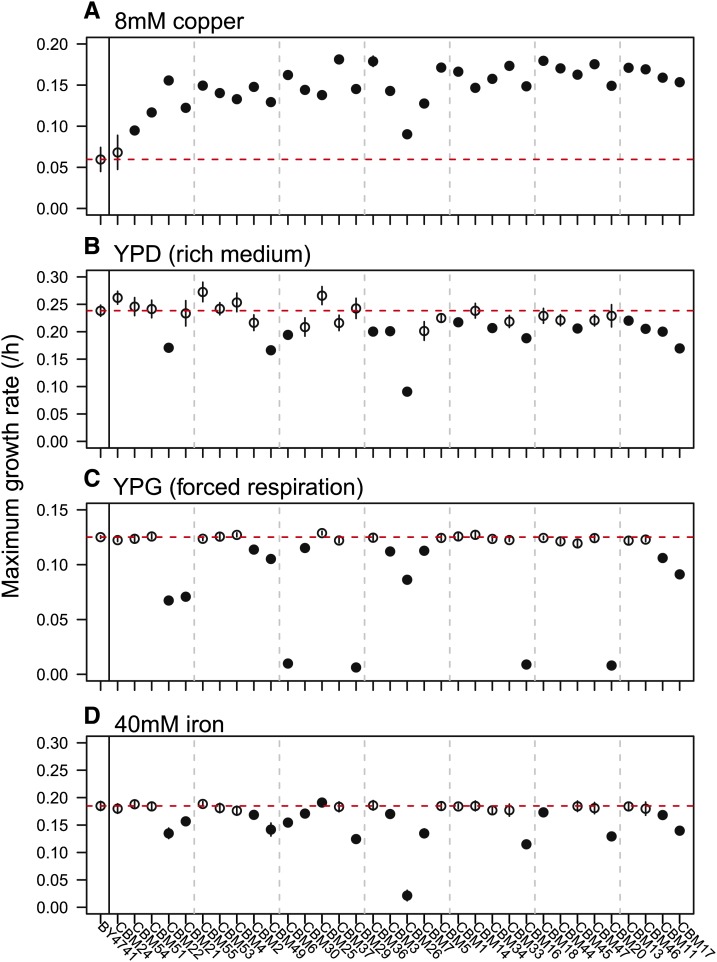
In addition to copper tolerance, we assayed maximum growth rates in copper, as well as YPD, YPG, and iron, using the Bioscreen C plate reader (Figure 3). Copper tended to be more inhibitory in the small volume plates used in the Bioscreen C, so we reduced copper levels to 8 mM CuSO4 (copper8). Growth rate in copper8 (Figure 3A) was significantly correlated with copper tolerance (Figure 2A, r = 0.69, t32 = 5.40, P < 0.0001). All lines except CBM24 grew significantly faster in copper8 than did BY4741, the ancestor (P < 0.05, Table S4). Copper tolerance did not correlate with growth in any of the three other environments examined (Figure 3, B–D; YPD: r = −0.31, t32 = −1.83, P = 0.077; YPG: r = −0.06, t32 = −0.34, P = 0.73; and iron: r = −0.09, t31 = −0.52, P = 0.61). No lines exhibited significantly increased growth in the rich medium, YPD, while about half had significantly decreased growth (Table S5). There was a negative, but not significant correlation between growth in YPD and growth in copper or copper tolerance (IC50). Three of the four slowest growing lines in YPD carried multiple aneuploid chromosomes (CBM17, 22, 26), but otherwise the slow growing lines spanned a range of genotypes and CUP1 copy numbers. There was greater variation in growth rates observed in YPG and in iron, with growth in these two environments being strongly correlated (r = 0.58, t31 = 3.98, P = 0.0004; YPG statistical results in Table S6, iron results in Table S2). The 15 CBM lines that grew significantly slower in YPG and iron included all lines with PMA1 mutations (CBM16, 20, 26, and 29), chrXVI aneuploidy (CBM7, 17, and 22), and chrII aneuploidy (CBM2, 3, 11, 16, 20, 21, 29, and 30), as well as all lines that lacked mtDNA (CBM16, 20, and 29).
Figure 3.
CBM lines have variable growth rates under different environmental conditions. Maximum growth rate (±1 SE) was assayed within the Bioscreen C in four different environments, each on a single day: (A) YPD + 8 mM CuSO4, (B) YPD, a standard laboratory rich medium, (C) YPG, a medium that forces respiration, and (D) YPD + 40 mM ferric citrate. Closed circles are lines that are significantly different from the wild type, BY4741 (red dashed line; see Table S2, Table S4, Table S5, and Table S6 for statistical information). Vertical gray dashed lines are for ease of comparison among the panels.
