Abstract
Both Staphylococcus aureus and Staphylococcus epidermidis can form biofilms on natural surfaces or abiotic surfaces, such as medical implants, resulting in biofilm-associated diseases that are refractory to antibiotic treatment. We previously reported a promising antibacterial compound (Compound 2) and its derivatives with bactericidal and anti-biofilm activities against both S. epidermidis and S. aureus. We have further evaluated the antibacterial activities of four Compound 2 derivatives (H2-38, H2-39, H2-74 and H2-81) against 163 clinical strains of S. epidermidis and S. aureus, including methicillin-susceptible and methicillin-resistant strains, as well as biofilm-forming and non-biofilm-forming strains. The four derivatives inhibited the planktonic growth of all of the clinical staphylococcal isolates, including methicillin-resistant S. aureus and methicillin-resistant S. epidermidis and displayed bactericidal activities against both immature (6 h) and mature (24 h) biofilms formed by the strong biofilm-forming strains. The derivatives, which all target YycG, will help us to develop new antimicrobial agents against multidrug-resistant staphylococci infections and biofilm-associated diseases.
Keywords: antibacterial, anti-biofilm activity, methicillin-resistant Staphylococcus aureus, minimal inhibitory concentration, MIC50, Staphylococcus epidermidis
INTRODUCTION
Staphylococci are major causes of hospital-acquired infections that are associated with high morbidity. Most stains of Staphylococcus aureus and Staphylococcus epidermidis that cause nosocomial infections are antibiotic-resistant.1,2 Methicillin-resistant S. aureus (MRSA) has emerged as a cause of potentially lethal infections, and it can increase morbidity, length of hospital stay and hospital costs.3,4 Moreover, the number of multidrug-resistant MRSA and methicillin-resistant S. epidermidis (MRSE) strains that exhibit resistance to antibiotics, such as aminoglycosides, macrolides and lincosamides, has been on the rise.5 In addition, the ability of S. epidermidis and S. aureus to form biofilms on natural and abiotic surfaces, such as medical implants, has made these pathogens a major cause of refractory biofilm-associated infections because the biofilm bacteria show phenotypic resistance to antibiotics.6,7
The bacteria embedded in a biofilm exist in a low metabolic state or at a stationary growth phase, and they are much less susceptible to the host immune system and to antimicrobial agents. Bacteria in a biofilm are 10–1000 times more resistant to the effects of antimicrobial agents.8,9 Bacterial biofilms account for more than 80% of all microbial infections in humans,10 and 60% of nosocomial infections are related to the formation of biofilms on implantable medical devices.11 The colonization of staphylococci on implanted medical devices and formation of biofilm can result in chronic infections that are difficult to eradicate, thereby causing increased trauma to the patient and increased cost of treatment.12,13
Conventional antimicrobial agents have limited effectiveness against biofilm-related infections,12 which increases the emergence of multidrug-resistant staphylococci. This and the increased use of implanted medical devices, such as vascular catheters and joint prostheses, together drive the need for developing new types of antibiotics to effectively combat multidrug-resistant and biofilm-associated diseases. Two-component systems (TCSs), which exist in most bacteria, play important roles in sensing and responding appropriately to a wide range of environmental signals, and they have been considered to be potential targets for antimicrobial therapy.14 The YycGF TCS, which was discovered in low G+C gram-positive bacteria, including S. aureus and S. epidermidis, is an essential regulatory system for cell wall metabolism and is highly conserved,15 thus making it an attractive drug target. YycG is a sensor histidine-kinase, and YycF is the cognate response regulator. Inhibitors targeting the YycG kinase or the YycF regulator have been reported for B. subtilis and S. aureus.16,17,18,19
We previously reported a compound (Compound 2) that targets the HK domain of the S. epidermidis YycG and exhibits bactericidal and anti-biofilm activities.20 The structure of Compound 2 was optimized by substituting different functional groups, and a series of derivatives were designed and synthesized. Several derivatives with more effective antibacterial activity were then screened out.21,22,23 In this study, 163 clinical strains of S. aureus and S. epidermidis were collected from three tertiary hospitals in eastern China and used to evaluate the anti-bacteria activities of the newly synthesized derivatives of Compound 2 (H2-38, 3-{5-{{3-(4-chlorophenyl)-2-[(4-chlorophenyl)imino]-4-oxothiazolidin-5-ylidene}methyl}furan-2-yl}benzoic acid; H2-39, 4-{5-{{3-(4-chlorophenyl)-2-[(4-chlorophenyl)imino]-4-oxothiazolidin-5-ylidene}methyl}furan-2-yl}benzoic acid; H2-74, 2-{4-{{3-(4-chlorophenyl)-2-[(4-chlorophenyl)imino]-4-oxothiazolidin-5-ylidene}methyl}phenoxy}acetic acid; and H2-81, 4-{5-{{3-(4-fluorophenyl)-2-[(4-phenyl)imino]-4-oxothiazolidin-5-ylidene}methyl}thiophene-2-yl}benzoic acid) against the clinical staphylococcal isolates under planktonic and biofilm growth conditions.
