Abstract
The immunological significance of autophagy in the tumor microenvironment remains unclear. To explore the relationship between autophagy and anti-tumor immune responses, we investigated the expression of autophagy-related proteins and infiltration of immune cells using immunohistochemistry (IHC). The expression of three representative autophagy components, LC3, Beclin-1 and p62/SQSTM1, as well as the number of dendritic cells (DC), T cells and NK cells were examined by IHC in 74 patients with oral squamous cell carcinoma (OSCC). The relationship between the expression of autophagy-associated molecules and various clinicopathological parameters was also evaluated. The expression of both LC3 and p62/SQSTM1 in the peripheral site significantly correlated with an increase in the infiltration of T cells. Furthermore, the expression of p62/SQSTM1 and Beclin-1 correlated with that of HLA class I and class II in tumor cells, respectively. In addition, several unfavorable clinicopathological parameters correlated with an increase in the expression of LC3 in the peripheral site. The correlation observed between LC3 or p62/SQSTM1 and the infiltration of T cells suggests that autophagy may actively mobilize immune cells toward the cancer bed. Meanwhile, the three autophagy-associated proteins examined were linked to malignant potential and an unfavorable prognosis.
Keywords: Autophagy, immune infiltrates, LC3, oral squamous cell carcinoma, p62/SQSTM1
Oral squamous cell carcinoma is a fatal malignancy and the sixth most common cancer worldwide. The 5-year survival rate of oral or pharyngeal cancer is approximately 50%, with over 24 000 new patients being diagnosed and nearly 4000 deaths in the USA each year.1 Although its treatment mainly depends on surgery, the survival rate of OSCC has remained unchanged for decades.2 Novel therapeutic modalities and new/reliable biomarkers are urgently needed for the treatment of OSCC.3
Intracellular degradation systems in eukaryotes employ two major pathways: the ubiquitin–proteasome system to selectively reduce ubiquitinated proteins, and autophagic clearance in autophagosomes, which debulks the aggregates of proteins and organelles such as mitochondria by fusing with lysosomes. Autophagy is a prosurvival function that degrades bulk proteins and organelles to maintain energy homeostasis under cellular stress and nutrient starvation.4 The role of autophagy is like a “double-edged sword” in malignant tumors, having opposite functions on cancer cell survival. Recent studies revealed that autophagy exhibited suppressive effects on chronic inflammation, thereby eliciting carcinogenesis in the early phase, whereas it facilitated cancer cell survival under hypoxic and starved conditions in the advanced stage of the disease.5–7 The findings of previous studies in which clinical samples from cancer patients were immunohistochemically stained indicated that strong relationships may exist between the expression of autophagy-associated molecules and clinicopathological parameters. Most of these studies regarded the autophagy components LC3, Beclin-1 and p62/SQSTM1 as prognostic factors. LC3, which is essential for the formation of autophagosomes, has been identified as the most specific and reliable marker of autophagy.8–10 Beclin-1 also plays a crucial and promoting role in the autophagic pathway by binding several factors, while interactions between Beclin-1 and the Bcl-2 family have been shown to inhibit autophagy.11–13 p62/SQSTM1 has been implicated in various cell signaling pathways, including autophagy-mediated protein degradation by binding with LC3. The autophagy-induced accumulation of ubiquitinated proteins has been shown to retain p62/SQSTM1 in cells, resulting in an increase in selective autophagy.14,15
Although numerous immunohistochemical studies have investigated autophagic proteins in human samples, the immunological aspect of autophagy in cancer cells remains unclear. Autophagy-dependent cross-presentation by DC and possible anticancer vaccines with purified autophagosomes were reported previously by Hu's group.16–18 Michaud et al.19 recently reported “immunologic cell death” via an autophagic pathway after chemotherapy in mice with calreticulin exposure. These findings indicated that autophagy may be closely related to immunological phenomena in tumor-bearing animals; however, few studies have examined the immunological significance of these autophagy components in a human tumor microenvironment. The present study is the first to demonstrate the relationship between autophagy and the immunogenicity of human cancer tissues in vivo. The aim of the present study is to elucidate the relationship between the expressions of the three autophagy-related proteins: LC3, Beclin-1 and p62/SQSTM1, and HLA molecule expression/immune infiltrates using immunohistochemistry (IHC) in OSCC patients. We also examined the relationship between these autophagy components and clinicopathological manifestations.
Materials and Methods
Patients
Seventy-four OSCC samples resected surgically at Gunma University Hospital between November 2000 and January 2012 were analyzed. All samples were primary tongue cancer specimens. Patients who received preoperative neoadjuvant chemotherapy were excluded. All surgical specimens were classified according to the World Health Organization classification by a pathologist who was blind to the clinical findings, and were diagnosed as squamous cell carcinomas. Pathological TN stages were established using the International System for Staging adopted by the American Joint Committee on Cancer and the Union Internationale Contre le Cancer. Clinicopathological variables, including age, sex, the primary tumor (pT), nodal metastasis (pN), the TNM stage, histological grade and lymphatic/vascular invasion, as well as p53/Ki-67 staining, were also evaluated. This study was approved by the Institutional Review Board of Gunma University, and was performed in line with the Declaration of Helsinki of 1996.
Immunohistochemistry staining
Surgical specimens were fixed in 10% formaldehyde and routinely processed for paraffin embedding. Serial histological sections (5-μm thick) were deparaffinized in xylene and hydrated in descending dilutions of ethanol. Antigen retrieval was achieved by autoclaving at 121°C for 20 min in citrate buffer (pH 6.0). Endogenous peroxidase was blocked by 3% H2O2, and the sections were then covered with 1% BSA/5% normal horse serum for 30 min at room temperature. The slides were incubated at 4°C overnight with the primary antibodies (Table1), followed by Labelled Polymer-HRP anti-mouse/rabbit (Dako, Glostrup, Denmark) for 45 min at room temperature. The reaction products were detected by 3.3′-diaminobenzidine (DAB; Dako), and then counterstained by Mayer's hematoxylin. After being dehydrated by ascending dilutions of ethanol, the slides were mounted with the non-aqueous mounting medium, DPX (Merck, Darmstadt, Germany).
