Abstract
Diffuse large B-cell lymphoma (DLBCL) displays striking heterogeneity at the clinical, genetic and molecular levels. Subtypes include germinal center B-cell-like (GCB) DLBCL and activated B-cell-like (ABC) DLBCL, according to microarray analysis, and germinal center type or non-germinal center type by immunohistochemistry. Although some reports have described genomic aberrations based upon microarray classification system, genomic aberrations based upon immunohistochemical classifications have rarely been reported. The present study aimed to ascertain the relationship between genomic aberrations and subtypes identified by immunohistochemistry, and to study the pathogenetic character of Chinese DLBCL. We conducted immunohistochemistry using antibodies against CD10, BCL6 and MUM1 in 59 samples of DLBCL from Chinese patients, and then performed microarray-based comparative genomic hybridization for each case. Characteristic genomic differences were found between GCB and non-GCB DLBCL from the array data. The GCB type was characterized by more gains at 7q (7q22.1, P < 0.05) and losses at 16q (P ≤ 0.05), while the non-GCB type was characterized by gains at 11q24.3 and 3q13.2 (P < 0.05). We found completely different mutations in BCL6+ and BCL6− non-GCB type DLBCL, whereby the BCL6− group had a higher number of gains at 1q and a loss at 14q32.13 (P ≤ 0.005), while the BCL6+ group showed a higher number of gains at 14q23.1 (P = 0.15) and losses at 6q (P = 0.07). The BCL6− group had a higher frequency of genomic imbalances compared to the BCL6+ group. In conclusion, the BCL6+ and BCL6− non-GCB type of DLBCL appear to have different mechanisms of pathogenesis.
Keywords: Array-comparative genomic hybridization, Chinese, diffuse large B-cell lymphoma, genomic profiling, lymphoma classification
Diffuse large B-cell lymphoma (DLBCL), as defined by the World Health Organization (WHO) classification, is a clinically, morphologically and genetically heterogeneous group of malignant proliferations of large lymphoid B-cells. Gene expression profiling has identified two major molecularly distinct forms of DLBCL, with gene expression patterns indicative of different stages of B-cell differentiation: the activated B-cell-like (ABC) subtype expresses the genes characteristic of activated B-cells and plasma cells, and the germinal center B-cell-like (GCB) subtype, which maintains the gene expression pattern of normal germinal center B-cells.(1,2)
Several groups have also attempted to classify and investigate DLBCL using immunohistochemistry.(3,4) Han et al. use CD10 and BCL6 as GCB B-cell markers, and multiple myeloma-1/interferon regulatory factor-4 (MUM1/IRF-4) as an activated or non-GCB B-cell marker. Our previous array-comparative genomic hybridization (CGH) study revealed distinct differences between the pattern of genomic aberrations in the ABC and GCB subtypes, as identified by expression profiling.(5) To date, there have been no reports of the genomic aberrations detected by array CGH in the GCB and non-GCB subtypes, as differentiated using immunohistochemistry. Immunohistochemistry has been reported to correctly identify 70–80% of the cases identified by expression profiling.(3,4) In this context, it is still not well known what relationship exists between genomic aberrations and the immunohistochemical characteristics of both subtypes of DLBCL.
BCL6 is a marker for germinal center cells, whereas MUM1/IRF4 is expressed on B-cells that are on the verge of exiting or have exited the germinal center and are committed to plasma cell differentiation.(6) However, some cases of DLBCL co-express BCL6 and MUM1. Hans et al. classify these cases as the non-GCB subtype, as some of these cases show the typical gene expression profile of ABC DLBCL;(3) however, other studies report that some of these cases have the characteristic gene expression profile of GCB DLBCL.(4) Therefore, it is important to understand how the immunohistochemical classification of both subtypes of DLBCL is associated with their genomic profiles.
Lymphoma is tumor type whose characteristics vary in different geographical areas. In this study, we used immunohistochemistry and array-CGH to unveil the pathogenetic characteristics of Chinese DLBCL cases, and, furthermore, investigated the genomic aberrations in the GCB and non-GCB subtypes, as classified by immunohistochemistry.
Materials and Methods
Patients and samples
Previously untreated patients diagnosed with de novo DLBCL between 1999 and 2006 were selected randomly from Xijing Hospital, Fourth Military Medical University, Xi'an, China. All patients provided informed consent in accordance with the Declaration of Helsinki, and the use of patient materials and information was approved by the institutional review boards of Xijing Hospital, Fourth Military Medical University, China.
The samples were immunostained with CD20 and 59 CD20-positive cases were selected for further study. All diagnoses were based on the criteria established in the WHO classification. The histological slides from all 59 cases were reviewed by two observers (Y. Guo and K. Ohshima).
Immunohistochemistry
Paraffin sections from each sample were immunostained with antibodies against CD10, BCL6 and MUM1 using the peroxidase–antiperoxidase method. The immunoreactive sites were visualized using the Dako Envision+ System peroxidase technique. The primary antibodies and pretreatments are shown in Table 1. CD10, BCL6 and MUM1 immunostaining were evaluated semi-quantitatively: the samples were classified as positive if more than 30% of the tumor cells were immunoreactive. The immunostaining results for CD10, BCL6 and MUM1 were used to subclassify the cases into GCB and non-GCB subtypes according to Hans' algorithms (Fig. 1).
Table 1.
