SUMMARY
Transcription factors and chromatin-remodeling complexes are key determinants of embryonic stem cell (ESC) identity. Here, we demonstrate that BRD4, a member of the bromodomain and extraterminal domain (BET) family of epigenetic readers, regulates the self-renewal ability and pluripotency of ESCs. BRD4 inhibition resulted in induction of epithelial-tomesenchymal transition (EMT) markers and commitment to the neuroectodermal lineage while reducing the ESC multidifferentiation capacity in teratoma as-says. BRD4 maintains transcription of core stem cell genes such as OCT4 and PRDM14 by occupying their super-enhancers (SEs), large clusters of regulatory elements, and recruiting to them Mediator and CDK9, the catalytic subunit of the positive transcription elongation factor b (P-TEFb), to allow Pol-II-dependent productive elongation. Our study describes a mechanism of regulation of ESC identity that could be applied to improve the efficiency of ESC differentiation.
INTRODUCTION
Embryonic stem cells (ESCs) rely on a small set of core pluripotency transcription factors (TFs) to preserve self-renewal capacity and cellular plasticity (Keramari et al., 2010; Mitsui et al., 2003; Pesce and Schöler, 2001). These cells possess a distinctive “open” chromatin structure with abundant posttranslational modifications on the N-terminal tails of histones, which allows them to sustain a high level of transcriptional activity compared to fully differentiated cells (Meshorer and Misteli, 2006). The conformational state of the chromatin functions as a signal inter preted by the transcriptional machinery to activate the expression of the core pluripotency TFs that maintain the ESC state (Ding et al., 2009; Fazzio et al., 2008; Li et al., 2012; Morey et al., 2012; Onder et al., 2012).
Bromodomain (BrD)-containing proteins function as key epigenome readers at the interface between chromatin remodeling and transcriptional regulation (Haynes et al., 1992). In humans, there are 61 predicted bromodomains in 46 proteins (Filippakopoulos et al., 2012; Sanchez and Zhou, 2009), including chromatin regulators of the SWI/SNF superfamily of DNA helicases (Tamkun et al., 1992), histone acetyltransferases (HATs) such as CREB-binding protein (Bannister and Kouzarides, 1996; Ogryzko et al., 1996) and TBP-associated factor 1 (Morinière et al., 2009), as well as the bromodomain and extraterminal domain (BET) family of transcriptional regulators (BRD2, BRD3, BRD4, and BRDT). BRD2, BRD3, and BRD4 are ubiquitously expressed, whereas BRDT expression is restricted to testis (Jones et al., 1997). BET proteins play multiple roles in transcription by binding to acetylated lysines and recruiting chromatin-modifying enzymes that function as context-dependent coactivators or corepressors (reviewed in Belkina and Denis, 2012). They serve as scaffolds for chromatin binding of HATs, histone deacetylases, and components of the SWI/SNF and Mediator complexes. In addition, BRD4 and BRDT interact with the positive transcription elongation factor b (P-TEFb) through their carboxy-terminal regions (Bisgrove et al., 2007). Recent studies have demonstrated important roles of BET proteins in development, inflammation, and cancer (reviewed in Belkina and Denis, 2012). Instrumental to these studies has been the availability of highly permeable, potent, and specific small-molecule inhibitors, which have allowed probing of BET functions in a variety of experimental systems (Segura et al., 2013; Filippakopoulos et al., 2010; Nicodeme et al., 2010; Zhang et al., 2012).
Our study reveals that BRD4 sustains ESC self-renewal and controls cell fate decisions by positively regulating the expression of pluripotency genes. BRD4 specifically governs the transcriptional elongation of stem cell transcripts by occupying their associated super-enhancers (SEs) and recruiting Mediator and CDK9 to those regulatory elements, thus emerging as a key regulator of the intricate gene expression network that maintains ESC identity.
RESULTS
BRD4 Is Required for Maintenance of ESC Identity
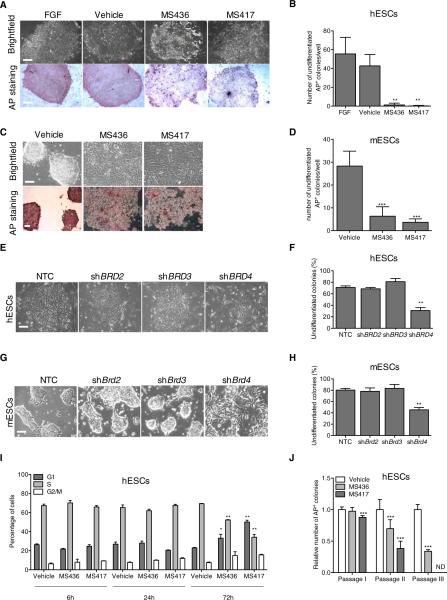
To investigate the role of BET proteins in the maintenance of ESC identity, we inhibited BET activity by pharmacological and genetic means. We used highly specific, cell-permeable small molecules to effectively block acetylated lysine binding by BET proteins (Borah et al., 2011; Zhang et al., 2012). MS436 is a broad-range diazobenzene compound with high affinity for some BrD-containing proteins (Figure S1A; Zhang et al., 2013). MS417 is a thienotriazolodiazepine BrD inhibitor with high affinity and specificity for the BrDs of BET proteins (Figure S1A and S1B) and is structurally related to previously reported BET inhibitors, JQ1 (Filippakopoulos et al., 2010) and GSK525762A (Nicodeme et al., 2010). Treatment of human ESCs (hESCs) with either MS436 or MS417 altered colony integrity and reduced alkaline phosphatase (AP) activity, compared to vehicle- or fibroblast growth factor (FGF)-treated cells (Figures 1A and 1B). Similar results were obtained in murine ESCs (mESCs) treated with BET inhibitors in the presence of leukemia inhibitory factor (LIF) (Figures 1C and 1D). To dissect the relative contributions of individual BET proteins to the observed morphological changes of ESCs following BET inhibition, we independently silenced BRD2, BRD3, and BRD4 by short hairpin RNA (shRNA)-mediated transduction in both hESCs and mESCs (Figures S2A and S2B). Depletion of BRD2 and BRD3 did not perturb ESC colony formation (Figures 1E–1H). In contrast, BRD4 silencing disrupted both mouse and human ESC colony integrity, yielding a flattened and dispersed morphology similar to that observed with BET inhibitors and characteristic of differentiating ESCs (Figures 1E– 1H). Similar results were obtained with an independent small interfering RNA (siRNA)-mediated knockdown of BRD4 in hESCs (Figures S2C–S2E). Importantly, BET inhibition and BRD4 suppression impaired ESC colony formation without overt induction of apoptosis (Figure S2F; data not shown), indicating that chemical or genetic inhibition of BRD4 is not detrimental to ESC viability. We also observed that prolonged compound treatment resulted in accumulation of ESCs in the G1 phase of the cell cycle at the expense of S phase and cell proliferation (Figures 1I, S2G, and S2H), which is consistent with the acquisition of a more-differentiated phenotype. Finally, BET inhibition reduced the ability of both human and murine ESCs to form colonies after repeated passaging (Figures 1J and S2I); compound-treated colonies that did form were only weakly positive for AP (Figure S2J). Collectively, these results demonstrate that BRD4 is a key regulator of ESC maintenance.
Figure 1. BRD4 Regulates ESC Identity.
(A) Representative bright field images (upper panel) of hESCs treated for 3 days with BET inhibitors compared to FGF- or vehicle-treated cells. Alkaline Phosphatase (AP) staining following treatment is shown in bottom panel.
(B) Quantification of AP-positive hESC colonies following FGF, vehicle, or compound treatment.
(C) Bright field images (upper panel) and AP staining (bottom panel) of mESCs treated with BET inhibitors for 3 days in the presence of LIF.
(D) Quantification of AP-positive mESC colonies following 3 days of compound treatment.
(E–H) Bright field images of hESC (E) and mESC (G) colonies following BRD2, BRD3, and BRD4 suppression. Percentage of undifferentiated hESCs and mESC colonies is shown in (F) and (H), respectively. NTC, nontargeting control.
(I) Cell-cycle analysis of hESCs following BET inhibitors treatment at the indicated time points as determined by FACS analysis.
(J) Self-renewal capacity of vehicle- and compound-treated hESCs passaged on mitotically inactivated MEFs in the presence of FGF as measured by AP staining. AP, alkaline phosphatase; ND, not detected. *p ≤ 0.05; **p ≤ 0.01; ***p ≤ 0.001. The scale bar represents 100 μm. Error bars represent SD. See also Figures S1 and S2.
