Abstract
Background
Corticosteroids exert their anti-inflammatory action by binding and activating the intracellular the glucocorticoid receptor (GR) hetero-complex.
Objective
Evaluate the genes HSPCB, HSPCA, STIP1, HSPA8, DNAJB1, PTGES3, FKBP5, and FKBP4 on corticosteroid response.
Methods
Caucasian asthmatics (382) randomized to once daily flunisolide or conventional inhaled corticosteroid therapy were genotyped. Outcome measures were baseline FEV1, % predicted FEV1, and % change in FEV1 after corticosteroid treatment. Multivariable analyses adjusted for age, gender, and height, were performed fitting the most appropriate genetic model based on quantitative mean derived from ANOVA models to determine if there was an independent effect of polymorphisms on change in FEV1 independent of baseline level.
Results
Positive recessive model correlations for STIP1 SNPs were observed for baseline FEV1 [rs4980524, p=0.009; rs6591838, p=0.0045; rs2236647, p=0.002; and rs2236648; p=0.013], baseline % predicted FEV1 [rs4980524, p=0.002; rs6591838, p=0.017; rs2236647, p=0.003; and rs2236648; p=0.008] ; % change in FEV1 at 4 weeks [rs4980524, p=0.044; rs6591838, p=0.016; rs2236647; p=0.01] and 8 weeks therapy [rs4980524, p=0.044; rs6591838, p=0.016; rs2236647; p=0.01]. Haplotypic associations were observed for baseline FEV1 and % change in FEV1 at 4 weeks therapy [p=0.05 and p=0.01, respectively]. Significant trends towards association were observed for baseline % predicted FEV1 and % change in FEV1 at 8 weeks therapy. Positive correlations between haplotypes and % change in FEV1 were also observed.
Conclusions
STIP1 genetic variations may play a role in regulating corticosteroid response in asthmatics with reduced lung function. Replication in a second asthma population is required to confirm these observations.
Clinical Implications
Identifying genes that regulate corticosteroid responses could allow a priori determination of individual responses to corticosteroid therapy, leading to more effective dosing and/or selection of drug therapies for treating asthma.
Keywords: corticosteroid, pharmacogenetics, glucocorticoid receptor, SNP, heat shock protein, heat shock organizing protein, immunophilin
Introduction
Corticosteroids are the most efficacious medication used for treating airway inflammation associated with asthma. In spite of the effectiveness of corticosteroids, however, there is considerable clinical evidence of wide variations in corticosteroid response among asthmatics (1;2). Severe asthmatics, for example, are generally treated with high doses of oral corticosteroids, yet their asthma symptoms persist in contrast to most mild and moderate asthmatics that are treated with lower doses of inhaled corticosteroids (ICS) (3-5). In addition, there also evidence that a small subset of subjects experience worsening of asthma symptoms after corticosteroid treatment (6). Understanding the genetic mechanisms that regulate corticosteroid response variability could allow a priori determination of an individual’s response to corticosteroid therapy, leading to more effective dosing and/or selection of single or combination drug therapies for treating asthma.
CRHR1 (corticotrophin-releasing hormone receptor 1) is the only gene that has been identified as a potential genetic predictor of lung function change in asthmatics treated with corticosteroids (7). Corticotrophin-releasing hormone receptor 1 is a key component of the molecular pathway that regulates the production of endogenous cortisol in response to stress. Genetic variations in CRHR1, therefore, are probably most useful for identifying individuals deficient in cortisol production and therefore more responsive to exogenously administered corticosteroids. An obvious candidate for corticosteroid response is the glucocorticoid receptor gene, NR3C1. Corticosteroids exert their action by binding cytoplasmic glucocorticoid receptors and inducing structural and conformational changes that activate the receptor. Once activated, glucocorticoid receptors translocate across the nuclear membrane, where receptor dimers bind glucocorticoid receptor response elements (GRE) in the regulatory regions of genes, causing increased or decreased gene expression. Several genetic variations have been found in NR3C1, some of which have been shown to affect the receptor’s ability to effectively bind steroids (8-13). These rare mutations, however, have been associated with familial corticosteroid resistance and thus do not account for the genetic variations that contribute to inter-individual corticosteroid resistance and response (9;14-17). In fact, two studies, one association study and one mutational analysis, conclude that NR3C1 is not a significant contributor to corticosteroid resistance (18;19).
The glucocorticoid receptor is not autonomous in its action, and is only one part of a large hetero-complex of proteins that cooperatively functions to activate the glucocorticoid receptor (20;21). In addition to the glucocorticoid receptor, the hetero-complex consists of several chaperone proteins, including heat shock protein 90 (Hsp90), heat shock protein 70 (Hsp70/Hsc70), heat shock protein 40 (Hsp40), heat shock organizing protein (Hop), p23, and the immunophilins FKBP51 and FKBP52. The assembly of this hetero-complex occurs in a series of ATP dependent steps and is highly regulated (20;22-25). Proper assembly and activation of the glucocorticoid receptor complex is thus critical for proper response to corticosteroid treatment. Therefore, to fully understand the pharmacogenetics of corticosteroid response, a comprehensive evaluation of all genes encoding hetero-complex components is required.
In this study, we hypothesize that variations in the genes encoding components of the glucocorticoid receptor complex may significantly contribute to variation in corticosteroid response in asthmatics. To test this hypothesis, we genotyped single nucleotide polymorphisms (SNPs) in eight glucocorticoid complex genes HSPCB (Hsp90 1α), HSPCA (Hsp90 1β), STIP1 (Hop), HSPA8 (Hsc70), DNAJB1 (Hsp40), PTGES3 (p23), FKBP5 (FKBP51), and FKBP4 (FKBP52) in an adult asthma population evaluated for corticosteroid response, as measured by change in FEV1. Based on these results, we have been able to determine that the variations in the gene STIP1 may be important in predicting corticosteroid responses in subjects with reduced lung function.
Methods
Study Population
The adult corticosteroid response used in this study was a multi-center clinical trial comparing once-daily high dose inhaled flunisolide versus standard inhaled corticosteroid therapy (7;26). DNA from 382 Caucasians who completed the study were genotyped.
Selection of Single Nucleotide Polymorphisms (SNPs)
SNPs were selected for HSPCB, HSPCA, STIP1, HSPA8, DNAJB1, PTGES3, FKBP5, and FKBP4 based on allele frequency and linkage disequilibrium (LD) information from the NCBI database dbSNP (http://www.ncbi.nlm.nih.gov/SNP/), HapMap (http://www.hapmap.org/), and on our own re-sequencing results (manuscript in preparation). Due to the small population sizes, SNPs with minor allele frequencies (MAF) <0.10 were not selected in order to increase our power to detect genetic associations. The gene NRC31 (glucocorticoid receptor) was previously genotyped and analyzed in this population (7).