List of the antibodies and pretreatment
| Antigen | Source | Clone | Dilution | Pretreatment |
|---|---|---|---|---|
| CD10 | Novocastra, Newcastle-upon-Tyne, UK | 56C6 | 1:40 | Microwave, 98°C, 10 min |
| Bcl-6 | Novocastra, Newcastle-upon-Tyne, UK | P1F6 | 1:20 | Microwave, 98°C, 20 min |
| MUM1 | Dako, Glostrup, Denmark | MUM1 p | 1:50 | Microwave, 95°C,20 min |
Fig. 1.
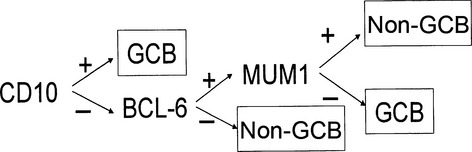
Decision tree for the immunostaining classification of diffuse large B-cell lymphoma (DLBCL). The germinal center B-cell-like (GCB) and non-GCB subtypes were classified according to the following standard: the cases were identified as GCB subtypes when the cases were CD10-positive. If they were both CD10-negative and BCL6-negative, then they belonged to the non-GCB subtype. If they were CD10-negative and BCL6-positive, the MUM1 immunostaining profile determined the subtype. The MUM1-positive cases were identified as non-GCB and MUM1-negative cases were identified as the GCB subtype.
DNA extraction from paraffin embedded sections
Between 10 and 15 5-μm-thick tissue sections were collected in 1.5-mL Eppendorff tubes, and deparaffinized in 1 mL of xylene followed by three ethanol washes (100, 85 and 70% ethanol). The sample was digested with 1 mL of TENS containing 2 mg/mL of proteinase K for 24 h at 55°C. Then, 1 mL of fresh TENS containing 2 mg/mL of proteinase K was added and incubated for another 48 h at 55°C. The sample was purified by phenol-chloroform extraction. The aqueous phase was precipitated with 2.5 volumes of cold 100% ethanol after the addition of 1:10 volume of 3 M sodium acetate (pH 5.3) and 40 mg of glycogen, centrifuged at 1700 × g at 4°C. After decantation and air-drying, DNA was re-suspended in 100–200 μL of TE at 37°C or room temperature overnight.
Array-comparative genomic hybridization
DNA preparation, labeling, array fabrication, hybridization and analysis were performed as described previously, except for the array glasses.(5,7) The array CGH used in this study was ACC Version 5.0. The array consisted of 2304 BAC/PAC clones from libraries RP11 and 13 for BAC clones and RP1, 3, 4 and 5 for PAC clones. These clones were obtained from the BAC/PAC Resource Center at the Children's Hospital Oakland Research Institute in Oakland, California (http://bacpac.chori.org/). The names of the clones and their chromosomal locations are listed together with the raw data in Supplementary Table S1. BAC and PAC clones were aligned with each of the chromosomes on the basis of data from Ensembl Genome Data Resources (release 40; http://www.ensembl.org/) or from the National Center for Biotechnology Information (Build 36; http://www.ncbi.nlm.nih.gov/). The locations of all the clones used for array CGH were confirmed by FISH. For the data analysis, 163 clones were excluded as they showed unreliable data for 17 unrelated normal individuals and those of sex chromosomes. Thus, a total of 2138 clones covering 3022 Mb (average resolution, 1.34 Mb) were subjected to further analysis. As >96% of the measured fluorescence log2 ratio values for each spot (2 × 2138 clones) ranged from +0.16 to −0.15, the thresholds for the log2 ratios of gains and losses were set at +0.16 and −0.15, respectively. Linearity between copy numbers and log2 ratio values was confirmed through the use of human fibroblast cell lines with different copy numbers of X chromosomes, as described previously.(7) Regions of low-level gain/amplification were defined as those with log2 ratios of +0.16 to +1.0, those suspected of containing a heterozygous loss/deletion as regions with log2 ratios of −1.0 to −0.15, those showing high-level gain/amplification as regions with a log2 ratio >+1.0, and those suspected of containing a homozygous loss/deletion as regions with a log2 ratio <−1.0. All data that showed recurrently altered genomic copy numbers were checked against the copy number polymorphism database (http://projects.tcag.ca/variation/).
Statistical methods
The distribution of the frequency of genomic alterations in different groups was investigated using the χ2-test. Probabilities of <0.05 were considered statistically significant.
Statistical analysis of array-comparative genomic hybridization
To investigate whether the differences in the genomic regions of the non-GCB and GCB patient groups were statistically significant, a dataset was constructed by defining genomic aberrations as copy number gains for log2 ratio thresholds of +0.2 or greater, and as copy number losses for thresholds of −0.2 or less. Clones showing a gain (log2 ratio ≥0.2) were inputted as “1” versus no-gain clones (log2 ratio <0.2) as “0” on an Excel (Microsoft, Redman, WA, USA) template for each case. Similarly, clones with losses (log2 ratio ≤0.2) were inputted as “1” versus no-loss clones (log2 ratio >0.2) as “0” on another Excel template for each case. The frequencies of gains or losses for each single clone were compared between the non-GCB and GCB groups.
Results
Classification of diffuse large B-cell lymphoma by CD10, BCL6 and MUM1 immunostaining
Positive expression of CD10 was observed in 29% (17/59) of the patients, BCL6 in 47% (28/59) of the patients and MUM1 in 51% (30/59) of the patients. In total, 26 of the 59 cases (44%) were classified as GCB and 33 (56%) were classified as non-GCB by the immunostaining analysis (Fig. 1). Of the 26 GCB cases, 10 cases (38%) expressed CD10 alone, nine cases (35%) expressed BCL6 alone; seven cases (27%) expressed both CD10 and BCL6, and only 1 case (0.4%) expressed CD10, BCL6 and MUM1. Of the 33 non-GCB cases, 17 cases (56%) expressed MUM1 alone, 12 cases (34%) expressed both MUM1 and BCL6, and four cases (10%) expressed none of these markers (Table 2).