Table 1.
Antibody sources, clones, and dilutions
| Antibody | Clone | Dilution | Vendeor |
|---|---|---|---|
| LC3B | 5F10 | 2.5 μg/mL (1:40) | Nanotools; Teningen, Germany |
| Beclin-1 | EPR1733Y | 1:50 | Abcam; Cambridge, UK |
| p62/SQSTM1 | Polyclonal | 2 μg/mL | Sigma-Aldrich; St. Louis, MO, USA |
| CD1a | O10 | Ready-to-use | Beckman Coulter; Brea, CA, USA |
| CD3 | Polyclonal | Ready-to-use | Dako; Glostrup, Denmark |
| CD56 | 123.C3.D5 | Ready-to-use | Thermo Scientific; Waltham, MA, USA |
| HLA class I heavy chain | HC-10/HC-A2 | 0.5 μg/mL each | * |
| HLA class II | LGII-612.14 | 1 μg/mL | * |
| p53 | DO7 | 1:50 | Dako |
| Ki-67 | MIB-1 | 1:300 | Dianova; Hamburg, Germany |
| Negative control | 11711 | 2.5 μg/mL | R&D Systems; Minneapolis, MN, USA |
Kind gifts from Dr. Soldano Ferrone, Massachusetts General Hospital, Harvard Medical School, Boston, MA, USA.
Evaluation of immunohistochemistry samples
Slides were evaluated by two independent investigators (K. S. and H. T.) in a blinded fashion using a light microscope, a Zeiss Axioscope (Carl Zeiss microscopy GmbH, Jena, Germany). The immunoreactivities of the three autophagy-related proteins were evaluated according to the criteria described by Choi et al. and Miracco et al.20,21 Staining for LC3 and p62/SQSTM1 was assessed on the basis of the proportion of stained cells and immunostaining intensity. The proportion of cells was graded as 0 (negative), 1 (<30% positive) and 2 (>30% positive), and staining intensity was graded as 0 (negative), 1 (low density of puncta, with diffuse cytoplasmic staining), 2 (moderate density of puncta) and 3 (high density of puncta). The scores for the proportion of stained cells and staining intensity were multiplied to provide a total score: negative (0–1) or positive (2–6). Nerve cells are regarded as the internal positive control for LC3. Staining intensity for Beclin-1 in the cytoplasm was graded as 0 (negative), 1 (weak), 2 (moderate) and 3 (strong). The scores for the proportion of cells and staining intensity were multiplied to provide a total score: negative (0–4) or positive (5–6). More than five areas of a representative field were counted at ×200 magnification for CD1a+ DC and CD56+ NK cells. The infiltration of CD3+ T cells in more than five ×400 high power fields (HPF) was graded as grade 1 (<10 positive cells/HPF), grade 2 (10–30/HPF), grade 3 (31–100/HPF) and grade 4 (>101/HPF), which was consistent with a previous study.22 The final scores of the immune infiltrates were defined as an average of the number of stained cells in each field, while “CD3 positive” was defined as CD3 grade 2 and higher. HLA class I heavy chain and HLA class II were scored as (+), (+/−) and (−) when the percentages of stained tumor cells in an entire lesion were >75%, 25–75% and <25%, respectively, according to the criteria established by the HLA and Cancer Component of the 12th International Histocompatibility Workshop.23 Two regions, the central and peripheral sites, were evaluated separately in each tumor tissue slide. The peripheral site was defined as a field that included cancer cells and adjacent non-cancerous cells at a magnification of ×200. When a tumor was too small to divide into the central and peripheral areas, the tumor was only evaluated as the peripheral site.
Based on previous studies,24,25 the expression of p53 in more than 10% of tumor cells was defined as positive expression. A highly cellular area of the stained sections was evaluated for ki-67. Approximately 1000 nuclei were counted on each slide, and proliferative activity was assessed as the percentage of Ki-67-stained nuclei (Ki-67 labeling index) in the sample.
Statistical analysis
Data were analyzed using the Statistical Package for Social Science version 22.0 (SPSS; IBM, Armonk, NY, USA) and Statcel 3 (OMS Publishing, Saitama, Japan). Mann–Whitney's U-test, the Kruskal–Wallis test, the chi-square-test for independence and Fisher's exact test were used to examine differences in continuous and categorical variables, respectively. Significance was defined when P < 0.05. Kaplan–Meier survival curves and log-rank statistics were used to evaluate disease-free survival and overall survival. Univariate/multivariate regression analyses were performed using the Cox proportional hazards model.
Results
Patient characteristics
The clinicopathological characteristics of all 74 patients are summarized in Table2. Postoperative adjuvant chemotherapy with the oral administration of S-1 (Taiho Pharmaceutical, Tokyo, Japan), tegafur or docetaxel was given to 9, 10 and 3 patients, respectively. The median follow-up duration was 915 days (range, 85–3452 days).
Table 2.
Clinicopathological features of patients
| Parameters | n = 74 (%) |
|---|---|
| Age | 33–92, median: 69 |
| Sex | |
| Male | 27 (36.5) |
| Female | 47 (63.5) |
| TNM | |
| T1/T2 | 63 (85.1) |
| T3/T4 | 11 (14.9) |
| N0 | 53 (71.6) |
| N+ | 21 (28.4) |
| M0 | 73 (98.6) |
| M1 | 1 (1.4) |
| Stage | |
| I/II | 47 (63.5) |
| III/IV | 27 (36.5) |
| Differentiation | |
| Well/moderate | 66 (89.2) |
| Poorly | 8 (10.8) |
| Invasion | |
| Lymphatic | 35 (47.3) |
| Vascular | 25 (33.8) |
| Adjuvant | |
| Chemotherapy | 22 (29.7) |
| Radiation +/− chemotherapy | 29 (39.2) |
| Recurrence | 27 (36.5) |
| Death | 22 (29.7) |
Expression of autophagy-associated proteins
Figure1(a) shows representative IHC staining of LC3, Beclin-1 and p62/SQSTM1, respectively, according to the scores for the staining intensity. LC3 and p62/SQSTM1 were expressed in puncta, corresponding to autophagosomes in the cytoplasm.26 Beclin-1 was mainly expressed in the cytoplasm. LC3, Beclin-1 and p62/SQSTM1 accumulated in 27 (36.5%), 27 (36.5%) and 24 (32.4%) of the 74 cases of OCCC examined, respectively. The detailed basic data of expressions of autophagy-related proteins are summarized in Table3. No significant differences were observed in the peripheral or central expression of these autophagy components.