BRD4 Positively Regulates the ESC Transcriptome
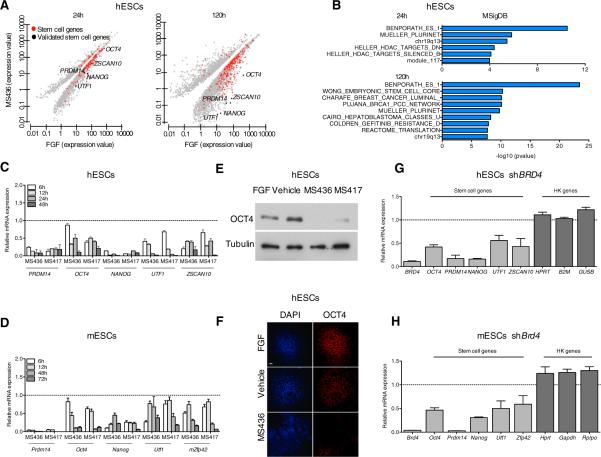
In deducing the mechanism by which BRD4 maintains the ESC state, we reasoned that BRD4 was likely to regulate the expression of genes required for ESC identity. Global gene-expression profiling by RNA sequencing of compound- versus FGF-treated hESCs confirmed the suppression of pluripotency transcripts (Table S1) at day 1 post-compound treatment, even more pronounced by day 5 (Figure 2A; Table S2). Gene set enrichment analysis of downregulated genes revealed enrichment in multiple ESC-related gene categories following compound treatment, indicating that the stem cell gene network is significantly perturbed by BET inhibition (Figure 2B). The expression of key determinants of ESC identity, including OCT4, NANOG, and PRDM14, was rapidly suppressed following compound treatment in human and mouse ESCs cultured even in the presence of FGF (hESCs; Figure 2C) or LIF (mESCs; Figure 2D), indicating that BET inhibition can counteract the extracellular signals that help to maintain ESCs in an undifferentiated state in vitro. Consistently, we found that OCT4 protein levels were reduced in hESCs following BET inhibition (Figures 2E and 2F). The rapid loss of expression of stem cell factors upon BET inhibition preceded the defects in cell cycle progression associated with the acquisition of a more differentiated phenotype (Figures 1I, S2G, and S2H), indicating that the former is not a result of the latter. Similar results were obtained in an established human induced pluripo-tent stem cell (iPSC) line (Papapetrou et al., 2009; Figure S3A) and in a pluripotent embryonic carcinoma cell line (NTERA-2; Figure S3B), indicating that the effects of BET inhibition on pluripotency gene expression are not restricted to ESC lines. Treatment of hESCs with the BET inhibitor JQ1 (Filippakopoulos et al., 2010) yielded comparable suppression of stem cell genes to MS417 treatment (Figure S3C). Moreover, RNAi-mediated BRD4 silencing led to reduced expression of key determinants of stem cell identity (e.g., OCT4, PRDM14, and NANOG) in human and mouse ESCs (Figures 2G and 2H), mimicking the effects of compound treatment. Interestingly, the expression of housekeeping genes was mostly unaffected upon BRD4 silencing (Figures 2G and 2H), suggesting a specific transcriptional regulation of stem cell genes by BRD4. The expression of stem cell genes was also suppressed in hESC, established iPSC, and NTERA lines following siRNA-mediated BRD4 depletion (Figures S3D–S3F). Our findings indicate that BRD4 specifically contributes to the transcriptional program that maintains the expression of many stem cell genes and ESC identity.
Figure 2. BRD4 Regulates the Expression of Stem Cell Genes.
(A) Scatter plots of gene expression at 24 hr and 120 hr in hESCs following MS436 treatment. Each point corresponds to a differentially expressed gene in compound-treated versus FGF-treated hESCs. Red dots indicate genes in the ESC gene categories (Table S1). Black dots represent stem cell genes validated throughout the manuscript. q value cutoff used throughout the analysis = 0.01.
(B) Gene set enrichment analysis of downregulated genes at 24 hr and 120 hr following BET inhibition, according to MSigDB.
(C and D) Time course expression of stem cell genes in hESCs (C) and mESCs (D), cultured in the presence of FGF (C) or LIF (D), upon compound treatment relative to vehicle-treated cells (dashed line), as detected by qRT-PCR.
(E) Effect of BET inhibitor treatment (3 days) on OCT4 protein levels as detected by immunoblotting. Tubulin is used as a loading control.
(F) Effect of compound treatment on hESC colony morphology (DAPI) and OCT4 protein expression as detected by immunostaining. The scale bar represents 100 μm.
(G and H) Relative mRNA expression of the indicated stem cell and housekeeping genes (HK) in hESCs (G) and mESCs (H) after BRD4 genetic depletion, as detected by qRT-PCR. Dashed lines represent nontargeting control (NTC)-transduced cells.
Error bars represent SD. See also Tables S1 and S2 and Figure S3.
Surprisingly, the expression of c-MYC, a positive regulator of proliferation and survival, was not downregulated after BET inhibition in either mouse or human ESCs (Figure S3G and S3H), despite being suppressed by compound treatment in cancer settings (Dawson et al., 2011; Zuber et al., 2011; Delmore et al., 2011). This suggests that c-MYC transcriptional regulation by BRD4 may be context dependent and, in the case of ESCs, BRD4 independent.
BRD4 Regulates Pluripotency of ESCs and Neuroectodermal Lineage Commitment
We next investigated whether BRD4 inhibition results in ESC differentiation together with suppression of stem cell genes and loss of self-renewal capacity. Compound treatment of ESCs modulated the expression of genes involved in epithelialto-mesenchymal transition (EMT) (Figures 3A and 3B), a defining feature of ESCs undergoing differentiation. Accompanying the emergence of an EMT signature, we observed an enrichment of neuroectodermal differentiation markers (e.g., NES, NCAD, and SOX1) relative to markers of other lineages in both hESCs (Figures 3B and 3C) and mESCs (Figures S4A and S4B), indicating lineage specificity associated with BET inhibition. Consistently, individual silencing of BRD4 mimicked the preferential regulation of EMT and neuroectodermal markers over other line-ages observed following pharmacologic BET inhibition in human (Figure 3D) and mouse ESCs (Figure S4C). BET inhibitor treat ment of ESCs significantly affected both the number and morphology of embryoid bodies (EBs) (Figures S5A and S5B), indicating that BRD4 activity is required for the formation of tridimensionally organized EBs. Moreover, BET inhibition resulted in suppression of stem cell genes (Figure S5C) and induction of neuroectodermal lineage genes over genes of other lineages in EBs formation assay (Figure S5D), highlighting the specific line-age commitment following BET inhibition.
Figure 3. BET Inhibition and BRD4 Suppression Lead to EMT Induction and Neuroectodermal Lineage Commitment of ESCs.
(A) Downregulation of the epithelial marker E-cadherin (ECAD) in FGF-, vehicle-, or compound-treated hESCs, as detected by immunofluorescence analysis after 5 days of compound treatment. The scale bar represents 100 μm.
(B) Expression over time of a panel of EMT, neuroectodermal, and mesendodermal markers in hESCs following BET inhibition, as detected by qRT-PCR. Dashed line represents expression in vehicle-treated cells.
(C) Gene set enrichment analysis of upregulated genes following 5 days of compound treatment in Gene Ontology terms compared to FGF-treated hESCs.
(D) Expression of EMT (red) and mesendodermal (green) genes following BRD4 depletion in hESCs, as detected by qRT-PCR. Dashed line represents NTC-transduced hESCs.
(E) Representative images of teratomas derived from NTC and shBrd4-transduced mESCs. The scale bar represents 1 cm.
(F) Weight distribution of NTC- and Brd4-depleted mESC-derived tumors. Horizontal line represents the median value.
(G) Representative micrographs of hematoxylin and eosin sections of NTC or shBrd4-mESCs-derived tumors. EC, ectoderm; EN, endoderm; M, mesoderm.
(H) Expression of Ki67 and NeuN in shBrd4-derived tumors compared to NTC-derived teratomas as detected by immunohistochemistry. The scale bar represents 200 μm.
(I) Oct4 expression levels following compound treatment in mESCs transduced with an empty vector or Oct4 cDNA-expressing vector, as detected by qRT-PCR.
(J) Expression by qRT-PCR of EMT and neuroectodermal genes (Ncad and Nestin) following BET inhibition in control and Oct4-expressing mESCs.
(K) Expression by qRT-PCR of mesendodermal genes (Brachyury and Cdx2) following BET inhibition in control and Oct4-expressing mESCs.
Error bars represent SD. *p ≤ 0.05. See also Table S3 and Figures S4 and S5.
A defining trait of ESC pluripotency is the ability to form teratomas in vivo. To determine if BRD4 regulates pluripotency in vivo, ESCs were efficiently transduced with either nontargeting control (NTC) or shBrd4 lentiviral vectors and injected subcutaneously into immunodeficient (Rag2−/−) mice. After 4 weeks, we found that Brd4 depletion significantly reduced both the size and number of engrafted teratomas (Figures 3E and 3F). Histological analysis of shBrd4- versus control-derived tumors revealed a reduced representation of meso- and endodermal germ layers relative to neuroectodermal lineage (Figure 3G; Table S3). This phenotype could not be explained merely by reduced proliferation as shown by the presence of Ki67-positive cells in Brd4-depleted tumors (Figure 3H, top panels). Furthermore, shBrd4-derived tumors displayed induction of neuroectodermal markers (i.e., NeuN), recapitulating the effects of BET inhibition in vitro in lineage commitment (Figure 3H, bottom panels). Together, these findings indicate that Brd4 is a critical mediator of ESC pluripotency in vivo.