Table 2.
Phenotype of germinal center B-cell-like (GCB) and non-GCB subtypes of diffuse large B-cell lymphoma (DLBCL)
| GCB type |
Non-GCB type |
||
|---|---|---|---|
| Phenotype | Cases (%) | Phenotype | Cases (%) |
| CD10+ (only) | 10 (38%) | Mum1+ | 17 (56%) |
| Bcl6+ (only) | 9 (35%) | Bcl6+, Mum1+ | 12 (34%) |
| CD10+, Bcl6+ | 7 (27%) | Bcl6–, Mum1– | 4 (10%) |
| Total | 26 | 33 | |
Array germinal center B-cell-like analysis of genomic aberrations in the non-germinal center B-cell-like and germinal center B-cell-like subtypes, as classified by immunostaining
Array CGH was successfully performed on 46 of the 59 DLBCL cases. Gains at 1q, 2p, 3q, 7, 11q, 12q, 17q and 18q, and losses at 1p, 2q, 6q, 8p and 16q were frequent (>40%) genomic aberrations among the Chinese DLBCL patients. Frequent genomic imbalances (copy number changes) in the non-GCB subtype were gains (>40%) at 1q, 3q, 6p21, 7q22–31.1, 9p24.1, 11q24, 12q, 17q21.1 and 18q and losses (>40%) at 1p36, 1q43, 6q, 8p21, 8q24, 10q, 15q11.2 and 16q12.2. Frequent genomic imbalances in the GCB subtype (>40%) were gains at 1q23, 2p16.1, 3q29, 6p21–23, 7, 8q24, 11q13.4, 11q22–q24, 12q15, 12q21–q24, 14q23.1, 16p13.3, 17q21.2, 17q24.1 and 21q22.1 and losses at 1p36.32, 2q14, 4p15.31, 6q and 16q. The non-GCB subtype was genomically characterized by more frequent gains at 11q24.3 and 3q13.2 (P < 0.05). The GCB group was genomically characterized by more frequent gains at 7q (7q22.1, P < 0.05), and losses at 16q (P < 0.05) (Figs 2and 3a–c, Table 3).
Fig. 2.
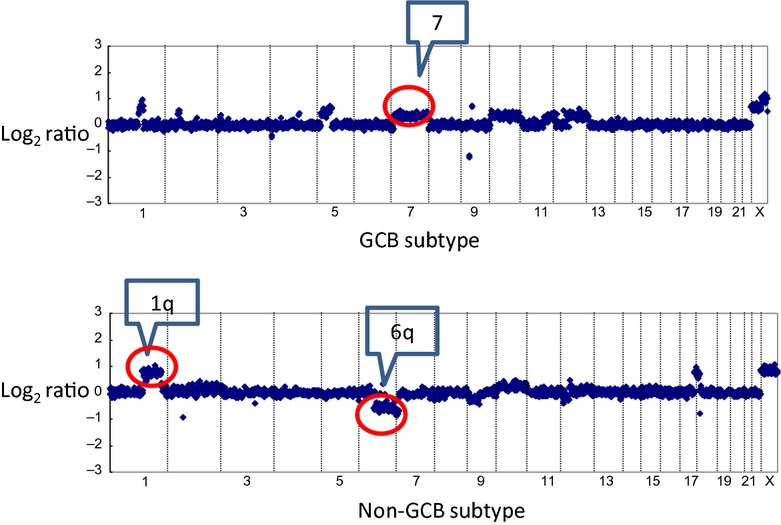
Representative array CGH profile of germinal center B-cell-like (GCB) and non-GCB diffuse large B cell lymphomas. Log2 ratios for all clones were plotted based on their chromosome position, with the vertical dotted bars representing the separation of chromosomes. Clones are ordered from chromosome 1 to 22 followed by X. The upper figure shows gain of 7q in a case of GCB diffuse large B-cell lymphoma (DLBCL). The lower figure shows a gain of 1q and a loss of 6q in a case of non-GCB DLBCL.
Fig. 3.