Figure 1.
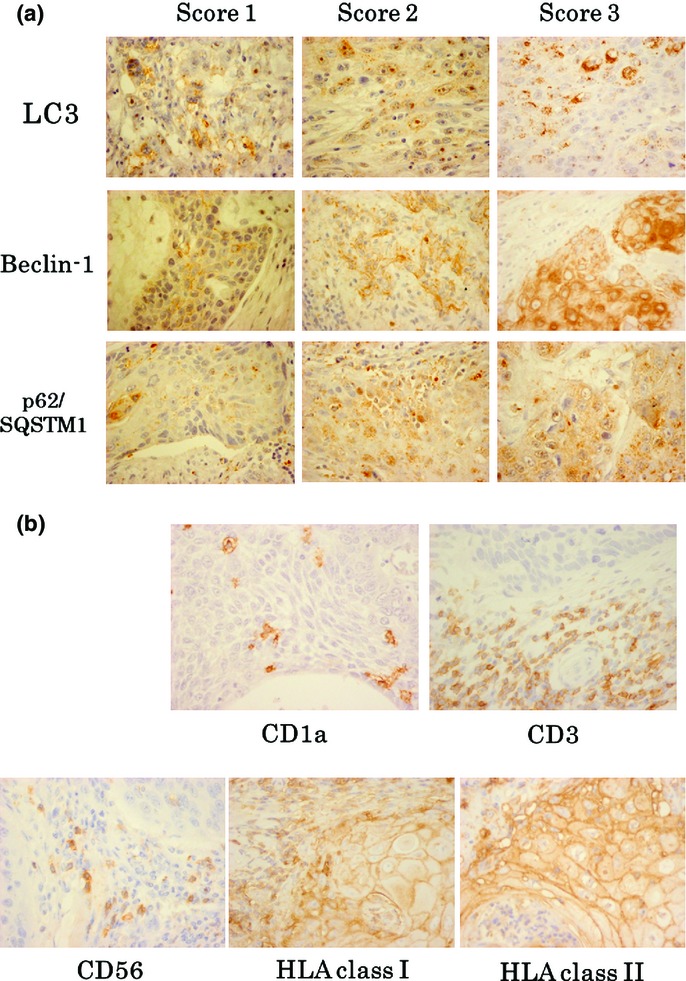
(a) Representative immunohistochemistry (IHC) staining of LC3, Beclin-1 and p62 (×400 magnification), according to the intensity scores. All pictures were taken under the same photographic condition. LC3 and p62 SQSTM1 were expressed in puncta corresponding to autophagosomes in the cytoplasm. Beclin-1 was mainly expressed in the cytoplasm. (b) Representative CD1a+ DC, CD3+ T cell, CD56+ NK cell, HLA class I heavy chain and class II stainings were presented under magnification of ×400. HLA class I heavy chain and class II were presented as a membranous pattern.
Table 3.
Expressions of autophagy-related proteins by immunohistochemistry
| Scores | Central site Number of cases (%) | Peripheral site Number of cases (%) |
|---|---|---|
| LC3 | ||
| 0 | 30 (46.9) | 33 (44.6) |
| 1 | 10 (15.6) | 14 (18.9) |
| 2 | 11 (17.2) | 11 (14.9) |
| 3 | 7 (10.9) | 5 (6.8) |
| 4 | 3 (4.7) | 5 (6.8) |
| 6 | 3 (4.7) | 6 (8.1) |
| Beclin-1 | ||
| 0 | 8 (13.3) | 13 (17.6) |
| 1 | 4 (6.7) | 5 (6.8) |
| 2 | 7 (11.7) | 17 (23.0) |
| 3 | 6 (10.0) | 2 (2.7) |
| 4 | 6 (10.0) | 11 (14.9) |
| 6 | 29 (48.3) | 26 (35.1) |
| p62/SQSTM1 | ||
| 0 | 28 (47.5) | 39 (52.7) |
| 1 | 8 (13.6) | 11 (14.9) |
| 2 | 18 (30.5) | 18 (24.3) |
| 3 | 1 (1.7) | 3 (4.1) |
| 4 | 3 (5.1) | 3 (4.1) |
| 6 | 1 (1.7) | 0 (0.0) |
Immunohistochemistry staining of tumor-infiltrating immune cells and HLA molecules
Representative CD1a+ DC, CD3+ T cell and CD56+ NK cell staining is shown in Figure1(b). These cells accumulated not only in the peritumoral “peripheral” site, but also in the tumor epithelial “central” site. The median values of these numbers/scores were: CD1a central, 16.4; CD1a peripheral, 20.5; CD3 central, 2.2; CD3 peripheral 3.0; CD56 central, 1.3; and CD56 peripheral, 6.6. The infiltration of CD3+ T cells was significantly greater in the peripheral site than in the central site of the tumor (P = 0.00560).
Figure1(b) also shows representative IHC staining of HLA class I heavy chain and HLA class II. These antigens presented a membranous pattern in IHC staining. HLA class I was expressed in 47.1% (+ and +/−) and class II was 23.7% (+ and +/−) of cells in the central site. In the peripheral site, HLA class I was expressed in 60.8% (+ and +/−), and class II was expressed in 35.1% (+ and +/−) of cells, which is consistent with previous findings.27,28 No correlations were observed between the expression of HLA molecules and the number of tumor-infiltrating immune cells, whereas the peripheral expression of HLA class II correlated with an increase in the accumulation of CD3+ T cells (P = 0.0480).