Next, we investigated the mechanisms by which BRD4 inhibition results in ESCs commitment toward the neuroectodermal lineage. Considering that, in addition to having a critical role in pluripotency, Oct4 also acts as a repressor of neuroectodermal specification (Thomson et al., 2011; Wang et al., 2012), we hypothesized that its downregulation following BRD4 inhibition may be responsible for the induction of neuroectodermal line-age. Therefore, ectopic Oct4 expression should prevent the induction of the neuroectodermal lineage upon compound treatment. Indeed, we found that ectopic Oct4 expression (Figure 3I) in ESCs treated with BET inhibitors restricted the induction of EMT and neuroectodermal markers (Figure 3J). Nevertheless, Oct4 upregulation in this context did not rescue the expression of stem cell genes and it was unable to induce the levels of mesoendodermal markers (Figure 3K; data not shown). In sum, these findings indicate that the neuroectodermal lineage commitment of ESCs following BRD4 inhibition is at least partially due to downregulation of Oct4 and imply that BRD4 controls both ESC identity and cell fate decisions via regulation of key pluripotency genes.
BRD4 Occupies and Transcriptionally Regulates Super-Enhancer-Associated Stem Cell Genes
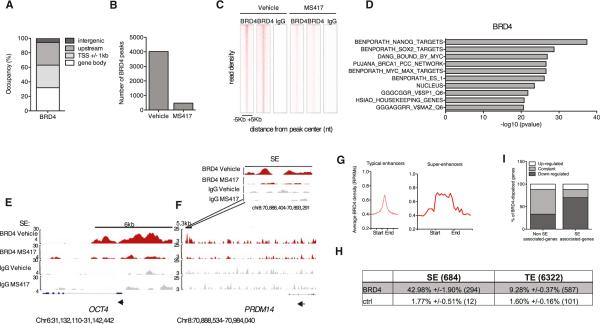
In order to dissect the mechanisms by which BRD4 regulates pluripotency gene expression, we performed chromatin immunoprecipitation sequencing (ChIP-seq) for BRD4 in vehicle- or compound-treated hESCs grown in the presence of FGF in two independent, highly reproducible (R2 = 0.815) biological replicates. Using stringent parameters (see Experimental Procedures section), we identified 4,026 BRD4-binding peaks that were common to both biological replicates. We first examined the distribution of BRD4 protein occupancy at defined regions relative to gene elements in vehicle-treated hESCs by classifying the identified peaks into (1) 1 kb transcription start site (TSS)-flanking regions of known transcripts; (2) gene body regions, excluding any overlapping regions with (1); or (3) upstream regions of 10 kb to 100 kb, excluding any overlapping regions with either (1) or (2). The remaining genomic loci were classified as intergenic regions. In vehicle-treated cells, BRD4 occupied gene bodies and TSSs (31.9% and 31.1%; Figure 4A) and was also present at upstream regions (31.1%; Figure 4A), suggesting a possible binding to regulatory elements in the genome in addition to promoters and gene bodies. BRD4 occupancy in hESCs was analyzed 6 hr after treatment to identify early compound-responsive elements. BET inhibition consistently resulted in robust and global displacement (defined as absence of peak in treatment; see Experimental Procedures) of BRD4 from the vast majority of the occupied genomic loci common to the two biological replicates (Figures 4B and 4C), indicating that our small-molecule compounds are potent displacers of BRD4 from chromatin. Gene set enrichment analysis of genes from which BRD4 was displaced following treatment revealed a significant enrichment in stem cell gene categories, enforcing the concept that BRD4 regulates the stem cell gene transcriptional network (Figure 4D; Table S4).
Figure 4. BRD4 Occupies and Regulates Super-Enhancer-Associated Genes.
(A) Genome-wide distribution of BRD4 in 1-kb-flanked TSS regions, gene bodies (excluding flanked TSS regions), upstream (up to 100 kb and excluding flanked TSSs and gene bodies), and intergenic regions (excluding all of the above) by ChIP-seq.
(B) Number of BRD4-binding peaks common to two biological replicates after 6 hr of vehicle or MS417 treatment.
(C) Heatmap representation of ChIP-seq binding for BRD4 peaks common to two independent biological replicates before and after BET inhibition, rank ordered form the most BRD4 to lowest BRD4. Immunoglobulin G (IgG) is shown as a negative control for enrichment.
(D) Gene set enrichment analysis of BRD4 target genes (e.g., genes from which BRD4 is displaced following BET inhibition [top ten categories are shown]).
(E and F) Genome browser representations of BRD4 and IgG ChIP-seq reads at stem cell genes [i.e., OCT4 (E) and PRDM14 (F)], in FGF-cultured hESCs treated with vehicle or MS417. Black bars indicate super-enhancers (SEs) (Hnisz et al., 2013). Arrows indicate direction of transcription. Insert represents a close up for BRD4 and IgG binding at PRDM14 SE before and after treatment. Chromosomal locations are indicated.
(G) Read density representation of global BRD4 occupancy at typical enhancer (TE) and SE elements. The x axis shows the center of TE regions flanked by 5 kb as well as the start and end of SE regions flanked by 5 kb of adjacent sequence. The y axis indicates reads per kilobase of transcript per million reads mapped (RPKMs).
(H) Table indicates the percentage and total number of BRD4-overlapping TEs and SEs at BRD4 peak signal distributions comparable between TEs and SEs. TEs were extended upstream and downstream from their centers such that their size equals the mean size of SEs. Control (ctrl) refers to the percent of peaks at SEs/TEs if the BRD4-displaced regions were randomly distributed along the genome.
(I) Histogram indicates the percentage of BRD4 target genes that are downregulated, constant, and upregulated in SE- and non-SE-associated gene categories upon compound treatment.
See also Tables S4 and S5 and Figure S7.
Recently, large enhancer domains called SEs have been identified at key cell identity genes (Whyte et al., 2013; Hnisz et al., 2013). We found that BRD4 localizes at SEs of stem cell genes, such as OCT4 and PRDM14, in vehicle-treated hESCs and is displaced from them following compound treatment (Figures 4E and 4F). Despite BRD4 peak signal intensity being similar in SEs and typical enhancers (TEs) in hESCs (Figure 4G), more BRD4-binding peaks were found in SEs than in TEs. Indeed, BRD4-binding peaks reside in 42.98% of SEs (294 out of 684 SEs) compared to 9.28% TEs (587 out of 6,322 TEs; Figure 4H), after controlling for BRD4 peak signal intensity and enhancer size. These data suggest that BRD4 may specifically bind and regulate SE-associated genes, which are key stem cell identity genes. In further support of preferential regulation of SE-associated genes by BRD4, integration of ChIP-seq data with changes in gene expression revealed that, of all BRD4 targets, those associated with SEs were mostly downregulated upon BRD4 displacement, whereas the majority of non-SE-associated genes remained constant (Figure 4I). These findings indicate that SE-associated genes are more dependent on BRD4 binding for their active transcription than genes that do not associate with SEs.
Expression of SE-Associated Genes Depends on BRD4-Mediated Recruitment of Mediator Complex at SEs
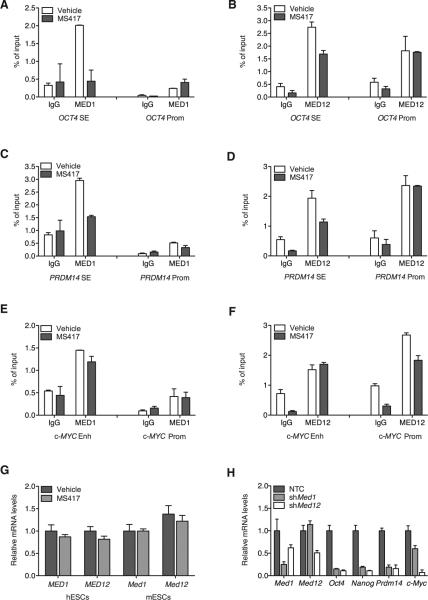
The Mediator complex is a transcriptional activator of the pluripotency gene network that represents a potentially important player in BRD4-mediated transcriptional regulation of SE-associated stem cell genes. We found that MED1 (Core Mediator) and MED12 (CDK8-module) subunits localize at both SEs and promoters of stem cell genes in hESCs (i.e., OCT4 and PRDM14; Figures 5A–5D). Notably, the binding of MED1 and MED12 at SEs, but not at promoters, was significantly reduced by BRD4 inhibition (Figures 5A–5D), suggesting that BRD4 may act as a docking site for the Mediator complex at SEs of stem cell genes, thus regulating their expression. Consistent with the observation that c-MYC was not downregulated following BRD4 inhibition (Figure S3G), we did not find a SE associated with c-MYC in hESCs. In this non-SE-associated gene, Mediator occupancy was not altered by compound treatment (Figures 5E and 5F). Importantly, displacement of Mediator from SEs upon compound treatment was not due to reduced levels of MED1 and MED12 following BRD4 inhibition in hESCs and mESCs (Figure 5G). Silencing of Mediator subunits resulted in suppression of stem cell genes, thus recapitulating the effects of BRD4 inhibition on the ESC transcriptional network (Figure 5H). The expression of c-Myc was also inhibited following Med1 and Med12 depletion (Figure 5H), suggesting that Mediator is a positive transcriptional regulator of c-MYC expression but independently of BRD4. Altogether, these data indicate that the expression of SE-associated stem cell genes relies on BRD4-dependent binding of Mediator-containing complexes at SEs.