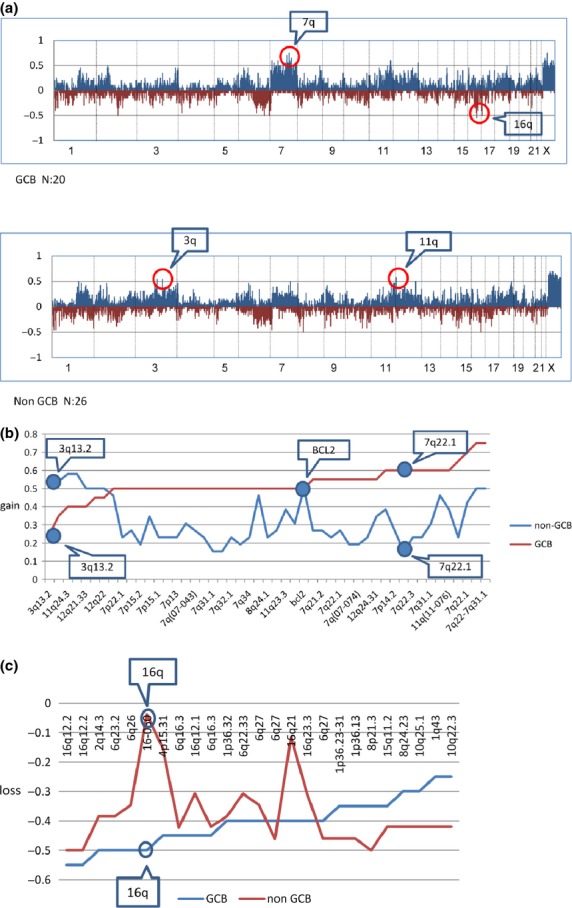
(a) Genome-wide frequency of the genomic imbalance in diffuse large B-cell lymphoma (DLBCL) subtypes: non-germinal center B-cell-like (GCB) group (26 cases) and GCB group (20 cases). Horizontal lines indicate 2213 BAC/PAC clones ordered from chromosomes 1 to 22 and X. Within each chromosome, clones are shown in order from the p telomere to the q telomere. Vertical lines indicate the frequency (%) of gains and losses. Non-GCB and GCB subtypes are shown to have different genomic imbalance characteristics. (b) Contrastive analysis of genomic gains of GCB and non-GCB subtypes with frequencies of >50%. Horizontal lines indicate BAC/PAC clones of genomic gains with high frequencies (>50%) in each subtype in order of increasing frequencies of gains in the GCB subtype. Vertical lines indicate the frequency (%) of gains. The figure shows that the frequencies of gains at specific loci in each subtype were different. (c) Contrastive analysis of genomic losses of GCB and non-GCB subtypes with frequencies of >40%. Horizontal lines indicate BAC/PAC clones of genomic losses with high frequencies (>40%) in each subtype in order of increasing frequency of losses in GCB samples. Vertical lines indicate the frequency (%) of losses. The frequencies of genomic losses with high frequencies (>40%) were different between the non-GCB and GCB subtypes.
Table 3.
Frequent regions of genomic imbalance in germinal center B-cell-like (GCB) or non-GCB diffuse large B-cell lymphoma (DLBCL)
| Gain |
Loss |
||||
|---|---|---|---|---|---|
| Chromosome band | GCB% (n/GCB) | Non-GCB% (n/Non-GCB) | Chromosome band | GCB% (n/GCB) | Non-GCB% (n/Non-GCB) |
| 11q24.3 | 0 (0/20) | 58 (15/26) | 1q43 | 25 (5/20) | 42 (11/26) |
| 3q13.2 | 5 (1/20) | 54 (14/26) | 8q24.23 | 30 (6/20) | 42 (11/26) |
| 7q22-7q31.1 | 5 (1/20) | 50 (13/26) | 10q25.1 | 30 (6/20) | 42 (11/26) |
| 11q24.3 | 15 (3/20) | 58 (15/26) | 1p36.23-31 | 35 (7/20) | 46 (12/26) |
| 1q23.3 | 25 (5/20) | 50 (13/26) | 1p36.13 | 35 (7/20) | 46 (12/26) |
| 12q22 | 25 (5/20) | 50 (13/26) | 8p21.3 | 35 (7/20) | 50 (13/26) |
| 12q21.33 | 30 (6/20) | 50 (13/26) | 15q11.2 | 35 (7/20) | 42 (11/26) |
| 3q23 | 45 (9/20) | 54 (14/26) | 1p36.32 | 40 (8/20) | 38 (10/26) |
| BCL2 | 50 (10/20) | 50 (13/26) | 6q22.33 | 40 (8/20) | 31 (8/26) |
| 7p21.1 | 55 (11/20) | 27 (7/26) | 6q27 | 40 (8/20) | 35 (9/26) |
| 7q21.2 | 55 (11/20) | 27 (7/26) | 16q21 | 40 (8/20) | 12 (3/26) |
| 7q34-7q35 | 55 (11/20) | 23 (6/26) | 16q23.3 | 40 (8/20) | 31 (8/26) |
| 12q24.31 | 55 (11/20) | 35 (9/26) | 6q27 | 40 (8/20) | 46 (12/26) |
| 7p14.2 | 60 (12/20) | 27 (7/26) | 4p15.31 | 45 (9/20) | 15 (4/26) |
| 7q22.1 | 60 (12/20) | 15 (4/26) | 6q16.3 | 45 (9/20) | 42 (11/26) |
| 7q22.3 | 60 (12/20) | 50 (13/26) | 16q12.1 | 45 (9/20) | 31 (8/26) |
| 7q31.1 | 60 (12/20) | 31 (8/26) | 6q16.3 | 45 (9/20) | 42 (11/26) |
| 7q31.2 | 65 (13/20) | 23 (6/26) | 2q14.3 | 50 (10/20) | 38 (10/26) |
| 6q23.2 | 50 (10/20) | 38 (10/26) | |||
| 6q26 | 50 (10/20) | 35 (9/26) | |||
| 16q | 50 (10/20) | 4 (1/26) | |||
| 16q12.2 | 55 (11/20) | 50 (13/26) | |||
Copy number change of X chromosome was excluded. n, case numbers in each sub group of DLBCL identified by immunostaining. %, percentage of positive cases in each sub group of DLBCL identified by immunostaining.
