Abstract
Irritable bowel syndrome (IBS) is a heterogeneous functional disorder with a multifactorial etiology that involves the interplay of both host and environmental factors. Among environmental factors relevant for IBS etiology, the diet stands out given that the majority of IBS patients report their symptoms to be triggered by meals or specific foods. The diet provides substrates for microbial fermentation, and, as the composition of the intestinal microbiota is disturbed in IBS patients, the link between diet, microbiota composition, and microbial fermentation products might have an essential role in IBS etiology. In this review, we summarize current evidence regarding the impact of diet and the intestinal microbiota on IBS symptoms, as well as the reported interactions between diet and the microbiota composition. On the basis of the existing data, we suggest pathways (mechanisms) by which diet components, via the microbial fermentation, could trigger IBS symptoms. Finally, this review provides recommendations for future studies that would enable elucidation of the role of diet and microbiota and how these factors may be (inter)related in the pathophysiology of IBS.
Introduction
With a prevalence of approximately 10–15%, irritable bowel syndrome (IBS) is one of the most common gastrointestinal (GI) disorders in the industrialized world (1). The high prevalence together with the reduced quality of life and associated co-morbidities in patients suffering from IBS put a significant negative burden on both patients and society (2). IBS is a heterogeneous functional disorder and commonly subtyped according to the prevailing bowel habit into IBS with constipation (IBS-C), IBS with diarrhea (IBS-D), mixed IBS with both constipation and diarrhea and unsubtyped IBS with neither constipation nor diarrhea using the Rome III criteria (3).
Although the etiology of IBS is incompletely understood, it is generally regarded as a multifactorial disorder involving the interplay of both host and environmental factors, including diet. The host factors include central factors, such as aberrant stress responses, psychiatric co-morbidity, and cognitive dysfunctions, whereas intestinal functions are also involved, e.g., dysmotility, visceral hypersensitivity, low-grade immune activation, altered barrier function, and the intestinal microbiota composition (4).
During recent years, perturbations in the intestinal microbiota are increasingly being linked to the pathophysiology of IBS. The increased risk of new onset IBS after an episode of gastroenteritis (5) and the association with prior antibiotic use (6) support the importance of the intestinal microbiota in IBS. Numerous studies have demonstrated altered microbial profiles in (specific subgroups of) IBS patients compared with healthy individuals (7, 8, 9, 10, 11). The intestinal microbiota strongly interacts with exogenous factors, in particular diet, which may also directly or indirectly provoke IBS symptoms.
In this review, we describe the current evidence regarding the impact of diet and the intestinal microbiota and their (inter)relation on the pathophysiology of IBS.
The Impact of Diet on the Intestinal Microbiota
The fetal intestine is thought to be sterile, although some recent studies indicate that colonization may already start before delivery, by bacterial transmission through the placental barrier (12, 13). The colonization during and upon delivery involves a succession of microbial populations, a seemingly chaotic process strongly influenced by birth mode and infant feeding. Vaginally born infants are initially colonized by maternal fecal and vaginal microbes, such as Bifidobacterium, Bacteroides, Prevotella, and Lactobacillus spp., whereas infants born by cesarean section are inoculated by typical skin bacteria and bacteria from the hospital's environment (14, 15).
The strong impact of diet on the indigenous microbial communities can already be observed even before weaning. Compared with formula-fed infants, the microbiota of breastfed infants is less diverse and more dominated by Bifidobacterium spp. (16, 17, 18). This is generally attributed to unique bioactive compounds such as human milk oligosaccharides that serve as metabolic substrates for a limited number of microbes (19), although breast milk itself also harbors a transferrable microbiota (20). A rapid diversification of the infants' intestinal microbiota and shift toward an adult-like microbiota occurs with the introduction of solid foods (21, 22); yet, the maturation of the microbiota appears to continue in parallel to physiological development of, for example, the gastrointestinal tract and the central nervous system, throughout childhood and adolescence (23, 24).
Once established, the intestinal microbiota appears to be relatively stable within individuals over time (25), whereas high variability between individuals is observed (26, 27, 28, 29).
Pronounced differences in the composition and functional capacity of the human microbiota between geographically distant populations point toward an important role of the diet in these inter-individual variations (26, 30, 31, 32). The microbiota in non-Western populations, consuming a diet high in plant-derived carbohydrates, has consistently been shown to be more diverse and enriched in Prevotella spp. at the expense of Bacteroides spp. as compared with Western populations consuming a diet high in animal protein, sugar, starch, and fat (reviewed in (33)). It remains to be determined to what extent the microbiota composition is shaped by the diet, as the impact of genetics, ethnic, and cultural factors, such as hygiene and living conditions, cannot be ruled out in such studies comparing distinct populations.
Nevertheless, habitual long-term dietary patterns have also been directly linked to intestinal microbial enterotypes. Especially, protein and animal fat intake has been associated with the Bacteroides enterotype, whereas a high carbohydrate intake was associated with the Prevotella enterotype (34). The effect of short-term dietary interventions on the microbiota composition appears to be only modest (34, 35), unless the intervention comprises an extreme switch in diet (36, 37, 38). David et al. (37) demonstrated that even within days after the transition to a diet entirely composed of animal or plant products, changes in microbial structure, metabolic activity, and gene expression can be observed. In subjects receiving the animal-based high fat and protein diet, the abundance of bile-tolerant bacteria (Alistipes, Bilophila and Bacteroides) together with branched short-chain fatty acids indicative of amino-acid fermentation increased, whereas levels of bacteria metabolizing dietary plant polysaccharides (Eubacterium rectale, Roseburia, Ruminococcus bromii) decreased. In agreement, switching to a diet high in resistant starch has been shown to increase Ruminococcus bromii and Eubacterium rectale related species (39), which are known for their saccharolytic properties. Altogether, these studies indicate that the intestinal microbial community structure as well as its function and metabolic output are influenced by our diet, especially in the case of clearly distinct dietary patterns.
Influence of Diet on IBS
Surveys on perceived food intolerance show that 64–89% of IBS patients report their symptoms to be triggered by meals or specific foods (40, 41, 42) The majority indicate that they limit or exclude certain foods without professional counseling (40, 42), thereby increasing the risk of inadequate dietary intake. Monsbakken et al. (40) found indications for inadequate dietary intake in 12% of IBS patients studied, whereas others found adequate or even increased intake of some nutrients (43, 44, 45). Overall, data on habitual dietary intake are limited and may depend on the study population included. Meal-related symptom aggravation is further supported by studies showing a postprandial worsening of pain using symptom diaries for 6 weeks (46) or rectal barostat (47, 48).
Foods often reported to provoke symptoms include wheat/grains, vegetables, milk products, fatty foods, spicy foods, coffee, and alcohol (40, 41, 42, 49) and are especially reported to be associated with abdominal pain and gas problems (41, 42). Overall, the percentage of responders on exclusion diets varies from 15 to 71% (50), but it has to be noted that most studies suffer from major methodological limitations and only a minority of subjects was found to react positively to double-blind food challenges (51, 52, 53).
Although placebo and possibly nocebo effects have to be considered in IBS, possible physiological mechanisms of (perceived) food intolerance include an exaggerated sensory and motor response and/or incomplete absorption, which may lead to symptoms in a susceptible host. High fat intake, for example, is associated with an exaggerated colonic motor response to eating and increased visceral sensitivity in IBS patients, whereas duodenal lipid infusions delay small intestinal transit. Randomized controlled trials adjusting fat intake are, however, limited (54). Poorly absorbed carbohydrates, like lactose, fructose, and galacto- or fructo-oligosaccharides (fructans), can result in luminal distension by osmotic effects and increased gas production due to microbial fermentation. Although this is a normal physiological phenomenon, it can result in symptoms in subjects with an altered microbiota, increased visceral sensitivity, and/or abnormal gas handling. A recent study by Yang et al. (55) found increased mucosal mast cell numbers, serum tumor necrosis factor-α, rectal sensitivity, and anxiety in lactose intolerant IBS-D patients, supporting that neuro-immune modulation of visceral function is a potential underlying mechanism. Perceived intolerance to lactose, fructose, and fructans is frequently reported and uncontrolled studies point to symptom improvement after removal of milk containing products or wheat from the diet (42). However, these studies should be interpreted with caution because of a large placebo and nocebo response in IBS. The overall evidence for an increased incidence of lactose intolerance in IBS patients was found to be weak and to be moderate—weak for the benefit of low lactose intake in these subjects (56).
