Abstract
Recent generation of induced pluripotent stem (iPSCs) has made a significant impact on the field of human regenerative medicine. Prior to the clinical application of iPSCs, testing of their safety and usefulness must be carried out using reliable animal models of various diseases. In order to generate iPSCs from common marmoset (CM; Callithrix jacchus), one of the most useful experimental animals, we have lentivirally transduced reprogramming factors, including POU5F1 (also known as OCT3/4), SOX2, KLF4, and c-MYC into CM fibroblasts. The cells formed round colonies expressing embryonic stem cell markers, however, they showed an abnormal karyotype denoted as 46, X, del(4q), +mar, and formed human dysgerminoma-like tumors in SCID mice, indicating that the transduction of reprogramming factors caused unexpected tumorigenesis of CM cells. Moreover, CM dysgerminoma-like tumors were highly sensitive to DNA-damaging agents, irradiation, and fibroblast growth factor receptor inhibitor, and their growth was dependent on c-MYC expression. These results indicate that DNA-damaging agents, irradiation, fibroblast growth factor receptor inhibitor, and c-MYC-targeted therapies might represent effective treatment strategies for unexpected tumors in patients receiving iPSC-based therapy.
Keywords: Common marmoset, FGFR, regenerating medicine, reprogramming factor, tumorigenesis
The development of a technology to generate iPSCs from differentiated somatic cells by the transduction of a set of transcription factors, OSKM, made a significant impact in the field of basic research for regenerative medicine, in light of their potential use as a cell source for transplantation therapy for various kinds of incurable diseases.(1,2)
However, the low efficiency of iPSC generation, the need to induce their efficient differentiation into specific cell types, and the risk of tumor formation in recipients transplanted with iPSC-derived functional cells have hindered the clinical application of iPSCs.(3) The transduction of transcription factors, including the oncogene c-MYC, insertional mutation of the genome caused by virus vectors, and genomic instability due to the stress of long-term culture for reprogramming, might contribute to tumor development when iPSC-derived functional cells are applied to clinical practice.(4) Although the technology used to generate iPSCs has improved with the use of OSK without M,(5) non-viral vectors,(6) other molecules such as mRNA or miRNA,(7) and chemicals,(8) tumor formation in recipients remains a major concern.(9) Despite these issues, reprogramming factor-related tumor cells have not been well-characterized to date.
The CM (Callithrix jacchus) has several advantages as an experimental laboratory primate, including ease of handling, being inexpensive to house and feed, and a high reproductive rate.(10) Therefore, CM and CM-derived iPSCs represent useful experimental tools for testing the clinical utility of iPSC-based regenerative medicine in vivo and in vitro.
In this study, we attempted to generate iPSCs from CM fibroblasts, and inadvertently produced immature malignant tumor cells. We therefore analyzed the biological characteristics of these cells in vitro and in vivo. The results may provide useful information for the development of strategies to deal with tumors unexpectedly formed in patients treated with iPSC-based therapies.
Materials and Methods
Cell culture, induction of reprogramming, and proliferation assay
Common marmoset ARCs, CM ESCs, and iPS A cells derived from fetal liver cells (provided by Erika Sasaki, KEIO-REKEN Research Center for Human Cognition, Keio University, Tokyo, Japan) were maintained in DMEM/F12 (Sigma-Aldrich, St. Louis, MO, USA) containing 20% Knockout Serum Replacement (Gibco, Carlsbad, CA, USA), 0.1 mM non-essential amino acid (Gibco), 1 mM L-glutamine (Nacalai Tesque, Kyoto, Japan), 1% antibiotic–antimycotics (Nacalai Tesque), 0.4 mM 2-mercaptoethanol (Sigma Aldrich), and 0.12% sodium hydroxide (Nacalai Tesque). The CM DGs were maintained in DMEM/F12 containing 10% FBS at 37°C in a 5% humidified CO2 atmosphere. Detailed descriptions of the cell culture, reprogramming method, and proliferation assay are provided in Figures 1 and 5.
Fig. 1.
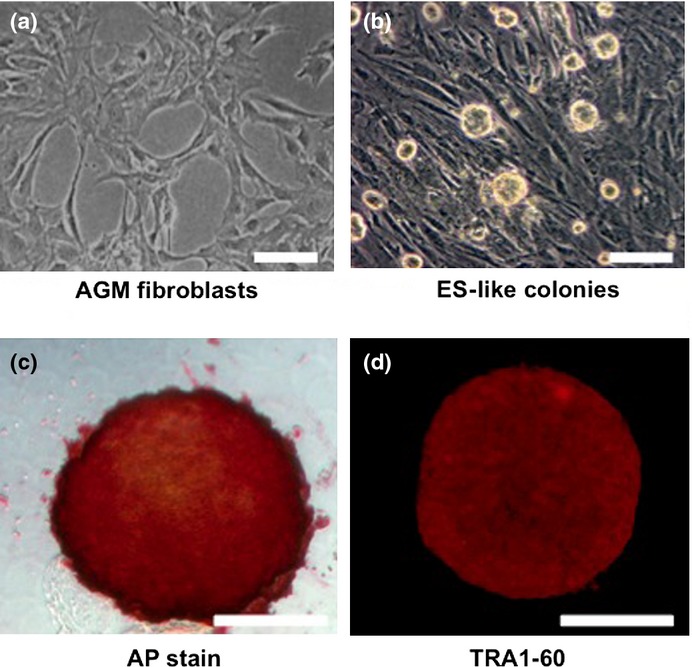
Characterization of aorta-gonado-mesonephros (AGM) fibroblast-derived colonies formed by transduction of reprogramming factors. Representative phase-contrast images of (a) AGM fibroblasts and (b) abnormally reprogrammed cells (ARCs) forming round-shaped colonies. (c) Representative image showing expression of alkaline phosphatase (AP) activity in ARCs. (d) Immunocytochemical staining showing expression of TRA1-60 in ARCs. Bar = 100 μm.