MATERIALS AND METHODS
Clinical bacterial strains and culture media
A total of 163 staphylococci, each from an individual patient, were retrospectively collected from three tertiary teaching hospitals in three provinces in eastern China (ZheJiang, Jiangsu and Shanghai) from July 2011 to January 2012. The isolates were identified by the VITEK 2 compact automated system (bioMerieux SA, Lyon, France) and stored in trypticase soy broth (Oxoid, Sydney, NSW, Australia) with 15% glycerol at −80 °C until they were used for the present study.24
S. aureus (ATCC2 5923), S. aureus (ATCC 49230) and S. epidermis (ATCC 35984) strains were tested by the broth microdilution minimum inhibitory concentration (MIC) test. The biofilm-forming strain, S. epidermis ATCC 35984, and the biofilm-negative strain, S. epidermis ATCC 12228, were used in the biofilm formation assay as controls. All staphylococci were cultured in tryptic soy broth medium (TSB; Oxoid Ltd, Basingstoke, UK). Mueller-Hinton broth (Oxoid Ltd) was used for the MIC assay.
Biofilm formation assay of the clinical staphylococcal isolates
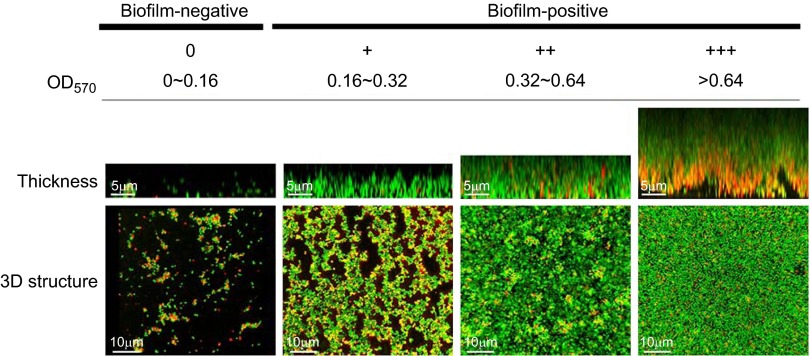
Biofilm formation of the clinical S. aureus and S. epidermis isolates was detected by a semi-quantitative assay in 96-well polystyrene microtiter plates. Overnight cultures of clinical strains were diluted 1:200 in TSB (Oxoid Ltd) containing 1% glucose and dispensed into 96-well plates (200 µL/well). After static incubation at 37 °C for 24 h, the plates were washed gently three times with phosphate-buffered saline to remove unattached bacteria, dried for 10 min at 60 °C, fixed with methanol and stained with 2% (w/v) crystal violet for 15 min at room temperature. Crystal violet staining was scanned at 570 nm using a 96-well plate spectrophotometer (DTX 880 Multimode Detector, Beckman Coulter, Brea, USA) to determine the optical density of the stained biofilms. The S. epidermidis (ATCC 35984) and S. epidermidis (ATCC 12228) strains were used as positive and negative biofilm-forming controls in the experiments, respectively.25 The clinical strains were divided into four groups according to their biofilm-forming ability as measured by OD values at 570 nm (OD570). The cutoff OD value (ODc) was defined as three standard deviations above the mean OD of the negative control. The OD value of a tested strain was expressed as follows: OD=average OD570−ODc. The strains were divided into the following groups: OD570≤ODc=no biofilm producer (−); ODc<OD570≤2×ODc=weak biofilm producer (+); 2×ODc<OD570≤4×ODc=moderate biofilm producer (++); and 4×ODc<OD570=strong biofilm producer (+++).26
MIC assay of the derivatives against clinical staphylococcal isolates
The derivatives of Compound 2 used in the experiment (H2-38, H2-39, H2-74 and H2-81) were designed by substituting functional groups while keeping the core thiazolidione structure intact. The compounds were synthesized by Professor Han's group at Nanjing University of Technology.
Broth micro-dilution testing was used to determine the MICs in accordance with the guidelines provided by the American Clinical and Laboratory Standards Institute.27 A range of different concentrations (0.8–200 µM) of derivative in Mueller-Hinton broth (Oxoid Ltd) was prepared and dispensed into a microtiter plate (100 µL per well), and 100 µL of bacteria (106 colony-forming units (CFU)/mL) was then added into each well resulting in final concentrations of the derivative from 0.4–100 µM. After a 12-h incubation at 37 °C, the MIC value was determined as the lowest concentration of the derivative to completely inhibit bacterial growth as observed by naked eye.28 The S. aureus (ATCC 25923), S. aureus (ATCC 49230), S. epidermis (ATCC 35984) and S. epidermis (ATCC 12228) strains were used as quality control strains. All tests were performed in triplicate and repeated three times.