Figure 5. Mediator Is Required for BRD4-Dependent Regulation of SE-Associated Genes.
(A–F) ChIP analysis of MED1 and MED12 binding at enhancer regions and promoters (prom) of SE-associated stem cell genes [OCT4 (A and B) and PRDM14 (C and D) and a non-SE-associated gene, c-MYC (E and F)] in hESCs following BET inhibition. Bar graphs represent the mean enrichment relative to input, and error bars reflect SD of a representative experiment run in triplicate.
(G) Expression levels of MED1 and MED12 by qRT-PCR following BET inhibition in hESCs and mESCs.
(H) Expression of the indicated genes in shMed1 and shMed12 mESCs compared to NTC-transduced cells as detected by qRT-PCR.
See also Figure S6H for primers design.
BRD4 Is Required for Productive Elongation of SE-Associated Genes
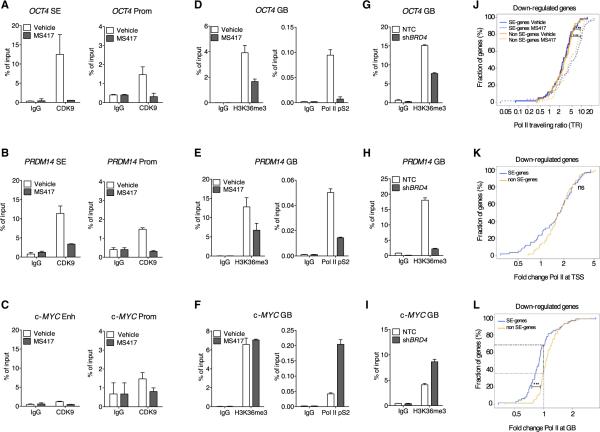
In addition to Mediator, BRD4 has been found to interact with CDK9, the catalytic subunit of the P-TEFb transcriptional elongation complex (Bisgrove et al., 2007; Dawson et al., 2011). To study whether BRD4 regulates the expression of SE-associated stem cell genes via productive transcriptional elongation, we performed ChIP assays for CDK9, as well as two elongation marks, the phosphorylated Pol II at Ser2 (pS2) and H3K36me3. We found that together with BRD4, CDK9 is more associated to SE of stem cell genes (e.g., OCT4 and PRDM14; Figures 6A and 6B) than to genes bearing regular enhancer elements (e.g., c-MYC; Figure 6C). BRD4 inhibition resulted in dramatic loss of CDK9 from the SEs and promoters (Figures 6A and 6B). Next, we investigated whether loss of BRD4 and CDK9 from SEs results in reduced levels of elongation marks in gene bodies (GBs) of SE-associated stem cell genes. H3K36me3 and Pol II pS2 were reduced following compound treatment at the gene bodies of OCT4 and PRDM14 (Figures 6D and 6E). Conversely, c-MYC, a non-SE-associated gene, did not display any loss of elongation marks (Figure 6F), consistent with its unreduced expression levels following BRD4 inhibition (Figure S3G). BRD4 silencing was sufficient to recapitulate the effects of BET inhibition on the transcriptional elongation of SE-associated stem cell genes and did not affect the levels of elongation marks at c-MYC gene body (Figures 6G–6I). The levels of active transcription and initiation marks, such as H3K4me3 and Pol II pS5, were mostly unaffected by BET inhibition at both SE and non-SE-associated genes (Figures S6A–S6C). Furthermore, levels of H4K5 acetylation, one of the docking sites for BET binding to chromatin (Filippakopoulos et al., 2012), remained unchanged upon compound treatment (Figures S6D–S6F). These findings suggest that BRD4 controls SE-associated stem cell gene expres sion by regulating their transcriptional elongation rather than transcriptional initiation or histone acetylation.
Figure 6. BRD4 Inhibition Impairs the Transcriptional Elongation of SE-Associated Stem Cell Genes.
(A–C) ChIP analysis of the elongation factor CDK9 at SEs and promoters of OCT4 (A), PRDM14 (B), and c-MYC enhancer and promoter (C) following compound treatment. IgG was used as control. Bar graphs represent the mean enrichment relative to input, and error bars reflect SD of a representative experiment run in triplicate.
(D–F) ChIP analysis of the elongation marks H3K36me3 and Pol II-pS2 on gene bodies (GBs) of SE-associated genes, OCT4 (D) and PRDM14 (E), and non-SE-associated gene, c-MYC (F), following compound treatment.
(G–I) ChIP analysis of the elongation marks H3K36me3 on GB of OCT4 (G), PRDM14 (H), and c-MYC (I) in BRD4-depleted cells compared to NTC-transduced hESCs.
(J) Traveling ratio distribution across SE- or non-SE-associated transcripts of similar expression before treatment and similarly downregulated after treatment. The initiating regions are defined as ±300 bp around the TSS, and the elongating regions are defined as +300 bp from the TSS to 3,000 bp after the transcript ends.
(K) Distribution of fold changes of Pol II densities (normalized by total number of reads per treatment) at TSSs following compound treatment in SE- and non-SE-downregulated transcripts.
(L) Distribution of fold changes of Pol II densities (normalized by total number of reads per treatment) at GBs following compound treatment in SE- and non-SE genes, downregulated following MS417 treatment. Dashed lines indicate the fraction of genes with reduced Pol II occupancy at gene bodies in the two gene categories.
ns, not significant; ***p ≤ 0.001. See also Figure S6.
Next, we investigated whether reduced elongation after BRD4 inhibition was mainly due to a defective Pol II pause release at the TSS and/or to loss of elongating Pol II from the gene body. For that, we analyzed genome-wide occupancy of Pol II in vehicle- or compound-treated hESCs and calculated the Pol II traveling ratio (Pol II at the TSS over the gene body) in SE and non-SE-associated genes that were downregulated following BRD4 inhibition. The analysis presented below was performed (1) on all SE versus non-SE genes and (2) on a subset of SE and non-SE genes of similar expression levels before treatment. Both analyses supported our conclusions, but for clarity, only the latter is presented. We found that the traveling ratios of both SE and non-SE genes significantly increase after compound treatment (p < 0.001; Wilcoxon test; Figure 6J). An increase in traveling ratio could be explained by Pol II stalling at the TSS, by a decrease in Pol II binding throughout the gene body with no increase at TSS, or by a combination of both features. We found that Pol II stalls at the TSS in a high percentage of both downregulated SE- and non-SE-associated genes following BRD4 inhibition (Figure 6K). Conversely, we found that a higher fraction of downregulated SE-associated genes displayed a significant Pol II loss from the gene body than down-regulated non-SE-associated genes upon BRD4 displacement (Figure 6L; snapshots of Pol II binding at OCT4 and PRDM14 in Figure S6G; primers design for ChIP experiments in Figures 5 and 6 are indicated in Figure S6H). This finding suggests that concomitant increase of Pol II stalling with loss of Pol II from the gene body is a unique feature of SE-associated genes and may be responsible for their defective transcriptional elongation following BRD4 inhibition.
We envision a model in which BRD4 recruits SE-associated complexes at SEs of stem cell genes, thus allowing the progression of a promoter-proximal Pol II through the gene body to achieve productive elongation (Figure 7A). Meanwhile, the transcriptional elongation of non-SE-associated genes remains mostly unperturbed by BRD4 inhibition (Figure 7B). Downregulation of the small percentage of non-SE-associated genes following BRD4 inhibition is mainly due to increased Pol II at the TSS rather than to loss of elongating Pol II throughout the gene body. Collectively, our findings support a specific regulation of transcriptional elongation of stem cell genes that relies on BRD4-dependent binding of Mediator and CDK9 to SEs to sustain ESC identity.
Figure 7. Model Depicting the Effects of BET Inhibition on the Binding of MED, BRD4, CDK9, and Pol II at SE-Associated Stem Cell Genes and Non-SE-Associated Genes.