Fig. 5.
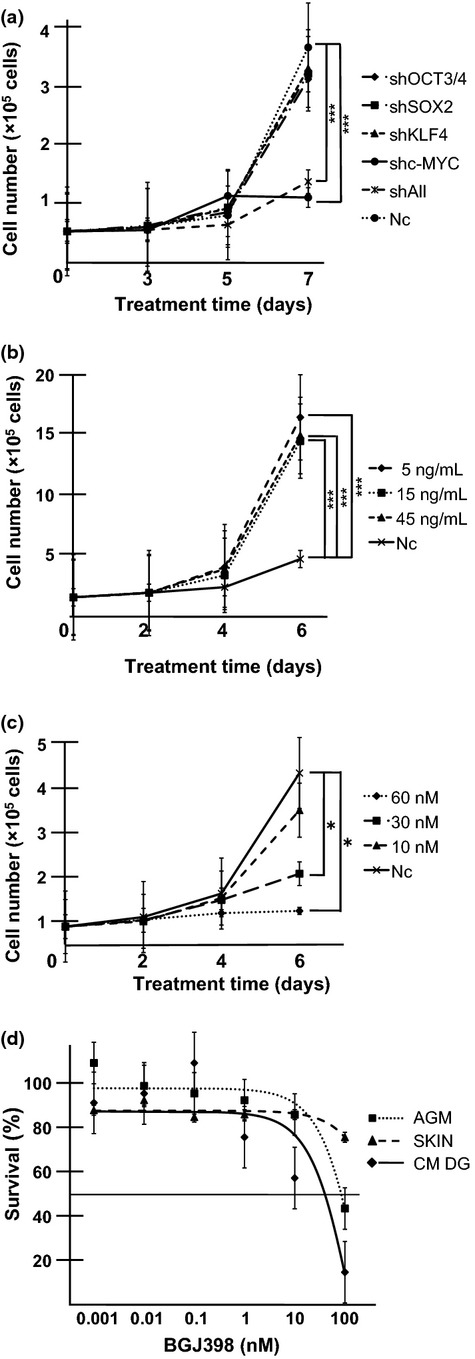
Dependence of common marmoset dysgerminoma-like (CM DG) cell growth on c-MYC and basic fibroblast growth factor (bFGF) signaling. (a) Inhibition of CM DG growth by knockdown of c-MYC. Cells (3 × 104) were seeded on 24-well plates and transduced with shRNA targeting OCT3/4, SOX2, KLF4, c-MYC, or all reprogramming factors (shAll). Cell growth curves were analyzed by cell counts at the indicated time points. Results are shown as means ± SD. ***P < 0.001. Nc, negative control (mock vector). (b) Growth rate of CM DGs was promoted by the addition of bFGF. Cells were cultured in the presence or absence (Nc) of bFGF. Cell numbers were counted at the indicated time points. Results are shown as means ± SD. ***P < 0.001. (c) FGFR inhibitor suppressed CM DG growth. Cells were cultured in the presence or absence (Nc) of the FGFR1-4 inhibitor BGJ398; bFGF was added at 5 ng/mL. Cell numbers were counted at the indicated time points. Results are shown as means ± SD. *P < 0.05. (d) CM DGs, aorta-gonado-mesonephros fibroblasts (AGM), and CM skin fibroblasts (SKIN) were treated with different concentrations of BGJ398 for 3 days, and the growth-inhibitory effects were analyzed by MTS assay. The IC50 for CM DGs was lower than those for parental AGM fibroblasts and control CM skin fibroblasts. Results are shown as means ± SD.
Plasmids and lentiviral vector production
Human OCT3/4, SOX2, KLF4, or c-MYC was inserted into CSIV-CMV-MCS-IRES2-Venus lentiviral vectors (kindly provided by Hiroyuki Miyoshi, Riken, Tsukuba, Japan). Short hairpin RNAs targeting OCT3/4, SOX2, and c-MYC were obtained from Addgene (Cambridge, MA, USA), and shRNA targeting KLF4 was obtained from Applied Biological Materials (Richmond, BC, Canada). Lentiviruses were produced as previously described.(11)
Microarray analysis
Total RNA from AGM fibroblasts, ARCs, and iPS A cells were isolated using the RNeasy Mini Kit (Qiagen, Valencia, CA, USA). RNA was reverse-transcribed, biotin-labeled, and hybridized for 16 h to a marmoset genome oligonucleotide custom array Marmo2 (in preparation)(12), which was subsequently washed and stained in a Fluidics Station 450 (Affymetrix, Santa Clara, CA, USA) according to the manufacturer's instructions. Detailed protocols of microarray analysis are provided in Figures 2 and S5.
Fig. 2.