Tetrad dissections to isolate single mutations
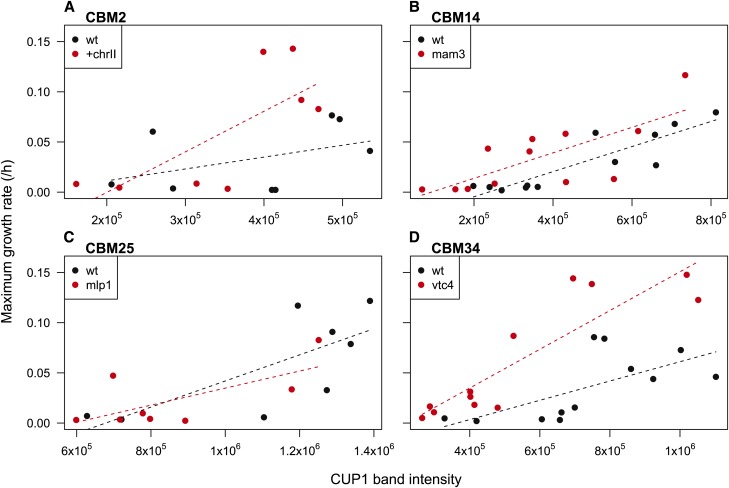
To examine the specific effect of common mutations, we crossed CBM2 (chrII aneuploidy), CBM14 (MAM3), CBM25 (MLP1 and ENA5), and CBM34 (VTC34) to BY4739 and dissected the resulting tetrads. As indicated above each panel in Figure 4, the four mutations and chrII aneuploidy segregated according to a 2:2 pattern (±), but segregation of CUP1 copy number was more variable (shown by colored circles). The variability in CUP1 inheritance may reflect noise in the assay (band densities on Southern blots, even though measured in triplicate; Figure S2B), or it may indicate changes in CUP1 over the course of the tetrad line construction. Indeed, previous work has shown copy number alterations were frequent (20%) during meiosis in lines heterozygous for CUP1 (Welch et al. 1991).
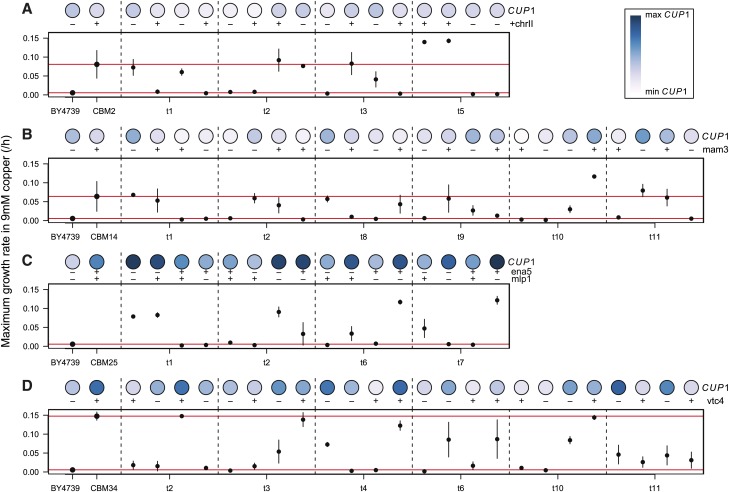
Figure 4.
Maximum growth rates of tetrad lines in YPD + 9 mM CuSO4. Tetrads were derived from four different CBM lines: (A) CBM2, (B) CBM14, (C) CBM25, and (D) CBM34. For each line, maximum growth rate was assayed within the Bioscreen C, with bars representing ±1 SE across 3 days. The darkness of the circle represents the line’s relative number of copies of CUP1, as assayed by Southern blot. Presence (+) or absence (−) of a segregating mutation is also noted. The maximum growth rate is shown for all tetrad lines, as well as for their two parents, BY4739 and the relevant CBM parent (red lines), except for the tetrads derived from CBM25 for which parental growth rate was not assayed (due to its initially being considered CBM22, see Materials and Methods).
Maximum growth rate in copper9 exhibited considerable variation among different spores from the same tetrad and rarely followed the strict 2:2 pattern expected if a single mutation underlay copper tolerance (Figure 4). The effect of each mutation on growth in copper is more easily seen in Figure 5, which shows linear fits through all of the tetrad data for a given CBM line (using the mean maximum growth rate across all replicates as a single data point for each spore). The interaction terms (seen as a difference in slope) were not significant, except for a marginally significant interaction between VTC4 and CUP1 (t20 = 2.044, P = 0.054, Figure 5D), and thus interactions were excluded from the main statistical analyses. For CBM14 and CBM34, the presence of a mutation in MAM3 and VTC4 (respectively), as well as CUP1 copy number, had significant positive effects on growth rate (CBM14, CUP1: t21 = 6.16, P < 0.0001, MAM3: t21 = 2.27, P = 0.034; and CBM34, CUP1: t21 = 6.16, P < 0.0001, VTC4: t21 = 4.41, P = 0.0002). By contrast, only CUP1 copy number had a significant effect on growth rate for CBM2 and CBM25 (CBM2, CUP1: t13 = 2.66, P = 0.020, +chrII: t13 = 1.73, P = 0.11; and CBM25, CUP1: t12 = 3.37, P = 0.0056, MLP1: t12 = −0.30, P = 0.77, ENA5: t12 = −0.018, P = 0.99).