Genomic imbalances in subgroups identified by immunostaining of germinal center B-cell-like diffuse large B-cell lymphoma
We examined the frequency of gains and losses in the subgroups identified by immunostaining of GCB DLBCL. The subgroups included CD10+only, BCL6+only, CD10+BCL6+ and CD10+BCL6+MUM1+. Frequent genomic imbalancein the CD10+only group were gains (>60%) at 7q, 11q23.3, 1q23.3, 2p16.1 and 3q28 and losses (≥50%) at 10q25.1, 6q16.3, 9p21.3, 16q12.2 and 16q24. Frequent genomic imbalances in the BCL6+ only group were gains (>60%) at 7, 2p14, 3q29, 8q24.1, 12q13.12 and BCL2, and losses (>60%) at 4p15.31, 6q22.33, 1p36.32, 1p31.1, 1q43, 3p12.1–p12.2, 4q13.1, 6q23.2. Frequent genomic imbalance (≥60%) in the BCL6+CD10+ group were gains at 2p16.2, 7, 11p11.2, 11q13.4, 11q22, 17q21.2, 17q24.1, 1q23.3, 1q25.1, 2p16.1, 3q23, 3q28, 6p21–6p23, 6q21, 8q24.21, 11p13, 11q13–11q14, 11q21–11q23, 12q21, 12q24.31, 14q23, 15q23, 16p13.3 and 17q25.3, and losses at 16q, 18q12.2d, 1p36.13, 2p24, 2q and 5p15.33. Genomic imbalances in the CD10+BCL6+MUM1+ group were gains at 1q, 2p12–2p16, 3q13.2, 6p21–6p25, 7, 8q12–8q13, 8q21–8q24, 11p13, 12q22, 12q24, 15q15.3, 17q, 18p11.32, 20q12, 20q13.2 and 22q, and losses at 1p34–36, 2q14.3, 2q35, 3q27–29, 4q34.3, 5p15.33, 6q14–6q16, 6q21-6q27, 11q21, 11q22.1, 11q22.3, 13q13–14, 16q12, 16q21, 17q25.3 and 22q13 (Table 4a,b). The similarities of the four subgroups were gains of 7 and losses at 16q.
Table 4.
Frequent regions of genomic imbalances in each subgroup of germinal center B-cell-like (GCB) or non-GCB diffuse large B-cell lymphoma (DLBCL) identified by immunostaining
| (a) Frequent regions of genomic gains in each sub group of GCB DLBCL identified by immunostaining | |||||||
|---|---|---|---|---|---|---|---|
| GCB gain | |||||||
| CD10+ only |
BCL6+ only |
CD10+BCL6+ |
CD10+BCL6+MUM1+ |
||||
| Chromosome band | % (n/CD10+only) | Chromosome band | % (n/BCL6+ only) | Chromosome band | % (n/CD10+BCL6+) | Chromosome band | % (n/CD10+BCL6+MUM1+) |
| 7q34–7q35 | 75 (6/8) | 7q22.1, 7q22–7q31.1 | 83 (5/6) | 2p16.2 | 80 (4/5) | 1q21–1q25, 1q31–1q32, 1q41–1q44 | 100 (1/1) |
| 11q23.3 | 75 (6/8) | 2p14 | 67 (4/6) | 7q22–7q31.1 | 80 (4/5) | 2p12–2p16 | 100 (1/1) |
| 1q23.3 | 63 (5/8) | 3q29 | 67 (4/6) | 11p11.2, 11q13.4, 11q22.1, 11q22.3, | 80 (4/5) | 3q13.2 | 100 (1/1) |
| 2p16.1 | 63 (5/8) | 7p11–7p14, 7q21–7q31 | 67 (4/6) | 17q21.2, 17q24.1, | 80 (4/5) | 6p25.3 | 100 (1/1) |
| 3q28 | 63 (5/8) | 8q24.1 | 67 (4/6) | 1q23.3, 1q25.1 | 60 (3/5) | 6p21–6p25 | 100 (1/1) |
| 7q22.1, 7q22–7q31.1, 7q31.1 – 7q34 | 63 (5/8) | 12q13.12 | 67 (4/6) | 2p16.1, | 60 (3/5) | 7p21–7p22 | 100 (1/1) |
| 11q23.3, 11q23.3 | 63 (5/8) | BCL2 | 67 (4/6) | 3q23, 3q28 | 60 (3/5) | 7p11–7p15, 7q11.23–7q21.1, 7q22–7q31, 7q36 | 100 (1/1) |
| 6p21–6p23, 6q21 | 60 (3/5) | 8q12–8q13, 8q21–8q24 | 100 (1/1) | ||||
| 7p22.1, 7p14–7p15, 7q31.2, 7q34 | 60 (3/5) | 11p13 | 100 (1/1) | ||||
| 8q24.21 | 60 (3/5) | 12q22, 12q24 | 100 (1/1) | ||||
| 11p13, 11q13– 11q14, 11q21–11q23 | 60 (3/5) | 15q15.3 | 100 (1/1) | ||||
| 12q21, 12q24.31 | 60 (3/5) | 17q12, 17q21.2, 17q25.3 | 100 (1/1) | ||||
| 14q23 | 60 (3/5) | 18p11.32 | 100 (1/1) | ||||
| 15q23 | 60 (3/5) | 20q12, 20q13.2 | 100 (1/1) | ||||
| 16p13.3 | 60 (3/5) | 22q | 100 (1/1) | ||||
| 17q25.3 | 60 (3/5) | ||||||