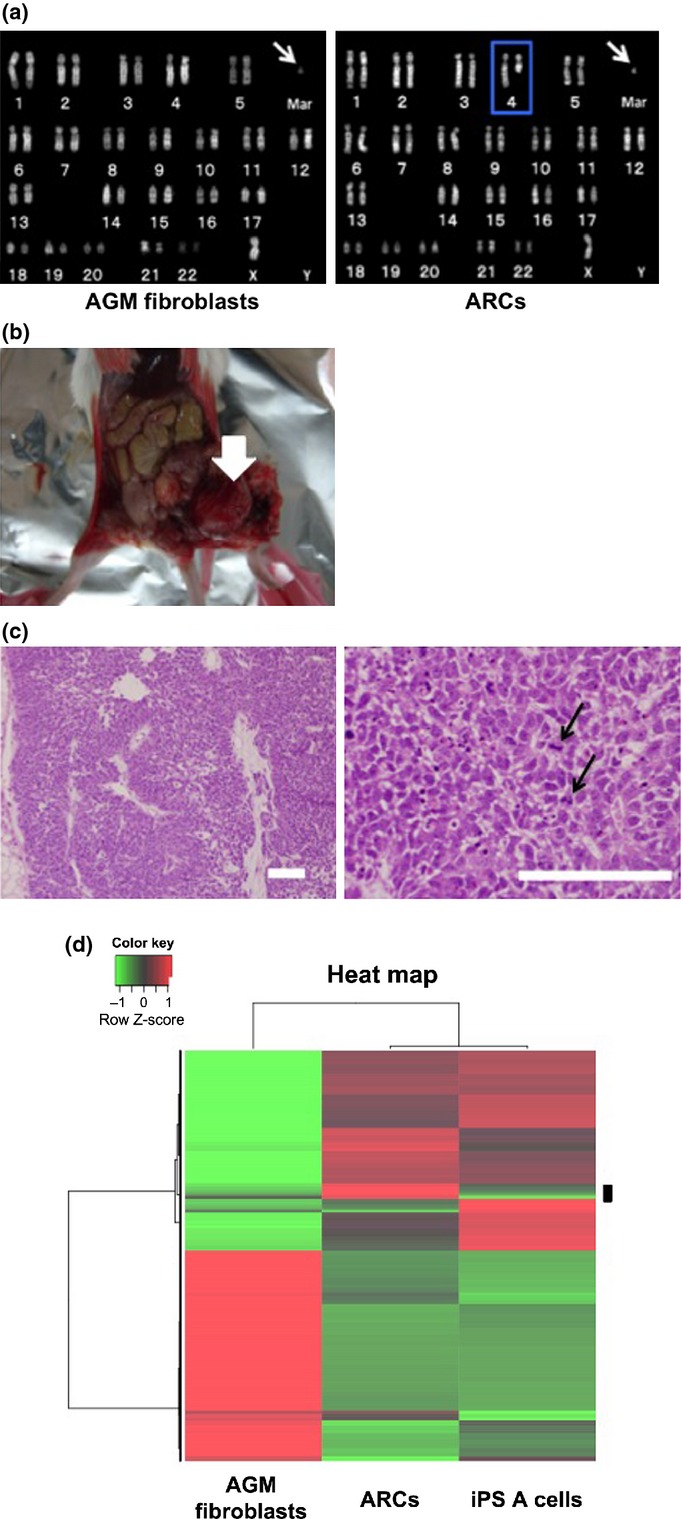
Chromosome abnormality and tumor-forming ability in abnormally reprogrammed cells (ARCs). (a) Karyotype analyses of aorta-gonado-mesonephros (AGM) fibroblasts (left panel) and ARCs (right panel). Arrows indicate marker chromosome. Blue outline indicates the deletion of 4q. Mar, marker chromosome. (b) Representative photograph of dysgerminoma-like tumor (arrow) formed by transplantation of ARCs into SCID mice. (c) Hematoxylin-eosin staining of dysgerminoma-like tumor tissues. Arrows in right panel indicate mitotic figures in tumor cells. Bar = 100 μm. (d) Microarray analysis. Gene expressions in AGM fibroblasts, ARCs, and normal induced pluripotent stem (iPS) A cells were analyzed by unsupervised hierarchical clustering. A heat map using probes showing differential expression levels in each cell line is shown. Red indicates upregulation; green indicates downregulation. The black bar on the right side of the heat map shows candidate differentially expressed probes in ARCs.
DNA-damaging treatments
The CM DGs were treated with 1 μg/mL MMC (Kyowa Hakko Kirin, Tokyo, Japan) or 10 μg/mL cisplatin (Sigma-Aldrich) for 1 h at 37°C. For irradiation, CM DGs were irradiated (20 Gy) using Gammacell 40 (Atomic Energy, Chalk River, Ontario, Canada). At 24 h after treatment, the cells were stained with propidium iodide (Nacalai Tesque), and the proportion of dead cells was analyzed as the sub-G1 population by flow cytometry (FACSCalibur; BD Biosciences, San Jose, CA, USA).
Statistical analysis
Statistical analyses were carried out with the graphpad prism 5.0d software package (GraphPad Software, La Jolla, CA, USA). Statistical analyses were carried out using a two-tailed unpaired Student's t-test or one-way anova followed by Tukey's multiple comparison test. P < 0.05 was considered statistically significant.
Additional information is provided in Supporting information.
Results
Characteristic of aorta-gonado-mesonephros fibroblast-derived colonies formed by transduction of reprogramming factors
To generate CM-derived iPSCs, reprogramming factors (OSKM) were transduced into AGM fibroblasts using lentiviral vectors (Fig. 1a). Then OSKM-transduced cells were transferred to mouse embryonic fibroblast feeder cells on day 7 post-infection, and cultured in medium for CM ESCs. We found that the cells formed sphere-like structures on day 17 post-infection (Fig. 1b). Moreover, these colonies showed AP activity (Fig. 1c), and expressed ESC markers such as TRA1-60, SALL1, LIN28, and DPPA4 (Figs 1d, S1). These results suggested that the reprogrammed AGM fibroblasts formed immature, round iPSC-like colonies.
Chromosome abnormality and tumor-forming ability in abnormally reprogrammed cells
Given that KLF4 and c-MYC are well-known oncogenes,(13,14) transduction with OSKM transcription factors may cause cell transformation and chromosome instability.(15) We carried out karyotype analysis of the colony-forming cells to determine if OSKM-transduced AGM fibroblasts exhibited chromosome instability. The normal karyotype of CM cells is 44 autosomes and two sex chromosomes (46, XX or 46, XY).(16) However, the round colony-forming cells contained 44 autosomes, one X chromosome, and an abnormal marker chromosome (mar), with deletions of chromosome 4q, and were therefore denoted as 46, X, del (4q), +mar (Fig. 2a, right panel). The karyotype of the parental AGM fibroblasts was 46, X, +mar (Fig. 2a, left panel). These results suggested that the reprogramming stress induced by OSKM might have caused the deletion of 4q, although the possibility that the stress of long-term in vitro culture might have resulted in chromosome instability could not be excluded. These colony-forming cells were named ARCs.