Relationship between autophagy components and immune infiltrates
Table4 shows the relationships between the autophagy-related proteins LC3, Beclin-1 and p62/SQSTM1, and the immune infiltrates DC, T cells, NK cells and HLA molecules in the central and peripheral sites of cancer tissue (by Mann–Whitney's U-test and the Kruskal–Wallis test). Table5 describes the detailed IHC data of the statistically significant parameters. The peripheral expression of LC3 was correlated with the number of CD3+ T cells in the peripheral site (Fig.2a), but not in the central site of tumor, in which hypoxia and nutrient starvation was apparent. The expression of p62/SQSTM1 in the peripheral site was also related to the peripheral infiltration of CD+ T cells (Fig.2a). Strong correlations were observed between the expression of p62/SQSTM1 and HLA class I (Fig.2b), as well as HLA class II and Beclin-1 (Fig.2c). As described above, the expression of the three autophagy components appeared to be associated with immunologically favorable phenomena, especially in the peripheral site of the tumor.
Table 4.
Relationship between autophagy components and immunological parameters
| Expressions | P-value | ||
|---|---|---|---|
| Central site of the tumor | LC3 expression | Versus CD1a+ infiltration | 0.625 |
| Versus CD3+ infiltration | 0.508 | ||
| Versus CD56+ infiltration | 0.875 | ||
| Versus HLA class I expression | 0.142 | ||
| Versus HLA class II expression | 0.676 | ||
| Beclin-1 expression | Versus CD1a+ infiltration | 0.646 | |
| Versus CD3+ infiltration | 0.357 | ||
| Versus CD56+ infiltration | 0.744 | ||
| Versus HLA class I expression | 0.792 | ||
| Versus HLA class II expression | 0.046 | ||
| p62/SQSTM1 expression | Versus CD1a+ infiltration | 0.221 | |
| Versus CD3+ infiltration | 0.104 | ||
| Versus CD56+ infiltration | 0.363 | ||
| Versus HLA class I expression | 0.013 | ||
| Versus HLA class II expression | 0.694 | ||
| Peripheral site of the tumor | LC3 expression | Versus CD1a+ infiltration | 0.129 |
| Versus CD3+ infiltration | 0.035 | ||
| Versus CD56+ infiltration | 0.145 | ||
| Versus HLA class I expression | 0.343 | ||
| Versus HLA class II expression | 0.115 | ||
| Beclin-1 expression | Versus CD1a+ infiltration | 0.185 | |
| Versus CD3+ infiltration | 0.165 | ||
| Versus CD56+ infiltration | 0.689 | ||
| Versus HLA class I expression | 0.925 | ||
| Versus HLA class II expression | 0.036 | ||
| p62/SQSTM1 expression | Versus CD1a+ infiltration | 0.259 | |
| Versus CD3+ infiltration | 0.008 | ||
| Versus CD56+ infiltration | 0.842 | ||
| Versus HLA class I expression | 0.003 | ||
| Versus HLA class II expression | 0.745 | ||
The bold values mean statistically significant (p<0.05).
Table 5.
Statistically significant data of the expressions of autophagic proteins and immunological parameters
| Expressions | P-value | |||||
|---|---|---|---|---|---|---|
| Central site of the tumor | HLA class II |
|||||
| (−) | (+/−) | (+) | ||||
| Beclin-1 | Negative | 26 | 2 | 2 | 0.0462 | |
| Positive | 15 | 8 | 2 | |||
| HLA class I | ||||||
| (−) | (+/−) | (+) | ||||
| p62/SQSTM1 | Negative | 22 | 11 | 1 | 0.0134 | |
| Positive | 7 | 11 | 5 | |||
| Peripheral site of the tumor | CD3 score |
|||||
| Number of cases | Median | SD | ||||
| LC3 | Negative | 46 | 2.8 | 0.66 | 0.0345 | |
| Positive | 27 | 3.2 | 0.69 | |||
| HLA class II |
||||||
| (−) | (+/−) | (+) | ||||
| Beclin-1 | Negative | 35 | 5 | 7 | 0.0362 | |
| Positive | 13 | 9 | 5 | |||
| CD3 score |
||||||
| Number of cases | Median | SD | ||||
| p62/SQSTM1 | Negative | 47 | 2.8 | 0.71 | 0.0084 | |
| Positive | 27 | 3.2 | 0.57 | |||
| HLA class I |
||||||
| (−) | (+/−) | (+) | ||||
| p62/SQSTM1 | Negative | 24 | 17 | 6 | 0.0029 | |
| Positive | 5 | 10 | 12 | |||
Figure 2.
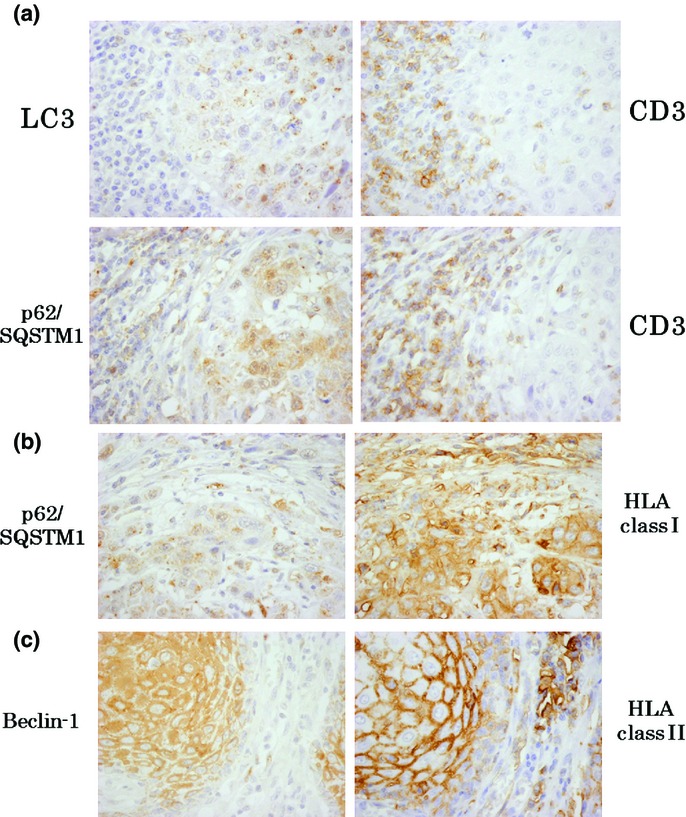
Representative serial immunohistochemistry (IHC) pictures of the expressions of molecules which showed statistically significant association each other. (a) LC3 and p62/SQSTM1 significantly correlated with CD3+ T cell infiltration. (b) p62/SQSTM1 was closely related to the expression of HLA class I. (c) Beclin-1 was significantly related to HLA class II expression.