Figure 5.
Impact of mutations on growth of tetrad lines in YPD + 9 mM CuSO4. Dots indicate maximum growth rates for tetrad lines carrying (red) or lacking (black) the mutation of interest: (A) chrII aneuploidy in CBM2, (B) MAM3 in CBM14, (C) MLP1 in CBM25, and (D) VTC4 in CBM34. For each line, linear model fits were performed with maximum growth rate as the response variable and CUP1 copy number and the allele status of the other mutation as the predictors. All fits are plotted including interaction terms between CUP1 copy number and the other gene of interest, where MLP1 was used as the gene of interest for CBM25 in C (no difference is seen when using ENA5).
The growth rate in copper9 may reflect an overall growth impact caused by the mutations, rather than a specific effect on growth in copper per se, interfering with our ability to detect improvement. To assess this possibility, we measured growth in the rich medium, YPD, and reran the models performed above, using growth rate in YPD as the response variable. The models for CBM14, CBM25, and CBM34 contained no significant terms (Table S7). However, the model for CBM2 indicated that the presence of an extra copy of chrII contributed to a significant decrease in growth rate in YPD (Table S7, Figure S4). We thus reran the copper9 linear growth models for CBM2, adjusting for growth rate in YPD by taking the difference. This model indicated that chrII aneuploidy indeed has a significant positive effect on growth in copper when controlling for its negative effect on growth in YPD (Table S8).
Together, these tetrad analyses indicate that CUP1 has a significant impact on growth in copper, as do the mutations in MAM3 and VTC4. In addition, chrII aneuploidy has a significantly more positive impact on growth in copper than expected based on its negative effect on growth in YPD.
Finally, from among all tetrads available for each line, we measured copper tolerance (IC50) for two spores that carried the mutation of interest (chrII aneuploidy, MAM3, MLP1, and VTC4) yet exhibited low CUP1 copy number (Figure S1). All mutants were found to have a significantly higher copper tolerance than either the BY4741 ancestor or BY4739 parent (Figure S5), despite their low CUP1 copy number. IC50 is likely to be a more sensitive assay of resistance to copper than maximum growth rate in a single copper level, suggesting that all of these mutations increase copper tolerance.
Reexamining the petite mutations
After the above analyses were conducted, we reexamined the lines that displayed small colonies on YPD plates (Table S1). To confirm that they were incapable of respiration, we assayed their growth on YPG plates. While 11 lines showed no growth, three lines (CBM9, 27, and 28) formed colonies, indicating that their mitochondria were still functional. These three lines were then whole-genome sequenced using population samples (Table S9). Line CBM9 carried a high-frequency SNP in PMA1, CBM27 carried a 3-bp deletion in PMA1, and CBM28 was aneuploid for chrIII, chrV, and chrVIII (a combination not seen in any other line). Levels of CUP1 were assayed as in Figure 1C and fell within the range of the BMN lines (CBM9, 0.69; CBM27, 0.83; and CBM28, 1.25), except for CBM28 once we account for its extra copy of chrVIII. Altogether, these lines provide additional confirmation of the role of PMA1 and chrVIII aneuploidy in the evolution of copper tolerance, although we infer that the mutational routes taken by these three lines were particularly deleterious in the absence of copper, given their small colony size on YPD plates.
Discussion
Depending on the different possible directions in which the environment may change, how rapidly can evolution happen and via how many different possible pathways? The advent of rapid sequencing technology has led to an increase in studies examining the repeatability of evolution at the level of the genotype, finding that the same genes often underlie parallel and convergent evolution in natural populations and during experimental evolution studies (Conte et al. 2012; Martin and Orgogozo 2013, see references within). Whether this repeatability reflects convergence over time to fitter genotypes or a limited scope of adaptive mutations along the way remains unknown. Also unknown is the relationship between the type of evolutionary challenge and the likelihood of parallel genetic changes (Stern 2013). This study aimed to contribute to our understanding of how the type of environmental challenge influences the genomic target size of the mutations selected during the very first steps of adaptation. We used the same mutation acquisition protocol as in our previous study on nystatin resistance (Gerstein et al. 2012) to obtain mutations in the presence of an inhibitory level of copper. We predicted that a broader range of genetic solutions would underlie copper adaptation, in contrast to the nystatin study that identified a narrow genetic solution (all 35 lines had mutations within four genes in the same pathway). Previous studies have suggested that xenobiotic environments (such as antimicrobial drugs) select for repeated genetic solutions (Martin and Orgogozo 2013). By contrast, copper is a very different kind of environmental stressor—it is essential for several different enzymatic processes in yeast (Graden and Winge 1997) and therefore cannot be blocked entirely from entering the cell. Copper is, however, toxic at high concentrations (Peña et al. 1999), and thus its concentration in the cell must be held in a delicate balance. As predicted, we identified a large number of mutations among our copper adaptation lines, with the level of genetic parallelism highly dependent on the type of mutation under investigation.