Effects of the derivatives on 6-h biofilms of clinical strains
The inhibitory effects of H2-38 and H2-81 on 6-h biofilms of the clinical staphylococcal isolates19 were evaluated by a semiquantitative plate assay.26 All of the tested strains were strong biofilm-forming strains, including eight MRSE, four methicillin-susceptible S. epidermidis (MSSE), five MRSA and two methicillin-susceptible S. aureus (MSSA) strains. The bacteria were statically incubated in polystyrene 96-well plates with TSB medium containing 0.25% glucose at 37 °C for 6 h. After the removal of planktonic cells, 12.5 µM H2-38 and H2-81 in fresh TSB were separately added to each well and incubated for another 16 h at 37 °C. The wells were then washed gently three times with phosphate-buffered saline, fixed with methanol, and stained with 2% (w/v) crystal violet. The OD570 of the wells was determined using a 96-well plate spectrophotometer (DTX880; Beckman Coulter, USA).
Visualization of 24-h biofilms by confocal laser scanning microscopy
Overnight cultures of the selected clinical strains diluted at a ratio of 1:200 were inoculated in cell culture dishes (WPI, Florida USA) and incubated at 37 °C for 24 h. After removal of non-adherent cells with phosphate-buffered saline, the biofilms were stained using the Live/Dead BacLight Viability Kit (BacLight; Molecular Probes, Eugene, Oregon, USA), which employs SYTO9 and propidium iodide (SYTO9/PI) and they were subsequently analyzed with a Leica confocal laser scanning microscope (TCS SP5; Leica, Heidelberg, Germany). A series of images was acquired at 1 µm intervals in the Z section to measure the biofilm thickness in microns. IMARIS 7.0.0 software (Bitplane, Zurich, Switzerland) was used to create a three-dimensional view of the formed biofilms. A minimum of six representative optical fields were randomly selected and observed per specimen.29
To evaluate antibacterial effects of the H2-38 and H2-81 derivatives on 24-h biofilms, six strains were randomly selected out of the 19 strong biofilm-forming strains (MRSA 1000234, MRSA 916054, MSSA 1815, MRSE 1020081, MRSE 915296 and MSSE 914111). Overnight cultures of the bacteria (diluted 1:200) were inoculated in cell culture dishes (23 mm diameter) with glass bottoms (WPI) and incubated at 37 °C for 24 h. After removal of the suspension cultures, four-fold MIC concentrations of the derivatives in fresh TSB were added and incubated at 37 °C for an additional 12 h. The biofilms were then stained with the Live/Dead BacLight Viability Kit and visualized by a Leica TCS SP5 confocal laser scanning microscope. Viable cells in the biofilms exhibited green fluorescence, and dead cells exhibited red fluorescence. IMARIS 7.0.0 software (Bitplane, Zurich, Switzerland) was used to create the three-dimensional structural images of biofilms, and the percentages of live bacteria in the total bacterial counts were calculated by ImageJ (Wayne Rasband, NIH, Bethesda, MD, USA).
Statistical analysis
The average biofilm thickness was calculated from the six views for each clinical isolate and reported as the mean with standard deviations. Fisher's exact test was conducted for the anti-staphylococcal activities of the derivatives with respect to the different strains.
RESULTS
Characteristics of the clinical staphylococcal isolates
The 163 clinical isolates comprising S. aureus (95) and S. epidermidis (68) were primarily isolated from blood, sputum and wound discharges (Supplementary Table S1). Of the isolates, 99 were methicillin-resistant accounting for 60.7% of the clinical staphylococcal isolates. The 95 S. aureus isolates included 40 MSSA and 55 MRSA strains. The 68 S. epidermidis isolates consisted of 44 MRSE and 24 MSSE strains. All methicillin-resistant staphylococcal strains were not susceptible to penicillin. The MRSA and MRSE strains showed much higher rates of multidrug resistance compared with the MSSA and MSSE strains. The 55 MRSA strains were characterized as follows: 36 (65.4%) were not susceptible to tetracycline and levofloxacin; 47 (85.4%) were resistant to erythromycin; 37 (67.3%) were resistant to gentamycin; 18 (32.7%) were resistant to rifampicin; and 19 (34.5%) were resistant to sulfadiazine and trimethoprim (SMZ-TMP) (Supplementary Table S2).
The microtiter plate assay of biofilm formation by the clinical isolates was done using the non-biofilm-forming S. epidermidis ATCC 12228 strain as a negative control, which gave an OD570 cutoff value (ODc) defined as 0.16. The clinical strains were classified into four groups as follows: non-biofilm producer (OD570 less than or equal to 0.16); weak biofilm producer (0.32≥OD570>0.16); moderate biofilm producer (0.64≥OD570>0.32); and strong biofilm producer (OD570>0.64). Of the 95 S. aureus isolates, 41 strains produced biofilms, and 7 isolates were strong biofilm producers. Of the 68 S. epidermidis isolates, 42 isolates produced biofilms, and 12 isolates were strong biofilm producers (Table 1). There was no statistically significant difference between the biofilm-forming abilities of the methicillin-resistant and methicillin-susceptible staphylococcal isolates (P>0.05).