(A) SE-associated stem cell genes depend on the cooperative binding of BRD4, MED, and CDK9 at their SEs for productive transcription elongation (Pol II pS2 and H3K36me3). BRD4 also localizes in the gene body, most likely to help the traveling of Pol II through nucleosomes. Compound treatment displaces, together with BRD4, MED, and CDK9, resulting in Pol II stalling at TSSs and reduced elongating Pol II in the gene body of stem cell genes.
(B) The vast majority of TE-associated genes does not rely on BRD4 binding for their elongation and are insensitive to BRD4 inhibition. Transcription factors and chromatin remodelers rather than BRD4 may be responsible for the expression of TE-associated genes.
DISCUSSION
The role of BET proteins as chromatin readers and transcriptional regulators of the ESC gene network remains unexplored. BET family members display similar structure and are conserved throughout evolution. When we compared the effect of BET inhibition on BRD2-, BRD3-, and BRD4-binding peaks, we found that compound treatment results in robust and global displacement of BRD2 and BRD3, as well as BRD4, from the vast majority of the occupied genomic loci (Figures 4B, 4C, and S7A–S7D), demonstrating that, by interfering with the binding of BET proteins to acetylated his-tones, small-molecule compounds act as potent displacers of all members of the BET family of proteins from the chromatin. Gene set enrichment analysis of BRD2 and BRD3 target genes displaced by compound treatment did not reveal a signifi-cant enrichment in stem cell gene cate gories (Figures S7E and S7F; Table S5), indicating that BRD4, not BRD2 or BRD3, is required to maintain ESC state. In agreement with this, BRD2 or BRD3 silencing did not affect ESC colony morphology (Figures 1E–1H) or the expression of stem cell genes (Figure S7G). Altogether, our data strongly suggest that, in the context of ESCs, BET inhibition effects are mostly due to BRD4 displacement and that, of all BET members, BRD4 acts as the most important regulator of ESC identity. Consistent with a role for BRD4 as a key mediator of ESCs identity, Brd4-null mouse embryos display postimplantation lethality that was attributed to degeneration of the inner cell mass (Houzelstein et al., 2002). Meanwhile, Brd2-null animals survive implantation and die at a later embryonic stage of developmental abnormalities unrelated to maintenance of the inner cell mass (Shang et al., 2009), confirming our results that Brd2 is not essential for ESC state maintenance. A Brd3 knockout mouse has not yet been reported, so role(s) of Brd3 in development remains unclear.
Self-renewal capacity of ESCs is tightly coupled to their proliferative potential. BET proteins, in particular BRD4, play essential roles in cell-cycle progression (You et al., 2009) and mitosis (Zhao et al., 2011), thus raising the possibility that BET inhibitor effects on ESC pluripotency could be explained by cell-cycle arrest and compromised cell viability. We do find that prolonged exposure to BET inhibitors and BRD4 depletion results in reduced proliferation of ESCs, which may contribute to the significantly smaller size of teratomas generated following Brd4 depletion. However, time course analyses of vehicle- and compound-treated ESCs demonstrated that changes in core pluripotency gene expression occurred within a few hours of BET inhibitor treatment, substantially preceding proliferation arrest and establishment of a differentiated state. These findings distinguish BRD4 as key regulator of cell identity of ESCs, rather than a mere cell-cycle regulator.
Our findings indicate that BRD4 chemical and genetic inhibition result in loss of the ESC state while driving lineage specification. The observed EMT and neuroectodermal commitment over other lineages pointed to the possibility that BRD4 may be exerting repressive functions on differentiation genes of this lineage. However, we did not find a significant enrichment of BRD4 in EMT or differentiation genes in our ChIP-seq data sets, suggesting that BRD4 does not directly repress neuroectodermal differentiation genes. Alternatively, transcriptional induction of EMT and differentiation genes following BRD4 inhibition could be explained by regulation of stem cell genes known to directly repress developmental genes and control germ layer fate selection (Thomson et al., 2011; Ma et al., 2011). Consistent with the role of Oct4 as an inhibitor of the neuroectodermal lineage, ectopic expression of Oct4 in ESCs limited the induction of neuroectodermal differentiation genes by BET inhibitors. These results suggest that the increased expression of neuroectodermal lineage markers upon BET displacement might be explained by the BET-dependent regulation of Oct4 (and possibly other neuroectodermal inhibitors).
Our study reveals that, relative to housekeeping genes, stem cell genes particularly rely on BRD4 occupancy for their expression. A unique chromatin conformation or pattern of histone modifications (Filippakopoulos et al., 2012) may favor BRD4-dependent recruitment of transcriptional complexes to stem cell genes over other gene categories. Recently, SEs have been identified at key cell identity genes in ESCs and other cell types (Hnisz et al., 2013; Whyte et al., 2013). Interestingly, our study reveals a disproportionate occupancy of BRD4 at SEs compared to TEs in hESCs. In addition, our discovery that, following BET inhibition, genes associated with SEs are much more frequently downregulated than non-SE-associated genes suggests that SE occupancy and recruitment of transcriptional coregulators may explain the specificity of stem cell gene regulation by BRD4. Master transcriptional regulators have also been described at SEs in ESCs (Hnisz et al., 2013). Protein-protein interactions of BRD4 with ESC transcription factors at SEs may also explain the BRD4-dependent specific regulation of stem cell genes. The multisubunit modular complex Mediator, known to regulate chromatin looping for gene activation (Poss et al., 2013), was enriched at SEs, and its depletion resulted in downregulation of SE-associated genes (Hnisz et al., 2013; Whyte et al., 2013). Because the Mediator complex lacks a DNA-binding domain, we hypothesize that BRD4, with its ability to bind acetylated lysines, recruits Mediator at SEs. Consistently, proteomics studies have revealed the direct interaction between BRD4 and Mediator complex (Dawson et al., 2011).
c-MYC is a proliferation-associated gene that regulates ESC self-renewal and survival. c-MYC was found to be a BRD4 target and modulated in response to BET inhibition in multiple cancer settings (Dawson et al., 2011; Zuber et al., 2011; Delmore et al., 2011; Lovén et al., 2013). Surprisingly, there is no SE associated to c-MYC in hESCs (Hnisz et al., 2013). Compound treatment did not affect the expression of c-MYC in ESCs and did not displace Mediator-containing complexes from its enhancer, suggesting a unique, BRD4-independent regulation of c-MYC in ESCs. However, our gene set enrichment analysis of BRD4 target genes identified c-MYC target genes as BRD4 bound and downregulated following compound treatment (data not shown), suggesting that, in ESCs, BRD4 may directly regulate the expression of MYC target genes that may rely on BRD4 binding to their SEs.
Notably, BRD4 was the BET family member found to be the most significantly enriched at SEs over TEs, compared to BRD2 and BRD3, reinforcing the hypothesis that SE-associated stem cell genes may depend more on BRD4 than BRD2 or BRD3 for their expression. The close similarity among the bromodo-mains of BET members suggests that their functional disparity in ESC maintenance, with BRD4 being essential, does not lie in the bromodomains but most likely in the extraterminal protein-protein interaction domain and the long unstructured C-terminal domain (CTD), unique to BRD4. In support of this concept, BRD4 was shown to be part of the super elongation complex in a model of mixed-lineage-leukemia-fusion leukemia (Dawson et al., 2011). In particular, the CTD of BRD4 was reported to interact and activate P-TEFb (CDK9/cyclin T1) to elicit full transcription (Bisgrove et al., 2007), whereas an atypical kinase domain residing between its bromodomains was shown to directly phosphorylate Pol II at position S2 to ensure productive elongation (Devaiah et al., 2012). Our study reveals that the P-TEFb subunit, CDK9, binds SEs and promoters of stem cell genes together with BRD4 and is displaced following BET inhibition. Because CDK9 activity depends on its acetylation status (Fu et al., 2007), BRD4 may bind preferentially to the acetylated form of CDK9 at SEs and sustain its kinase activity during transcriptional elongation. BRD4 has been described as a Pol II pause release factor necessary for gene expression (Anand et al., 2013). Interestingly, the evidence that an increased Pol II pausing index in the majority of downregulated SE-associated stem cell genes is due to concomitant stalled Pol II at the TSSs and loss of Pol II from the gene body suggests that both events contribute to the nonproductive elongation of stem cell genes following BRD4 inhibition. One intriguing possibility is that loss of elongating Pol II from the gene body of SE-associated stem cell genes following BRD4 displacement might be due to lack of the previously reported BET chaperone activity through acetylated nucleosomes (LeRoy et al., 2008).
We present a model in which BRD4 binds SEs, recruits Mediator and CDK9 at those regulatory elements, and allows Pol II binding throughout the gene body of SE-associated pluripotency genes for their productive elongation. This study provides evidence that the expression of stem cell genes relies on a distinctly productive transcriptional elongation and places BRD4 at the center of a hub that recruits the transcriptional machinery to locus-specific open chromatin, thus connecting these two main regulators of stem cell identity and cell fate decisions.