| (b) Frequent regions of genomic losses in each subgroup of GCB DLBCL identified by immunostaining | |||||||
| GCB loss | |||||||
| CD10+ only |
BCL6+ only |
CD10+BCL6+ |
CD10+BCL6+MUM1+ |
||||
| Chromosome band | % (n/CD10+ only) | Chromosome band | % (n/BCL6+ only) | Chromosome band | % (n/CD10+BCL6+) | Chromosome band | % (n/CD10+BCL6+MUM1+) |
| 10q25.1 | 50 (4/8) | 4p15.31 | 83 (5/6) | 16q12, 16q22– 16q24 | 60 (3/5) | 1p36,1p35, 1p34.3 | 100 (1/1) |
| 6q16.3 | 50 (4/8) | 6q22.33 | 67 (4/6) | 18q12.2d | 60 (3/5) | 2q14.3, 2q35 | 100 (1/1) |
| 9p21.3 | 50 (4/8) | 1p36.32, 1p31.1, 1q43 | 67 (4/6) | 1p36.13 | 60 (3/5) | 3q27.2, 3q28, 3q29 | 100 (1/1) |
| 16q12.2 | 50 (4/8) | 3p12.1–p12.2 | 67 (4/6) | 2p24 | 60 (3/5) | 4q34.3 | 100 (1/1) |
| 16q24 | 50 (4/8) | 4q13.1 | 67 (4/6) | 2q14.3, 2q21– 2q24 | 60 (3/5) | 5p15.33 | 100 (1/1) |
| 6q23.2 | 67 (4/6) | 5p15.33 | 60 (3/5) | 6q14–6q16, 6q21–6q27 | 100 (1/1) | ||
| 8p22 | 67 (4/6) | 6q26, 6q27 | 60 (3/5) | 11q21, 11q22 | 100 (1/1) | ||
| 16p13.2, 16q12, 16q23.3 | 67 (4/6) | 8p21.3, 8q24.23 | 60 (3/5) | 13q13.1–13q13.3, 13q14 | 100 (1/1) | ||
| 16q12, 16q21 | 100 (1/1) | ||||||
| 17q25.3 | 100 (1/1) | ||||||
| 22q13 | 100 (1/1) | ||||||
| (c) Frequent regions of genomic gains in each subgroup of Non-GCB DLBCL identified by immunostaining | |||||||
| Non-GCB gain | |||||||
| MUM1+ only |
BCL6+MUM1+ |
||||||
| Chromosome band | % (n/MUM1+only) | Chromosome band | % (n/BCL6+MUM1+) | ||||
| 1q31 | 69 (9/13) | 3q13.2 | 64 (7/11) | ||||
| 1q24.2, 1q24.3, 1q25.1, 1q31.2, 1q31.3-q32.1 | 62 (8/13) | 11q24.3 | 64 (7/11) | ||||
| 1q22, 1q23.3, 1q25.1, 1q25.3, 1q31.1, 1q31.2, 1q31.3, 1q32.1 | 54 (7/13) | 3q12.3 | 55 (6/11) | ||||
| 3q29 | 54 (7/13) | 3q23 | 55 (6/11) | ||||
| 7q22-7q31.1 | 54 (7/13) | 7q22-7q31.1 | 55 (6/11) | ||||
| 9p24.2, 9p24.1, 9q34.3 | 54 (7/13) | 11-075-1 | 55 (6/11) | ||||
| 11q24.1, 11q24.3, | 54 (7/13) | 11q24.3 | 55 (6/11) | ||||
| 12p11.21, 12q12, 12q15, 12q21.33, 12q22, 12q24.31 | 54 (7/13) | 14q23.1c | 55 (6/11) | ||||
| 18q21 | 54 (7/13) | ||||||
| (d) Frequent regions of genomic losses in each subgroup of Non-GCB DLBCL identified by immunostaining | |||||||
| Non-GCB loss | |||||||
| MUM1 only |
BCL6+MUM1+ |
||||||
| Chromosome band | % (n/MUM1 only) | Chromosome band | % (n/BCL6+MUM1+) | ||||
| 14q32.13 | 54 (7/13) | 6q16.3 | 64 (7/11) | ||||
| 1p36.32, 1p36.23-31, 1p36.13 | 54 (7/13) | 6q12 | 55 (6/11) | ||||
| 2q21.1 | 54 (7/13) | 6q23.2, 6q23.3, 6q24.2, 6q25.3, 6q13, 6q15, 6q16.1, 6q16.2, 6q22.31, 6q22.33, | 45 (5/11) | ||||
| 8p21.3 | 54 (7/13) | 2q14.3 | 45 (5/11) | ||||
| 9p21.3 | 46 (6/13) | 8p21.3 | 45 (5/11) | ||||
| 14q24.3 | 46 (6/13) | 16q12.2 | 45 (5/11) | ||||
| 5p15 | 46 (6/13) | ||||||
| 5p15.32 | 46 (6/13) | ||||||
| 15q12, 15q26.1 | 46 (6/13) | ||||||
| 1p35.1, 1p34.3, 1q43 | 46 (6/13) | ||||||
| 2q12.2, 2q14.1 | 46 (6/13) | ||||||
| 6q21, 6q27 | 46 (6/13) | ||||||
| 8p23, 8q24.23 | 46 (6/13) | ||||||
| 10q22.3, 10q25.1 | 46 (6/13) | ||||||
| 15q11.2 | 46 (6/13) | ||||||
| 16q12.2 | 46 (6/13) | ||||||
| 16q23.3 | 46 (6/13) | ||||||
| 20q13.33 | 46 (6/13) | ||||||
Tables 4a–d: Copy number change of X chromosome was excluded. n, case numbers in each subgroup of GCB or non-GCB DLBCL identified by immunostaining. %, percentage of positive cases in each subgroup of GCB or non-GCB DLBCL identified by immunostaining.