Table 1. The biofilm-forming abilities of the clinical staphylococcal isolates.
| Strainsa | Biofilm-negativeb (number (%)) | Biofilm-positiveb (number (%)) | ||
|---|---|---|---|---|
| - | + | ++ | +++ | |
| MRSA | 32 (57.6) | 15 (27.3) | 3 (6.0) | 5 (9.1) |
| MSSA | 22 (54.1) | 13 (33.3) | 3 (8.3) | 2 (4.3) |
| MRSE | 16 (37.2) | 11 (25.7) | 9 (20.0) | 8 (17.1) |
| MSSE | 10 (41.6) | 7 (29.2) | 3 (12.5) | 4 (16.7) |
Biofilm formation by the clinical isolates was assessed by semiquantitative assay with microtiter plates.
The clinical strains were classified into four groups according to their abilities to form biofilms as measured by OD570. The cutoff OD value (ODc) was 0.16. The four groups were as follows: no biofilm producer (−), OD570≤0.16; weak biofilm producer (+), 0.16<OD570≤0.32; moderate biofilm producer (++), 0.32<OD570≤0.64; and strong biofilm producer (+++), OD570>0.64.
Confocal microscopy measurements of biofilm thickness showed the following results: the strong biofilm producers formed dense biofilms with thicknesses ranging from 9 to 14 µm; the moderate biofilm producers produced biofilms that were 5–8 µm thick; and the weak biofilm producers produced biofilms that were 2–4 µm thick. With those strains that did not produce biofilms, only scattered microcolonies at the bottom of the dishes were observed (Figure 1).
Figure 1.
Representative biofilms of the clinical staphylococcal strains. The clinical staphylococcal strains were classified into four groups according to their biofilm-forming ability. After a 24-h incubation, the biofilms were observed by confocal microscopy with Live/Dead staining. A series of images were acquired at 1 µm intervals along the Z section, and three-dimensional biofilm architectures were constructed by IMARIS 7.0.0.
Antimicrobial efficacy of H2 derivatives against clinical MRSA and MRSE isolates
Four of the newly synthesized derivatives of Compound 2 (H2-38, H2-39, H2-74 and H2-81) had strong antimicrobial efficacy against staphylococcus (S. epidermis ATCC 35984, S. aureus ATCC 25923 and S. aureus ATCC 49230),23 and their anti-bacterial activities against 163 clinical staphylococcal isolates (99 of which were methicillin-resistant) were evaluated in this experiment. Most MIC values of the four derivatives were in the range of 0.75–25 µM. Both the methicillin-resistant and methicillin-susceptible strains of S. aureus and S. epidermidis were susceptible to the four derivatives (Table 2).
Table 2. MIC values of the four derivatives against the clinical staphylococcal isolates.
| Strains | Derivativesa | The range of MICb (μM) | MIC50c (μM) | MIC90c (μM) |
|---|---|---|---|---|
| MRSE (44) | H2-38 | 0.75–12.5 | 3.13 | 6.25 |
| H2-39 | 3.125–12.5 | 3.13 | 6.25 | |
| H2-74 | 3.125–12.5 | 6.25 | 12.5 | |
| H2-81 | 0.75–6.25 | 3.13 | 6.25 | |
| MSSE (24) | H2-38 | 0.75–6.25 | 3.13 | 6.25 |
| H2-39 | 1.5–12.5 | 3.13 | 6.25 | |
| H2-74 | 3.13–25 | 6.25 | 12.5 | |
| H2-81 | 0.75–12.5 | 3.13 | 6.25 | |
| MRSA (55) | H2-38 | 0.75–12.5 | 6.25 | 12.5 |
| H2-39 | 1.5–25 | 6.25 | 12.5 | |
| H2-74 | 1.5–25 | 6.25 | 12.5 | |
| H2-81 | 0.75–12.5 | 3.13 | 6.25 | |
| MSSA (40) | H2-38 | 0.75–12.5 | 3.13 | 6.25 |
| H2-39 | 1.5–12.5 | 3.13 | 6.25 | |
| H2-74 | 3.125–100 | 6.25 | 12.5 | |
| H2-81 | 0.75–12.5 | 3.13 | 6.25 |
Stock solutions of the compounds were prepared in 0.1% DMSO.
MIC represents the minimal inhibitory concentration of the derivatives against the test strains as determined by broth micro-dilution testing on a microtiter plate according to the standards of the American Clinical and Laboratory Standards Institute.
The MIC50 is the concentration below which 50% of the clinical isolate MIC values lie, and MIC90 is the concentration below which 90% of the isolate MIC values lie.