Statistical Methods
Hardy-Weinberg equilibrium (HWE) tests were performed on all genotyped polymorphisms using the GDA computer programs (27) and SAS/Genetics (2002). Linkage disequilibrium (LD), in terms of D’ and r2, were calculated using the program Haploview (28). The empirical p-values of both the HWE and LD tests were based on 10,000 replicate samples. ANOVAs were performed for quantitative pulmonary function measurements to test for association. Multivariable analyses of the associations were performed, adjusting for age, gender, and height, and were performed fitting the most appropriate genetic model based upon the results of the mean quantitative trait derived from the ANOVA models to determine if there was an independent effect of polymorphisms on change in FEV1 independent of baseline level. Association tests for haplotypes were performed using HAPLO.SCORE (29). Genetic association significance was assessed by performing 10,000 permutations of the single-locus genotypes among individuals in the sample to simulate the null distribution.
Genotyping
Genotyping was performed using the iPlex MassARRAY genotyping platform (Sequenom Inc., San Diego, CA, USA. Controls and blanks were included for quality and error checking. For all genes, the genotyping success rate was >95%.
Results
Study Population Characteristics
The characteristics for the study population are shown in Table I. The original study consisted of 515 moderate to severe asthmatic adults in ages 16-75, of which 58% were female, and 87% were self-reported as Caucasian (white) (7;26). Inclusion criteria included a history of asthma and documentation within the previous year of at least 12% improvement in FEV1 with administration of the short acting β-agonist, albuterol. Exclusion criteria included a history of significant pulmonary disease other than asthma, smoking in excess of 10 pack-years, recent upper respiratory infection, and recent asthma exacerbations requiring hospitalization or oral prednisone usage. Subjects were followed by phone contact on a weekly basis and had follow-up spirometry at 4 and 8 weeks. This study was conducted by Forest Pharmaceuticals and all subjects signed informed consent for the study and for genetic testing. The study protocol was approved by the Institutional Review Board at the Brigham and Women’s Hospital, Harvard University.
Table I. Characteristics of Study Population(26).
| Flunisolide (n=416) |
Non Flunisolide** (n=99) |
|
|---|---|---|
| White (US Caucasian) | 362 | 88 |
| Black (African American) | 26 | 8 |
| Other* | 28 | 3 |
| Age [Y (mean ±SD)] | 39.9 ± 13.7 | 40.7 ± 13.5 |
| Sex (% Male) | 41.8 | 41.4 |
| Asthma duration [Y (mean ±SD)] | 20.7 ± 14.2 | 21.1 ± 12.7 |
| Pack years (mean ±SD) | 5.6 ±7.7 | 7.4 ±7.8 |
| Subjects completing 8-week study | 348 | 91 |
Asian, Hispanic, Native American
triamcinolone, beclomethasone, or fluticasone propionate
The primary outcome measure was % change in FEV1 after 8 weeks, with a secondary analysis of the intermediary % change in FEV1 at 4 weeks. In this study, subjects were randomized 4:1 flunisolide versus other corticosteroid therapy (triamcinolone, beclomethasone, or fluticasone propionate). The mean change in the FEV1 was the same in both treatment groups (p = 0.30), therefore all results were combined in our analyses. The adjusted measurement % predicted FEV1 was derived based from Hankinson values(30). The mean baseline FEV1 as % predicted for this population was 72.2 ± 16.2%. The mean % change in FEV1 in this population was 7.0± 19.3% after 8 weeks treatment with corticosteroids. Multivariable analyses genetic ANOVA models to determine if there was an independent effect of genetic variations on change in FEV1 independent of baseline level. Of the 515 subjects, 439 completed the study (348 flunisolide and 91 non-flunisolide), of which 382 were Caucasian. Due to the small number of subjects in the minor ethnic groups and potential for population stratification, this study was confined to the Caucasians subset. A separate analysis of a random panel of 59 SNPs across the genome in this population indicated no evidence of population stratification (p > 0.05 for dichotomizations of baseline FEV1 and FEV1 change into highest and lowest quartiles).
STIP1 SNPs Are Associated with Baseline Lung Function Measures
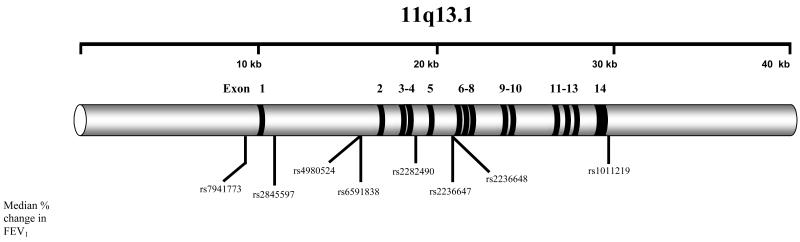
We observed the greatest associations for measures of baseline lung function in the gene STIP1 (Figure 1). All genotyped STIP1 SNPs tested were in HWE. Four STIP1 SNPs, rs4980524 (intron 1), rs6591838 (intron 1), rs2236647 (intron 5), and rs2236648 (intron 5), were consistently correlated with trends in baseline FEV1 (p=0.03, p=0.02, p=0.009, and p=0.043, respectively) (Table II). The largest difference for baseline FEV1 was measured for SNP rs 6591838 genotypes (AA baseline FEV1= 2.51±0.76; GG baseline FEV1= 1.97±0.49). Three SNPs, rs4980524, rs2236647, and rs2236648, were also correlated with trends in % predicted FEV1 (p=0.009, p=0.013, and p=0.03, respectively), again with the largest difference observed for rs6591838 between AA (72.31% ±12.71) and GG (63.83%±10.92) genotypes. Genetic modeling confirmed a strong recessive effect for each SNP for trends in both baseline FEV1 (p-value range 0.002-0.013) and % predicted FEV1 (p-value range 0.002-0.017), with the rare alleles correlating with lower lung function. Minor allele frequencies (MAFs) between rs498024 (MAF=0.41), rs236647 (MAF=0.43), and rs2236648 (MAF=0.42) were similar, and r2 values between these three SNPs were >0.90, thus it could not be determined which SNP was most concordant with the trend effects. SNP rs6591838 was not in strong LD with rs4980524, rs236647, or rs2236648 (r2 ≤ 0.46). No significant associations were observed between baseline lung function measures and SNPs in the remaining seven genes (Supplementary Tables SI-SVII).