Increased copy number of the CUP1 locus through tandem duplication or aneuploidy of chrVIII was by far the most common mutation, seen in 27 of the 34 copper adaptation lines. CUP1 exists as a tandem repeat in the S288C genome, and adaptation under copper stress has previously been shown to select for amplification of this locus (Fogel and Welch 1982; Adamo et al. 2012). Indeed, Covo et al. (2014) recently demonstrated that moving CUP1 onto other chromosomes can efficiently select for disomy under copper stress. The number of CUP1 copies varies among naturally isolated wild and vineyard strains (between 1 and 18 copies among 14 wild strains, Zhao et al. 2014, and 4 and 18 copies among 15 Italian vineyard strains, Stroobants et al. 2008). As CUP1 is present in the ancestral genome as a tandem repeat, it is likely prone to alterations in copy number as a consequence of unequal crossover, gene conversion, or single-strand annealing (Zhang et al. 2013). Furthermore, CUP1 amplification seems to incur few pleiotropic costs, as seen by the lack of an observed effect of CUP1 copy number on growth rate in YPD among our tetrad lines (Table S7).
Chromosomal aneuploidy also repeatedly arose within our lines. Twelve of the 34 lines were aneuploid, and each of these 12 lines contained either chrVIII aneuploidy (5 lines) or chrII aneuploidy (8 lines). Chromosomal aneuploidy seems to be a common route to adaptation for fungal species reproducing asexually in a diverse array of environmental stressors such as drug resistance (Selmecki et al. 2009; Sionov et al. 2010), high temperature (Yona et al. 2012), and salt (Dhar et al. 2011). Aneuploidy is an intriguing beneficial mutation, as it has the potential to affect many genes simultaneously, yet has a much higher reversion rate than other types of mutations. Whether such a high degree of aneuploidy would have been seen had our strains evolved for longer remains an open question. It may be that aneuploidy serves primarily as a stop-gap adaptation until other beneficial mutations appear in the genome with fewer costs (Yona et al. 2012). In our experiment, chrVIII aneuploidy may have been positively selected for its effect on CUP1 copy number, as seen by Fogel and Welch (1982). Similarly, chrII aneuploidy had a more beneficial effect in copper (Figure 5, Table S8) than expected, based on its low growth in YPD (Table S7), perhaps because it amplified genes contributing to copper tolerance, such as SCO1 and SCO2 (nonadjacent gene duplicates on chrII), which function in the delivery of copper to cytochrome c oxidase in the mitochondrial inner membrane. ChrII aneuploidy was also repeatedly observed by Covo et al. (2014) when investigating chromosomes gained in response to copper stress. Whether the other aneuploid chromosomes had an effect on copper tolerance remains unknown. Repeated attempts to sporulate CBM22 (aneuploid for chrVIII and chrXVI) and CBM26 (aneuploid for chrI, chrV, and chrVIII) failed to yield tetrads.
The major genetic contributors to copper tolerance besides CUP1 were four genes involved in maintaining plasma membrane potential (PMA1), vacuolar transport (VTC1 and VTC4), and mitochondrial morphology (MAM3). These genes each bore several independent protein-coding mutations in different lines, which is highly unlikely in the absence of selection. Indeed, of the seven lines that did not exhibit CUP1 amplification (relative to the range of BMN lines), six involved mutations in these genes (three in PMA1, one in VTC1, and two in VTC4), with the remaining low-CUP1 line exhibiting little copper tolerance (CBM24).
The types of mutations observed in VTC4 and VTC1 point to selection for loss of function in these genes, with several mutations inducing stop codons or frame shifts (Table 1). Furthermore, deletion of VTC1 and VTC4, as well as MAM3, significantly increased copper tolerance (Figure S3). By contrast, PMA1 codes for a plasma membrane H+-ATPase that regulates cytoplasmic pH, and yeast are inviable when this gene is deleted. High levels of copper have previously been shown to have a deleterious effect on plasma membrane organization (Fernandes et al. 2000), while strongly stimulating plasma membrane ATPase activity (Fernandes and Sá-Correia 2001). It thus seems likely that the mutations identified in PMA1 alter, rather than inactivate, this protein, suggesting a gain (or fine tuning) of function. Consistent with this view, none of the PMA1 mutations involved stop codons or frameshifts (neither the four listed in Table 1 nor the two additional PMA1 mutations among the lines initially categorized as petites, Table S9).