H2-38 and H2-81 inhibited the growth of all the 163 staphylococcal isolates (68 S. epidermidis strains and 95 S. aureus strains) at concentrations ≤12.5 µM. H2-39 and H2-74 inhibited the growth of 158 isolates (68 S. epidermidis strains and 95 S. aureus strains) at concentrations ≤12.5 µM, and four isolates (MRSA 14320, MRSA 1000718, MRSA 0916054 and MSSA 15760) exhibited a MIC of 25 µM. Only one S. aureus (MSSA 15760) isolate showed a higher MIC than the other strains (with H2-74, MIC=100 µM) (Table 3). There were no obvious differences between the MIC50 values of the four derivatives, but when comparing the MIC90 values, the anti-staphylococcal activities of H2-38 and H2-81 were greater than those of H2-39 and H2-74 (Table 2). H2-38 and H2-81 were used for further evaluating the anti-biofilm efficacy of the derivatives against the clinical staphylococcal strains.
Table 3. The MIC distribution for the four derivatives and the clinical staphylococcal isolates.
| Derivatives | MICa | |||||
|---|---|---|---|---|---|---|
| ≤12.5 (μM) | 25–50 (μM) | >50 (μM) | ||||
| SEb | SAb | SE | SA | SE | SA | |
| H2-38 | 68 | 95 | 0 | 0 | 0 | 0 |
| H2-39 | 68 | 91 | 0 | 4 | 0 | 0 |
| H2-74 | 67 | 91 | 1 | 3 | 0 | 1 |
| H2-81 | 68 | 95 | 0 | 0 | 0 | 0 |
MIC represents the minimal inhibitory concentration of the derivatives against the test strains as determined by broth micro-dilution testing on a microtiter plate according to the standards of the American Clinical and Laboratory Standards Institute.
SE represents S. epidermidis (68 clinical stains) and SA represents S. aureus (95 clinical strains).
Anti-biofilm efficacy against clinical staphylococcal strains
The microtiter plate assay showed that the minimal biofilm eradication concentrations of H2-38 and H2-81 against immature (6 h) biofilms of S. epidermidis ATCC 35984 were 12.5 µM and 6.3 µM, respectively. At a concentration of 6.25 µM, immature biofilms of all of the tested isolates (eight MRSE, four MSSE, five MRSA and two MSSA) were inhibited by both derivatives.
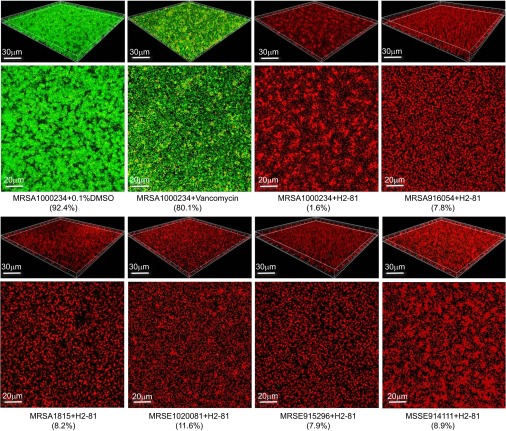
Confocal microscopy with Live/Dead staining showed that at a concentration of 4×MIC (6.25 µM) both H2-81 and H2-38 had bactericidal activity against mature (24 h) biofilms of all six strong biofilm-forming clinical strains (two MRSA, one MSSA, two MRSE and one MSSE). The number of viable cells in the biofilm was determined by ImageJ software. With H2-81 treatment, the percentages of viable cells in the biofilms of the six strains were reduced to 1.6%–11.6% (Table 4 and Figure 2). With H2-38 treatment, the viable cells were decreased to 3.5%–16.2% (Table 4). For the controls, 75.4%–80.8% of the cells were viable in the biofilm treated with vancomycin (128 µg/mL), and 69.8% of the cells were viable in the biofilm treated with the prototype of Compound 2. Using 0.1% dimethyl sulfoxide (DMSO) as a negative control, 92.4% of the cells were viable in the biofilms.
Table 4. The percentages of viable cells in the biofilms of the six strains treated with H2-81 and H2-38.
| Strainsa | |||||||
|---|---|---|---|---|---|---|---|
| Treatmenta | MRSA 1000234 | MRSA 916054 | MSSA 1815 | MRSE 1020081 | MRSE 915296 | MSSE 914111 | |
| Viable cells/total (%) |
H2-81 | 1.6 | 7.8 | 8.2 | 11.6 | 7.9 | 8.9 |
| H2-38 | 3.5 | 10.4 | 7.5 | 16.2 | 8.6 | 12.3 | |
| Vancomycin | 80.1 | 75.6 | 77.8 | 75.4 | 80.8 | 79.6 | |
| DMSOb |
92.4 |
89.1 |
88.5 |
91.8 |
91.4 |
93.6 |
|
| MIC (μM) | H2-81 | 1.5 | 12.5 | 1.5 | 3.1 | 3.1 | 1.5 |
| H2-38 | 1.5 | 12.5 | 1.5 | 6.3 | 3.1 | 1.5 | |
The six strains were selected randomly from 19 strong biofilm producers (MRSA 1000234, MRSA 916054, MSSA 1815, MRSE 1020081, MRSE 915296 and MSSE 914111).
Stock solutions of the compounds were prepared in 0.1% DMSO. Biofilms treated with 0.1% DMSO were used as the negative control.