Figure 1. Diagram of STIP1 and Location of Genotyped SNPs.
Table II. Genetic Association of STIP1 SNPs with Baseline Lung Function Measures.
| Baseline FEV1 | Baseline % Predicted FEV1 | |||||||
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| N | Mean (SD) | Univariate ANOVA p-value | Multivariate Recessive* | Mean (SD) | Univariate ANOVA p-value | Multivariate Recessive* | ||
| SNP | ||||||||
| AA | 126 | 2.54 ±0.79 | 72.62 ±12.57 | |||||
| rs4980524 | AC | 195 | 2.51 ±0.56 | 0.03 | 0.009 | 72.71 ±12.84 | 0.009 | 0.002 |
| CC | 57 | 2.27 ±0.60 | 67.09 ±11.85 | |||||
| AA | 229 | 2.51 ±0.76 | 72.31 ±12.71 | |||||
| rs6591838 | AG | 127 | 2.49 ±0.68 | 0.018 | 0.0045 | 71.81 ±12.85 | 0.055 | 0.017 |
| GG | 14 | 1.97 ±0.49 | 63.83 ±10.92 | |||||
| CC | 118 | 2.54 ±0.76 | 72.92 ±12.82 | |||||
| rs2236647 | CT | 193 | 2.55 ±0.69 | 0.009 | 0.002 | 72.95 ±12.78 | 0.013 | 0.003 |
| TT | 60 | 2.25 ±0.54 | 67.67 ±11.85 | |||||
| CC | 120 | 2.51 ±0.77 | 72.31 ±16.76 | |||||
| rs2236648 | CT | 198 | 2.53 ±0.70 | 0.043 | 0.013 | 72.85 ±12.80 | 0.03 | 0.008 |
| TT | 57 | 2.28 ±0.60 | 67.80 ±11.55 | |||||
Recessive model used based upon results from univariate analysis.
STIP1 SNPs Are Associated with Change in FEV1 After Corticosteroid Treatment
Significant correlations for % change in FEV1 were observed for STIP1 SNPs rs6591838 and rs2236647 at 4 weeks (p=0.025 and p=0.04, respectively) and SNPs rs6591838 and rs1011219 (3′ non-coding region) at 8 week for (p=0.014 and p=0.005, respectively; Table III). The largest % change in FEV1 (20.70%±28.29) occurred with SNP rs6591838 at the 8 week time point for the rare GG genoytpe. Genetic modeling confirmed a consistent recessive effect at 4 weeks for SNPs rs4980524, rs6591838, and rs2236647 (p-value range 0.01-0.044) and at 8 weeks for SNPs rs4980524, rs6591838, rs2236647, and rs1011219 (p-value range 0.0014-0.044). The significance observed for SNP rs1011219 was driven by two individuals homozygous for the A allele, who had large negative changes (−50% and −23.7%) in FEV1 at 8 weeks. No significant associations were observed between change in FEV1 and SNPs in the remaining seven genes.
Table III. Genetic Association of STIP1 SNPs with % Change in FEV1.
| % Change FEV1 4 Weeks | % Change FEV1 8 Weeks | ||||||||
|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||
| N | Mean (SD) | Univariate ANOVA p-value | Multivariate Recessive* | N | Mean (SD) | Univariate ANOVA p-value | Multivariate Recessive* | ||
| SNP | |||||||||
| AA | 118 | 5.10 ±17.16 | 124 | 5.60 ±15.26 | |||||
| rs4980524 | AC | 187 | 5.40 ±19.00 | 0.13 | 0.044 | 193 | 6.54 ±19.87 | 0.12 | 0.044 |
| CC | 55 | 11.03 ±24.03 | 55 | 11.91 ±25.44 | |||||
| AA | 214 | 4.57 ±16.82 | 227 | 5.50 ±17.63 | |||||
| rs6591838 | AG | 123 | 7.32 ±22.07 | 0.025 | 0.016 | 123 | 7.43 ±14.97 | 0.014 | 0.016 |
| GG | 14 | 18.41 ±26.15 | 14 | 20.70 ±28.29 | |||||
| CC | 110 | 5.23 ±17.52 | 116 | 5.55 ±15.51 | |||||
| rs2236647 | CT | 186 | 4.95 ±18.40 | 0.04 | 0.01 | 191 | 6.02 ±19.49 | 0.11 | 0.01 |
| TT | 58 | 12.05 ±23.99 | 58 | 11.68 ± 24.68 | |||||
| CC | 112 | 5.23 ±17.67 | 118 | 5.80 ±16.29 | |||||
| rs2236648 | CT | 190 | 5.60 ±19.30 | 0.25 | 0.10 | 196 | 6.74 ±19.88 | 0.3 | 0.13 |
| TT | 55 | 10.22 ±23.50 | 55 | 10.70 ±24.40 | |||||
| AA | 1 | −15.93 | 2 | −36.85 ±18.59 | |||||
| rs1011219 | AG | 85 | 6.75 ±22.58 | 0.5 | 0.26 | 83 | 8.36 ±21.14 | 0.005 | 0.0014 |
| GG | 272 | 6.00 ±18.14 | 285 | 6.92 ±18.74 | |||||
Recessive model used based upon results from univariate analysis.
STIP1 Haplotypes Are Correlated with Measures of Lung Function
Six STIP1 haplotypes were derived for the Caucasians using eight SNPs (Tables IV and V) SNPs rs7941773 and rs1011219 were in identity with each other (r2=1), as were the SNP pair rs4980524 and rs2236648. Overall global significance (comparison between all haplotypes; p=0.05) was observed for baseline FEV1 (Table IV), with haplotypes 1 (p=0.025; Hap-score=−2.24) and 6 (p=0.01;Hap-score=2.57) attaining individual significance. Haplotype 1 (freq = 0.03) and haplotype 6 (freq = 0.56) differ only at SNP rs2236647 (C/T). A trend towards significance was observed for % predicted FEV1 (p=0.09) (Table IV), with haplotypes 2 (p= 0.03 ; Hap-score=−2.18) and 6 (p=0.03; Hap-score=2.16) attaining individual significance. Based on the above observations, a three SNP sliding window sub-haplotype analysis was employed to better discern the contribution of each SNP or gene region towards the primary haplotype associations. For baseline FEV1, the largest correlations were observed with the three SNP sub-haplotypes in the 3′ portion of STIP1 (three SNP windows 4D, 4E, and 4F), all of which contained the SNP rs2236647. The lowest p-values (0.008) were observed for two rare three SNP sub-haplotypes in window 4E (rs2282490/rs2236647/rs2236648; T/T/C) and window 4F (rs2236647/rs2236648/rs1011219; T/C/G). As mentioned above, SNP rs2236647 differentiated the highest and lowest scoring eight SNP haplotypes (1 and 6) for FEV1 and correlated with high (C allele) and low (T allele) trends in baseline FEV1 measurements in the single SNP analysis (Table II). In contrast to baseline FEV1, the highest correlations for % predicted FEV1 were observed in the 5′ portion of STIP1 (three SNP windows 4A and 4B; Table IV), where the A allele of SNP rs4980524 correlated with the highest scoring three SNP sub-haplotypes. Based on single SNP analysis, the A allele of rs4980524 correlated with high measures of % predicted FEV1 (Table II).