Evolving from previous studies on lactose and fructose intolerance, a more generalized approach of intolerance to poorly absorbed and rapidly fermented carbohydrates has been introduced: FODMAPS, including fermentable oligo-, di- and monosaccharides and polyols. Both retrospective and prospective open studies have shown reduced symptoms after introduction of a low-FODMAP diet in IBS patients with suspected or proven fructose or lactose malabsorption (57, 58). These findings are confirmed by randomized controlled trials, showing a significant improvement of overall symptoms, abdominal pain and bloating, in patients with bloating and/or diarrhea (59) and in small (n = 15−30) unselected IBS patient groups (60, 61). One study showed recurrence of symptoms on re-challenge with fructose and/or fructans (62). Involvement of gas production is supported by findings that IBS patients produced more hydrogen relative to healthy controls and lacked an increased methane production on a high vs. low-FODMAP diet (60). However, it should be noted that the long-term benefit of a low-FODMAP approach in large (unselected) patient groups is unclear, especially considering its invasiveness, which requires strict guidance by a dietician, and the potential risks of, for example, reduced fiber intake and changes in microbiota composition and activity (59, 63). Furthermore, randomized controlled trials are needed to prove whether it is superior to other dietary interventions (e.g., lactose or fructose reduction) or general dietary advice (e.g., the NICE guidelines). Finally, the FODMAP levels vary between studies, countries, and products, which should be taken into account when interpreting study results and considering low-FODMAP diets.
Foods can also evoke symptom onset by immune activation or altered neuro-endocrine responses (42). Food allergy or intolerance associated with IgE-mediated immune responses is uncommon and evidence for IgG/IgG4-mediated hypersensitivity is inconclusive (42, 64). However, in subsets of IBS patients, increased numbers of T-lymphocytes, mast cells, eosinophils, and/or enteroendocrine cells have been found (65, 66, 67), but their exact role in food intolerance and symptom generation is unclear.
Interest in the role of gluten intolerance (and the possible benefit of gluten-free diets) is increasing. A systematic meta-analysis found a pooled prevalence for celiac disease up to 4% among IBS patients (68). The prevalence of non-celiac gluten sensitivity among unselected IBS patients is unknown and complicated by clear diagnostic criteria and overlap in symptoms. Although symptom reduction after a gluten-free diet (69) and worsening of symptoms after a gluten challenge (70) have been reported, the recent study Biesiekierski et al. (71) found no symptom induction by giving pure gluten to IBS patients following a gluten- and FODMAP-free diet. These findings suggest that not gluten but fructans and/or other components might contribute to symptoms in perceived “wheat” intolerance. Furthermore, Carroccio et al. (72) showed that the majority of IBS patients diagnosed with wheat sensitivity had multiple food sensitivities.
Dietary supplementation studies focus on probiotics, prebiotics, synbiotics, and fiber intake. A recent meta-analysis on the use of probiotics shows significant effects on subjective global symptom improvement, but study heterogeneity is statistically significant and no conclusions can be drawn on which individual species or strains are most beneficial (73). Studies on prebiotics and synbiotics are limited and insufficient to draw conclusions. Evidence for the benefit of dietary fiber was found for soluble fiber intake only (74), but it should be noted that overall study quality was moderate and more studies are needed on its effects in IBS subgroups.
Although several food components are associated with symptom generation, detailed analyses on the effect of food intake on symptom generation and underlying mechanisms are limited. In addition, many studies suffer from methodological limitations, in part inherent to the complexity of food research.
Microbiota Alterations in IBS
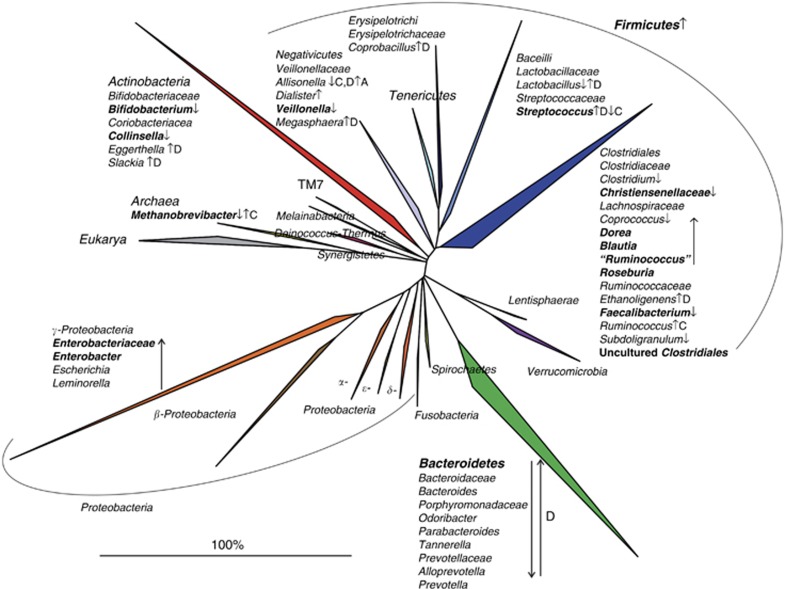
Following the first comprehensive analysis reporting a distinctive intestinal microbiota composition in IBS patients (75), several studies have identified differences between the microbiota of IBS patients, or subgroups thereof, and healthy controls (reviewed in (7)). Recently, clinical guidance regarding the modulation of intestinal microbiota in IBS was provided by the Rome Team Working Group (76), which concluded that there is good evidence supporting the concept that the intestinal microbiota is perturbed in patients with IBS. However, despite a growing consensus regarding an association between the intestinal microbiota and IBS, results of current studies lack general consensus and a specific microbial signature in IBS remains elusive (Figure 1). Lack of detailed phenotypic characterization of patients, small sample sizes, and the cross-sectional study designs (providing only a single snapshot of the microbiota composition) in the majority of studies may all contribute to the fact that the markers of the IBS microbiota reported in several studies (marked in bold on Figure 1) are not reproducibly detected in all cohorts. Moreover, exogenous factors, including diet, are most often not taken into account, despite the demonstrated effect of diet on the microbiota and the potential alterations in diet associated with IBS. Finally, it is noteworthy that Jeffrey and colleagues identified distinct subsets of IBS patients, not corresponding to the traditional IBS subtypes, with an altered or normal-like microbiota composition (8), indicating that the disturbed microbiota might be relevant for the pathology of part of the IBS patients. Moreover, those with a normal-like microbiota had more adverse psychological factors, suggesting that central factors may predominate over microbiological factors in some but not all IBS patients.
Figure 1.
Phylogenetic tree of the human intestinal microbiota with indicated microbial groups that are significantly altered in irritable bowel syndrome (IBS) patients relative to controls. The increase and decrease in microbial groups in IBS patients are marked by arrows facing up and down, respectively, and in bold if reproduced by at least two independent studies. If microbial groups are altered only in diarrhea or constipation predominant IBS patients, letter D or C in superscript follows the arrows, respectively. The figure is generated on the basis of the data published in (8, 9, 10, 11, 75, 77, 78, 79, 82, 85, 98, 116).
Several recent comprehensive studies of the microbiota in IBS have reported an increase in the relative abundance of Firmicutes, mainly Clostridium cluster XIVa and Ruminococcaceae, together with a reduction in the relative abundance of Bacteroidetes (7, 8, 9, 77). Bifidobacteria have been shown to be depleted in both fecal and mucosal samples of IBS patients (9, 78, 79, 80, 81), which adds to the trend toward a modest beneficial effect of Bifidobacterium supplementation, in terms of the improvement of global IBS symptoms and pain scores as reported in a meta-analysis of probiotic trials.(73) A lower diversity and a higher instability of the (Firmicutes fraction or function of the) microbiota in IBS patients relative to controls have been reported (82, 83, 84, 85), although these findings need confirmation in studies with sufficient depth of the microbiota analysis.