Figure 2.
The bactericidal activities of H2-81 on 24-h biofilms of the clinical staphylococcal strains. The six strong biofilm-forming clinical strains were incubated in Fluoro Dishes at 37 °C for 24 h, and the biofilms were treated with H2-81 at concentrations of 4×MIC (6.25 µM) for 12 h. The biofilms were visualized by confocal laser scanning microscopy with the Live/Dead viability stain (SYTO9/PI). In the three-dimensional structural images of biofilms, the viable cells exhibit green fluorescence, whereas dead cells exhibit red fluorescence. The percentages of live bacteria of the total bacterial counts as shown in brackets were calculated by ImageJ software.
DISCUSSION
The increasing prevalence of antibiotic-resistant bacterial infections has become one of the greatest threats to human health. The clinical significance of staphylococci is closely related to their ability to form biofilms because biofilms are phenotypic resistance to antibiotics and often result in long-lasting infections.30 Most antibiotics are active against planktonic bacteria but not against biofilms, and they fail to cure biofilm-associated infections.31 The challenge of developing therapeutics to treat staphylococcal biofilm infections is that several characteristics of biofilms promote their antibiotic resistance, including the slow growth rate of bacteria in a biofilm, the high bacterial density and the binding of antibiotics to the biofilm slime.32
New types of antibiotics are urgently needed to combat the growing threat from MRSA and biofilm-forming strains that are resistant to conventional antibiotics.33,34 The YycFG TCS and its orthologs (renamed as WalK/WalR)35 are found in most low G+C gram-positive bacteria and play important roles in bacterial murein biosynthesis, cell wall metabolism and biofilm formation. TCSs have been regarded as potential targets for the development of new antibiotics, and inhibitors targeting YycG and YycF have been described. We previously reported that Compound 2, which targets the HK domain of the S. epidermidis YycG protein, inhibits S. epidermidis growth and biofilm formation.20 The structure of Compound 2 was optimized by retaining the intact thiazolidione core structure and altering the functional groups. A total of 91 derivatives were synthesized. Four of these derivatives (H2-38, H2-39, H2-74 and H2-81) exhibited high antibacterial activity and were selected to evaluate their efficacy against clinical staphylococcus strains.
In the present study, the 163 staphylococcal isolates were collected from three hospitals as follows: 99 (60.7%) isolates were methicillin-resistant (MRSA and MRSE); 106 isolates (65.0%) were resistant to erythromycin; 55 isolates (33.7%) were resistant to SMZ-TMP; 68 isolates (41.7%) were resistant to gentamycin; and 27 isolates (16.5%) were resistant to rifampicin. A total of 43.1% of S. aureus isolates and 61.7% of S. epidermidis isolates were biofilm producers. We evaluated the efficacy of the derivatives against the clinical strains. The four derivatives inhibited the planktonic growth of all isolates, including the MRSA and MRSE isolates. These derivatives displayed bactericidal activities against both immature (6 h) biofilms and mature (24 h) biofilms produced by the strong biofilm-forming strains, thus demonstrating that they can act at both the early and late stages of biofilm formation.
Combinations of antibiotics can enhance the effect of individual antimicrobials through synergic interactions and are considered to be a new therapeutic option for the treatment of staphylococcal biofilm-associated infections.36 However, it is important to choose the correct antibiotics for combination therapy. Rifampin is currently the most common constituent of antibiotic combinations against staphylococcal biofilms, and other frequently used antimicrobials are vancomycin and fusidic acid.32,37 In a preceding study, we demonstrated a synergistic effect between two of the derivatives (H2-74 and H2-81) and vancomycin or cefazolin on the S. epidermidis 3 5984 strain,23 thereby suggesting that these derivatives can be used alone or in combination with other antibiotics to combat staphylococcal biofilms.
In summary, the newly synthesized derivatives of Compound 2 exhibit substantial anti-staphylococcal activity against both methicillin-susceptible and methicillin-resistant clinical strains. Thus, these results warrant further investigation to develop a YycG inhibitor as a candidate drug that will be effective against multidrug-resistant staphylococci infections and biofilm-associated diseases.
Acknowledgments
This work was supported by the National Science and Technology Major Project of China (2012ZX10003008-010, 2012ZX10004401 and 2012ZX09301002-001) and the National Natural Science Foundation of China (81271791, 21072095 and 81101214).