Table IV. STIP1 Haplotype Association with Baseline FEV1 and Baseline % Predicted FEV1.
| Baseline FEV1 Global** p = 0.05 |
Baseline % pred FEV1 Global** p = 0.09 |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| rs7941773 | rs2845597 | rs4980524 | rs6591838 | rs2282490 | rs2236647 | rs2236648 | rs1011219 | Haplotype Frequency |
Hap-Score | p-value | Hap-Score | p-value | |
| 1 | C | G | A | A | T | T | C | G | 0.03 | −2.24 | 0.025 | −0.16 | 0.87 |
| 2 | C | G | C | G | T | T | T | G | 0.07 | −1.67 | 0.09 | −2.18 | 0.03 |
| 3 | C | A | C | A | C | T | T | G | 0.19 | −1.06 | 0.29 | −1.57 | 0.12 |
| 4 | C | A | C | G | T | T | T | G | 0.03 | −1.01 | 0.31 | 0.85 | 0.39 |
| 5 | T | A | C | G | T | T | T | A | 0.11 | 0.58 | 0.56 | 0.47 | 0.64 |
| 6 | C | G | A | A | T | C | C | G | 0.56 | 2.57 | 0.01 | 2.16 | 0.03 |
|
| |||||||||||||
| c | G | C | 0.07 | −1.72 | 0.09 | −2.72 | 0.007 | ||||||
| 3A | C | A | C | 0.22 | −1.31 | 0.19 | −1.03 | 0.30 | |||||
| T | A | C | 0.12 | 0.64 | 0.52 | 0.31 | 0.76 | ||||||
| c | G | A | 0.60 | 1.65 | 0.10 | 2.13 | 0.034 | ||||||
|
| |||||||||||||
| G | C | G | 0.07 | −1.76 | 0.08 | −2.75 | 0.006 | ||||||
| 3B | A | C | A | 0.19 | −0.60 | 0.54 | −1.19 | 0.23 | |||||
| A | C | G | 0.14 | −0.36 | 0.71 | 0.45 | 0.65 | ||||||
| G | A | A | 0.60 | 1.72 | 0.09 | 2.11 | 0.035 | ||||||
|
| |||||||||||||
| C | G | T | 0.21 | −1.13 | 0.25 | −1.39 | 0.17 | ||||||
| 3C | C | A | C | 0.19 | −0.82 | 0.41 | −1.03 | 0.30 | |||||
| A | A | T | 0.60 | 1.78 | 0.08 | 2.13 | 0.033 | ||||||
|
| |||||||||||||
| A | T | T | 0.03 | −2.11 | 0.035 | −1.49 | 0.13 | ||||||
| 3D | G | T | T | 0.21 | −1.24 | 0.21 | −1.15 | 0.25 | |||||
| A | C | T | 0.19 | −0.83 | 0.41 | −0.04 | 0.97 | ||||||
| A | T | C | 0.57 | 2.47 | 0.01 | 2.19 | 0.03 | ||||||
|
| |||||||||||||
| T | T | c | 0.03 | −2.66 | 0.008 | −1.54 | 0.12 | ||||||
| 3E | C | T | T | 0.19 | −0.90 | 0.37 | −1.25 | 0.21 | |||||
| T | T | T | 0.21 | −0.84 | 0.40 | −0.53 | 0.60 | ||||||
| T | C | c | 0.56 | 2.40 | 0.017 | 2.03 | 0.04 | ||||||
|
| |||||||||||||
| T | c | G | 0.03 | −2.66 | 0.008 | −2.34 | 0.02 | ||||||
| 3F | T | T | G | 0.28 | −2.10 | 0.035 | −1.26 | 0.21 | |||||
| T | T | A | 0.12 | 0.84 | 0.40 | 0.72 | 0.47 | ||||||
| C | C | G | 0.56 | 2.39 | 0.017 | 2.05 | 0.04 | ||||||
Trends towards significance in italics.
Based on Hap-Score comparisons between six haplotypes.
Table V. STIP1 Haplotype Association with % Change in FEV1.