Most if not all studies that compare microbial profiles in IBS patients and healthy controls are associative and cannot distinguish cause from consequence. However, evidence of a potential causal role for the GI microbiota is supported by a fecal transplantation experiment in which visceral hypersensitivity, presumed to underpin abdominal pain in a subgroup of IBS patients, could be transferred via the microbiota of IBS patients to previously germ-free rats (86). Furthermore, germ-free mice colonized with the microbiota from IBS-D patients have been shown to exhibit a faster GI transit and impaired intestinal permeability as compared with mice gavaged with the microbiota from healthy humans (87). The mechanisms through which the microbiota exerts these effects are not fully understood, but the fact that supernatants from colon biopsies of IBS patients, with increased levels of histamine and proteases, excite human submucosal neurons could indicate a brain–gut axis connection (88). Furthermore, an increase in fecal serine proteases has been linked with symptom development in IBS patients (89, 90). Although the origin of both histamine and proteases could be both human and microbial, it is noteworthy that some of the Firmicutes bacteria that were found to be increased in abundance in IBS patients (8, 9) are known to secrete large amounts of extracellular proteases (91). Alternatively, the predominantly endogenous fecal proteases might not be degraded by microbes because of the accelerated transit (as seen in IBS-D patients) or because of the disturbed metabolic activity of the microbiota (92). Serine protease inhibitors, the antagonists of serine proteases, are produced by many bacteria, including bifidobacteria (93), which may contribute to the possible benefit of bifidobacterial supplementation.
Dysregulated intestinal immune function, chronic low-grade mucosal inflammation, and increased mucosal permeability and barrier dysfunction have all been suggested as putative pathogenic mechanisms in IBS, in which the intestinal microbiota might have a role (reviewed in (76, 94)). Moreover, the bidirectional interactions between the intestines and the central nervous system, which have an important role in the pathogenesis of IBS, have suggested to be modulated by the microbiota (reviewed in (95)). Yet, there is a lack of data on the exact mechanisms through which the host-microbiota interactions underlie pathophysiology and generate symptoms. Identifying these mechanisms is further complicated by the fact that the majority of GI microbes remain uncharacterized (96). Indeed, most putative microbial markers of IBS are among uncultured bacteria. For example, uncultured bacteria related to Ruminococcus torques are significantly enriched in IBS patients in several cohorts (9, 11, 75), and their abundance positively correlates with bowel symptoms (9, 10, 97). It is intriguing that this bacterial group has been shown to be suppressed following a multispecies probiotic treatment that alleviated IBS symptoms (98). Similarly, uncultured bacteria within the Clostridiales order are reproducibly detected in significantly depleted abundance in IBS (9, 10) but also in ulcerative colitis (99). Focus on the function of these uncultured bacteria should be of major interest for future studies.
Microbiota–Diet Interactions in IBS
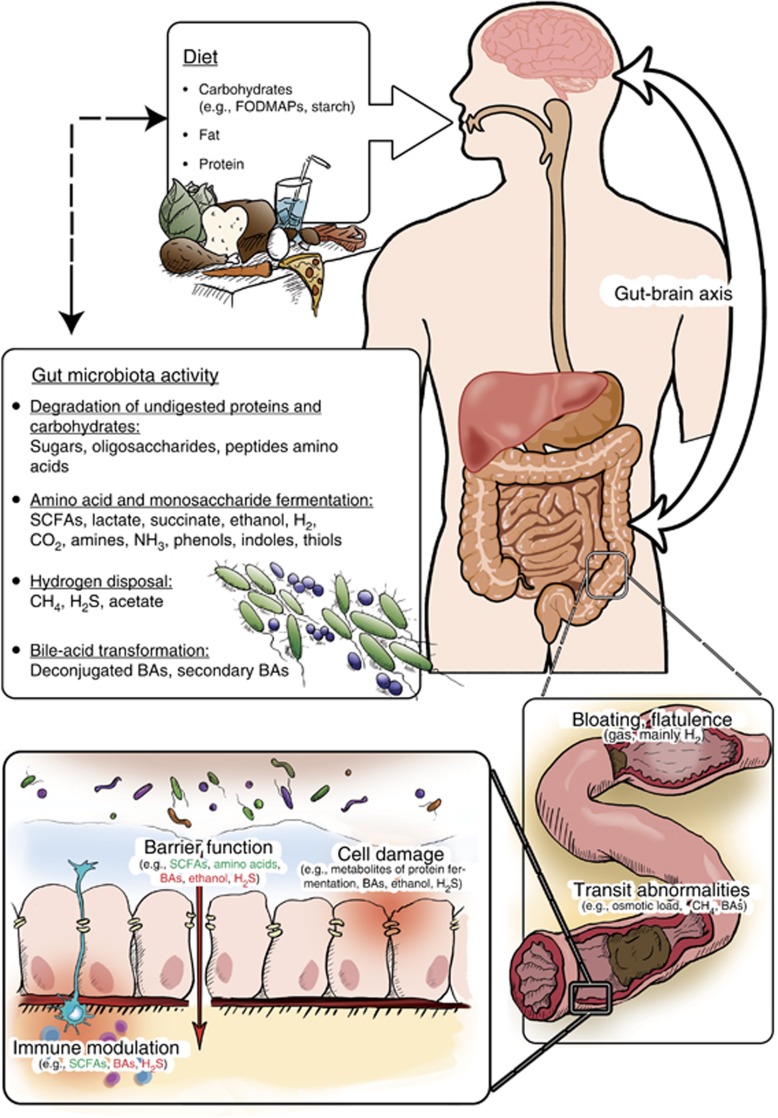
Intestinal microbes have an important role in the digestion of dietary components, resulting in metabolites that may directly or indirectly contribute to IBS symptoms (Figure 2).
Figure 2.
Non-exclusive listing of dietary metabolites that can be regulated by microbial activity and contribute to irritable bowel syndrome (IBS) symptoms. The impact of metabolites affecting different domains of intestinal health is depicted; however, it should be noted that the effects of some metabolites depend on their concentration or further conversion. Note: for immune modulation and barrier function, metabolites with positive and negative effects are colored in green and red, respectively. SCFAs, short-chain fatty acids; BAs, bile acids.
Colonic fermentation of carbohydrates mainly results in short-chain fatty acids (SCFAs, including acetate, propionate, and butyrate) and variable amounts of hydrogen and carbon dioxide. Several low-abundant microbial groups can subsequently dispose the produced hydrogen into acetate, hydrogen sulfide, and methane. The fermentation of protein residues, primarily in the distal colon, produces a variety of metabolites, including ammonia, organic acids, heterocyclic amides, and phenolic and indolic compounds, that are toxic and detrimental to gut health (100). Moreover, fat and digested proteins increase the excretion of bile acids that are subject to extensive bacterial transformation in the intestinal tract. This could be of particular relevance as bile acids inhibit many bacteria and may modulate microbiota composition and also have the potential to affect major pathophysiological factors in IBS, including GI motility, secretion, and immune function (101, 102). Indeed, in a recent large-scale study, increased colonic bile acid exposure was demonstrated in a subset of, predominantly non-constipated, IBS patients correlating with diarrhea and accelerated colonic transit (101).
An association between the intestinal microbiota and protein metabolism has been established by a significant correlation between the abundance of several post-infectious (PI)-IBS microbial markers and host amino-acid metabolism (10). Among harmful products of protein fermentation (100, 103), hydrogen sulfide might be relevant for compromising intestinal health as it directly impairs epithelial metabolism (104) and acts as a gut–brain signaling molecule (105). Hydrogen sulfide can be converted to thiosulfate and further oxidized to tetrathionate during inflammation. The latter supports the growth of Salmonella (106) and other tetrathionate utilizing pathogens from the Gamma-Proteobacteria class (107), many of which have been associated with bowel symptoms of IBS patients (9, 10, 82).