Relationship between autophagy-related proteins and clinicopathological features
We also investigated the relationship between the expression of autophagy components and clinicopathological parameters using the chi-square-test for independence (Table6). Lymphatic invasion by tumors was positively associated with the expression of LC3 and Beclin-1, whereas vascular invasion was closely related to the expression of p62/SQSTM1. The peritumoral expression of LC3 was also positively correlated with the clinical stage, T-factor and Ki-67 labeling index. Well/moderately differentiated tumor tissue was more closely associated with the positive expression of Beclin-1 and p62/SQSTM1 than poorly differentiated tumor tissue. These results indicated that the expression of autophagy-related proteins appeared to be related to unfavorable prognostic factors.
Table 6.
Relationship between autophagy components and clinicopathological features
| LC3 P-value | Beclin-1 P-value | p62/SQSTM1 P-value | |
|---|---|---|---|
| Central site | |||
| Differentiation | 0.896 | 0.137 | 0.241 |
| Lymphatic invasion | 0.0214 | 0.00342 | 0.119 |
| Vascular invasion | 0.189 | 0.106 | 0.0116 |
| Nodal status | 0.359 | 0.245 | 0.285 |
| Stage | 0.0309 | 0.0815 | 0.0476 |
| T-factor | 0.0549 | 0.878 | 0.433 |
| Recurrence | 0.315 | 0.0815 | 0.223 |
| Ki-67 | 0.164 | 0.933 | 0.642 |
| p53 | 0.190 | 0.326 | 0.378 |
| Peripheral site | |||
| Differentiation | 0.503 | 0.00644 | 0.0426 |
| Lymphatic invasion | 0.00258 | 0.0682 | 0.118 |
| Vascular invasion | 0.142 | 0.567 | 0.0127 |
| Nodal status | 0.271 | 0.614 | 0.590 |
| Stage | 0.00980 | 0.0534 | 0.281 |
| T-factor | 0.00680 | 0.172 | 0.503 |
| Recurrence | 0.564 | 0.353 | 0.281 |
| Ki-67 | 0.0187 | 0.583 | 0.166 |
| p53 | 0.210 | 0.465 | 0.210 |
The bold values mean statistically significant (p<0.05).
Survival and univariate/multivariate regression analyses
Overall/progression-free survival rates were compared between the positive and negative expression of LC3, Beclin1 and p62/SQSTM1 in the central and peripheral areas. No significant differences were observed in survival by the log-rank test, except for shorter overall survival with the positive expression of LC3 in the peripheral area (P = 0.030), as shown in Figure3. We also examined the relationship between overall/progression-free survival rates and the number (divided into higher/lower groups by median) of CD1a+ dendritic cells, CD3+ T cells or CD56+ NK cells, as well as the expression of HLA class I/class II by the log-rank test; however, no correlations were detected among these parameters. Univariate regression analysis revealed that lymphatic invasion, vascular invasion, lymph node metastasis, an advanced clinical stage, recurrence and the peripheral expression of LC3 correlated with overall survival, while lymphatic invasion, lymph node metastasis, recurrence and a high Ki-67 labeling index correlated with progression-free survival, as indicated in Table7. Multivariate regression analysis revealed that vascular invasion, recurrence and the peripheral expression of LC3 correlated with overall survival, but not progression-free survival (Table7).
Figure 3.
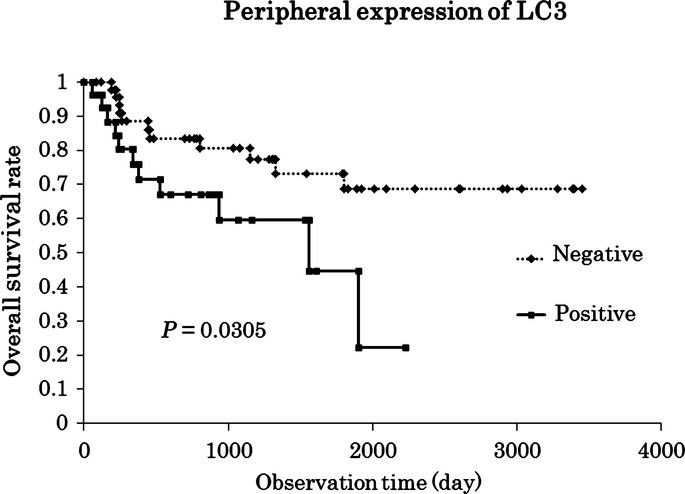
The log-rank test revealed that the positive expression of LC3 in the peripheral site of the tumor led to a significantly shorter survival. Multivariate regression analysis also identified the peripheral expression of LC3 as an independent prognostic factor (P = 0.034).
Table 7.
Univariate and multivariate regression analyses of survival
| Different variables | Overall survival Univariate P-value | Multivariate P-value | Progression free survival Univariate P-value | Multivariate P-value |
|---|---|---|---|---|
| Poorly differentiated | 0.092 | 0.053 | ||
| Lymphatic invasion | 0.008 | 0.807 | 0.013 | 0.306 |
| Vascular invasion | 0.003 | 0.028 | 0.100 | |
| Lymph node metastasis | 0.049 | 0.585 | 0.005 | 0.073 |
| Positive surgical margin | 0.553 | 0.568 | ||
| Advanced clinical stage | 0.018 | 0.592 | 0.142 | |
| Large T-factor | 0.115 | 0.842 | ||
| Recurrence | <0.001 | 0.002 | <0.001 | 0.875 |
| High Ki-67 labelling index | 0.065 | 0.012 | 0.157 | |
| p53 positive | 0.163 | 0.817 | ||
| Expression in the peripheral site | ||||
| LC3 | 0.034 | 0.030 | 0.517 | |
| Beclin-1 | 0.919 | 0.708 | ||
| p62/SQSTM1 | 0.901 | 0.160 | ||
| CD1a+ | 1.000 | 1.000 | ||
| CD3+ | 0.856 | 0.981 | ||
| CD56+ | 1.000 | 1.000 | ||
| HLA class I | 0.778 | 0.894 | ||
| HLA class II | 0.901 | 0.683 | ||
The bold values mean statistically significant (p<0.05).