Genomic imbalances in BCL6+ and BCL6 non-germinal center B-cell-like diffuse large B-cell lymphoma
We examined the frequency of gains and losses in the BCL6+ non-GCB and BCL6− non-GCB groups. Frequent genomic imbalances (>40%) in the BCL6+ group were gains at 2p, 3q, 4p14, 6p12–p23, 7p11, 7p15, 7q, 11q21–q24, 14q23.1, 17q21.2, 18q21 and Bcl2, and losses at 2q14.3, 6q, 8p21.3 and 16q12.2. The most frequent genomic imbalances (>40%) in the BCL6− group were gains at 1q22–q24, 1q32, 2p16.1, 2q33.2, 3q, 4q13.3–q21.1, 6p21–p22, 6q21, 7p22.1, 7p15.2, 7q22–q31.1, 9p21–p24, 9q34, 11q13.4, 11q23–q24, 12p21, 12q, 15q15, 16p11, 17q, 18q12.1, 18q21–32, 19p13, 19q13 and 20q12, and losses at 1p36, 1q, 2q, 4p16, 5p15, 6q,7q36, 8p21, 8p23, 8q24, 9p24, 9p21, 10q, 13q34, 14q, 15q, 16q12.2, 16q23.2, 17p11.2, 20q11 and 20q13. Comparison of the genomic imbalances between the BCL6− and BCL6+ non-GCB DLBCL cases revealed a higher number of gains at 1q (1q24.2, 1q24.3, 1q25,1) (P < 0.05) and loss at 14q32.13 (P < 0.05) in the BCL6− group and a higher number of gains at 14q23.1 (P = 0.15) and losses at 6q (6q13, 6q22.31, 6q23.2, 6q24.2, 6q12) (P = 0.068) in the BCL6+ group. BCL6− non-GCB DLBCL showed a much higher number of genomic imbalances of gains and losses with high frequencies (>40%) compared to the BCL6+ non-GCB type (gains, BCL6−/BCL6+: 23/12; losses, BCL6−/BCL6+: 41/20). In the BCL6+ non-GCB group, the frequency of gains at 3q27 was 27%, compared to 40% in the Bcl6− non-GCB group. Overall, the BCL6− and BCL6+ cases had different patterns of genomic aberrations (Fig. 4a–c, Table 4c,d).
Fig. 4.
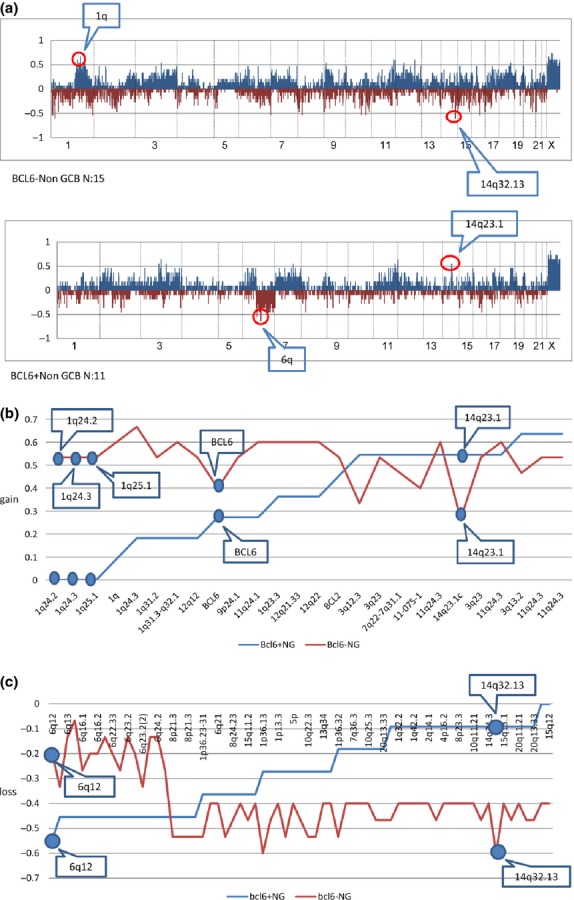
(a) Genome-wide frequencies of genomic imbalance in BCL6+ non-germinal center B-cell-like (GCB) and BCLl6− non-GCB groups. Horizontal lines indicate 2213 BAC/PAC clones in order from chromosomes 1 to 22 and X. Within each chromosome, clones are shown in order from the p telomere to the q telomere. Vertical lines indicate the frequency (%) of gains and losses. The genomic imbalance characteristics of BCL6+ non-GCB (11 cases) and BCL6− non-GCB (15 cases) groups were different. (b) Contrastive analysis of genomic gains of BCL6+ and BCL6− non-GCB diffuse large B-cell lymphoma (DLBCL) (NG) groups with frequency over 50% (plus BCL6) in each group. Horizontal lines indicate BAC/PAC clones of genomic gains with high frequencies (>50%) in each group in order of increasing frequency of gains in the BCL6+ non-GCB group. Vertical lines indicate the frequency (%) of gains. (c) Contrastive analysis of genomic losses of BCL6+ and BCL6− non-GCB DLBCL (NG) groups with frequencies of >40% (plus BCL6). The frequencies of genomic losses with high frequencies (>40%) differed between the BCL6+ non-GCB and BCL6− non-GCB groups.