FODMAPS, indigestible carbohydrates, can be fermented by intestinal microbes, resulting in increased gas production and an osmotic effect that can provoke IBS symptoms in a sensitive host (108), although similar changes may go unperceived in normal subjects (109). Carbohydrate utilizing GI bacteria such as Dorea spp., major gas producing bacteria in the human intestine (110), were reported to be significantly increased in abundance in IBS patients (9, 11). Overproduction of gas, specifically hydrogen, has been associated with IBS symptoms (111), especially abdominal pain and flatulence. In patients with IBS-D, whose colon was found to be less able to accommodate the increased intestinal volume (112), overproduction of gas could be a trigger for increased colonic wall tension. In general, hydrogen produced in the gut is most efficiently removed by the methanogenic archaea (113) that appear to be depleted in IBS-D patients (9, 10) and increased in IBS with constipation patients (reviewed in (114)) Although there is substantial evidence for implication of methane in IBS symptomology, most data are based upon methane breath testing. Hence, future studies, especially those addressing the diet-microbiota connection in IBS, should quantify methanogens to provide more direct evidence.
Alternative pathway for gas elimination is the conversion into acetate by acetogenic bacteria, Blautia spp. (115). Blautia spp. are highly prevalent and dominant intestinal bacteria, which have been found to be elevated in IBS patients (9, 79). Intriguingly, the major fermentation end-product—acetate—has been reported as significantly increased in IBS patients (116), although this was not confirmed in another study (117). This may relate to the fact that most studies measure stool concentrations without taking the total stool volume into account, thereby potentially concealing overall increased acetate production as a result of dilution due to increased stool volume.
SCFAs produced upon the microbial fermentation of undigested carbohydrates and reduced levels thereof may also contribute to symptom generation in IBS. SCFAs have some documented health benefits, in particular butyrate, which is an important energy source for colonocytes, and among others inhibits inflammation and enhances barrier function (118). Moreover, intraluminal administration of butyrate into the distal colon has been shown to decrease visceral sensitivity in healthy humans (119). However, excessive intake of fermentable carbohydrates and high SCFA levels can also increase the osmotic load and thereby lead to diarrhea (120).
Data on SCFAs in IBS are limited and inconsistent, showing no alterations, increased or decreased levels of SCFAs relative to controls (61, 116, 117, 121). Furthermore, altered SCFAs profiles might merely reflect diet rather than a feature of the condition. Given that the major products of FODMAPs fermentation are SCFAs, and that some major microbial SCFA-producers may be altered in IBS, the connection between FODMAPs, SCFAs, microbiota, and IBS symptoms calls for further elucidation.
Few studies have examined the impact of dietary interventions, in particular FODMAP restriction, on the microbiota in IBS patients. A low-FODMAP diet has been linked to reduced bifidobacterial counts (59), which seems a paradox given their potential health benefit. A recent study reported a reduction in total bacterial abundance following the introduction of a low-FODMAP diet, as well as a reduction in absolute counts but not relative abundance of specific bacterial groups, including bifidobacteria as compared with habitual diet (63). However, as compared with a diet high in FODMAP content, the low-FODMAP diet appeared to be associated with marked lower relative abundances of butyrate-producing bacteria and A. muciniphila, and a significantly higher abundance of R. torques, adding more paradoxical data on the effects of low-FODMAP diet (63).
Extensive analyses of microbiota composition, functionality, and fermentation products in relation to FODMAP restriction and symptom generation are currently, however, lacking.
The direct impact of diet-microbiota interactions on IBS symptoms has been demonstrated in an elegant study on the impact of a diet rich in fermentable substrates. The introduction of this flatulogenic diet significantly increased the gas volume and the number of gas evacuations, as well as abdominal symptoms and digestive discomfort in both healthy subjects and flatulent patients (122). Yet, upon dietary challenge, patients' microbiota developed instability in composition, exhibiting variations in the Bacteroidetes and Firmicutes phyla and reduction in microbial diversity, whereas healthy subjects' microbiota remained stable. Hence, the extent of dysbiosis in IBS patients likely depends on their diet. In a small study among children with IBS, the introduction of a low fermentable substrate diet significantly decreased abdominal pain severity and frequency in some but not all children. Responders to this dietary intervention appeared to differ in intestinal microbiota composition at baseline from nonresponders, indicating that the efficacy of dietary interventions might be influenced by the patient's microbiota (123). To prove the validity, these pilot results will have to be replicated in larger studies.
Conclusion, Recommendations for Future Research
Both experimental and observational studies provide good evidence to conclude that microbial alterations frequently observed in IBS patients potentially drive or perpetuate gastrointestinal symptoms. Yet, the strong interaction between distinct dietary patterns and the intestinal microbial communities is likely one of the explanations for the inconclusive findings among studies comparing the microbiota composition in IBS patients and healthy subjects.
More well-designed studies on the effect of food intake on symptom generation and the underlying mechanisms are warranted. The potential benefit of FODMAP restriction seems promising but requires further study and dietary guidance is essential given the complexity and risk of inadequate nutrient intake. In addition, more evidence on long-term outcome and well-performed challenge or re-introduction trials are warranted as important adaptions may occur. The incorporation of microbiota characterization in such studies would both provide insight into the impact of these dietary interventions on the microbiota, as well as offer the opportunity to examine whether the microbiota can predict response.
Given that IBS patients comprise a heterogeneous group both in the type of bowel symptoms and food components to which they might respond, and that the microbiota is extremely complex and variable between individuals, it is clear that defining the correlations between IBS, microbiota, and diet is not an easy task. However, high-throughput molecular methods enable reproducible and in-depth analysis of the intestinal microbiota, providing the necessary tool for testing these correlations. The major challenge remains to include the conscientious collection of dietary information in future studies in this field. Moreover, future studies should include multiple microbiota analysis at various time points in a large number of well-phenotyped patients to increase our insight in the contribution of intestinal microbiota perturbations to the diverse and fluctuating symptom pattern in IBS patients.
Acknowledgments
We are thankful to Mayk Lucchesi for figure designing and formatting.
Guarantor of the article: John Penders, PhD.
Specific author contributions: The initial drafting of the manuscript was prepared by Mirjana Rajilić-Stojanović, Daisy M. Jonkers, Robin C. Spiller, Anne Salonen, Kurt Hanevik, Jonna Jalanka, Chaysavanh Manichanh, Elena Philippou, Gerard Clarke, and John Penders. Editing and draft revisions were done by Mirjana Rajilić-Stojanović, Daisy M. Jonkers, Anne Salonen, Kurt Hanevik, Jeroen Raes, Jonna Jalanka, Willem M. de Vos, Chaysavanh Manichanh, Natasa Golic, Paul Enck, Elena Philippou, Fuad A. Iraqi, Gerard Clarke, Robin C. Spiller, and John Penders. All authors have approved the final draft.
Financial Support: This work was supported by collaboration and network activities promoted under the frame of the international network GENIEUR (Genes in Irritable Bowel Syndrome Europe), which is currently funded by the COST program (BM1106, www.GENIEUR.eu).
Potential competing interests: Willem M. de Vos served as a member of the Scientific Advisory Boards of NIHS and Johnson & Johnson and has filed patent applications on IBS diagnostics. Paul Enck has received research funds from SymbioPharm, Germany, and has received consultancy fees and travel reimbursement from SymbioPharm, Germany, and Danone, France. Robin Spiller has received research funding from Lesaffre and Ironwood and free drug for clinical trial from Norgine and Falk Pharma. He has also acted on Advisory Boards for Almirall, Astellas, Yuhan Corporation, and Danone. Jeroen Raes served on advisory boards for Johnson & Johnson, GSK Vaccines, and 23andMe. The Alimentary Pharmabiotic Centre is a research center funded by Science Foundation Ireland (SFI; grant numbers SFI/12/RC/2273, 02/CE/B124 and 07/CE/B1368), through the Irish Government's National Development Plan. The Centre has conducted studies in collaboration with several companies including GSK, Pfizer, Wyeth, and Mead Johnson. Gerard Clarke is also supported by the Health Research Board (HRB) through a Health Research Award (grant no HRA_POR/2011/23) and by a NARSAD Young Investigator Grant from the Brain and Behavior Research Foundation (grant Number 20771). The content of this article was neither influenced nor constrained by this support.