| 4 weeks % change FEV1 Global** p = 0.012 |
8 weeks % change FEV1 Global** p = 0.07 |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| rs7941773 | rs2845597 | rs4980524 | rs6591838 | rs2282490 | rs2236647 | rs2236648 | rs1011219 | Haplotype Frequency |
Hap-Score | p-value | Hap-Score | p-value | |
| 6 | C | G | A | A | T | C | C | G | 0.56 | −1.93 | 0.053 | −1.96 | 0.05 |
| 5 | T | A | C | G | T | T | T | A | 0.11 | 0.06 | 0.95 | −0.05 | 0.96 |
| 3 | C | A | C | A | C | T | T | G | 0.19 | 0.03 | 0.98 | −0.02 | 0.98 |
| 1 | C | G | A | A | T | T | C | G | 0.03 | −0.02 | 0.98 | 0.14 | 0.89 |
| 4 | C | A | C | G | T | T | T | G | 0.03 | −0.36 | 0.71 | 0.21 | 0.84 |
| 2 | C | G | C | G | T | T | T | G | 0.07 | 2.29 | 0.02 | 2.93 | 0.0034 |
|
| |||||||||||||
| C | G | A | 0.60 | −1.76 | 0.08 | −1.87 | 0.06 | ||||||
| 4A | C | A | C | 0.22 | −0.08 | 0.93 | −0.18 | 0.86 | |||||
| T | A | C | 0.11 | 0.49 | 0.62 | 0.15 | 0.88 | ||||||
| C | G | C | 0.07 | 2.86 | 0.004 | 3.67 | 0.0002 | ||||||
|
| |||||||||||||
| G | A | A | 0.60 | −1.77 | 0.08 | −1.87 | 0.06 | ||||||
| 4B | A | C | A | 0.19 | −0.03 | 0.97 | −0.21 | 0.84 | |||||
| A | C | G | 0.14 | 0.41 | 0.67 | 0.19 | 0.85 | ||||||
| G | C | G | 0.07 | 2.86 | 0.004 | 3.64 | 0.0003 | ||||||
|
| |||||||||||||
| A | A | T | 0.60 | −1.78 | 0.08 | −1.87 | 0.06 | ||||||
| 4C | C | A | C | 0.19 | 0.02 | 0.98 | −0.25 | 0.80 | |||||
| C | G | T | 0.21 | 2.04 | 0.04 | 2.50 | 0.01 | ||||||
|
| |||||||||||||
| A | T | C | 0.57 | −1.67 | 0.09 | −1.77 | 0.08 | ||||||
| 4D | A | C | T | 0.19 | −0.01 | 0.99 | −0.34 | 0.73 | |||||
| A | T | T | 0.03 | −0.08 | 0.93 | 0.27 | 0.79 | ||||||
| G | T | T | 0.21 | 2.02 | 0.044 | 2.34 | 0.02 | ||||||
|
| |||||||||||||
| T | C | C | 0.56 | −1.90 | 0.06 | −1.93 | 0.054 | ||||||
|
| |||||||||||||
| 4E | C | T | T | 0.19 | 0.02 | 0.98 | −0.34 | 0.73 | |||||
| T | T | C | 0.03 | 0.93 | 0.35 | 1.19 | 0.23 | ||||||
| T | T | T | 0.21 | 1.56 | 0.12 | 1.93 | 0.054 | ||||||
|
| |||||||||||||
| C | C | G | 0.56 | −1.91 | 0.056 | −1.94 | 0.053 | ||||||
| 4F | T | T | A | 0.11 | 0.04 | 0.96 | −0.38 | 0.70 | |||||
| T | C | G | 0.03 | 0.92 | 0.36 | 1.19 | 0.24 | ||||||
| T | T | G | 0.28 | 1.43 | 0.15 | 1.72 | 0.09 | ||||||
Trends towards significance in italics.
Based on Hap-Score comparisons between six haplotypes.
STIP1 Haplotypes Are Correlated with Change in FEV1 After Corticosteroid Treatment
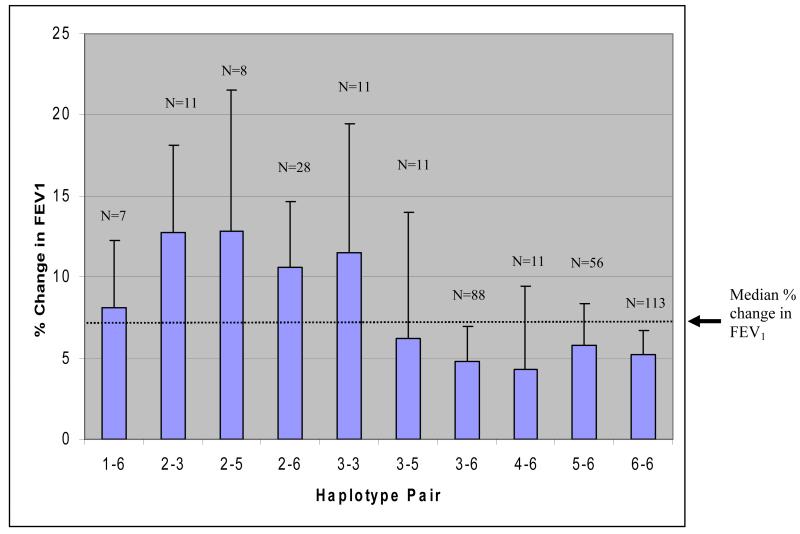
Eight SNP haplotype and three SNP sliding window haplotype analyses were also performed for 4 week and 8 week % change in FEV1 after corticosteroid therapy (Table V). Global significance (p=0.01) was observed at 4 weeks % change in FEV1 while a trend towards significance (p=0.07) was observed at 8 weeks % change in FEV1. Haplotype 2 (freq = 0.07) was individually significant for 4 week (p=0.02) and 8 week (p=0.0034) % change in FEV1 while haplotype 6 (freq = 0.56) trended towards significance for both 4 week (p=0.053) and 8 week (p=0.05) % change in FEV1. Based on single SNP analysis of rs2236647 for 4 and 8 week % change in FEV1 (C allele corresponding to lowest change in FEV1; Table III), haplotype 6 correlates with individuals with the lowest % change in FEV1. The three SNP sub-haplotypes (C/G/C; rs7941773/rs2845597/rs4980524; window 5A) and (G/C/G; rs2845597/rs4980524/rs6591838; window 5B) had the strongest correlation with % change in FEV1 at both 4 weeks (p=0.004 and p=0.004, respectively) and at 8 weeks (p=0.0002 and p=0.0003, respectively). These three SNP sub-haplotypes only occur in the eight SNP haplotype 2. Based on single SNP analyses of rs4980524 (A allele corresponding to lowest % change in FEV1) and rs6591838 (G allele corresponding with the highest % change in FEV1) in Table III, it can be logically deduced that each of these three SNP sub-haplotypes and the eight SNP haplotype 2 correlate directly with the largest change in FEV1 after both 4 and 8 week therapy, however these haplotypes account for only 7% of the total haplotypes. A comparison of haplotype pairs (Fig 2; number of haplotype pairs N) showed that subjects inheriting one copy of haplotype 2 had mean increases in % change in FEV1 >10% after 8 weeks of therapy. Only one other haplotype pair 3/3 had a mean increase in % change in FEV1 >10%. With the exception of haplotype pairs 1/6 and 2/6, a majority of subjects inheriting one copy of haplotype 6 consistently had mean % change in FEV1 less than the mean % change (6.97%).
Figure 2. Correlation Between Haplotype Pairs and % Change in FEV1 After 8 Weeks of Flunisolide Therapy in Caucasians.
The median % change in FEV1 for all haplotype pairs (6.97%) is indicated by the dotted horizontal line. N = number of subjects possessing each haplotype pair.