In addition, 25 genes bore a single mutation in a single line in our experiment (Table 1). These unique mutations were no more likely to be nonsynonymous than expected, based on the mutational spectrum and no more likely to occur within exons than expected. That said, several of the unique mutations caused stop codons (4/25), a marginally significant excess. Furthermore, 12/18 of the deletion lines tested for this subset of genes were found to have significantly altered copper tolerance when compared to BY4741 (Figure S3), suggesting that at least some of the uniquely hit genes may be playing a role in copper tolerance. Alternatively, many of the mutations in singly hit genes may have been neutral but spread via hitchhiking, a pervasive phenomenon in other batch culture experiments (Lang et al. 2013).
Beyond genetic changes, epigenetic changes may have contributed to copper adaptation. This possibility was not formally investigated in our study. We can, however, conclude that epigenetic change was not the primary cause of adaptation, given that plausible causative mutations, involving either CUP1 or the four multiply hit genes, occurred in every line except one (Table 1, Figure 2B). The exception, CBM24, was the least copper tolerant line, bore only two intergenic mutations, and remains a possible candidate for epigenetic adaptation, although our genomic analysis may not have found all rearrangements or changes in hard-to-align regions.
One question of interest is why our copper-adapted lines carried so many more mutations, on average, than the nystatin-adapted lines studied previously (Gerstein et al. 2012). In no case did we see a BMN line that carried two mutations thought to be adaptive; all lines carried one and only one mutation in an ERG gene. By contrast, 21 of the 34 CBM lines carried more than one mutation for which we have evidence of a beneficial effect in copper (including the four multiply hit genes, chrII aneuploidy, and CUP1 levels above the BMN range when including chrVIII aneuploidy; 15 of 34 lines if we exclude chrII aneuploidy). The greater contribution of multiple beneficial changes to copper adaptation cannot be explained by a larger pool of large-effect beneficial mutations or a higher mutation rate, because it took longer to observe growth in copper (7–14 days) than in nystatin (4–7 days). One possibility is that many of the adaptive mutations may not have been beneficial enough on their own to generate detectable growth; instead, they may have allowed a line to persist for longer or to expand slightly in population size, facilitating the appearance of subsequent large-effect mutations. An alternative possibility is that positive epistasis among mutations more strongly favored the spread of secondary mutations. Our tetrad analysis provides some evidence for this possibility. Figure 5 shows that the effect of VTC4 on growth rate rises with CUP1 copy number among tetrads of CBM34, a marginally significant positive interaction (t20 = 2.044, P = 0.054). Similarly, the benefits of chrII aneuploidy also appear mild at low CUP1 levels and rise with increasing CUP1 copy number among CBM2 tetrads, although this interaction is not significant (t12 = 1.61, P = 0.13). In accordance with either of these explanations (mutations facilitating subsequent adaptation or positive epistasis), VTC4 mutations occurred more often among the CBM lines with higher CUP1 copy numbers (Table 1, Figure 2B).
This study provides an in depth analysis of how a eukaryotic organism, like yeast, takes its first few evolutionary steps toward tolerating an inhibitory but essential element, copper. Our genome-wide analysis of 34 strains found that adaptation often involved a common step (especially amplification of CUP1), but that routes less taken were also available. These alternate routes often involved chromosomal aneuploidy of chrII or chrVIII and four genes with roles in a wide variety of cellular functions—vacuolar transporters, mitochondrial morphology, and cytoplasmic pH regulation. Compared to our previous study of nystatin resistance (Gerstein et al. 2012), the variety of genes allowing growth in copper suggests that altered environments with more widespread effects on a cell may also provide a broader genetic basis for evolutionary recovery. Previous longer-term experimental evolution studies with microbes (e.g., Conrad et al. 2009; Miller et al. 2011; Tenaillon et al. 2012; Wong et al. 2012; Herron and Doebeli 2013; Lang et al. 2013; Kryazhimskiy et al. 2014) have found a high degree of parallel evolution at the gene level, and our results suggest that this can be the case even among the very first mutations selected. Whether adaptive mutations are likely to recur depends, in theory, on the effect size of the mutations as well as their mutation rate. In our case, amplification of CUP1 is both a relatively large effect mutation and easily acquired, contributing to its highly repeated nature, but it was also aided by the beneficial effects of many other less repeated mutations. In short, adaptation to copper is both more and less repeatable than adaptation to nystatin, with adaptation via CUP1 representing the route most commonly taken, but with mutations affecting a variety of other cellular processes providing a diversity of less traveled paths toward copper adaptation.