References
- Lovell RM, Ford AC. Global prevalence of and risk factors for irritable bowel syndrome: a meta-analysis. Clin Gastroenterol Hepatol. 2012;10:712–721. doi: 10.1016/j.cgh.2012.02.029. [DOI] [PubMed] [Google Scholar]
- Simrén M, Svedlund J, Posserud I, et al. Health-related quality of life in patients attending a gastroenterology outpatient clinic: functional disorders versus organic diseases. Clin Gastroenterol Hepatol. 2006;4:187–195. doi: 10.1016/s1542-3565(05)00981-x. [DOI] [PubMed] [Google Scholar]
- Longstreth GF, Thompson WG, Chey WD, et al. Functional bowel disorders. Gastroenterology. 2006;130:1480–1491. doi: 10.1053/j.gastro.2005.11.061. [DOI] [PubMed] [Google Scholar]
- Camilleri M, Lasch K, Zhou W. Irritable bowel syndrome: methods, mechanisms, and pathophysiology. The confluence of increased permeability, inflammation, and pain in irritable bowel syndrome. Am J Physiol Gastrointest Liver Physiol. 2012;303:G775–G785. doi: 10.1152/ajpgi.00155.2012. [DOI] [PubMed] [Google Scholar]
- Thabane M, Kottachchi DT, Marshall JK. Systematic review and meta-analysis: The incidence and prognosis of post-infectious irritable bowel syndrome. Aliment Pharmacol Ther. 2007;26:535–544. doi: 10.1111/j.1365-2036.2007.03399.x. [DOI] [PubMed] [Google Scholar]
- Villarreal AA, Aberger FJ, Benrud R, et al. Use of broad-spectrum antibiotics and the development of irritable bowel syndrome. WMJ. 2012;111:17–20. [PubMed] [Google Scholar]
- Salonen A, de Vos WM, Palva A. Gastrointestinal microbiota in irritable bowel syndrome: present state and perspectives. Microbiology. 2010;156:3205–3215. doi: 10.1099/mic.0.043257-0. [DOI] [PubMed] [Google Scholar]
- Jeffery IB, O'Toole PW, Öhman L, et al. An irritable bowel syndrome subtype defined by species-specific alterations in faecal microbiota. Gut. 2012;61:997–1006. doi: 10.1136/gutjnl-2011-301501. [DOI] [PubMed] [Google Scholar]
- Rajilić-Stojanović M, Biagi E, Heilig HGHJ, et al. Global and deep molecular analysis of microbiota signatures in fecal samples from patients with irritable bowel syndrome. Gastroenterology. 2011;141:1792–1801. doi: 10.1053/j.gastro.2011.07.043. [DOI] [PubMed] [Google Scholar]
- Jalanka-Tuovinen J, Salojärvi J, Salonen A, et al. Faecal microbiota composition and host-microbe cross-talk following gastroenteritis and in postinfectious irritable bowel syndrome. Gut. 2014;63:1737–1745. doi: 10.1136/gutjnl-2013-305994. [DOI] [PubMed] [Google Scholar]
- Saulnier DM, Riehle K, Mistretta T-A, et al. Gastrointestinal microbiome signatures of pediatric patients with irritable bowel syndrome. Gastroenterology. 2011;141:1782–1791. doi: 10.1053/j.gastro.2011.06.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funkhouser LJ, Bordenstein SR. Mom knows best: the universality of maternal microbial transmission. PLoS Biol. 2013;11:e1001631. doi: 10.1371/journal.pbio.1001631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aagaard K, Ma J, Antony KM, et al. The placenta harbors a unique microbiome. Sci Transl Med. 2014;6:237ra65. doi: 10.1126/scitranslmed.3008599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez-Bello MG, Costello EK, Contreras M, et al. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc Natl Acad Sci USA. 2010;107:11971–11975. doi: 10.1073/pnas.1002601107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penders J, Thijs C, Vink C, et al. Factors influencing the composition of the intestinal microbiota in early infancy. Pediatrics. 2006;118:511–521. doi: 10.1542/peds.2005-2824. [DOI] [PubMed] [Google Scholar]
- Penders J, Gerhold K, Stobberingh EE, et al. Establishment of the intestinal microbiota and its role for atopic dermatitis in early childhood J Allergy Clin Immunol 2013132601–607.e8. [DOI] [PubMed] [Google Scholar]
- Azad MB, Konya T, Maughan H, et al. Gut microbiota of healthy Canadian infants: profiles by mode of delivery and infant diet at 4 months. CMAJ. 2013;185:385–394. doi: 10.1503/cmaj.121189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fallani M, Young D, Scott J, et al. Intestinal microbiota of 6-week-old infants across Europe: geographic influence beyond delivery mode, breast-feeding, and antibiotics. J Pediatr Gastroenterol Nutr. 2010;51:77–84. doi: 10.1097/MPG.0b013e3181d1b11e. [DOI] [PubMed] [Google Scholar]
- Marcobal A, Barboza M, Froehlich JW, et al. Consumption of human milk oligosaccharides by gut-related microbes. J Agric Food Chem. 2010;58:5334–5340. doi: 10.1021/jf9044205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jost T, Lacroix C, Braegger C, et al. Assessment of bacterial diversity in breast milk using culture-dependent and culture-independent approaches. Br J Nutr. 2013;110:1253–1262. doi: 10.1017/S0007114513000597. [DOI] [PubMed] [Google Scholar]
- Palmer C, Bik EM, Digiulio DB, et al. Development of the human infant intestinal microbiota. PLoS Biol. 2007;5:e177. doi: 10.1371/journal.pbio.0050177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koenig JE, Spor A, Scalfone N, et al. Succession of microbial consortia in the developing infant gut microbiome. Proc Natl Acad Sci USA. 2011;108 (Suppl 1:4578–4585. doi: 10.1073/pnas.1000081107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nylund L, Satokari R, Salminen S, et al. Intestinal microbiota during early life—impact on health and disease. Proc Nutr Soc. 2014;73:457–469. doi: 10.1017/S0029665114000627. [DOI] [PubMed] [Google Scholar]
- Borre YE, O'Keeffe GW, Clarke G, et al. Microbiota and neurodevelopmental windows: implications for brain disorders. Trends Mol Med. 2014;20:509–518. doi: 10.1016/j.molmed.2014.05.002. [DOI] [PubMed] [Google Scholar]
- Rajilić-Stojanović M, Heilig HGHJ, Tims S, et al. Long-term monitoring of the human intestinal microbiota composition. Environ Microbiol. 2013;15:1146–1159. doi: 10.1111/1462-2920.12023. [DOI] [PubMed] [Google Scholar]
- Yatsunenko T, Rey FE, Manary MJ, et al. Human gut microbiome viewed across age and geography. Nature. 2012;486:222–227. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costello EK, Lauber CL, Hamady M, et al. Bacterial community variation in human body habitats across space and time. Science. 2009;326:1694–1697. doi: 10.1126/science.1177486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoetendal EG, Akkermans AD, de Vos WM. Temperature gradient gel electrophoresis analysis of 16S rRNA from human fecal samples reveals stable and host-specific communities of active bacteria. Appl Environ Microbiol. 1998;64:3854–3859. doi: 10.1128/aem.64.10.3854-3859.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding T, Schloss PD. Dynamics and associations of microbial community types across the human body. Nature. 2014;509:357–360. doi: 10.1038/nature13178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin A, Bik EM, Costello EK, et al. Distinct distal gut microbiome diversity and composition in healthy children from Bangladesh and the United States. PLoS ONE. 2013;8:e53838. doi: 10.1371/journal.pone.0053838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnorr SL, Candela M, Rampelli S, et al. Gut microbiome of the Hadza hunter-gatherers. Nat Commun. 2014;5:3654. doi: 10.1038/ncomms4654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Filippo C, Cavalieri D, Di Paola M, et al. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc Natl Acad Sci. 2010;107:14691–14696. doi: 10.1073/pnas.1005963107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salonen A, de Vos WM. Impact of diet on human intestinal microbiota and health. Annu Rev Food Sci Technol. 2014;5:239–262. doi: 10.1146/annurev-food-030212-182554. [DOI] [PubMed] [Google Scholar]