Discussion
Many studies have examined the immunohistochemical expression of autophagy components such as LC3, Beclin-1 and p62/SQSTM1 in various malignancies,20,21,29–31 including head and neck cancer.32–36 However, the findings reported are controversial: Tang et al., Choi et al., Chen et al., Guo et al. and Fujii et al. showed that the strong expression of autophagy-related proteins, as confirmed by immunohistochemistry, correlated with unfavorable clinicopathological parameters, which is consistent with the results of the present study;20,30–32,36 however, other studies report a contrasting conclusion.21,33,35 Even among head and neck cancer patients, interpretations of the relationship between autophagy and prognosis were diametrically opposite.32–36 These discrepancies may be attributed to investigators evaluating positive signals in tumor tissues, for example, in the peripheral or central area. Because most of these studies did not describe which part they evaluated in tumor tissue, variations in the findings reported were expected. In principle, malignant tumors are structurally heterogeneous, especially between the central and peripheral sites. Only one study by Fujii et al. compared the expression of the autophagy marker LC3 between the central and peripheral areas in pancreatic cancer. They report that the expression of LC3 in the peripheral area not only correlated with a poor prognosis and short disease-free period, but also corresponded to the expression of the hypoxia marker carbonic anhydrase IX.31 More oxygen and nutrition may be required in the peritumoral invasive margin than in the central site of the tumor. In the present study, no significant differences were observed in the expression of any of the autophagy-related proteins examined between the central and peripheral sites, and most of our important positive data were obtained from the peripheral site. For example, the infiltration/expression of all immune cells and HLA molecules was slightly higher in the peripheral site than in the central site (only that of CD3+ T cells was significant). A positive correlation between the expression of LC3 or p62/SQSTM1 and infiltration CD3+ T cells was only detected in the peripheral site. Moreover, the expression of LC3 in the peripheral site correlated with unfavorable prognostic factors, such as an advanced clinical stage, greater tumor size, high Ki-67 labeling index and shorter overall survival, and was identified as an independent prognostic factor by multivariate regression analysis. Thus, these results indicated that autophagy in the peripheral, but not central, site of tumor tissue reflected malignant potential and poor prognosis. The following paragraph focused on data obtained from the peripheral site.
To date, a few reports have implied that autophagy in tumor cells was closely related to tumor antigen presentation and tumor-specific cytotoxic T cell (CTL) induction in in vitro and animal experiments. Demachi-Okamura et al.37 reveal that autophagy in cancer cells contributed to MHC class I processing and presentation. Moreover, according to Hu and colleagues, autophagosomes containing tumor antigens derived from cancer cells were recognized by DC to induce cross-presentation. This autophagy-dependent cross-presentation of tumor antigens by DC suggested possible tumor-derived autophagosome vaccines to drive T cell proliferation.16,17 Based on these findings, we designed the present study to confirm that notion in an in vivo setting. In fact, this study is the first to elucidate the relationship between autophagy and immunological phenomena in the tumor microenvironment in cancer patients. Our novel in vivo findings by immunohistochemistry suggest that autophagy in tumor cells may induce not only an increase in tumor cell antigenicity, but also anti-tumor T cell responses, supporting the previous in vitro and animal studies, while our present study did not range over functional activities of autophagy components. We detected a positive correlation between the expression of p62/SQSTM1 and HLA class I, which was essential for the presentation of antigens from antigen-presenting cells, such as DC, to CD8+ CTL. A correlation was also observed between the expression of Beclin-1 and HLA class II binding to CD4+ helper T cells. We revealed that an increase in the expression of LC3 and LC3-binding protein p62/SQSTM1 correlated with the number of CD3+ T cells around the tumor. Therefore, autophagy appeared to induce the expression of HLA molecules, followed by the recognition of cancer cells by tumor-infiltrating lymphocytes (TIL) in the tumor microenvironment of cancer patients. Moreover, in vivo immunohistochemical findings as well as the present results revealed a positive correlation between the expression of HLA class I and tumor-infiltrating CTL38,39 or between the expression of HLA class II and TIL.40,41 Therefore, our results suggested a direct relationship between autophagy in cancer cells and the accumulation of T cells due to an increase in the expression of HLA molecules in patients with OSCC.
The accumulation of T cells in the peritumoral area was previously correlated with a good prognosis.42,43 Thus, the mobilization of T cells by autophagy in cancer cells, as shown in our present study, may have a favorable impact on the survival of cancer cells. Meanwhile, in our study, the expression of autophagy-related proteins correlated with unfavorable prognostic factors such as lymphatic/vascular invasion, an advanced clinical stage and shorter overall survival. Although we did not obtain any direct evidence to explain this contradiction in autophagic functions between T cell induction and poor clinicopathological representations, some pathogeneses can be speculated as follows. TIL in OSCC are known to exhibit signaling abnormalities, apoptosis and reduced proliferation.22 In the present study, autophagy components appeared to increase the peritumoral infiltration of T cells; however, the functions of these T cells may be defective, resulting in a poor prognosis. Moreover, the subpopulation of CD3+ T cells infiltrating around the tumor may also be important: these CD3+ T cells may contain regulatory T cells (Tregs). Several studies report that Tregs were enriched in the TIL of head and neck cancer patients and also that they secreted immunologically inhibitory cytokines such as interleukin (IL)-10 and transforming growth factor-beta1 (TGF-β1).44,45 Therefore, qualitative deteriorations and variations in subpopulations of CD3+ T cells mobilized to the cancer bed may have caused discrepancies in autophagic functions in the tumor microenvironment.
In summary, the expression of autophagy components in the peripheral invasive margin of cancer tissue was found to reflect clinicopathological features. We elucidated the relationship between autophagy and immunological phenomena in the tumor microenvironment in cancer patients for the first time. The correlation observed between LC3 or p62/SQSTM1 and T cell infiltration suggests that autophagy may actively mobilize immune cells toward the cancer bed. Furthermore, autophagy components may contribute to increases in tumor antigenicity, whereas these three autophagy-associated proteins may also be related to malignant potential and an unfavorable prognosis. Further studies are required to elucidate the functional roles of these components in anti-tumor immunity.