Acknowledgments
We are grateful to Sherry Li, Anna Van Tol, William Li, Xinya Wang, and Angelica Lillico-Ouachour for assistance in the lab; Barry Williams for helpful discussions regarding the genes involved in copper adaptation; Chris Walkey for advice on sporulation; Krystina Ho for advice on tetrad dissection; Patricia Schulte and Tim Healy for assistance running and analyzing qPCR results; Vivien Measday and Corey Nislow for assistance with the deletion lines; Martin Kircher and Edward Fox for discussions regarding the mutagenic properties of copper; and to two reviewers for their helpful comments on the manuscript. This work was supported by the National Science and Engineering Research Council of Canada (A.C.G. and S.P.O.), the Killam Trusts (A.C.G.), and a University of British Columbia graduate fellowship to J.O.
Footnotes
Supporting information is available online at http://www.genetics.org/lookup/suppl/doi:10.1534/genetics.114.171124/-/DC1.
Genomic fastq files have been deposited in the National Center for Biotechnology Information–Sequence Read Archive database under the accession code PRJNA261735. The remaining raw data and statistical analyses have been deposited in the Dryad Digital Repository (http://doi.org/10.5061/dryad.5gp25).
Communicating editor: S. I. Wright
Literature Cited
- Abramoff M., Magalhaes P., Ram S., 2004. Image processing with ImageJ. Biophotonics International 11: 36–42. [Google Scholar]
- Adamo G. M., Lotti M., Tamás M. J., Brocca S., 2012. Amplification of the CUP1 gene is associated with evolution of copper tolerance in Saccharomyces Cerevisiae. Microbiol. 158: 2325–35. [DOI] [PubMed] [Google Scholar]
- Alexander R. P., Fang G., Rozowsky J., Snyder M., Gerstein M. B., 2010. Annotating non-coding regions of the genome. Nat. Rev. Genet. 11: 559–571. [DOI] [PubMed] [Google Scholar]
- Balakrishnan R., Park J., Karra K., Hitz B. C., Binkley G., et al. , 2012. YeastMine—an integrated data warehouse for Saccharomyces cerevisiae data as a multipurpose tool-kit. Database 2012: bar062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell G., Collins S., 2008. Adaptation, extinction and global change. Evol. Appl. 1: 3–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Ari G., Zenvirth D., Sherman A., David L., Klutstein M., et al. , 2006. Four linked genes participate in controlling sporulation efficiency in budding yeast. PLoS Genet. 2: e195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burch C. L., Chao L., 1999. Evolution by small steps and rugged landscapes in the RNA virus phi6. Genetics 151: 921–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou H. H., Chiu H. C., Delaney N. F., Segre D., Marx C. J., 2011. Diminishing returns epistasis among beneficial mutations decelerates adaptation. Science 332: 1190–1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad T. M., Joyce A. R., Applebee M. K., Barrett C. L., Xie B., et al. , 2009. Whole-genome resequencing of Escherichia coli K-12 MG1655 undergoing short-term laboratory evolution in lactate minimal media reveals flexible selection of adaptive mutations. Genome Biol. 10: R118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conte G. L., Arnegard M. E., Peichel C. L., Schluter D., 2012. The probability of genetic parallelism and convergence in natural populations. Proc. Biol. Sci. 279: 5039–5047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covo S., Puccia C. M., Argueso J. L., Gordenin D. A., Resnick M. A., 2014. The sister chromatid cohesion pathway suppresses multiple chromosome gain and chromosome amplification. Genetics 196: 373–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutschbauer A. M., Davis R. W., 2005. Quantitative trait loci mapped to single-nucleotide resolution in yeast. Nat. Genet. 37: 1333–1340. [DOI] [PubMed] [Google Scholar]
- Dhar R., Sägesser R., Weikert C., Yuan J., Wagner A., 2011. Adaptation of Saccharomyces cerevisiae to saline stress through laboratory evolution. J. Evol. Biol. 24: 1135–1153. [DOI] [PubMed] [Google Scholar]
- Entian K.-D., Schuster T., Hegemann J., Becher D., Feldmann H., et al. , 1999. Functional analysis of 150 deletion mutants in Saccharomyces cerevisiae by a systematic approach. Mol. Gen. Genet. 262: 683–702. [DOI] [PubMed] [Google Scholar]
- Fernandes A. R., Prieto M., Sá-Correia I., 2000. Modification of plasma membrane lipid order and H+-ATPase activity as part of the response of Saccharomyces cerevisiae to cultivation under mild and high copper stress. Arch. Microbiol. 173: 262–268. [DOI] [PubMed] [Google Scholar]
- Fernandes A. R., Sá-Correia I., 2001. The activity of plasma membrane H(+)-ATPase is strongly stimulated during Saccharomyces cerevisiae adaptation to growth under high copper stress, accompanying intracellular acidification. Yeast 18: 511–521. [DOI] [PubMed] [Google Scholar]