- Wu GD, Chen J, Hoffmann C, et al. Linking long-term dietary patterns with gut microbial enterotypes. Science. 2011;334:105–108. doi: 10.1126/science.1208344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lappi J, Salojärvi J, Kolehmainen M, et al. Intake of whole-grain and fiber-rich rye bread versus refined wheat bread does not differentiate intestinal microbiota composition in Finnish adults with metabolic syndrome. J Nutr. 2013;143:648–655. doi: 10.3945/jn.112.172668. [DOI] [PubMed] [Google Scholar]
- Cotillard A, Kennedy SP, Kong LC, et al. Dietary intervention impact on gut microbial gene richness. Nature. 2013;500:585–588. doi: 10.1038/nature12480. [DOI] [PubMed] [Google Scholar]
- David LA, Maurice CF, Carmody RN, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505:559–563. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salonen A, Lahti L, Salojarvi J, et al. Impact of diet and individual variation on intestinal microbiota composition and fermentation products in obese men. ISME J. 2014;8:2218–2230. doi: 10.1038/ismej.2014.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker AW, Ince J, Duncan SH, et al. Dominant and diet-responsive groups of bacteria within the human colonic microbiota. ISME J. 2011;5:220–230. doi: 10.1038/ismej.2010.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monsbakken KW, Vandvik PO, Farup PG. Perceived food intolerance in subjects with irritable bowel syndrome—etiology, prevalence and consequences. Eur J Clin Nutr. 2006;60:667–672. doi: 10.1038/sj.ejcn.1602367. [DOI] [PubMed] [Google Scholar]
- Simrén M, Mansson A, Langkilde AM, et al. Food-related gastrointestinal symptoms in the irritable bowel syndrome. Digestion. 2001;63:108–115. doi: 10.1159/000051878. [DOI] [PubMed] [Google Scholar]
- Hayes PA, Fraher MH, Quigley EM. Irritable bowel syndrome: the role of food in pathogenesis and management. Gastroenterol Hepatol. 2014;10:164–174. [PMC free article] [PubMed] [Google Scholar]
- Böhn L, Störsrud S, Simrén M. Nutrient intake in patients with irritable bowel syndrome compared with the general population. Neurogastroenterol Motil. 2013;25:23–30. doi: 10.1111/nmo.12001. [DOI] [PubMed] [Google Scholar]
- Saito YA, Locke GR, Weaver AL, et al. Diet and functional gastrointestinal disorders: a population-based case control study. Am J Gastroenterol. 2005;100:2743–2748. doi: 10.1111/j.1572-0241.2005.00288.x. [DOI] [PubMed] [Google Scholar]
- Williams EA, Nai X, Corfe BM. Dietary intakes in people with irritable bowel syndrome. BMC Gastroenterol. 2011;11:9. doi: 10.1186/1471-230X-11-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragnarsson G, Bodemar G. Pain is temporally related to eating but not to defaecation in the irritable bowel syndrome (IBS). Patients' description of diarrhea, constipation and symptom variation during a prospective 6-week study. Eur J Gastroenterol Hepatol. 1998;10:415–421. doi: 10.1097/00042737-199805000-00011. [DOI] [PubMed] [Google Scholar]
- Simrén M, Agerforz P, Bjornsson ES, et al. Nutrient-dependent enhancement of rectal sensitivity in irritable bowel syndrome (IBS) Neurogastroenterol Motil. 2007;19:20–29. doi: 10.1111/j.1365-2982.2006.00849.x. [DOI] [PubMed] [Google Scholar]
- Ludidi S, Conchillo JM, Keszthelyi D, et al. Does meal ingestion enhance sensitivity of visceroperception assessment in irritable bowel syndrome Neurogastroenterol Motil 20122447–53.e3. [DOI] [PubMed] [Google Scholar]
- Böhn L, Storsrud S, Tornblom H, et al. Self-reported food-related gastrointestinal symptoms in IBS are common and associated with more severe symptoms and reduced quality of life. Am J Gastroenterol. 2013;108:634–641. doi: 10.1038/ajg.2013.105. [DOI] [PubMed] [Google Scholar]
- Niec AM, Frankum B, Talley NJ. Are adverse food reactions linked to irritable bowel syndrome. Am J Gastroenterol. 1998;93:2184–2190. doi: 10.1111/j.1572-0241.1998.00531.x. [DOI] [PubMed] [Google Scholar]
- Atkins FM. A critical evaluation of clinical trials in adverse reactions to foods in adults. J Allergy Clin Immunol. 1986;78:174–182. doi: 10.1016/0091-6749(86)90010-2. [DOI] [PubMed] [Google Scholar]
- Young E, Stoneham MD, Petruckevitch A, et al. A population study of food intolerance. Lancet. 1994;343:1127–1130. doi: 10.1016/s0140-6736(94)90234-8. [DOI] [PubMed] [Google Scholar]
- Carroccio A, Brusca I, Mansueto P, et al. Fecal assays detect hypersensitivity to cow's milk protein and gluten in adults with irritable bowel syndrome Clin Gastroenterol Hepatol 20119965–971.e3. [DOI] [PubMed] [Google Scholar]
- Feinle-Bisset C, Azpiroz F. Dietary lipids and functional gastrointestinal disorders. Am J Gastroenterol. 2013;108:737–747. doi: 10.1038/ajg.2013.76. [DOI] [PubMed] [Google Scholar]
- Yang J, Fox M, Cong Y, et al. Lactose intolerance in irritable bowel syndrome patients with diarrhoea: the roles of anxiety, activation of the innate mucosal immune system and visceral sensitivity. Aliment Pharmacol Ther. 2014;39:302–311. doi: 10.1111/apt.12582. [DOI] [PubMed] [Google Scholar]
- McKenzie YA, Alder A, Anderson W, et al. British Dietetic Association evidence-based guidelines for the dietary management of irritable bowel syndrome in adults. J Hum Nutr Diet. 2012;25:260–274. doi: 10.1111/j.1365-277X.2012.01242.x. [DOI] [PubMed] [Google Scholar]
- Shepherd SJ, Gibson PR. Fructose malabsorption and symptoms of irritable bowel syndrome: guidelines for effective dietary management. J Am Diet Assoc. 2006;106:1631–1639. doi: 10.1016/j.jada.2006.07.010. [DOI] [PubMed] [Google Scholar]
- de Roest RH, Dobbs BR, Chapman BA, et al. The low FODMAP diet improves gastrointestinal symptoms in patients with irritable bowel syndrome: a prospective study. Int J Clin Pract. 2013;67:895–903. doi: 10.1111/ijcp.12128. [DOI] [PubMed] [Google Scholar]
- Staudacher HM, Lomer MC, Anderson JL, et al. Fermentable carbohydrate restriction reduces luminal bifidobacteria and gastrointestinal symptoms in patients with irritable bowel syndrome. J Nutr. 2012;142:1510–1518. doi: 10.3945/jn.112.159285. [DOI] [PubMed] [Google Scholar]