Discussion
In this study, we report that genetic variations in the gene STIP1 may have value in identifying asthmatics that are more responsive to corticosteroid therapy. Most specifically, we have found that alleles for four STIP1 SNPs consistently correlate with levels of baseline lung function (baseline FEV1 and % predicted FEV1) and with improvement in lung function (% change in FEV1) after 4 and 8 weeks of corticosteroid therapy. In addition, we have also been able to identify haplotypes and haplotype pairs that also correlate with baseline lung function measures and improvement in lung function after corticosteroid therapy. None of the polymorphisms tested in this study were coding, thus it is difficult to assess the potential functional effects of STIP1 based on our observations. In addition, because we observed genetic association with multiple SNPs across the gene, it is difficult to pinpoint a specific region of the gene contributing most to the observed associations. We were, however, able to identify consistent trends in lung function measures and changes in FEV1 after corticosteroid therapy and able to identify SNPs most correlated with these haplotype trends utilizing eight and three SNP haplotype analyses. In spite of these haplotypic observations, however, we still cannot conclude whether the SNPs tested in this study or unknown SNPs in LD are responsible for the observed effects.
Based on the location of the strongest genotypic and haplotypic associations, our results suggest that a region between and inclusive of intron 1 (SNP rs4980524) and intron 5 (SNP rs2236647) may contain key genetic variants or combinations of genetic variants which control STIP1 expression. STIP1 is composed of 14 exons dispersed over ~18kb of genomic DNA, and there are at least eight different forms of STIP1 mRNA sequences deposited in GenBank. One of these mRNAs (Accession # AK225736), is truncated, containing only exons 1-5 and a short portion of intron 5 directly flanking exon 5. This small isoform of Hop produced by this truncated mRNA has not been characterized, however elimination of exons 6-14 would delete the important C-terminal tetratricopeptide repeat (TPR) domain (20;31;32) in Hop. This C-terminal TPR functions as the binding domain for the EEVD peptide sequence in the C-terminus of Hsp90 or Hsp70. Hypothetically, our association results could be indicative of genetic variation that regulates splicing between exons 5 and 6, which in turn could generate novel Hop isoforms which could have effects on assembly of the Hsp90-Hop-Hsp70 complex. We have re-sequenced STIP1 in a small screening panel of asthmatics and unaffected subjects in order to verify existing and identify potentially uncharacterized polymorphisms in the intron 1-intron 5 region (manuscript in preparation). We have not, however, re-sequenced STIP1 specifically in individuals with wide ranges of corticosteroid response, results from which could reveal risk/protective genetic variants closely associated with the production of STIP1 splice variants.
In light of the function of STIP and the importance of the TPR domains in Hop for controlling the assembly of the GR hetero-complex (20;33-36), the differences in lung function changes observed between genotypic groups is not surprising. The product of STIP1, the heat shock organizing protein (Hop), plays a critical role in assembly of the GR hetero-complex by specifically binding ADP bound conformations hsp90 and mediating its association to hsp70, through TPR interactions, prior to incorporation into the GR hetero-complex (20). The incorporation of the hsp70/hsp90 into the GR hetero-complex allows effective binding of corticosteroids into the open cleft of the GR. Without activation of the GR hetero-complex through hsp70/hsp90 association, efficient binding of corticosteroids would be attenuated, thus depriving the cytoplasm of activated GR hetero-complexes available for transport into the cell nucleus. Failure of activated GR hetero-complexes transport would result in dysregulation of genes controlling the inflammatory pathway. While this breakdown in GR hetero-complex activation may be explained by defects in STIP1 expression, we cannot completely conclude that our observations are pharmacogenetic in nature. Since we observed a strong correlation of STIP1 SNPs and haplotypes with baseline lung function measures, it is possible that STIP1 genetic variations simply correlate with subjects with poor lung function. Considering this possibility, in the case of asthmatics tested in this study, we would be observing individuals with lower baseline lung function measures who have more capacity for lung function improvement when treated with corticosteroids compared to subjects with higher baseline lung function measures. If this last possibility is true, STIP1 genetic variations could also be a contributor to asthma risk. In fact, we have performed additional analyses of STIP1 in two additional asthma population, neither of which have corticosteroid response data, that indicate that STIP1 is associated with varying levels of bronchial hyperresponsiveness (BHR) as measured by PC20 (manuscript in preparation).
There were several limitations to this study. First, we do not have confirmatory data from a replicate corticosteroid response population that strongly supports our observations for STIP1. The availability of corticosteroid response populations with similar study designs is limited, and those that are available are insufficiently powered or lack DNA for genetic studies. Second, we limited our genotyping to SNPs located approximately ~1 kb flanking the exonic regions of STIP1, thus our assessment of SNPs in the putative promoter region, intronic regions, and the 3′ UTR of the gene was not comprehensive. This study was also limited in that the subset of African Americans was insufficient to effectively evaluate genetic association. While we could have combined genotyping data for SNPs with similar genotypic frequencies between Caucasian and African American populations, the population stratification and admixture effects could have introduced significant errors into our analysis. We also must point out that because our initial screening included 60 SNPs from nine genes, our results could be affected by random associations caused by multiple comparisons. We did, however, try to control for this potential confounder by performing a permutation analysis on all data sets. Finally, while baseline FEV1 is a value affected by factors such as age, race, weight, and sex, which could affect our observations, our confirmatory association with the adjusted value % predicted FEV1 (Table II) and the consistent trends in lung function measures by genotype provide support that the observed changes in FEV1 after corticosteroid therapy are significant observations.
The ability to predict an individual’s response to corticosteroid therapy could have tremendous implications for increasing the effectiveness of treating inflammatory diseases such as asthma. This study, however, illustrates several issues in identifying and using genetic variants to test corticosteroid responsiveness. First, the pathway regulating corticosteroid response is complicated. In order to accurately assess the pharmacogenetics of corticosteroid response, the effects of more than one gene will have to be considered, and may require consideration of gene-gene interactions. Second, unless detailed DNA sequencing is performed on a patient by patient basis, we may never know the genetic contributions of rare genetic variants on corticosteroid response between individuals. The assessment of rare genetic variants becomes more problematic when considering that many of rare genetic variants may be specific to only one ethnicity. Third, the availability of comparable corticosteroid response populations is paramount for replication. While corticosteroid response studies have been performed (37-39), the differences in study design and the confounding effects of co-administered medications complicate final data interpretation. In addition, older corticosteroid response studies may have not had the foresight for performing pharmacogenetic analyses, therefore DNA was not collected. Finally, the availability of tools to measure the functional effects of genetically associated non-coding variants is still limited. Therefore, while these non-coding genetic variations may eventually be useful in predicting drug responsiveness, the inability to define functional roles for these non-coding genetic variations leaves questions as to their effects on lung function and corticosteroid response.