Footnotes
Note: Supplementary Information for this article can be found on Emerging Microbes and Infections' website (http://www.nature.com/emi/)
Supplementary Information
References
- Manunga J, Jr, Olak J, Rivera C, Martin M. Prevalence of methicillin-resistant Staphylococcus aureus in elective surgical patients at a public teaching hospital: an analysis of 1039 patients. Am Surg. 2012;78:1096–1099. [PubMed] [Google Scholar]
- Haddadin AS, Fappiano SA, Lipsett PA. Methicillin resistant Staphylococcus aureus (MRSA) in the intensive care unit. Postgrad Med J. 2002;78:385–392. doi: 10.1136/pmj.78.921.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morell EA, Balkin DM. Methicillin-resistant Staphylococcus aureus: a pervasive pathogen highlights the need for new antimicrobial development. Yale J Biol Med. 2010;83:223–233. [PMC free article] [PubMed] [Google Scholar]
- Kateete DP, Namazzi S, Okee M, et al. High prevalence of methicillin resistant Staphylococcus aureus in the surgical units of Mulago hospital in Kampala, Uganda. BMC Res Notes. 2011;4:326. doi: 10.1186/1756-0500-4-326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haaber J, Cohn MT, Frees D, Andersen TJ, Ingmer H. Planktonic aggregates of Staphylococcus aureus protect against common antibiotics. PloS One. 2012;7:e41075. doi: 10.1371/journal.pone.0041075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto M. Staphylococcal infections: mechanisms of biofilm maturation and detachment as critical determinants of pathogenicity. Annu Rev Med. 2013;64:175–188. doi: 10.1146/annurev-med-042711-140023. [DOI] [PubMed] [Google Scholar]
- Cue D, Lei MG, Lee CY. Genetic regulation of the intercellular adhesion locus in staphylococci. Front Cell Infect Microbiol. 2012;2:38. doi: 10.3389/fcimb.2012.00038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mah TF, O'Toole GA. Mechanisms of biofilm resistance to antimicrobial agents. Trends Microbiol. 2001;9:34–39. doi: 10.1016/s0966-842x(00)01913-2. [DOI] [PubMed] [Google Scholar]
- Lewis K. Riddle of biofilm resistance. Antimicrob Agents Chemother. 2001;45:999–1007. doi: 10.1128/AAC.45.4.999-1007.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou W, Sun X, Wang Z, Zhang Y. Biofilm-forming capacity of Staphylococcus epidermidis, Staphylococcus aureus, and Pseudomonas aeruginosa from ocular infections. Invest Ophthalmol Vis Sci. 2012;53:5624–5631. doi: 10.1167/iovs.11-9114. [DOI] [PubMed] [Google Scholar]
- Darouiche RO. Treatment of infections associated with surgical implants. N Engl J Med. 2004;350:1422–1429. doi: 10.1056/NEJMra035415. [DOI] [PubMed] [Google Scholar]
- Walters MC 3rd, Roe F, Bugnicourt A, Franklin MJ, Stewart PS Contributions of antibiotic penetration, oxygen limitation, and low metabolic activity to tolerance of Pseudomonas aeruginosa biofilms to ciprofloxacin and tobramycin. Antimicrob Agents Chemother. 2003;47:317–323. doi: 10.1128/AAC.47.1.317-323.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aendekerk S, Diggle SP, Song Z, et al. The MexGHI-OpmD multidrug efflux pump controls growth, antibiotic susceptibility and virulence in Pseudomonas aeruginosa via 4-quinolone-dependent cell-to-cell communication. Microbiology. 2005;151(Pt 4):1113–1125. doi: 10.1099/mic.0.27631-0. [DOI] [PubMed] [Google Scholar]
- Dubrac S, Msadek T. Tearing down the wall: peptidoglycan metabolism and the WalK/WalR (YycG/YycF) essential two-component system. Adv Exp Med Biol. 2008;631:214–228. doi: 10.1007/978-0-387-78885-2_15. [DOI] [PubMed] [Google Scholar]
- Dubrac S, Bisicchia P, Devine KM, Msadek T. A matter of life and death: cell wall homeostasis and the WalKR (YycGF) essential signal transduction pathway. Mol Microbiol. 2008;70:1307–1322. doi: 10.1111/j.1365-2958.2008.06483.x. [DOI] [PubMed] [Google Scholar]
- Okada A, Igarashi M, Okajima T, et al. Walkmycin B targets WalK (YycG), a histidine kinase essential for bacterial cell growth. J Antibiot. 2010;63:89–94. doi: 10.1038/ja.2009.128. [DOI] [PubMed] [Google Scholar]
- Watanabe T, Hashimoto Y, Yamamoto K, et al. Isolation and characterization of inhibitors of the essential histidine kinase, YycG in Bacillus subtilis and Staphylococcus aureus. J Antibiot. 2003;56:1045–1052. doi: 10.7164/antibiotics.56.1045. [DOI] [PubMed] [Google Scholar]
- Yamamoto K, Kitayama T, Minagawa S, et al. Antibacterial agents that inhibit histidine protein kinase YycG of Bacillus subtilis. Biosci Biotechnol Biochem. 2001;65:2306–2310. doi: 10.1271/bbb.65.2306. [DOI] [PubMed] [Google Scholar]