EXPERIMENTAL PROCEDURES
Cell Culture
mESCs were a kind gift from Dr. Ihor Lemischka (Mount Sinai School of Medicine) to I.A. They were passaged and maintained on gelatin-coated dishes or in coculture with mitomycin C (MITC)-treated mouse embryo fibroblasts (MEF) (GlobalStem) in presence of LIF (ESGRO-LIF) under previously described conditions (Schaniel et al., 2009). H9 hESCs were purchased from Wicell Institute and maintained in coculture with MITC-treated MEFs in the presence of FGF (10 ng/ml; R&D) under conditions described by the supplier. hESCs were plated on Matrigel (BD Pharmingem)-coated plates in the presence of conditioned medium from MITC-treated MEFs, FGF (10 ng/ml; R&D Systems), and ROCK inhibitor (10 μM; Stemgent) for 48 hr before compound treatment to allow hESCs to resume growth following single-cell suspension with Accutase (Innovative Cell Technologies). NTERA-2 cl.D1, 293T cells were purchased from ATCC and grown under standard tissue culture conditions. Human iPSCs, a kind gift of Drs. Mark J. Tomishima and Michel Sadelain (Papapetrou et al., 2009), were cultured under standard hESC conditions. MS436 was used at 10 μM, and MS417 was used at 250 nM concentration throughout the study. DMSO was used as a vehicle control for compounds. –(+) JQ1 enantiomers were used at 500 nM concentration for 6 hr.
Alkaline Phosphatase Staining
Cells were fixed with 4% formaldehyde and stained according to the manufacturer's protocol (Alkaline Phosphatase Detection kit; Millipore).
Self-Renewal Assay
mESCs were trypsinized (Trypsin; GIBCO), and hESCs were treated with Accutase and plated on MITC-MEFs-coated 6-well plates (1.5 × 105 cells per well) in complete ESC media containing LIF (for mESCs) or FGF (for hESCs) in the presence of vehicle or compounds. Cells were passaged every 48 hr for five passages (mESCs) and every 7 days for three passages (hESCs), and the same number of cells was replated. Compounds were replaced every day. At each passage, cells were fixed and stained in duplicates for alkaline phosphatase (AP). For every condition, cells were imaged and the number of alkaline phosphatase positive colonies per well was quantified.
Embryoid Bodies Assay
mESCs were plated in 12-well low attachment plates (Nunc; 300 cells per well) in mESC media in the absence of LIF, supplemented with vehicle or BET inhibitors. After 48 hr, cells were imaged and the number of embryoid bodies in each well was counted (n = 12) and collected for gene expression analysis.
Apoptosis and Cell Cycle Analysis
We seeded 1.1 × 105 mESCs in 6-well gelatin-coated plates in complete medium with LIF. hESCs were seeded in 6-well Matrigel-coated plates in complete medium with FGF. The next day, media was replaced with complete medium in the presence of vehicle or BET inhibitors. Compounds were replaced every day. For apoptosis analysis, on each time point, cells were brought to suspension and stained for annexin V:phycoerythrin (PE) and 7-aminoactinomycin D according to the manufacturer protocol (Annexin V: PE Apoptosis Detection Kit BD Biosciences; 559763). As a positive control for the staining, mESCs were treated with 500 μM H2O2 for 24 hr. For cell cycle analysis, on each time point, cells were brought to suspension, washed with PBS, and resus-pended in 70% ethanol for 30 min at 4°C. Cells were spun and resuspended in 100 μl of fluorescence-activated cell sorting (FACS) buffer with RNase and phosphatidylinositol stained for 30 min at 37°C. A BD LSR II cytometer was used for data acquisition and Flowjo and Modfit LT software for analysis.
Proliferation Assays
We seeded 4 × 103 ESCs per well in gelatin or Matrigel-coated 96-well plates (n = 5/condition) in presence of LIF (mESCs) or FGF (hESCs). After 24 hr, medium was changed to mESC media containing LIF or hESC media containing FGF and supplemented with vehicle or BET inhibitors. At each time point, cells were fixed in 100% methanol and stored in PBS at 4°C. At the end of the experiment, cells were stained with 0.5% crystal violet. Crystals were dissolved with 15% acetic acid, and optical density was read at 590 nm.
Western Blot
hESCs were harvested in lysis buffer (125 mM Tris [pH 6.8], 4% SDS, and 20% glycerol). Cell lysates (25–30 μg of total proteins) were resolved on 4%–20% Tris-glycine SDS-PAGE gels (Invitrogen) and transferred to nitrocellulose membranes. Membranes were blocked for 1 hr with 5% nonfat milk and probed with primary antibodies overnight at 4°C (OCT4 [Abcam; ab93085] and tubulin [Sigma]). Membranes were incubated with horseradish-peroxidaseconjugated secondary antibodies for 1 hr before developing with enhanced chemiluminescence plus western blotting detection kit (GE Healthcare).
Immunofluorescence Analysis
hESCs grown on coverslips on low-density, MITC-treated MEFs were analyzed 5 days after FGF, vehicle, or MS436 treatment. Cells were fixed for 10 min with 4% paraformaldehyde, washed three times with PBS, permeabilized with PBS-T (PBS-0.02% and Tween-20), blocked with 1% BSA in PBS, and incubated 1 hr at room temperature with OCT4 or ECAD (Abcam; ab1416) antibodies. Samples were washed three times with PBS and incubated for 45 min with Alexa Fluor (Molecular Probes) secondary antibodies. After three washes, samples were mounted in Vectashield (Vector Laboratories) containing DAPI and imaged with a Leica TCS SP5 II confocal microscope.
siRNA Transfection
hESCs, iPSCs, and NTERA-2 cells were transfected with siGENOME Non-Targeting siRNA no. 1 (D-001210-01-20) or siGENOME SMART POOL BRD4 (M-004937-02-0010; Dharmacon) using Lipofectamine 2000 (Invitrogen) according to suggested manufacturer procedures. hESCs and iPSCs were plated on MEFs-coated dishes in the presence of FGF and analyzed 5 days posttransfection.
Lentiviral Production and ESC Transduction
Lentiviruses were produced in 293T cells, and supernatants were collected at 24, 48, and 72 hr. Viral particles were concentrated by ultracentrifugation and stored at –80°C. A multiplicity of infection ranging from 10 to 20 was used to infect ESCs. mESCs were detached and infected with viral supernatant in the presence of polybrene (Sigma) at 8 μg/ml concentration. mESCs incubated with viral supernatants for 2 hr in suspension were seeded in gelatin-coated dishes and left to recover before puromycin (Sigma; 1 μg/ml) selection. mESCs were analyzed 5 days postselection. hESCs were detached with Accutase to obtain single-cell suspensions, incubated with viral supernatants for 2 hr in suspension, and seeded on Matrigel-coated dishes in the presence of FGF, MITC-MEFs-conditioned medium, and Rock inhibitor. hESCs were analyzed 2 weeks posttransduction for gene expression and colony morphology.
Plasmids
pGIPZ plasmids targeting human RNA were purchased from Open Bio-systems. shRNA sequences were subcloned to pH1-IRES-Hygro-GFP reporter vector (a gift from Dr. Ruben Hoya Arias; Ivanova et al., 2006). For the data shown in Figures 6G–6I, the shBRD4 sequence was subcloned to a pTRIPZ inducible lentiviral vector. pLKO.1 plasmids targeting murine Brd2, Brd3, Brd4, Med1, Med12, and a nontargeting control were purchased from Thermo Scientific. See Supplemental Experimental Procedures for sequences. pLVX-IRES-Oct4tdTomato and pLVX-IRES-tdTomato vector (Figures 3I–3K) were a kind gift of M. Stadtfeld.
Teratoma Formation Assay
All mouse experiments were conducted under the Guidelines of the NYU Institutional Animal Care and Use Committee, protocol no. 120405. mESCs were infected with lentiviruses containing nontargeting control vector or shRNA-targeting Brd4. Infected cells were selected with puromycin (1 μg/ml) for 48 hr and injected subcutaneously in the flanks of RAG2-deficient mice (Taconic; 1.5 × 106 cells/flank). After 4 weeks, mice were sacrificed and teratomas were extracted, weighed, fixed in 10% formalin, embedded in paraffin, sectioned, and stained with hematoxylin and eosin. The following primary antibodies were used for immunohistochemistry: Ki67 (Master Diagnostica) and NeuN (Millipore).
Quantitative RT-PCR
Total RNA was extracted using RNeasy QIAGEN kit according to the manufacturer's instructions. It was then subjected to DNase treatment and retrotranscription. Real-time PCR was performed using SYBR green fluorescence (Roche). hsa-GAPDH and mmu-18S were used as internal standards.
RNA-Sequencing Procedure and Analysis
Deep sequencing of RNA from hESCs FGF or MS436 treated at day 1 and day 5 was performed as described (Rajadhyaksha et al., 2010). See Supplemental Experimental Procedures for detailed information on samples analysis.