To examine the ability of ARCs to differentiate into three germ layers like ESCs, we carried out an in vitro differentiation assay based on the protocol for human ESC differentiation.(17,18) Unlike human ESCs, ARCs did not differentiate into neural progenitors, cardiomyocytes, or hepatic cells (Fig. S2, Video S1).
We carried out in vivo differentiation assays by injecting ARCs into the testes of SCID mice. Approximately 6 weeks after injection, 11 of 18 mice injected with ARCs showed tumor formation (Fig. 2b), whereas no mice injected with AGM fibroblasts showed tumor formation (0/3, data not shown). Staining with H&E revealed that the tumors were relatively homogenous, with high cellular density, necrosis, and pleomorphism, indicating their malignant phenotype (Fig. 2c). In addition, tumors were composed of nests and sheets of uniform round or polygonal cells with abundant, clear to faintly eosinophilic cytoplasm with well-demarcated cytoplasmic borders and a delicate network of thin-walled blood vessels in the tumor nests (Fig. 2c, right panel). Furthermore, immunohistochemical analyses revealed that the tumor cells were focally and weakly immunopositive for vimentin, and immunonegative for the differentiation markers cytokeratin, S100, desmin, α-smooth muscle actin, and neuron-specific enolase (data not shown). Tumor tissues also expressed c-KIT, but not CD30 or CD45 (Fig. S3). These molecular expression profiles implied that the tumor was equivalent to human malignant dysgerminoma, rather than other types of immature tumors such as embryonal carcinoma, yolk sac tumor, or teratoma.(19,20) The tumor was named CM DG.
We next carried out soft agar assays to determine if ARCs were transformed and showed anchorage-independent growth as a result of ectopic expression of reprogramming factors. The ARCs were cultured in 0.5% agarose-containing medium for 20 days, and the number of colonies was counted. The ARCs formed many colonies, compared with parental AGM fibroblasts (Fig. S4a and data not shown). These results strongly suggested that ARCs were transformed during reprogramming, and acquired the capacity for anchorage-independent growth. To clarify the contribution of reprogramming factors that could transform AGM fibroblasts, we transduced various combinations of these factors into AGM fibroblasts, and examined if the transduced cells were transformed by the colony formation assay on mouse embryonic fibroblasts, AP staining assay, and soft agar assay. The iPSC-like colonies were found in OSKM- and OSM-transduced cells (OSKM, 30 ± 3/5000; OSM, 6 ± 0/5000), but they were not found at all when OSK, OS, OM, SM, O, S, K, or M were transduced (Fig. S4b). AP activity was found in both OSKM- and OSM-transduced cells, although OSM-transduced cells showed weaker AP activity than OSKM-transduced cells (Fig. S4c). In soft agar assay, the anchorage-independent growth was found in both OSKM- and OSM-transduced cells (Fig. S4d; OSKM, 160 ± 23/1000; OSM, 163 ± 10/1000). These results indicated that the simultaneous expression of OCT3/4, SOX2, and c-MYC was at least required for the transformation of AGM fibroblasts, while KLF4 did not play a major role in the transformation of AGM fibroblasts.
Tomioka et al. reported the establishment of CM iPSCs (iPS A cells) showing normal karyotype.(12) To characterize the gene expression in ARCs, we carried out microarray analyses using mRNA from ARCs, iPS A cells, and AGM fibroblasts. According to the clustering pattern and the heat map, 171 probes that showed higher expression levels in ARCs as compared to other cells were selected as candidate differentially expressed genes in ARCs (Fig. 2d). Moreover, we focused on the genes specifically highly expressed in ARCs compared to those in iPS A cells, and the top seven genes highly expressed in ARCs were selected (ZFHX4, PCDH19, NFIX, HOXC8, STMN2, SERPINA3, and CXORF67). Then we validated these data by semiquantitative RT-PCR analyses, and five genes (ZFHX4, NFIX, HOXC8, STMN2, and CXORF67) were confirmed to be more expressed in ARCs than those in controls (Fig. S5). The high expression of these five genes might be characteristics of ARCs.
Characteristic of CM DGs
We then surgically removed CM DGs and cultured them in vitro to examine their biological characteristics. The CM DGs could grow infinitely in a semifloating state in the culture dish, and showed continuous expression of Venus fluorescent protein (Fig. 3a,b). We generated five CM DG cell lines (CMY401, CMY402a, CMY402b, CMY403a, and CMY403b) from five independent tumors formed by the injection of ARCs into SCID mice, and found that all four transduced reprogramming factors were integrated into their genomes (Fig. S6a). Both endogenous and exogenous reprogramming factors were expressed in these cell lines (Fig. 3c,d).
Fig. 3.