- Ong DK, Mitchell SB, Barrett JS, et al. Manipulation of dietary short chain carbohydrates alters the pattern of gas production and genesis of symptoms in irritable bowel syndrome. J Gastroenterol Hepatol. 2010;25:1366–1373. doi: 10.1111/j.1440-1746.2010.06370.x. [DOI] [PubMed] [Google Scholar]
- Halmos EP, Power VA, Shepherd SJ, et al. A diet low in FODMAPs reduces symptoms of irritable bowel syndrome Gastroenterology 201414667–75.e5. [DOI] [PubMed] [Google Scholar]
- Shepherd SJ, Parker FC, Muir JG, et al. Dietary triggers of abdominal symptoms in patients with irritable bowel syndrome: randomized placebo-controlled evidence. Clin Gastroenterol Hepatol. 2008;6:765–771. doi: 10.1016/j.cgh.2008.02.058. [DOI] [PubMed] [Google Scholar]
- Halmos EP, Christophersen CT, Bird AR, et al. Diets that differ in their FODMAP content alter the colonic luminal microenvironment. Gut. 2014;64:93–100. doi: 10.1136/gutjnl-2014-307264. [DOI] [PubMed] [Google Scholar]
- Ligaarden SC, Lydersen S, Farup PG. IgG and IgG4 antibodies in subjects with irritable bowel syndrome: a case control study in the general population. BMC Gastroenterol. 2012;12:166. doi: 10.1186/1471-230X-12-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiller RC, Jenkins D, Thornley JP, et al. Increased rectal mucosal enteroendocrine cells, T lymphocytes, and increased gut permeability following acute Campylobacter enteritis and in post-dysenteric irritable bowel syndrome. Gut. 2000;47:804–811. doi: 10.1136/gut.47.6.804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker MM, Warwick A, Ung C, et al. The role of eosinophils and mast cells in intestinal functional disease. Curr Gastroenterol Rep. 2011;13:323–330. doi: 10.1007/s11894-011-0197-5. [DOI] [PubMed] [Google Scholar]
- Matricon J, Meleine M, Gelot A, et al. Review article: associations between immune activation, intestinal permeability and the irritable bowel syndrome. Aliment Pharmacol Ther. 2012;36:1009–1031. doi: 10.1111/apt.12080. [DOI] [PubMed] [Google Scholar]
- Ford AC, Chey WD, Talley NJ, et al. Yield of diagnostic tests for celiac disease in individuals with symptoms suggestive of irritable bowel syndrome: systematic review and meta-analysis. Arch Intern Med. 2009;169:651–658. doi: 10.1001/archinternmed.2009.22. [DOI] [PubMed] [Google Scholar]
- Wahnschaffe U, Schulzke JD, Zeitz M, et al. Predictors of clinical response to gluten-free diet in patients diagnosed with diarrhea-predominant irritable bowel syndrome Clin Gastroenterol Hepatol 20075844–850.quiz 769.. [DOI] [PubMed] [Google Scholar]
- Biesiekierski JR, Newnham ED, Irving PM, et al. Gluten causes gastrointestinal symptoms in subjects without celiac disease: a double-blind randomized placebo-controlled trial Am J Gastroenterol 2011106508–514.quiz 515.. [DOI] [PubMed] [Google Scholar]
- Biesiekierski JR, Peters SL, Newnham ED, et al. No effects of gluten in patients with self-reported non-celiac gluten sensitivity after dietary reduction of fermentable, poorly absorbed, short-chain carbohydrates Gastroenterology 2013145320–328.e1-3. [DOI] [PubMed] [Google Scholar]
- Carroccio A, Mansueto P, D'Alcamo A, et al. Non-celiac wheat sensitivity as an allergic condition: personal experience and narrative review Am J Gastroenterol 20131081845–1852.quiz 1853. [DOI] [PubMed] [Google Scholar]
- Ford AC, Quigley EM, Lacy BE, et al. Efficacy of prebiotics, probiotics, and synbiotics in irritable bowel syndrome and chronic idiopathic constipation: systematic review and meta-analysis. Am J Gastroenterol. 2014;109:1547–1561. doi: 10.1038/ajg.2014.202. [DOI] [PubMed] [Google Scholar]
- Moayyedi P, Quigley EM, Lacy BE, et al. The effect of fiber supplementation on irritable bowel syndrome: a systematic review and meta-analysis. Am J Gastroenterol. 2014;109:1367–1374. doi: 10.1038/ajg.2014.195. [DOI] [PubMed] [Google Scholar]
- Kassinen A, Krogius-Kurikka L, Mäkivuokko H, et al. The fecal microbiota of irritable bowel syndrome patients differs significantly from that of healthy subjects. Gastroenterology. 2007;133:24–33. doi: 10.1053/j.gastro.2007.04.005. [DOI] [PubMed] [Google Scholar]
- Simrén M, Barbara G, Flint HJ, et al. Intestinal microbiota in functional bowel disorders: a Rome foundation report. Gut. 2013;62:159–176. doi: 10.1136/gutjnl-2012-302167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogius-Kurikka L, Lyra A, Malinen E, et al. Microbial community analysis reveals high level phylogenetic alterations in the overall gastrointestinal microbiota of diarrhoea-predominant irritable bowel syndrome sufferers. BMC Gastroenterol. 2009;9:95. doi: 10.1186/1471-230X-9-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerckhoffs APM, Samsom M, van der Rest ME, et al. Lower Bifidobacteria counts in both duodenal mucosa-associated and fecal microbiota in irritable bowel syndrome patients. World J Gastroenterol. 2009;15:2887–2892. doi: 10.3748/wjg.15.2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malinen E, Rinttilä T, Kajander K, et al. Analysis of the fecal microbiota of irritable bowel syndrome patients and healthy controls with real-time PCR. Am J Gastroenterol. 2005;100:373–382. doi: 10.1111/j.1572-0241.2005.40312.x. [DOI] [PubMed] [Google Scholar]
- Balsari A, Ceccarelli A, Dubini F, et al. The fecal microbial population in the irritable bowel syndrome. Microbiologica. 1982;5:185–194. [PubMed] [Google Scholar]
- Si JM, Yu YC, Fan YJ, et al. Intestinal microecology and quality of life in irritable bowel syndrome patients. World J Gastroenterol. 2004;10:1802–1805. doi: 10.3748/wjg.v10.i12.1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll IM, Shen XJ, Keku TO, et al. Characterization of the fecal microbiota in patients with diarrhea predominant irritable bowel syndrome. Gastroenterology. 2008;134:A-681. [Google Scholar]
- Durbán A, Abellán JJ, Jiménez-Hernández N, et al. Instability of the faecal microbiota in diarrhoea-predominant irritable bowel syndrome. FEMS Microbiol Ecol. 2013;86:581–589. doi: 10.1111/1574-6941.12184. [DOI] [PubMed] [Google Scholar]
- Mättö J, Maunuksela L, Kajander K, et al. Composition and temporal stability of gastrointestinal microbiota in irritable bowel syndrome—a longitudinal study in IBS and control subjects. FEMS Immunol Med Microbiol. 2005;43:213–222. doi: 10.1016/j.femsim.2004.08.009. [DOI] [PubMed] [Google Scholar]
- Maukonen J, Satokari R, Mättö J, et al. Prevalence and temporal stability of selected clostridial groups in irritable bowel syndrome in relation to predominant faecal bacteria. J Med Microbiol. 2006;55:625–633. doi: 10.1099/jmm.0.46134-0. [DOI] [PubMed] [Google Scholar]
- Crouzet L, Gaultier E, Del'Homme C, et al. The hypersensitivity to colonic distension of IBS patients can be transferred to rats through their fecal microbiota. Neurogastroenterol Motil. 2013;25:e272–e282. doi: 10.1111/nmo.12103. [DOI] [PubMed] [Google Scholar]
- De Palma G.Transfer of anxiety and gut dysfunction from IBS patients to gnotobiotic mice through microibota transplatation. CDDW 2014; 2014: A183.