Acknowledgments
This work was supported in part by Grants-in-Aid for Young Scientists (24791820 to K.S. and 25861525 to M. T.) and a Grant-in-Aid for Scientific Research (C) (26670736 to K.C.) from the Ministry of Education, Culture, Sports, Science and Technology, Japan. We thank Dr Soldano Ferrone for providing monoclonal antibodies and Dr Toshiaki Hikino for his technical assistance with immunohistochemical staining.
Disclosure Statement
The authors have no conflict of interest to declare.
References
- Siegel R, Naishandham D, Jemal A. Cancer statistics, 2012. CA Cancer J Clin. 2012;62:10–29. doi: 10.3322/caac.20138. [DOI] [PubMed] [Google Scholar]
- Shah JP, Gil Z. Current concepts in management of oral cancer surgery. Oral Oncol. 2009;45:394–401. doi: 10.1016/j.oraloncology.2008.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bello IO, Soini Y, Salo T. Prognostic evaluation of oral tongue cancer: means, markers and prospectives (I) Oral Oncol. 2010;46:630–5. doi: 10.1016/j.oraloncology.2010.06.006. [DOI] [PubMed] [Google Scholar]
- Mizushima N, Komatsu M. Autophagy: renovation of cells and tissues. Cell. 2011;147:728–41. doi: 10.1016/j.cell.2011.10.026. [DOI] [PubMed] [Google Scholar]
- Mathew R, Karp CM, Beaudoin B, et al. Autophagy suppresses tumorigenesis through elimination of p62. Cell. 2009;137:1062–75. doi: 10.1016/j.cell.2009.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degenhardt K, Mathew R, Beaudoin B, et al. Autophagy promotes tumor cell survival and restricts necrosis, inflammation, and tumorigenesis. Cancer Cell. 2006;10:51–64. doi: 10.1016/j.ccr.2006.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White E. Deconvoluting the context-dependent role for autophagy in cancer. Nat Rev Cancer. 2012;12:401–10. doi: 10.1038/nrc3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N, Yamamoto A, Matsui M, et al. In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol Biol Cell. 2004;15:1101–11. doi: 10.1091/mbc.E03-09-0704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky DJ, Cuervo AM, Seglen PO. Methods for monitoring autophagy from yeast to human. Autophagy. 2007;3:181–206. doi: 10.4161/auto.3678. [DOI] [PubMed] [Google Scholar]
- Mizushima N, Yoshimori T, Levine B. Methods in mammalian autophagy research. Cell. 2010;140:313–26. doi: 10.1016/j.cell.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang XH, Jackson S, Seaman M, et al. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature. 1999;402:672–6. doi: 10.1038/45257. [DOI] [PubMed] [Google Scholar]
- Pattingre S, Tassa A, Qu X, et al. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell. 2005;122:927–39. doi: 10.1016/j.cell.2005.07.002. [DOI] [PubMed] [Google Scholar]
- Zhou F, Yang Y, Xing D. Bcl-2 and Bcl-xL play important roles in the crosstalk between autophagy and apoptosis. FEBS J. 2011;278:403–13. doi: 10.1111/j.1742-4658.2010.07965.x. [DOI] [PubMed] [Google Scholar]
- Komatsu M, Waguri S, Koike M, et al. Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell. 2007;131:1149–63. doi: 10.1016/j.cell.2007.10.035. [DOI] [PubMed] [Google Scholar]
- Komatsu M, Kageyama S, Ichimura Y. p62/SQSTM1/A170: physiology and pathology. Pharmacol Res. 2012;66:457–62. doi: 10.1016/j.phrs.2012.07.004. [DOI] [PubMed] [Google Scholar]
- Li Y, Wang LX, Yang G, et al. Efficient cross-presentation depends on autophagy in tumor cells. Cancer Res. 2008;68:6889–95. doi: 10.1158/0008-5472.CAN-08-0161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Wang LX, Pang P, et al. Tumor-derived autophagosome vaccine: mechanism of cross-presentation and therapeutic efficacy. Clin Cancer Res. 2011;17:7047–57. doi: 10.1158/1078-0432.CCR-11-0951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twitty CG, Jensen SM, Hu HM, et al. Tumor-derived autophagosome vaccine: induction of cross-protective immune responses against short-lived proteins through a p62-dependent mechanism. Clin Cancer Res. 2011;17:6467–81. doi: 10.1158/1078-0432.CCR-11-0812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaud M, Martins I, Sukkurwala AQ, et al. Autophagy-dependent anticancer immune responses induced by chemotherapeutic agents in mice. Science. 2011;334:1573–7. doi: 10.1126/science.1208347. [DOI] [PubMed] [Google Scholar]
- Choi J, Jung W, Koo JS. Expression of autophagy-related markers beclin-1, light chain 3A, light chain 3B and p62 according to the molecular subtype of breast cancer. Histopathology. 2013;62:275–86. doi: 10.1111/his.12002. [DOI] [PubMed] [Google Scholar]
- Miracco C, Cevenini G, Franchi A, et al. Beclin 1 and LC3 autophagic gene expression in cutaneous melanocytic lesions. Hum Pathol. 2010;41:503–12. doi: 10.1016/j.humpath.2009.09.004. [DOI] [PubMed] [Google Scholar]
- Reichert TE, Strauss L, Wagner EM, et al. Signaling abnormalities, apoptosis, and reduced proliferation of circulating and tumor-infiltrating lymphocytes in patients with oral carcinoma. Clin Cancer Res. 2002;8:3137–45. [PubMed] [Google Scholar]
- Garrido F, Cabrera T, Accolla RS. HLA and cancer. In: Charron D, et al., editors. HLA. Paris: EDK; 1997. pp. 445–52. [Google Scholar]