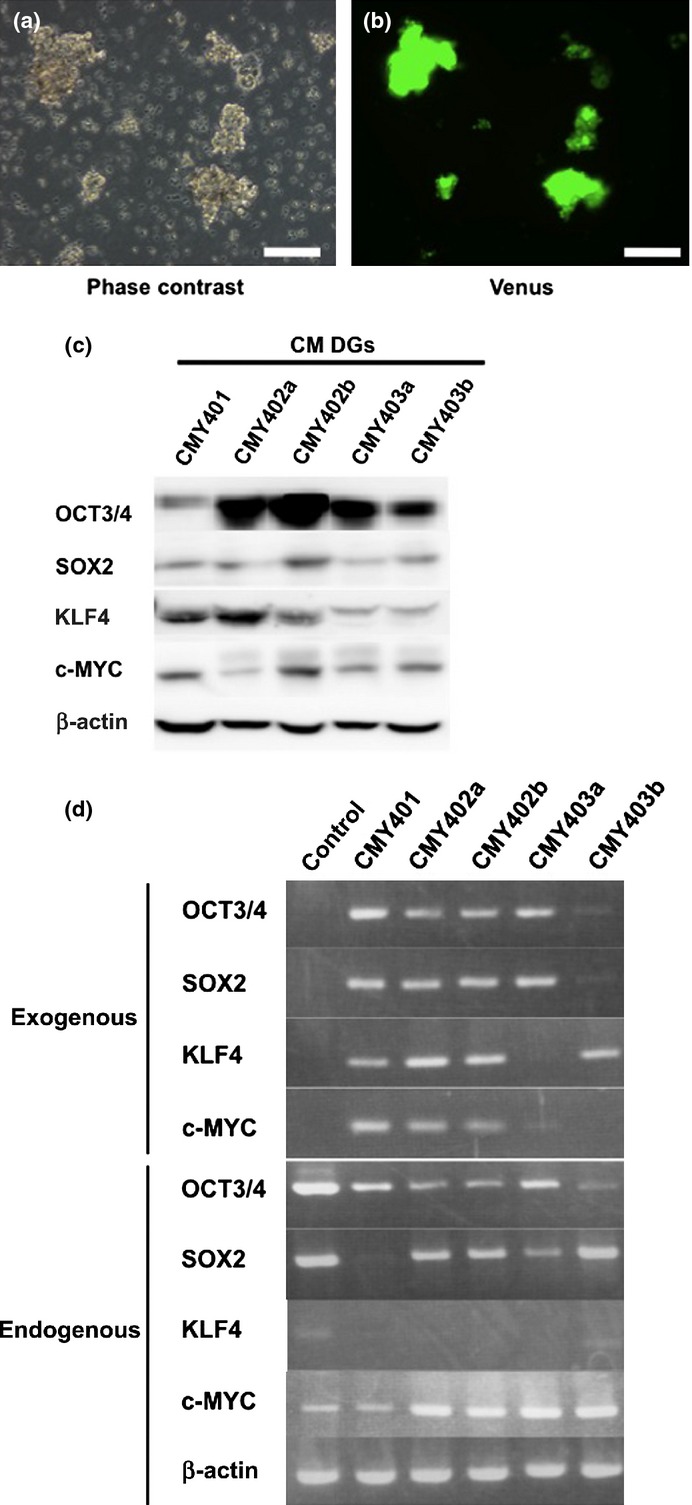
Characterization of common marmoset dysgerminoma-like (CM DG) cells in culture. (a) Representative phase-contrast image of CM DGs. (b) Immunofluorescent image of Venus expression in CM DGs. Bar = 100 μm. (c) Western blot analysis showing expression of reprogramming factors in CM DG cell lines. (d) RT-PCR analysis showing the expression of endogenous or exogenous reprogramming factors in CM DGs. Cj11 (CM embryonic stem cell line) was used as control.
Effects of DNA-damaging agents and irradiation on CM DGs
Dysgerminomas are generally sensitive to cisplatin and irradiation.(21,22) We therefore examined the effects of DNA-damaging agents such as MMC and cisplatin on CM DGs. Three CM DG cell lines (CMY402a, CMY402b, and CMY403a) were treated with MMC for 1 h, and the proportion of dead cells was analyzed by flow cytometry at 24 h after treatment. The percentage of cells with a sub-G1 DNA content was taken as a measure of dead cells in the population. The proportion of dead cells in MMC-treated CM DG cultures was significantly higher than that in controls (MMC-treated AGM fibroblasts) (Figs 4a, S7). Similar results were obtained when these three cell lines were treated with cisplatin or irradiation (Figs 4b,c, S8, S9). These results suggested that CM DGs were more sensitive to DNA damage than their parental AGM fibroblasts.
Fig. 4.
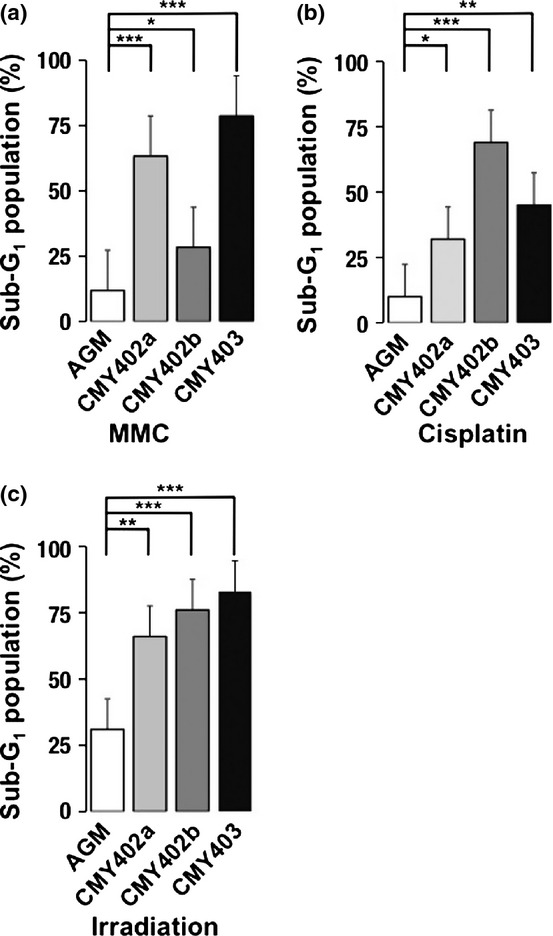
Effects of DNA-damaging agents and irradiation on common marmoset dysgerminoma-like cells (CM DGs). The cells were treated with (a) mitomycin C (MMC), (b) cisplatin, or (c) irradiation, and the proportions of sub-G1 populations in aorta-gonado-mesonephros (AGM) fibroblasts or CM DG cell lines were analyzed by FACS. Results are shown as means ± SD. *P < 0.05; **P < 0.01; ***P < 0.001.