ChIP and ChIP-Seq
Chromatin isolated from FGF-maintained hESCs, vehicle or MS417 treated (at 250 nM concentration for 6 hr), was subjected to ChIP and ChIP-seq protocols (Ntziachristos et al., 2012). Chromatin from hESCs transduced with a pTRIPZ vector expressing NTC or shBRD4 sequence was isolated upon 72 hr of doxycycline induction (2 μg/μl) and subjected to ChIP for H3K36me3. See Supplemental Experimental Procedures for protocol details and quantitative RT-PCR (qRT-PCR) primer sequences.
Computational Analysis of RNA-Seq and ChIP-Seq Data Sets
See Supplemental Experimental Procedures for detailed information on data sources and computational tools used for RNA-seq and ChIP-seq analysis, peak characterization, generation of Pol II occupancy profiles, transcriptional elongation, and super-enhancer data analysis.
Supplementary Material
Highlights.
BRD4 regulates ESC self-renewal and expression of pluripotency genes
Inhibition of BRD4 results in commitment to the neuroectodermal lineage
BRD4 occupies SEs of core stem cell genes and recruits active transcription complexes
BRD4 controls transcriptional elongation of SE-associated pluripotency genes
ACKNOWLEDGMENTS
We thank Lisa Koetz for experimental help, Jenny Xiang (Cornell University Genomics Core lab) for RNA-seq library preparation, the NYU Experimental Pathology Core Facility (Director, Cindy Loomis) for tissue processing, and the NYU Genome Technology Center (Director, Adriana Heguy) for ChIP sequencing (Cancer Center Support Grant NIH/NCI 5P30CA016087-32). We thank Weijia Zhang for initial bioinformatics analysis on RNA-seq data sets and Farbod Darvishian for an independent histological analysis of tissues. We thank Matthias Stadtfeld for sharing reagents. We thank Denes Hnisz for sharing data sets and for discussion. We thank Avital Gaziel-Sovran, Doug Hanniford, Lisa Dailey, Matthew Murtha, Martin J. Walsh, Pedro Rocha, Voigt Philipp, Matthias Stadtfeld, and Markus Schober for critical reading of the manuscript. Chemical compounds developed by M.-M.Z are in a patent application. P.N. is supported by Lady Tata Memorial Trust for Leukemia, American Society of Hematology, and the National Cancer Institute of the NIH (K99CA188293). R.D.M was supported by Human Frontier Science Project (HFSP) fellowship and is currently supported by a New York Stem Cell Foundation Druckenmiller postdoctoral fellowship. A.T. performed part of this work while at the Computational Biology Center, IBM Research, Yorktown Heights, NY. I.A.'s laboratory is supported by the NIH (RO1CA133379, RO1CA105129, RO1CA149655, and RO1GM088847) and the Leukemia & Lymphoma Society (TRP program grants), M.-M.Z.'s laboratory by the NIH (R01HG004508 and R01CA87658), and E.H.'s laboratory by the NIH (R01CA155234, R01CA163891, and R21AR062239). This research was supported by The New York Stem Cell Foundation.
Footnotes
AUTHOR CONTRIBUTIONS
R.D.M., B.F.-C., and V.L. performed and analyzed the experiments; P.N., S.K.Y., C.D.L., and Y.Y. contributed to experimental work; M.C. provided histological analyses; G.Z., E.R., G.G.-N., M.O., and M.-M.Z. provided small-molecule inhibitors; A.T. performed and supervised bioinformatic analyses conducted by I.D.; B.F.-C., V.L., and I.A. provided critical reading and editing of the manuscript; and R.D.M. and E.H. conceived the research project, analyzed the data, and wrote the manuscript.
ACCESSION NUMBERS
The National Center for Biotechnology Information Gene Expression Omnibus accession number for the RNA-seq and ChIP-seq data reported in the study is GSE60171.
SUPPLEMENTAL INFORMATION
Supplemental Information includes Supplemental Experimental Procedures, seven figures, and five tables and can be found with this article online at http://dx.doi.org/10.1016/j.celrep.2014.08.055.
REFERENCES
- Anand P, Brown JD, Lin CY, Qi J, Zhang R, Artero PC, Alaiti MA, Bullard J, Alazem K, Margulies KB, et al. BET bromodomains mediate transcriptional pause release in heart failure. Cell. 2013;154:569–582. doi: 10.1016/j.cell.2013.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannister AJ, Kouzarides T. The CBP co-activator is a histone acetyltransferase. Nature. 1996;384:641–643. doi: 10.1038/384641a0. [DOI] [PubMed] [Google Scholar]
- Belkina AC, Denis GV. BET domain co-regulators in obesity, inflammation and cancer. Nat. Rev. Cancer. 2012;12:465–477. doi: 10.1038/nrc3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisgrove DA, Mahmoudi T, Henklein P, Verdin E. Conserved P-TEFb-interacting domain of BRD4 inhibits HIV transcription. Proc. Natl. Acad. Sci. USA. 2007;104:13690–13695. doi: 10.1073/pnas.0705053104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borah JC, Mujtaba S, Karakikes I, Zeng L, Muller M, Patel J, Moshkina N, Morohashi K, Zhang W, Gerona-Navarro G, et al. A small molecule binding to the coactivator CREB-binding protein blocks apoptosis in cardiomyocytes. Chem. Biol. 2011;18:531–541. doi: 10.1016/j.chembiol.2010.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson MA, Prinjha RK, Dittmann A, Giotopoulos G, Bantscheff M, Chan WI, Robson SC, Chung CW, Hopf C, Savitski MM, et al. Inhibition of BET recruitment to chromatin as an effective treatment for MLL-fusion leukaemia. Nature. 2011;478:529–533. doi: 10.1038/nature10509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delmore JE, Issa GC, Lemieux ME, Rahl PB, Shi J, Jacobs HM, Kastritis E, Gilpatrick T, Paranal RM, Qi J, et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell. 2011;146:904–917. doi: 10.1016/j.cell.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devaiah BN, Lewis BA, Cherman N, Hewitt MC, Albrecht BK, Robey PG, Ozato K, Sims RJ, 3rd, Singer DS. BRD4 is an atypical kinase that phosphorylates serine2 of the RNA polymerase II carboxy-terminal domain. Proc. Natl. Acad. Sci. USA. 2012;109:6927–6932. doi: 10.1073/pnas.1120422109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding L, Paszkowski-Rogacz M, Nitzsche A, Slabicki MM, Heninger AK, de Vries I, Kittler R, Junqueira M, Shevchenko A, Schulz H, et al. A genome-scale RNAi screen for Oct4 modulators defines a role of the Paf1 complex for embryonic stem cell identity. Cell Stem Cell. 2009;4:403–415. doi: 10.1016/j.stem.2009.03.009. [DOI] [PubMed] [Google Scholar]
- Fazzio TG, Huff JT, Panning B. An RNAi screen of chromatin proteins identifies Tip60-p400 as a regulator of embryonic stem cell identity. Cell. 2008;134:162–174. doi: 10.1016/j.cell.2008.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filippakopoulos P, Qi J, Picaud S, Shen Y, Smith WB, Fedorov O, Morse EM, Keates T, Hickman TT, Felletar I, et al. Selective inhibition of BET bromodomains. Nature. 2010;468:1067–1073. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filippakopoulos P, Picaud S, Mangos M, Keates T, Lambert JP, Barsyte- Lovejoy D, Felletar I, Volkmer R, Müller S, Pawson T, et al. Histone recognition and large-scale structural analysis of the human bromodomain family. Cell. 2012;149:214–231. doi: 10.1016/j.cell.2012.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu J, Yoon HG, Qin J, Wong J. Regulation of P-TEFb elongation complex activity by CDK9 acetylation. Mol. Cell. Biol. 2007;27:4641–4651. doi: 10.1128/MCB.00857-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes SR, Dollard C, Winston F, Beck S, Trowsdale J, Dawid IB. The bromodomain: a conserved sequence found in human, Drosophila and yeast proteins. Nucleic Acids Res. 1992;20:2603. doi: 10.1093/nar/20.10.2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hnisz D, Abraham BJ, Lee TI, Lau A, Saint-André V, Sigova AA, Hoke HA, Young RA. Super-enhancers in the control of cell identity and disease. Cell. 2013;155:934–947. doi: 10.1016/j.cell.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houzelstein D, Bullock SL, Lynch DE, Grigorieva EF, Wilson VA, Beddington RS. Growth and early postimplantation defects in mice deficient for the bromodomain-containing protein Brd4. Mol. Cell. Biol. 2002;22:3794–3802. doi: 10.1128/MCB.22.11.3794-3802.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanova N, Dobrin R, Lu R, Kotenko I, Levorse J, DeCoste C, Schafer X, Lun Y, Lemischka IR. Dissecting self-renewal in stem cells with RNA interference. Nature. 2006;442:533–538. doi: 10.1038/nature04915. [DOI] [PubMed] [Google Scholar]