- Kaira K, Sunose Y, Arakawa K, et al. Prognostic significance of L-type amino-acid transporter 1 expression in surgically resected pancreatic cancer. Br J Cancer. 2012;107:632–8. doi: 10.1038/bjc.2012.310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck AC, Schirrmeister HH, Guhlmann CA, et al. Ki-67 immunostaining in pancreatic cancer and chronic active pancreatitis: does in vivo FDG uptake correlate with proliferative activity? J Nucl Med. 2001;42:721–5. [PubMed] [Google Scholar]
- Ladoire S, Chaba K, Martins I, et al. Immunohistochemical detection of cytoplasmic LC3 puncta in human cancer specimens. Autophagy. 2012;8:1175–84. doi: 10.4161/auto.20353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogino T, Bandoh N, Hayashi T, et al. Association of tapasin and HLA class I antigen down-regulation in primary maxillary sinus squamous cell carcinoma lesions with reduced survival of patients. Clin Cancer Res. 2003;9:4043–51. [PubMed] [Google Scholar]
- Meissner M, Whiteside TL, van Kuik-Romein P, et al. Loss of interferon-gamma inducibility of the MHC class II antigen processing pathway in head and neck cancer: evidence for post-transcriptional as well as epigenetic regulation. Br J Dermatol. 2008;158:930–40. doi: 10.1111/j.1365-2133.2008.08465.x. [DOI] [PubMed] [Google Scholar]
- Baspinar S, Bircan S, Yavuz G, et al. Beclin 1 and bcl-2 expressions in bladder urothelial tumors and their association with clinicopathological parameters. Pathol Res Pract. 2013;209:418–23. doi: 10.1016/j.prp.2013.04.006. [DOI] [PubMed] [Google Scholar]
- Guo GF, Jiang WQ, Zhang B, et al. Autophagy-related proteins Beclin-1 and LC3 predict cetuximab efficacy in advanced colorectal cancer. World J Gastroenterol. 2011;17:4779–86. doi: 10.3748/wjg.v17.i43.4779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii S, Mitsunaga S, Yamazaki M, et al. Autophagy is activated in pancreatic cancer cells and correlates with poor patient outcome. Cancer Sci. 2008;99:1813–9. doi: 10.1111/j.1349-7006.2008.00893.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang JY, Hsi E, Huang YC, et al. High LC3 expression correlates with poor survival in patients with oral squamous cell carcinoma. Hum Pathol. 2013;44:2558–62. doi: 10.1016/j.humpath.2013.06.017. [DOI] [PubMed] [Google Scholar]
- Wang Y, Wang C, Tang H, et al. Decrease of autophagy activity promotes malignant progression of tongue squamous cell carcinoma. J Oral Pathol Med. 2013;42:557–64. doi: 10.1111/jop.12049. [DOI] [PubMed] [Google Scholar]
- Huang L, Wang S, Li SS, et al. Prognostic significance of Beclin-1 expression in laryngeal squamous cell carcinoma. Pathol Oncol Res. 2013;19:771–7. doi: 10.1007/s12253-013-9642-0. [DOI] [PubMed] [Google Scholar]
- Jiang L, Huang S, Li W, et al. Expression of autophagy and ER stress-related proteins in primary salivary adenoid cystic carcinoma. Pathol Res Pract. 2012;208:635–41. doi: 10.1016/j.prp.2012.07.008. [DOI] [PubMed] [Google Scholar]
- Chen Y, Li X, Wu X, et al. Autophagy-related proteins LC3 and Beclin-1 impact the efficacy of chemoradiation on esophageal squamous cell carcinoma. Pathol Res Pract. 2013;209:562–7. doi: 10.1016/j.prp.2013.06.006. [DOI] [PubMed] [Google Scholar]
- Demachi-Okamura A, Torikai H, Akatsuka Y, et al. Autophagy creates a CTL epitope that mimics tumor-associated antigens. PLoS ONE. 2012;7:e47126. doi: 10.1371/journal.pone.0047126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogino T, Shigyo H, Ishii H, et al. HLA class I antigen down-regulation in primary laryngeal squamous cell carcinoma lesions as a poor prognostic marker. Cancer Res. 2006;66:9281–9. doi: 10.1158/0008-5472.CAN-06-0488. [DOI] [PubMed] [Google Scholar]
- Tsuchikawa T, Ikeda H, Cho Y, et al. Association of CD8+ T cell infiltration in oesophageal carcinoma lesions with human leucocyte antigen (HLA) class I antigen expression and survival. Clin Exp Immunol. 2011;164:50–6. doi: 10.1111/j.1365-2249.2010.04311.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leffers N, Fehrmann RS, Gooden MJ, et al. Identification of genes and pathways associated with cytotoxic T lymphocyte infiltration of serous ovarian cancer. Br J Cancer. 2010;103:685–92. doi: 10.1038/sj.bjc.6605820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaida MM, Welsch T, Herpel E, et al. MHC class II expression in pancreatic tumors: a link to intratumoral inflammation. Virchows Arch. 2012;460:47–60. doi: 10.1007/s00428-011-1175-x. [DOI] [PubMed] [Google Scholar]
- Mei Z, Liu Y, Liu C, et al. Tumour-infiltrating inflammation and prognosis in colorectal cancer: systematic review and meta-analysis. Br J Cancer. 2014;110:1595–605. doi: 10.1038/bjc.2014.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung AC, Guihard S, Krugell S, et al. CD8-alpha T-cell infiltration in human papillomavirus-related oropharyngeal carcinoma correlates with improved patient prognosis. Int J Cancer. 2013;132:E26–36. doi: 10.1002/ijc.27776. [DOI] [PubMed] [Google Scholar]
- Albers AE, Ferris RL, Kim GG, et al. Immune responses to p53 in patients with cancer: enrichment in tetramer+ p53 peptide-specific T cells and regulatory T cells at tumor sites. Cancer Immunol Immunother. 2005;54:1072–81. doi: 10.1007/s00262-005-0670-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauss L, Bergmann C, Szczepanski M, et al. A unique subset of CD4+ CD25highFoxp3+ T cells secreting interleukin-10 and transforming growth factor-beta1 mediates suppression in the tumor microenvironment. Clin Cancer Res. 2007;13:4345–54. doi: 10.1158/1078-0432.CCR-07-0472. [DOI] [PubMed] [Google Scholar]