We carried out inverse PCR analyses to identify the integration sites of the lentiviral vectors expressing reprogramming factors in CM DGs. OCT 3/4-, SOX2-, KLF4-, and c-MYC-expressing lentiviral vectors were integrated into 5, 12, 5, and 9 genomic sites, respectively (Fig. S6b, Table S1). The possibility that multiple integrations of lentiviral vectors into the genome caused chromosome instability, leading to the formation of CM DGs, could therefore not be excluded.
Dependence of CM DGs growth on c-MYC and bFGF signalings
To address the question of whether proliferation of CM DGs was dependent on reprogramming factors, we observed the proliferation rate after suppression of each reprogramming factor by shRNA (Fig. S10). Suppression of c-MYC or all four reprogramming factors greatly inhibited the proliferation of CM DGs, indicating that the growth of CM DGs was highly dependent on c-MYC (Fig. 5a).
The proliferation of human ESCs is known to be promoted by bFGF signaling.(23) We examined the possibility that the growth of CM DGs might also be enhanced by bFGF signaling by analyzing the proliferation of CM DGs cultured in medium with or without bFGF. The growth of CM DGs was highly dependent on bFGF (Fig. 5b). Consistent with these results, BGJ398, an inhibitor for FGFR 1 to 4, remarkably inhibited the growth of CM DGs in a dose-dependent manner (Fig. 5c). Moreover, FACS analyses revealed that the sub-G1 population, representing dead cells, was increased in the presence of BGJ398 (Fig. S11). It should be emphasized that the IC50 of BGJ398 (59 nM) was lower for CM DGs than for their parental AGM fibroblasts and control CM skin fibroblasts (Fig. 5d), indicating higher sensitivity of CM DGs. These results suggested that the growth of CM DGs was dependent on bFGF signaling, and therefore FGFR inhibitor could be used to control the growth of the reprogramming factor-induced tumor.
Discussion
In this study we investigated the characteristics of ARCs and CM DGs generated in the reprogramming process of CM AGM fibroblasts by Yamanaka factors.
A normal iPSC line of iPS A cells, showed the expression of ES markers, pluripotency, and flattened morphology, like human iPSCs.(20) In contrast, ARCs showed sphere-like structures, like mouse iPSCs.(1) This morphological difference between iPS A cells and ARCs might be useful to select “true” iPSCs derived from CM, although the underlying molecular mechanisms responsible for this morphological difference remain unknown.
We found, by microarray analyses, that the gene expression pattern in ARCs was more similar to that in iPS A cells than that in AGM fibroblasts, suggesting that reprogramming processes have been done in ARCs by the transduction of reprogramming factors. We also found that genes such as ZFHX4, NFIX, HOXC8, STMN2, and CXORF67 were highly expressed in ARCs. It should be noted that, among these, HOXC8 is known to be a transcriptional factor related to tumorigenesis.(24) Therefore, these candidates of markers might be useful to predict the tumorigenic potential of iPSCs. Further evaluation is required to confirm our hypothesis.
The original AGM fibroblasts had an abnormal marker chromosome (mar; Fig. 2a, left panel). Although tumor formation was not evident caused in SCID mice (data not shown), this chromosome instability might also be one of the inducers of carcinogenesis during the reprogramming process. Thus, needless to say, to generate “safe” iPSCs, validation of the karyotype of the original cells is needed. Moreover, ARCs lost chromosome 4q and X or Y, and possessed an abnormal marker chromosome (mar). Various tumor suppressors including human tumor suppressor gene 1, large tumor suppressor 1 and P36 transformed follicular lymphoma gene have been identified on chromosome 4q in CM cells (Table S2), suggesting that loss of these tumor suppressors might have induced the transformation of CM AGM fibroblasts during reprogramming, although the possibility that translocation of chromosome 4q occurred during the reprogramming process caused the transformation of cells could not be excluded.
It is also possible that the continuous activation of ectopically-transduced transcription factors, including the oncogene c-MYC, might have contributed to cell transformation, as described previously.(25,26) Indeed, CM DGs overexpressed c-MYC, and their growth was highly dependent on c-MYC expression, suggesting that downregulation of c-MYC might represent a possible strategy for inhibiting the growth of reprogramming factor-related tumors.
Insertional mutation caused by the integration of lentiviral vectors into the genome might also have promoted cell transformation. Lentiviral vectors expressing reprogramming factors were integrated into at least 31 different genomic sites in CM DGs, some of which were in the vicinity of protein-encoding genes. Moreover, the expression of reprogramming factors transduced by lentiviral vectors continued for over a year in ARCs (data not shown). A safer method, without genome integration, is therefore required for the delivery of reprogramming factors to somatic cells to generate iPSCs applicable for transplantation therapies. Although Sendai virus vectors or transfection of DNA or mRNA may be safer methods,(27) these are lengthy processes that can take more than 1 month to obtain iPSCs,(28) which could also cause stress and lead to genomic instability and subsequent tumor formation. More sophisticated, safer, and more rapid methods of reprogramming might be desirable.
Common marmoset DGs resembled human dysgerminomas in terms of both their pathology and sensitivity to irradiation and DNA-damaging agents.(21,22) In addition, the growth of CM DGs was significantly inhibited by an FGFR1-4 inhibitor. Therefore irradiation, chemotherapy, and FGFR1-4 inhibitors might be effective strategies for controlling human dysgerminomas, and also for tumors that develop in patients treated with iPSC-based therapies.