- Fisher R. A., 1930. The Genetical Theory of Natural Selection, Oxford University Press, Oxford. [Google Scholar]
- Fogel S., Welch J. W., 1982. Tandem gene amplification mediates copper resistance in yeast. Proc. Natl. Acad. Sci. USA 79: 5342–5346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fogel S., Welch J. W., Cathala G., Karin M., 1983. Gene amplification in yeast: CUP1 copy number regulates copper resistance. Curr. Genet. 7: 347–355. [DOI] [PubMed] [Google Scholar]
- Fogle C. A., Nagle J. L., Desai M. M., 2008. Clonal interference, multiple mutations and adaptation in large asexual populations. Genetics 180: 2163–2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstein A. C., Lo D. S., Otto S. P., 2012. Parallel genetic changes and non-parallel gene-environment interactions underlie drug resistance in yeast. Genetics 192: 241–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstein A. C., Kuzmin A., Otto S. P., 2014. Loss of heterozygosity facilitates passage through Haldane’s sieve. Nat. Genet. 5: 3819. [DOI] [PubMed] [Google Scholar]
- Giaever G., Chu A. M., Ni L., Connelly C., Riles L., et al. , 2002. Functional profiling of the Saccharomyces cerevisiae genome. Nature 418: 387–391. [DOI] [PubMed] [Google Scholar]
- Gompel N., Prud’homme B., 2009. The causes of repeated genetic evolution. Dev. Biol. 332: 36–47. [DOI] [PubMed] [Google Scholar]
- Gompel N., Prud’homme B., Wittkopp P. J., Kassner V. A., Carroll S. B., 2005. Chance caught on the wing: cis-regulatory evolution and the origin of pigment patterns in Drosophila. Nature 433: 481–487. [DOI] [PubMed] [Google Scholar]
- Gould S., 1989. Wonderful Life: The Burgess Shale and the Nature of History, Norton, New York. [Google Scholar]
- Graden J. A., Winge D. R., 1997. Copper-mediated repression of the activation domain in the yeast Mac1p transcription factor. Proc. Natl. Acad. Sci. USA 94: 5550–5555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herron M. D., Doebeli M., 2013. Parallel evolutionary dynamics of adaptive diversification in escherichia coli. PLoS Biol. 11: e1001490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huse H. K., Kwon T., Zlosnik J. E. A., Speert D. P., Marcotte E. M., et al. , 2010. Parallel evolution in Pseudomonas aeruginosa over 39,000 generations in vivo. MBio 1: e00199–e001910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kryazhimskiy S., Rice D., Jerison E., Desai M., 2014. Global epistasis makes adaptation predictable despite sequence-level stochasticity. Science 344: 1519–1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kvitek D. J., Sherlock G., 2011. Reciprocal sign epistasis between frequently experimentally evolved adaptive mutations causes a rugged fitness landscape. PLoS Genet. 7: e1002056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang G. I., Rice D. P., Hickman M. J., Sodergren E., Weinstock G. M., et al. , 2013. Pervasive genetic hitchhiking and clonal interference in forty evolving yeast populations. Nature 500: 571–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., et al. , 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25: 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losos J. B., 1992. The evolution of convergent structure in Caribbean Anolis communities. Syst. Biol. 41: 403–420. [Google Scholar]
- Lynch M., Sung W., Morris K., Coffey N., Landry C., 2008. A genome-wide view of the spectrum of spontaneous mutations in yeast. Proc. Natl. Acad. Sci. USA 105: 9272–9277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manceau M., Domingues V. S., Linnen C. R., Rosenblum E. B., Hoekstra H. E., 2010. Convergence in pigmentation at multiple levels: mutations, genes and function. Philos. Trans. R. Soc. Lond. B Biol. Sci. 365: 2439–2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin A., Orgogozo V., 2013. The loci of repeated evolution: a catalog of genetic hotspots of phenotypic variation. Evolution 67: 1235–1250. [DOI] [PubMed] [Google Scholar]
- Miller C., Joyce P., Wichman H., 2011. Mutational effects and population dynamics during viral adaptation challenge current models. Genetics 187: 185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peña M., Lee J., Thiele D., 1999. A delicate balance: homeostatic control of copper uptake and distribution. J. Nutr. 129: 1251–1260. [DOI] [PubMed] [Google Scholar]
- Prá D., Franke S. I. R., Giulian R., Yoneama M. L., Dias J. F., et al. , 2008. Genotoxicity and mutagenicity of iron and copper in mice. Biometals 21: 289–297. [DOI] [PubMed] [Google Scholar]