Supplementary Material
Acknowledgements
We want to thank all the participants of the adult asthma study. We want to also thank Dr. Elizabeth Ampleford, Huashi Li, Abdoulaye Diallo, and Catherine Brewer for additional technical support.
Sources of funding: This work was supported by NIH grants NHLBI HL077916 to GAH, NHLBI HL69197 and HL076285 to ERB, and NHLBI HL65899 to STW.
Abbreviations
- FEV1
forced expiratory volume in 1 second
- Freq
frequency
- GR
glucocorticoid receptor
- GRE
glucocorticoid response element
- HWE
Hardy-Weinberg equilibrium
- ICS
inhaled corticosteroid
- LD
linkage disequilibrium
- MAF
minor allele frequency
- PC20
provocative concentration causing a 20% drop in FEV1
- SNP
single nucleotide polymorphism
- 3′ UTR
3′ untranslated region
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- (1).Barnes PJ. Corticosteroids: the drugs to beat. Eur J Pharmacol. 2006;533(1-3):2–14. doi: 10.1016/j.ejphar.2005.12.052. [DOI] [PubMed] [Google Scholar]
- (2).Tantisira KG, Drazen JM. Pharmacogenetics. In: Silverman EK, Shapiro SD, Lomas DA, Weiss ST, editors. Respiratory Genetics. Hodder Education; London, England: 2005. pp. 191–216. [Google Scholar]
- (3).Busse WW, Banks-Schlegel S, Wenzel SE. Pathophysiology of severe asthma. J Allergy Clin Immunol. 2000;106(6):1033–42. doi: 10.1067/mai.2000.111307. [DOI] [PubMed] [Google Scholar]
- (4).Moore WC, Bleecker ER, Curran-Everett D, Erzurum SC, Ameredes BT, Bacharier L, et al. Characterization of the severe asthma phenotype by the National Heart, Lung, and Blood Institute’s Severe Asthma Research Program. J Allergy Clin Immunol. 2007;119(2):405–13. doi: 10.1016/j.jaci.2006.11.639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Wenzel S. Severe asthma: epidemiology, pathophysiology and treatment. Mt Sinai J Med. 2003;70(3):185–90. [PubMed] [Google Scholar]
- (6).Malmstrom K, Rodriguez-Gomez G, Guerra J, Villaran C, Pineiro A, Wei LX, et al. Montelukast/Beclomethasone Study Group Oral montelukast, inhaled beclomethasone, and placebo for chronic asthma. A randomized, controlled trial. Ann Intern Med. 1999;130(6):487–95. doi: 10.7326/0003-4819-130-6-199903160-00005. [DOI] [PubMed] [Google Scholar]
- (7).Tantisira KG, Lake S, Silverman ES, Palmer LJ, Lazarus R, Silverman EK, et al. Corticosteroid pharmacogenetics: association of sequence variants in CRHR1 with improved lung function in asthmatics treated with inhaled corticosteroids. Hum Mol Genet. 2004;13(13):1353–9. doi: 10.1093/hmg/ddh149. [DOI] [PubMed] [Google Scholar]
- (8).Bray PJ, Cotton RG. Variations of the human glucocorticoid receptor gene (NR3C1): pathological and in vitro mutations and polymorphisms. Hum Mutat. 2003;21(6):557–68. doi: 10.1002/humu.10213. [DOI] [PubMed] [Google Scholar]
- (9).Brufsky AM, Malchoff DM, Javier EC, Reardon G, Rowe D, Malchoff CD. A glucocorticoid receptor mutation in a subject with primary cortisol resistance. Trans Assoc Am Physicians. 1990;103:53–63. [PubMed] [Google Scholar]
- (10).Hillmann AG, Ramdas J, Multanen K, Norman MR, Harmon JM. Glucocorticoid receptor gene mutations in leukemic cells acquired in vitro and in vivo. Cancer Res. 2000;60(7):2056–62. [PubMed] [Google Scholar]
- (11).Hawkins GA, Amelung PJ, Smith RS, Jongepier H, Howard TD, Koppelman GH, et al. Identification of polymorphisms in the human glucocorticoid receptor gene (NR3C1) in a multi-racial asthma case and control screening panel. DNA Seq. 2004;15(3):167–73. doi: 10.1080/10425170410001704517. [DOI] [PubMed] [Google Scholar]
- (12).DeRijk RH, Schaaf M, de Kloet ER. Glucocorticoid receptor variants: clinical implications. J Steroid Biochem Mol Biol. 2002;81(2):103–22. doi: 10.1016/s0960-0760(02)00062-6. [DOI] [PubMed] [Google Scholar]
- (13).Rivers C, Levy A, Hancock J, Lightman S, Norman M. Insertion of an amino acid in the DNA-binding domain of the glucocorticoid receptor as a result of alternative splicing. J Clin Endocrinol Metab. 1999;84(11):4283–6. doi: 10.1210/jcem.84.11.6235. [DOI] [PubMed] [Google Scholar]
- (14).Bray PJ, Cotton RG. Variations of the human glucocorticoid receptor gene (NR3C1): pathological and in vitro mutations and polymorphisms. Hum Mutat. 2003;21(6):557–68. doi: 10.1002/humu.10213. [DOI] [PubMed] [Google Scholar]
- (15).Karl M, Lamberts SW, Detera-Wadleigh SD, Encio IJ, Stratakis CA, Hurley DM, et al. Familial glucocorticoid resistance caused by a splice site deletion in the human glucocorticoid receptor gene. J Clin Endocrinol Metab. 1993;76(3):683–9. doi: 10.1210/jcem.76.3.8445027. [DOI] [PubMed] [Google Scholar]
- (16).Koper JW, Stolk RP, de Lange P, Huizenga NA, Molijn GJ, Pols HA, et al. Lack of association between five polymorphisms in the human glucocorticoid receptor gene and glucocorticoid resistance. Hum Genet. 1997;99(5):663–8. doi: 10.1007/s004390050425. [DOI] [PubMed] [Google Scholar]
- (17).Vottero A, Kino T, Combe H, Lecomte P, Chrousos GP. A novel, C-terminal dominant negative mutation of the GR causes familial glucocorticoid resistance through abnormal interactions with p160 steroid receptor coactivators. J Clin Endocrinol Metab. 2002;87(6):2658–67. doi: 10.1210/jcem.87.6.8520. [DOI] [PubMed] [Google Scholar]