Acknowledgments
We thank Hiroyuki Miyoshi (Riken, Tsukuba, Japan) for providing lentiviral vectors, Norihiko Kinoshita (Kyushu University, Fukuoka, Japan) and the Laboratory for Technical Support (Medical Institute of Bioregulation, Kyushu University) for their technical assistance, Michiko Ushijima for administrative assistance, and members of Prof. Kenzaburo Tani's laboratory for constructive criticisms. This work was supported by grants from the Project for Realization of Regenerative Medicine (K.T., 08008010) and Kakenhi (T.M., 23590465) from the Ministry of Education, Culture, Sports, Science and Technology, Japan.
Glossary
- AGM
aorta-gonado-mesonephros
- AP
alkaline phosphatase
- ARC
abnormally reprogrammed cell
- bFGF
basic fibroblast growth factor
- CM
common marmoset
- DG
human dysgerminoma-like cell
- ESC
embryonic stem cell
- FGFR
fibroblast growth factor receptor
- iPSC
induced pluripotent stem cell
- K
KLF4
- M
c-MYC
- MMC
mitomycin C
- O
OCT3/4
- S
SOX2
Disclosure Statement
The authors have no conflicts of interest.
Supporting Information
Additional supporting information may be found in the online version of this article:
Fig. S1 Expression of embryonic stem cell (ESC) markers in abnormally reprogrammed cells (ARCs).
Fig. S2 Impaired differentiation of abnormally reprogrammed cells (ARCs).
Fig. S3 Expression of c-KIT, CD30, and CD45 in common marmoset dysgerminoma-like cells.
Fig. S4 Colony formation of aorta-gonado-mesonephros fibroblasts by the transduction of OCT3/4, SOX2, KLF4, and c-MYC (OSKM) and OSM.
Fig. S5 Validation of genes showing upregulation in abnormally reprogrammed cells (ARCs) compared to normal induced pluripotent stem (iPS) A cells in microarray analysis.
Fig. S6 Integration of reprogramming genes into the genome of common marmoset dysgerminoma-like cells (CM DGs).
Fig. S7 Fluorescence-activated cell sorter analyses to reveal effects of mitomycin C treatment on common marmoset dysgerminoma-like cell lines.
Fig. S8 Fluorescence-activated cell sorter analyses to reveal effects of cisplatin treatment on common marmoset dysgerminoma-like cell lines.
Fig. S9 Fluorescence-activated cell sorter analyses to reveal effects of irradiation on common marmoset dysgerminoma-like cell lines.
Fig. S10 Knockdown of OCT3/4, SOX2, KLF4, or c-MYC by shRNA in common marmoset dysgerminoma-like cell lines.
Fig. S11 Induction of cell death in common marmoset dysgerminoma-like cells by BGJ398.
Table S1 Lentiviral vector integration sites in common marmoset (CM) dysgerminoma-like cells.
Table S2 Human homologs of candidate tumor suppressors located on chromosome 4q in common marmoset (CM).
Video S1 In vitro differentiation assay to assess the ability of abnormally reprogrammed cells to differentiate into cardiomyocytes.
Data S1 Materials and Methods.
References
- 1.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–76. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 2.Okita K, Yamanaka S. Induced pluripotent stem cells: opportunities and challenges. Philos Trans R Soc Lond B Biol Sci. 2011;366:2198–207. doi: 10.1098/rstb.2011.0016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Okita K, Ichisaka T, Yamanaka S. Generation of germline-competent induced pluripotent stem cells. Nature. 2007;448:313–U1. doi: 10.1038/nature05934. [DOI] [PubMed] [Google Scholar]
- 4.Ohm JE, Mali P, Van Neste L, et al. Cancer-related epigenome changes associated with reprogramming to induced pluripotent stem cells. Cancer Res. 2010;70:7662–73. doi: 10.1158/0008-5472.CAN-10-1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wernig M, Meissner A, Cassady JP, Jaenisch R. c-Myc is dispensable for direct reprogramming of mouse fibroblasts. Cell Stem Cell. 2008;2:10–2. doi: 10.1016/j.stem.2007.12.001. [DOI] [PubMed] [Google Scholar]
- 6.Okita K, Nakagawa M, Hong HJ, Ichisaka T, Yamanaka S. Generation of mouse induced pluripotent stem cells without viral vectors. Science. 2008;322:949–53. doi: 10.1126/science.1164270. [DOI] [PubMed] [Google Scholar]
- 7.Judson RL, Babiarz JE, Venere M, Blelloch R. Embryonic stem cell-specific microRNAs promote induced pluripotency. Nat Biotechnol. 2009;27:459–61. doi: 10.1038/nbt.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lin TX, Ambasudhan R, Yuan X, et al. A chemical platform for improved induction of human iPSCs. Nat Methods. 2009;6:805–U24. doi: 10.1038/nmeth.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Madonna R. Human-induced pluripotent stem cells. in quest of clinical applications. Mol Biotechnol. 2012;52:193–203. doi: 10.1007/s12033-012-9504-0. [DOI] [PubMed] [Google Scholar]
- 10.Lunn SF. Systems for collection of urine in the captive common marmoset, Callithrix-Jacchus. Lab Anim. 1989;23:353–6. doi: 10.1258/002367789780745935. [DOI] [PubMed] [Google Scholar]
- 11.Marumoto T, Tashiro A, Friedmann-Morvinski D, et al. Development of a novel mouse glioma model using lentiviral vectors. Nat Med. 2009;15:110–6. doi: 10.1038/nm.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tomioka I, Maeda T, Shimada H, et al. Generating induced pluripotent stem cells from common marmoset (Callithrix jacchus) fetal liver cells using defined factors, including Lin28. Genes Cells. 2010;15:959–69. doi: 10.1111/j.1365-2443.2010.01437.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Foster KW, Frost AR, McKie-Bell P, et al. Increase of GKLF messenger RNA and protein expression during progression of breast cancer. Cancer Res. 2000;60:6488–95. [PubMed] [Google Scholar]
- 14.Gustafson WC, Weiss WA. Myc proteins as therapeutic targets. Oncogene. 2010;29:1249–59. doi: 10.1038/onc.2009.512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vogelstein B. Genetic instabilities in human cancers. Biophys J. 1999;76:A135–A. doi: 10.1038/25292. [DOI] [PubMed] [Google Scholar]
- 16.Sasaki E, Hanazawa K, Kurita R, et al. Establishment of novel embryonic stem cell lines derived from the common marmoset (Callithrix jacchus. Stem Cells. 2005;23:1304–13. doi: 10.1634/stemcells.2004-0366. [DOI] [PubMed] [Google Scholar]
- 17.Li XJ, Du ZW, Zarnowska ED, et al. Specification of motoneurons from human embryonic stem cells. Nat Biotechnol. 2005;23:215–21. doi: 10.1038/nbt1063. [DOI] [PubMed] [Google Scholar]
- 18.Liao JY, Marumoto T, Yamaguchi S, et al. Inhibition of PTEN tumor suppressor promotes the generation of induced pluripotent stem cells. Mol Ther. 2013;21:1242–50. doi: 10.1038/mt.2013.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.1998. pp. 239–65. Tumor of the ovary, Maldeveloped Gonads, Fallopian tube, and Broad Ligament.