- Prud’homme B., Gompel N., Rokas A., Kassner V. A., Williams T. M., et al. , 2006. Repeated morphological evolution through cis-regulatory changes in a pleiotropic gene. Nature 440: 1050–1053. [DOI] [PubMed] [Google Scholar]
- Robinson M. D., Grigull J., Mohammad N., Hughes T. R., 2002. FunSpec: a web-based cluster interpreter for yeast. BMC Bioinformatics 3: 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen D., de Visser J., Gerrish P., 2002. Fitness effects of fixed beneficial mutations in microbial populations. Curr. Biol. 12: 1040–1045. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., and D. W. Russell, 2001 Molecular Cloning: A Laboratory Manual, Ed. 3. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Schluter D., Clifford E. A., Nemethy M., McKinnon J. S., 2004. Parallel evolution and inheritance of quantitative traits. Am. Nat. 163: 809–822. [DOI] [PubMed] [Google Scholar]
- Schneeberger K., 2014. Using next-generation sequencing to isolate mutant genes from forward genetic screens. Nature Publishing Group 15: 662–676. [DOI] [PubMed] [Google Scholar]
- Selmecki A. M., Dulmage K., Cowen L. E., Anderson J. B., Berman J., 2009. Acquisition of aneuploidy provides increased fitness during the evolution of antifungal drug resistance. PLoS Genet. 5: e1000705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sionov E., Lee H., Chang Y. C., Kwon-Chung K. J., 2010. Cryptococcus neoformans overcomes stress of azole drugs by formation of disomy in specific multiple chromosomes. PLoS Pathog. 6: e1000848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern D. L., 2000. Perspective: evolutionary developmental biology and the problem of variation. Evolution 54: 1079–1091. [DOI] [PubMed] [Google Scholar]
- Stern D. L., 2013. The genetic causes of convergent evolution. Nat. Rev. Genet. 14: 751–764. [DOI] [PubMed] [Google Scholar]
- Stroobants A., Delroisse J.-M., Delvigne F., Delva J., Portetelle D., et al. , 2008. Isolation and biomass production of a Saccharomyces cerevisiae strain binding copper and zinc ions. Appl. Biochem. Biotechnol. 157: 85–97. [DOI] [PubMed] [Google Scholar]
- Tenaillon O., Rodriguez-Verdugo A., Gaut R. L., McDonald P., Bennett A. F., et al. , 2012. The molecular diversity of adaptive convergence. Science 335: 457–461. [DOI] [PubMed] [Google Scholar]
- Teste M.-A., Duquenne M., Francois J. M., Parrou J.-L., 2009. Validation of reference genes for quantitative expression analysis by real-time RT-PCR in Saccharomyces cerevisiae. BMC Mol. Biol. 10: 99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tkeshelashvili L., McBride T., Spence K., Loeb L., 1991. Mutation spectrum of copper-induced DNA damage. J. Biol. Chem. 266: 6401–6406. [PubMed] [Google Scholar]
- Weirauch M. T., Yang A., Albu M., Cote A. G., Montenegro-Montero A., et al. , 2014. Determination and inference of eukaryotic transcription factor sequence specificity. Cell 158: 1431–1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch J., Maloney D., Fogel S., 1991. Gene conversions within the Cup1r region from heterologous crosses in Saccharomyces cerevisiae. Mol. Gen. Genet. 229: 261–266. [DOI] [PubMed] [Google Scholar]
- Wenger J. W., Piotrowski J., Nagarajan S., Chiotti K., Sherlock G., et al. , 2011. Hunger artists: yeast adapted to carbon limitation show trade-offs under carbon sufficiency. PLoS Genet. 7: e1002202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong A., Rodrigue N., Kassen R., 2012. Genomics of adaptation during experimental evolution of the opportunistic pathogen pseudomonas aeruginosa. PLoS Genet. 8: e1002928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods R., 1971. Nystatin-resistant mutants of yeast: alterations in sterol content. J. Bacteriol. 108: 69–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yampolsky L. Y., Stoltzfus A., 2005. The exchangeability of amino acids in proteins. Genetics 170: 1459–1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yona A. H., Manor Y. S., Herbst R. H., Romano G. H., Mitchell A., et al. , 2012. Chromosomal duplication is a transient evolutionary solution to stress. Proc. Natl. Acad. Sci. USA 109: 21010–21015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Zeidler A. F. B., Song W., Puccia C. M., Malc E., et al. , 2013. Gene copy-number variation in haploid and diploid strains of the yeast Saccharomyces cerevisiae. Genetics 193: 785–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Strope P. K., Kozmin S. G., McCusker J. H., Dietrich F. S., et al. , 2014. Structures of naturally evolved cup1 tandem arrays in yeast indicate that these arrays are generated by unequal non-homologous recombination. G3 4: 2259–2269. [DOI] [PMC free article] [PubMed] [Google Scholar]