- Hilliard JJ, Goldschmidt RM, Licata L, Baum EZ, Bush K. Multiple mechanisms of action for inhibitors of histidine protein kinases from bacterial two-component systems. Antimicrob Agents Chemother. 1999;43:1693–1699. doi: 10.1128/aac.43.7.1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin Z, Zhang J, Xu B, et al. Structure-based discovery of inhibitors of the YycG histidine kinase: new chemical leads to combat Staphylococcus epidermidis infections. BMC Microbiol. 2006;6:96. doi: 10.1186/1471-2180-6-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang RZ, Zheng LK, Liu HY, et al. Thiazolidione derivatives targeting the histidine kinase YycG are effective against both planktonic and biofilm-associated Staphylococcus epidermidis. Acta Pharmacol Sin. 2012;33:418–425. doi: 10.1038/aps.2011.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan B, Huang R, Zheng L, et al. Thiazolidione derivatives as novel antibiofilm agents: design, synthesis, biological evaluation, and structure-activity relationships. Eur J Med Chem. 2011;46:819–824. doi: 10.1016/j.ejmech.2010.12.014. [DOI] [PubMed] [Google Scholar]
- Liu H, Zhao D, Chang J, et al. Efficacy of novel antibacterial compounds targeting histidine kinase YycG protein. Appl Microbiol Biotechnol. 2014;98:6003–6013. doi: 10.1007/s00253-014-5685-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenover FC, Tickler IA, Goering RV, Kreiswirth BN, Mediavilla JR, Persing DH. Characterization of nasal and blood culture isolates of methicillin-resistant Staphylococcus aureus from patients in United States Hospitals. Antimicrob Agents Chemother. 2012;56:1324–1330. doi: 10.1128/AAC.05804-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neill E, Pozzi C, Houston P, et al. Association between methicillin susceptibility and biofilm regulation in Staphylococcus aureus isolates from device-related infections. J Clin Microbiol. 2007;45:1379–1388. doi: 10.1128/JCM.02280-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanovic S, Vukovic D, Hola V, et al. Quantification of biofilm in microtiter plates: overview of testing conditions and practical recommendations for assessment of biofilm production by staphylococci. APMIS. 2007;115:891–899. doi: 10.1111/j.1600-0463.2007.apm_630.x. [DOI] [PubMed] [Google Scholar]
- Jiang L, Wang F, Han F, Prinyawiwatkul W, No HK, Ge B. Evaluation of diffusion and dilution methods to determine the antimicrobial activity of water-soluble chitosan derivatives. J Appl Microbiol. 2013;114:956–963. doi: 10.1111/jam.12111. [DOI] [PubMed] [Google Scholar]
- Peterson JF, Pfaller MA, Diekema DJ, Rinaldi MG, Riebe KM, Ledeboer NA. Multicenter comparison of the Vitek 2 antifungal susceptibility test with the CLSI broth microdilution reference method for testing caspofungin, micafungin, and posaconazole against Candida spp. J Clin Microbiol. 2011;49:1765–1771. doi: 10.1128/JCM.02517-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ning Y, Hu X, Ling J, et al. Candida albicans survival and biofilm formation under starvation conditions. Int Endod J. 2013;46:62–70. doi: 10.1111/j.1365-2591.2012.02094.x. [DOI] [PubMed] [Google Scholar]
- Liduma I, Tracevska T, Bers U, Zilevica A. Phenotypic and genetic analysis of biofilm formation by Staphylococcus epidermidis. Medicina (Kaunas) 2012;48:305–309. [PubMed] [Google Scholar]
- Olson ME, Ceri H, Morck DW, Buret AG, Read RR. Biofilm bacteria: formation and comparative susceptibility to antibiotics. Can J Vet Res. 2002;66:86–92. [PMC free article] [PubMed] [Google Scholar]
- Saginur R, Stdenis M, Ferris W, et al. Multiple combination bactericidal testing of staphylococcal biofilms from implant-associated infections. Antimicrob Agents Chemother. 2006;50:55–61. doi: 10.1128/AAC.50.1.55-61.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elhani D.Does the emergence of antibiotic resistance announce the return of the dark ages Ann Biol Clin (Paris) 201169637–646.French. [DOI] [PubMed] [Google Scholar]
- Woodford N, Wareham DW. UK Antibacterial Antisense Study Group. Tackling antibiotic resistance: a dose of common antisense. J Antimicrob Chemother. 2009;63:225–229. doi: 10.1093/jac/dkn467. [DOI] [PubMed] [Google Scholar]
- Dubrac S, Boneca IG, Poupel O, Msadek T. New insights into the WalK/WalR (YycG/YycF) essential signal transduction pathway reveal a major role in controlling cell wall metabolism and biofilm formation in Staphylococcus aureus. J Bacteriol. 2007;189:8257–8269. doi: 10.1128/JB.00645-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monzon M, Oteiza C, Leiva J, Amorena B. Synergy of different antibiotic combinations in biofilms of Staphylococcus epidermidis. J Antimicrob Chemother. 2001;48:793–801. doi: 10.1093/jac/48.6.793. [DOI] [PubMed] [Google Scholar]
- Kiedrowski MR, Horswill AR. New approaches for treating staphylococcal biofilm infections. Ann NY Acad Sci. 2011;1241:104–121. doi: 10.1111/j.1749-6632.2011.06281.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.