- 20.Ulbright TM. Germ cell tumors of the gonads: a selective review emphasizing problems in differential diagnosis, newly appreciated, and controversial issues. Mod Pathol. 2005;18(Suppl 2):S61–79. doi: 10.1038/modpathol.3800310. [DOI] [PubMed] [Google Scholar]
- 21.Brewer M, Gershenson DM, Herzog CE, Mitchell MF, Silva EG, Wharton JT. Outcome and reproductive function after chemotherapy for ovarian dysgerminoma. J Clin Oncol. 1999;17:2670–5. doi: 10.1200/JCO.1999.17.9.2670. [DOI] [PubMed] [Google Scholar]
- 22.Thoeny RH, Dockerty MB, Hunt AB, Childs DS., Jr A study of ovarian dysgerminoma with emphasis on the role of radiation therapy. Surg Gynecol Obstet. 1961;113:692–8. [PubMed] [Google Scholar]
- 23.Bendall SC, Stewart MH, Menendez P, et al. IGF and FGF cooperatively establish the regulatory stem cell niche of pluripotent human cells in vitro. Nature. 2007;448:1015–U3. doi: 10.1038/nature06027. [DOI] [PubMed] [Google Scholar]
- 24.Li Y. HOXC8-dependent cadherin 11 expression facilitates breast cancer cell migration through trio and rac. Genes Cancer. 2011;2:880–8. doi: 10.1177/1947601911433129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nakagawa M, Koyanagi M, Tanabe K, et al. Generation of induced pluripotent stem cells without Myc from mouse and human fibroblasts. Nat Biotechnol. 2008;26:101–6. doi: 10.1038/nbt1374. [DOI] [PubMed] [Google Scholar]
- 26.Gordan JD, Thompson CB, Simon MC. HIF and c-Myc: sibling rivals for control of cancer cell metabolism and proliferation. Cancer Cell. 2007;12:108–13. doi: 10.1016/j.ccr.2007.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Seki T, Yuasa S, Fukuda K. Derivation of induced pluripotent stem cells from human peripheral circulating T cells. Curr Protoc Stem Cell Biol. 2011:11–14. doi: 10.1002/9780470151808.sc04a03s18. Chapter 4: Unit4A.3. [DOI] [PubMed] [Google Scholar]
- 28.Buganim Y, Faddah DA, Cheng AW, et al. Single-cell expression analyses during cellular reprogramming reveal an early stochastic and a late hierarchic phase. Cell. 2012;150:1209–22. doi: 10.1016/j.cell.2012.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Expression of embryonic stem cell (ESC) markers in abnormally reprogrammed cells (ARCs).
Fig. S2 Impaired differentiation of abnormally reprogrammed cells (ARCs).
Fig. S3 Expression of c-KIT, CD30, and CD45 in common marmoset dysgerminoma-like cells.
Fig. S4 Colony formation of aorta-gonado-mesonephros fibroblasts by the transduction of OCT3/4, SOX2, KLF4, and c-MYC (OSKM) and OSM.
Fig. S5 Validation of genes showing upregulation in abnormally reprogrammed cells (ARCs) compared to normal induced pluripotent stem (iPS) A cells in microarray analysis.
Fig. S6 Integration of reprogramming genes into the genome of common marmoset dysgerminoma-like cells (CM DGs).
Fig. S7 Fluorescence-activated cell sorter analyses to reveal effects of mitomycin C treatment on common marmoset dysgerminoma-like cell lines.
Fig. S8 Fluorescence-activated cell sorter analyses to reveal effects of cisplatin treatment on common marmoset dysgerminoma-like cell lines.
Fig. S9 Fluorescence-activated cell sorter analyses to reveal effects of irradiation on common marmoset dysgerminoma-like cell lines.
Fig. S10 Knockdown of OCT3/4, SOX2, KLF4, or c-MYC by shRNA in common marmoset dysgerminoma-like cell lines.
Fig. S11 Induction of cell death in common marmoset dysgerminoma-like cells by BGJ398.
Table S1 Lentiviral vector integration sites in common marmoset (CM) dysgerminoma-like cells.
Table S2 Human homologs of candidate tumor suppressors located on chromosome 4q in common marmoset (CM).
Video S1 In vitro differentiation assay to assess the ability of abnormally reprogrammed cells to differentiate into cardiomyocytes.
Data S1 Materials and Methods.