- Buhner S, Li Q, Vignali S, et al. Activation of human enteric neurons by supernatants of colonic biopsy specimens from patients with irritable bowel syndrome. Gastroenterology. 2009;137:1425–1434. doi: 10.1053/j.gastro.2009.07.005. [DOI] [PubMed] [Google Scholar]
- Gecse K, Roka R, Ferrier L, et al. Increased fecal serine-protease activity in diarrheic IBS patients: a colonic lumenal factor impairing colonic permeability and sensitivity. Gut. 2008;57:591–599. doi: 10.1136/gut.2007.140210. [DOI] [PubMed] [Google Scholar]
- Tooth D, Garsed K, Singh G, et al. Characterisation of faecal protease activity in irritable bowel syndrome with diarrhoea: origin and effect of gut transit. Gut. 2014;63:753–760. doi: 10.1136/gutjnl-2012-304042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macfarlane GT, Allison C, Gibson SA, et al. Contribution of the microflora to proteolysis in the human large intestine. J Appl Bacteriol. 1988;64:37–46. doi: 10.1111/j.1365-2672.1988.tb02427.x. [DOI] [PubMed] [Google Scholar]
- Tooth D, Garsed K, Singh G, et al. Characterisation of faecal protease activity in irritable bowel syndrome with diarrhoea: origin and effect of gut transit. Gut. 2014;63:753–760. doi: 10.1136/gutjnl-2012-304042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov D, Emonet C, Foata F, et al. A serpin from the gut bacterium Bifidobacterium longum inhibits eukaryotic elastase-like serine proteases. J Biol Chem. 2006;281:17246–17252. doi: 10.1074/jbc.M601678200. [DOI] [PubMed] [Google Scholar]
- Lee KN, Lee OY. Intestinal microbiota in pathophysiology and management of irritable bowel syndrome. World J Gastroenterol. 2014;20:8886–8897. doi: 10.3748/wjg.v20.i27.8886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer EA, Savidge T, Shulman RJ. Brain-gut microbiome interactions and functional bowel disorders. Gastroenterology. 2014;146:1500–1512. doi: 10.1053/j.gastro.2014.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajilić-Stojanović M, Smidt H, de Vos WM. Diversity of the human gastrointestinal tract microbiota revisited. Environ Microbiol. 2007;9:2125–2136. doi: 10.1111/j.1462-2920.2007.01369.x. [DOI] [PubMed] [Google Scholar]
- Malinen E, Krogius-Kurikka L, Lyra A, et al. Association of symptoms with gastrointestinal microbiota in irritable bowel syndrome. World J Gastroenterol. 2010;16:4532–4540. doi: 10.3748/wjg.v16.i36.4532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyra A, Krogius-Kurikka L, Nikkila J, et al. Effect of a multispecies probiotic supplement on quantity of irritable bowel syndrome-related intestinal microbial phylotypes. BMC Gastroenterol. 2009;10:110. doi: 10.1186/1471-230X-10-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajilić-Stojanović M, Guarner F, Shanahan F, et al. Phylogenetic analysis of dysbiosis in ulcerative colitis during remission. Inflamm Bowel Dis. 2013;19:481–488. doi: 10.1097/MIB.0b013e31827fec6d. [DOI] [PubMed] [Google Scholar]
- Smith EA, Macfarlane GT. Dissimilatory amino acid metabolism in human colonic bacteria. Anaerobe. 1997;3:327–337. doi: 10.1006/anae.1997.0121. [DOI] [PubMed] [Google Scholar]
- Bajor A, Törnblom H, Rudling M, et al. Increased colonic bile acid exposure: a relevant factor for symptoms and treatment in IBS. Gut. 2014;64:84–92. doi: 10.1136/gutjnl-2013-305965. [DOI] [PubMed] [Google Scholar]
- Hylemon PB, Zhou H, Pandak WM, et al. Bile acids as regulatory molecules. J Lipid Res. 2009;50:1509–1520. doi: 10.1194/jlr.R900007-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajilić-Stojanović M. Function of the microbiota. Best Pract Res Clin Gastroenterol. 2013;27:5–16. doi: 10.1016/j.bpg.2013.03.006. [DOI] [PubMed] [Google Scholar]
- Jørgensen J, Mortensen P. Hydrogen sulfide and colonic epithelial metabolism. Dig Dis Sci. 2001;46:1722–1732. doi: 10.1023/a:1010661706385. [DOI] [PubMed] [Google Scholar]
- Schicho R, Krueger D, Zeller F, et al. Hydrogen sulfide is a novel prosecretory neuromodulator in the guinea-pig and human colon. Gastroenterology. 2006;131:1542. doi: 10.1053/j.gastro.2006.08.035. [DOI] [PubMed] [Google Scholar]
- Thiennimitr P, Winter SE, Winter MG, et al. Intestinal inflammation allows Salmonella to use ethanolamine to compete with the microbiota. Proc Natl Acad Sci USA. 2011;108:17480–17485. doi: 10.1073/pnas.1107857108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissfeld AS, Sonnenwirth AC. Rapid isolation of Yersinia spp. from feces. J Clin Microbiol. 1982;15:508–510. doi: 10.1128/jcm.15.3.508-510.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staudacher HM, Irving PM, Lomer MC, et al. Mechanisms and efficacy of dietary FODMAP restriction in IBS. Nat Rev Gastroenterol Hepatol. 2014;11:256–266. doi: 10.1038/nrgastro.2013.259. [DOI] [PubMed] [Google Scholar]
- Murray K, Wilkinson-Smith V, Hoad C, et al. Differential effects of FODMAPs (fermentable oligo-, di-, mono-saccharides and polyols) on small and large intestinal contents in healthy subjects shown by MRI. Am J Gastroenterol. 2014;109:110–119. doi: 10.1038/ajg.2013.386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taras D, Simmering R, Collins MD, et al. Reclassification of Eubacterium formicigenerans Holdeman and Moore 1974 as Dorea formicigenerans gen. nov., comb. nov., and description of Dorea longicatena sp. nov., isolated from human faeces. Int J Syst Evol Microbiol. 2002;52:423–428. doi: 10.1099/00207713-52-2-423. [DOI] [PubMed] [Google Scholar]
- King TS, Elia M, Hunter JO. Abnormal colonic fermentation in irritable bowel syndrome. Lancet. 1998;352:1187–1189. doi: 10.1016/s0140-6736(98)02146-1. [DOI] [PubMed] [Google Scholar]
- Pritchard SE, Marciani L, Garsed KC, et al. Fasting and postprandial volumes of the undisturbed colon: normal values and changes in diarrhea-predominant irritable bowel syndrome measured using serial MRI. Neurogastroenterol Motil. 2014;26:124–130. doi: 10.1111/nmo.12243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pimentel M, Gunsalus RP, Rao SSC, et al. Methanogens in human health and disease. Am J Gastroenterol Suppl. 2012;1:28–33. [Google Scholar]
- Triantafyllou K, Chang C, Pimentel M. Methanogens, methane and gastrointestinal motility. J Neurogastroenterol Motil. 2014;20:31–40. doi: 10.5056/jnm.2014.20.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Finegold SM, Song Y, et al. Reclassification of Clostridium coccoides, Ruminococcus hansenii, Ruminococcus hydrogenotrophicus, Ruminococcus luti, Ruminococcus productus and Ruminococcus schinkii as Blautia coccoides gen. nov., comb. nov., Blautia hansenii comb. nov., Blautia hydrogenotrophica comb. nov., Blautia luti comb. nov., Blautia producta comb. nov., Blautia schinkii comb. nov. and description of Blautia wexlerae sp. nov., isolated from human faeces. Int J Syst Evol Microbiol. 2008;58:1896–1902. doi: 10.1099/ijs.0.65208-0. [DOI] [PubMed] [Google Scholar]
- Tana C, Umesaki Y, Imaoka A, et al. Altered profiles of intestinal microbiota and organic acids may be the origin of symptoms in irritable bowel syndrome. Neurogastroenterol Motil. 2010;22:512–519. doi: 10.1111/j.1365-2982.2009.01427.x. [DOI] [PubMed] [Google Scholar]
- Treem WR, Ahsan N, Kastoff G, et al. Fecal short-chain fatty acids in patients with diarrhea-predominant irritable bowel syndrome: in vitro studies of carbohydrate fermentation. J Pediatr Gastroenterol Nutr. 1996;23:280–286. doi: 10.1097/00005176-199610000-00013. [DOI] [PubMed] [Google Scholar]
- Hamer HM, Jonkers D, Venema K, et al. Review article: the role of butyrate on colonic function. Aliment Pharmacol Ther. 2008;27:104–119. doi: 10.1111/j.1365-2036.2007.03562.x. [DOI] [PubMed] [Google Scholar]
- Vanhoutvin SALW, Troost FJ, Kilkens TOC, et al. The effects of butyrate enemas on visceral perception in healthy volunteers. Neurogastroenterol Motil. 2009;21:952–e76. doi: 10.1111/j.1365-2982.2009.01324.x. [DOI] [PubMed] [Google Scholar]
- Fritz E, Hammer HF, Lipp RW, et al. Effects of lactulose and polyethylene glycol on colonic transit. Aliment Pharmacol Ther. 2005;21:259–268. doi: 10.1111/j.1365-2036.2005.02244.x. [DOI] [PubMed] [Google Scholar]
- Mortensen PB, Andersen JR, Arffmann S, et al. Short-chain fatty acids and the irritable bowel syndrome: the effect of wheat bran. Scand J Gastroenterol. 1987;22:185–192. doi: 10.3109/00365528708991878. [DOI] [PubMed] [Google Scholar]
- Manichanh C, Eck A, Varela E, et al. Anal gas evacuation and colonic microbiota in patients with flatulence: effect of diet. Gut. 2014;63:401–408. doi: 10.1136/gutjnl-2012-303013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chumpitazi BP, Hollister EB, Oezguen N, et al. Gut microbiota influences low fermentable substrate diet efficacy in children with irritable bowel syndrome. Gut Microbes. 2014;5:165–175. doi: 10.4161/gmic.27923. [DOI] [PMC free article] [PubMed] [Google Scholar]