- Jones MH, Numata M, Shimane M. Identification and characterization of BRDT: A testis-specific gene related to the bromodomain genes RING3 and Drosophila fsh. Genomics. 1997;45:529–534. doi: 10.1006/geno.1997.5000. [DOI] [PubMed] [Google Scholar]
- Keramari M, Razavi J, Ingman KA, Patsch C, Edenhofer F, Ward CM, Kimber SJ. Sox2 is essential for formation of trophectoderm in the preimplantation embryo. PLoS ONE. 2010;5:e13952. doi: 10.1371/journal.pone.0013952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeRoy G, Rickards B, Flint SJ. The double bromodomain proteins Brd2 and Brd3 couple histone acetylation to transcription. Mol. Cell. 2008;30:51–60. doi: 10.1016/j.molcel.2008.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Li L, Pandey R, Byun JS, Gardner K, Qin Z, Dou Y. The histone acetyltransferase MOF is a key regulator of the embryonic stem cell core transcriptional network. Cell Stem Cell. 2012;11:163–178. doi: 10.1016/j.stem.2012.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovén J, Hoke HA, Lin CY, Lau A, Orlando DA, Vakoc CR, Bradner JE, Lee TI, Young RA. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell. 2013;153:320–334. doi: 10.1016/j.cell.2013.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Z, Swigut T, Valouev A, Rada-Iglesias A, Wysocka J. Sequence-specific regulator Prdm14 safeguards mouse ESCs from entering extraembryonic endoderm fates. Nat. Struct. Mol. Biol. 2011;18:120–127. doi: 10.1038/nsmb.2000. [DOI] [PubMed] [Google Scholar]
- Meshorer E, Misteli T. Chromatin in pluripotent embryonic stem cells and differentiation. Nat. Rev. Mol. Cell Biol. 2006;7:540–546. doi: 10.1038/nrm1938. [DOI] [PubMed] [Google Scholar]
- Mitsui K, Tokuzawa Y, Itoh H, Segawa K, Murakami M, Takahashi K, Maruyama M, Maeda M, Yamanaka S. The homeoprotein Nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell. 2003;113:631–642. doi: 10.1016/s0092-8674(03)00393-3. [DOI] [PubMed] [Google Scholar]
- Morey L, Pascual G, Cozzuto L, Roma G, Wutz A, Benitah SA, Di Croce L. Nonoverlapping functions of the Polycomb group Cbx family of proteins in embryonic stem cells. Cell Stem Cell. 2012;10:47–62. doi: 10.1016/j.stem.2011.12.006. [DOI] [PubMed] [Google Scholar]
- Morinière J, Rousseaux S, Steuerwald U, Soler-López M, Curtet S, Vitte AL, Govin J, Gaucher J, Sadoul K, Hart DJ, et al. Cooperative binding of two acetylation marks on a histone tail by a single bromodomain. Nature. 2009;461:664–668. doi: 10.1038/nature08397. [DOI] [PubMed] [Google Scholar]
- Nicodeme E, Jeffrey KL, Schaefer U, Beinke S, Dewell S, Chung CW, Chandwani R, Marazzi I, Wilson P, Coste H, et al. Suppression of inflammation by a synthetic histone mimic. Nature. 2010;468:1119–1123. doi: 10.1038/nature09589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ntziachristos P, Tsirigos A, Van Vlierberghe P, Nedjic J, Trimarchi T, Flaherty MS, Ferres-Marco D, da Ros V, Tang Z, Siegle J, et al. Genetic inactivation of the polycomb repressive complex 2 in T cell acute lymphoblastic leukemia. Nat. Med. 2012;18:298–301. doi: 10.1038/nm.2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogryzko VV, Schiltz RL, Russanova V, Howard BH, Nakatani Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- Onder TT, Kara N, Cherry A, Sinha AU, Zhu N, Bernt KM, Cahan P, Marcarci BO, Unternaehrer J, Gupta PB, et al. Chromatin-modifying enzymes as modulators of reprogramming. Nature. 2012;483:598–602. doi: 10.1038/nature10953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papapetrou EP, Tomishima MJ, Chambers SM, Mica Y, Reed E, Menon J, Tabar V, Mo Q, Studer L, Sadelain M. Stoichiometric and temporal requirements of Oct4, Sox2, Klf4, and c-Myc expression for efficient human iPSC induction and differentiation. Proc. Natl. Acad. Sci. USA. 2009;106:12759–12764. doi: 10.1073/pnas.0904825106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesce M, Schöler HR. Oct-4: gatekeeper in the beginnings of mammalian development. Stem Cells. 2001;19:271–278. doi: 10.1634/stemcells.19-4-271. [DOI] [PubMed] [Google Scholar]
- Poss ZC, Ebmeier CC, Taatjes DJ. The Mediator complex and transcription regulation. Crit. Rev. Biochem. Mol. Biol. 2013;48:575–608. doi: 10.3109/10409238.2013.840259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajadhyaksha AM, Elemento O, Puffenberger EG, Schierberl KC, Xiang JZ, Putorti ML, Berciano J, Poulin C, Brais B, Michaelides M, et al. Mutations in FLVCR1 cause posterior column ataxia and retinitis pigmentosa. Am. J. Hum. Genet. 2010;87:643–654. doi: 10.1016/j.ajhg.2010.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez R, Zhou MM. The role of human bromodomains in chromatin biology and gene transcription. Curr. Opin. Drug Discov. Devel. 2009;12:659–665. [PMC free article] [PubMed] [Google Scholar]
- Schaniel C, Ang YS, Ratnakumar K, Cormier C, James T, Bernstein E, Lemischka IR, Paddison PJ. Smarcc1/Baf155 couples selfrenewal gene repression with changes in chromatin structure in mouse embryonic stem cells. Stem Cells. 2009;27:2979–2991. doi: 10.1002/stem.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segura MF, Fontanals-Cirera B, Gaziel-Sovran A, Guijarro MV, Hanniford D, Zhang G, González-Gomez P, Morante M, Jubierre L, Zhang W, et al. BRD4 sustains melanoma proliferation and represents a new target for epigenetic therapy. Cancer Res. 2013;73:6264–6276. doi: 10.1158/0008-5472.CAN-13-0122-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang E, Wang X, Wen D, Greenberg DA, Wolgemuth DJ. Double bromodomain-containing gene Brd2 is essential for embryonic development in mouse. Dev. Dyn. 2009;238:908–917. doi: 10.1002/dvdy.21911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamkun JW, Deuring R, Scott MP, Kissinger M, Pattatucci AM, Kaufman TC, Kennison JA. brahma: a regulator of Drosophila homeotic genes structurally related to the yeast transcriptional activator SNF2/ SWI2. Cell. 1992;68:561–572. doi: 10.1016/0092-8674(92)90191-e. [DOI] [PubMed] [Google Scholar]
- Thomson M, Liu SJ, Zou LN, Smith Z, Meissner A, Ramanathan S. Pluripotency factors in embryonic stem cells regulate differentiation into germ layers. Cell. 2011;145:875–889. doi: 10.1016/j.cell.2011.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Oron E, Nelson B, Razis S, Ivanova N. Distinct lineage specification roles for NANOG, OCT4, and SOX2 in human embryonic stem cells. Cell Stem Cell. 2012;10:440–454. doi: 10.1016/j.stem.2012.02.016. [DOI] [PubMed] [Google Scholar]
- Whyte WA, Orlando DA, Hnisz D, Abraham BJ, Lin CY, Kagey MH, Rahl PB, Lee TI, Young RA. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013;153:307–319. doi: 10.1016/j.cell.2013.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You J, Li Q, Wu C, Kim J, Ottinger M, Howley PM. Regulation of aurora B expression by the bromodomain protein Brd4. Mol. Cell. Biol. 2009;29:5094–5103. doi: 10.1128/MCB.00299-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, Liu R, Zhong Y, Plotnikov AN, Zhang W, Zeng L, Rusinova E, Gerona-Nevarro G, Moshkina N, Joshua J, et al. Down-regulation of NF-κB transcriptional activity in HIV-associated kidney disease by BRD4 inhibition. J. Biol. Chem. 2012;287:28840–28851. doi: 10.1074/jbc.M112.359505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, Plotnikov AN, Rusinova E, Shen T, Morohashi K, Joshua J, Zeng L, Mujtaba S, Ohlmeyer M, Zhou MM. Structure-guided design of potent diazobenzene inhibitors for the BET bromodomains. J. Med. Chem. 2013;56:9251–9264. doi: 10.1021/jm401334s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao R, Nakamura T, Fu Y, Lazar Z, Spector DL. Gene bookmarking accelerates the kinetics of post-mitotic transcriptional re-activation. Nat. Cell Biol. 2011;13:1295–1304. doi: 10.1038/ncb2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuber J, Shi J, Wang E, Rappaport AR, Herrmann H, Sison EA, Magoon D, Qi J, Blatt K, Wunderlich M, et al. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature. 2011;478:524–528. doi: 10.1038/nature10334. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.