- (18).Koper JW, Stolk RP, de Lange P, Huizenga NA, Molijn GJ, Pols HA, et al. Lack of association between five polymorphisms in the human glucocorticoid receptor gene and glucocorticoid resistance. Hum Genet. 1997;99(5):663–8. doi: 10.1007/s004390050425. [DOI] [PubMed] [Google Scholar]
- (19).Huizenga NA, de LP, Koper JW, de Herder WW, Abs R, Kasteren JH, et al. Five patients with biochemical and/or clinical generalized glucocorticoid resistance without alterations in the glucocorticoid receptor gene. J Clin Endocrinol Metab. 2000;85(5):2076–81. doi: 10.1210/jcem.85.5.6542. [DOI] [PubMed] [Google Scholar]
- (20).Pratt WB, Morishima Y, Murphy M, Harrell M. Chaperoning of glucocorticoid receptors. Handb Exp Pharmacol. 2006;(172):111–38. doi: 10.1007/3-540-29717-0_5. [DOI] [PubMed] [Google Scholar]
- (21).Pratt WB, Toft DO. Regulation of signaling protein function and trafficking by the hsp90/hsp70-based chaperone machinery. Exp Biol Med (Maywood ) 2003;228(2):111–33. doi: 10.1177/153537020322800201. [DOI] [PubMed] [Google Scholar]
- (22).Chen S, Smith DF. Hop as an adaptor in the heat shock protein 70 (Hsp70) and hsp90 chaperone machinery. J Biol Chem. 1998;273(52):35194–200. doi: 10.1074/jbc.273.52.35194. [DOI] [PubMed] [Google Scholar]
- (23).Johnson BD, Schumacher RJ, Ross ED, Toft DO. Hop modulates Hsp70/Hsp90 interactions in protein folding. J Biol Chem. 1998;273(6):3679–86. doi: 10.1074/jbc.273.6.3679. [DOI] [PubMed] [Google Scholar]
- (24).Dittmar KD, Hutchison KA, Owens-Grillo JK, Pratt WB. Reconstitution of the steroid receptor.hsp90 heterocomplex assembly system of rabbit reticulocyte lysate. J Biol Chem. 1996;271(22):12833–9. doi: 10.1074/jbc.271.22.12833. [DOI] [PubMed] [Google Scholar]
- (25).Dittmar KD, Banach M, Galigniana MD, Pratt WB. The role of DnaJ-like proteins in glucocorticoid receptor.hsp90 heterocomplex assembly by the reconstituted hsp90.p60.hsp70 foldosome complex. J Biol Chem. 1998;273(13):7358–66. doi: 10.1074/jbc.273.13.7358. [DOI] [PubMed] [Google Scholar]
- (26).Bielory L, Piccone F, Rabinowitz P, Rossoff L, Winder J, Incaudo G, et al. Multicentre, randomised, parallel-group study of the efficacy and tolerability of flunisolide administered once daily via AeroChamber (R) in the treatment of mild to moderate asthma. Clinical Drug Investigation. 2000;19(2):93–101. [Google Scholar]
- (27).Weir BS. Genetic Data Analysis II: Methods for Discrete Population Genetic Data. Sinauer Associations, Inc. Publishers; Sunderland, MA: 1996. [Google Scholar]
- (28).Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21(2):263–5. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- (29).Schaid DJ, Rowland CM, Tines DE, Jacobson RM, Poland GA. Score tests for association between traits and haplotypes when linkage phase is ambiguous. Am J Hum Genet. 2002;70(2):425–34. doi: 10.1086/338688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Hankinson JL, Odencrantz JR, Fedan KB. Spirometric reference values from a sample of the general U.S. population. Am J Respir Crit Care Med. 1999;159(1):179–87. doi: 10.1164/ajrccm.159.1.9712108. [DOI] [PubMed] [Google Scholar]
- (31).Cortajarena AL, Regan L. Ligand binding by TPR domains. Protein Sci. 2006;15(5):1193–8. doi: 10.1110/ps.062092506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Odunuga OO, Longshaw VM, Blatch GL. Hop: more than an Hsp70/Hsp90 adaptor protein. Bioessays. 2004;26(10):1058–68. doi: 10.1002/bies.20107. [DOI] [PubMed] [Google Scholar]
- (33).Longshaw VM, Chapple JP, Balda MS, Cheetham ME, Blatch GL. Nuclear translocation of the Hsp70/Hsp90 organizing protein mSTI1 is regulated by cell cycle kinases. J Cell Sci. 2004;117(Pt 5):701–10. doi: 10.1242/jcs.00905. [DOI] [PubMed] [Google Scholar]
- (34).Odunuga OO, Longshaw VM, Blatch GL. Hop: more than an Hsp70/Hsp90 adaptor protein. Bioessays. 2004;26(10):1058–68. doi: 10.1002/bies.20107. [DOI] [PubMed] [Google Scholar]
- (35).Smith DF, Sullivan WP, Marion TN, Zaitsu K, Madden B, McCormick DJ, et al. Identification of a 60-kilodalton stress-related protein, p60, which interacts with hsp90 and hsp70. Mol Cell Biol. 1993;13(2):869–76. doi: 10.1128/mcb.13.2.869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Pratt WB, Toft DO. Regulation of signaling protein function and trafficking by the hsp90/hsp70-based chaperone machinery. Exp Biol Med (Maywood ) 2003;228(2):111–33. doi: 10.1177/153537020322800201. [DOI] [PubMed] [Google Scholar]
- (37).Lemanske RF, Jr., Sorkness CA, Mauger EA, Lazarus SC, Boushey HA, Fahy JV, et al. Inhaled corticosteroid reduction and elimination in patients with persistent asthma receiving salmeterol: a randomized controlled trial. JAMA. 2001;285(20):2594–603. doi: 10.1001/jama.285.20.2594. [DOI] [PubMed] [Google Scholar]
- (38).Lazarus SC, Boushey HA, Fahy JV, Chinchilli VM, Lemanske RF, Jr., Sorkness CA, et al. Long-acting beta2-agonist monotherapy vs continued therapy with inhaled corticosteroids in patients with persistent asthma: a randomized controlled trial. JAMA. 2001;285(20):2583–93. doi: 10.1001/jama.285.20.2583. [DOI] [PubMed] [Google Scholar]
- (39).Deykin A, Wechsler ME, Boushey HA, Chinchilli VM, Kunselman SJ, Craig TJ, et al. Combination therapy with a long-acting beta-agonist and a leukotriene antagonist in moderate asthma. Am J Respir Crit Care Med. 2007;175(3):228–34. doi: 10.1164/rccm.200601-112OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.