Discussion
Comparison of subtype classification methods
Our previous study proved that distinct differences exist in the pattern of genomic imbalances in the ABC and GCB subtypes, as classified by gene expression profiling.(5) In this study we also found specific genomic imbalances in the non-GCB and GCB subtypes, based upon Hans' immunohistochemistry classification. The non-GCB group was genomically characterized by more frequent gains at 11q24.1 and 3q (P < 0.05). The GCB group was genomically characterized by more frequent gains at 7q (7q22.1, P < 0.05) and losses at 16q (P < 0.05). The results from the current study are not absolutely identical to those of our former study(5) and another study.(8) In our former study, the ABC group was genomically characterized by more frequent gains at 3p23–q28, 18q11.2–q23 and 19q13.41–q13.43, and losses at 6q22.31–q24.1 and 9p21.3, whereas the GCB group was genomically characterized by more frequent gains at 1q21.1–q23.3, 1q31.1–q42.13, 2p15–p16.1, 7q22.1–q36.2 and 12q13.1–q14. Comparing the two studies, the gains of 18q11.2–q23 and 19q13.41–q13.43 and loss of 9p21.3 in the ABC group of the former study are not so frequent in the non-GCB group of the current study. In contrast, gains of 1q31.1–q42.13 in the GCB group of the former study are not so frequent in the GCB group of the current study. The characteristic frequent gains at 11q24.3 of the non-GCB group and frequent losses at 16 of the GCB group in our current study were not so frequent in similar ABC and GCB groups in our former study. However, the most distinct differences in the genomic aberrations in the GCB and non-GCB subtypes in this study (e.g. gain at 3q in non-GCB and gain at seven in GCB) were the same as in our previous study. Using comparative genomic hybridization, Bea et al. also report that the gain at 3q was a characteristic genomic imbalance of the non-GCB subtype.(8) The discrepancy between the precise genomic aberrations observed in each subtype, as classified by different methods, may be due to the different identification methods or differences in the patient population (Japanese and Chinese) in each study.
We tried to make clinical study between genomic alteration and treatment of DLBCL, but unfortunately the samples we used were old and we lost contact with some patients, so there was no follow-up. Therefore, we failed to determine the distinct genomic alteration's clinical significance. This should be an important aspect in future study.
Subgroups of germinal center B-cell-like diffuse large B-cell lymphoma identified by immunostaining
We analyzed the genomic imbalance of subgroups of GCB DLBCL by immunostaining. The GCB DLBCL can be divided into four subgroups: CD10+ only, BCL6+ only, CD10+BCL6+ and CD10+BCL6+MUM1+. Comparing the genomic imbalance in each group, we found that the GCB DLBCL is a heterogeneous disease that contains many different frequent gains and losses in each subgroup. Although each subgroup contains a small case number, it showed recurrent genomic imbalances with high frequency. In particular, in CD10+BCL6+ subgroup it contains 25 recurrent genomic gains in over 60% of cases. It seems that each subgroup stands for special molecular pathways. Each subgroup contains only a small number of cases, and a conclusion requires analysis based on more cases. Regardless, the four subgroups share common gains of seven and losses of 16q, which may be associated with their germinal center B cell-derived character.
Relationship of BCL6 expression with genomic profiling
BCL6 is a zinc-finger protein that acts as a transcriptional repressor and is expressed in germinal center B-cells and a subset of CD4+ T cells. Unlike normal germinal center B-cells, in which the expression of BCL6 and MUM1 are mutually exclusive, DLBCL tumor cells can co-express both proteins.(6) MUM1 is involved in the late stages of B-cell differentiation and T-cell activation, and is deregulated in DLBCL. As BCL6 negatively regulates NF-κB activation,(9) it is conceivable that anti-apoptotic effects in lymphoma cells caused by NF-κB activation might be suppressed by BCL6. This hypothesis could explain the better prognosis observed in GCB-like DLBCL. We were interested to investigate which genomic aberrations existed in cases co-expressing BCL6 and MUM1, and found that the genomic characteristics of BCL6+ and BCL6− in non-GCB DLBCL were different, suggesting they belong to different subtypes of disease. We did not find significant differences in the frequency of 3q27 gains (the genomic site of BCL6) between the two groups. It appears that the expression of BCL6 has no relationship with its degree of amplification. We also found that BCL6− non-GCB DLBCL had a much higher frequency of genomic imbalances (gains and losses) compared with BCL6+ non-GCB DLBCL. This observation may also explain why BCL6 expression alone can predict a good outcome in DLBCL, even in non-GCB.(10,11) Because cases of DLBCL co-expressing BCL6 and MUM1 are not rare, the identification of these cases needs to be modified in the future.
In conclusion, we have investigated the pathogenetic characteristics of different subtypes of DLBCL in Chinese patients. By comparing immunoprofiling and array-CGH profiling, we observed that distinct chromosomal aberrations are correlated with each subtype of DLBCL. Cases of non-GCB DLBCL that co-express BCL6 and MUM1 have different genomic aberration profiles than single MUM1-positive non-GCB DLBCL, which suggests that different oncogenetic mechanisms of lymphoma may occur in these diseases.
Acknowledgments
This work was supported in part by grants from the Ministry of Health, Labor and Welfare, the Ministry of Education, Culture, Sports, Science and Technology, the Japan Society for the Promotion of Science, the Foundation of Promotion of Cancer Research, the Wella Award from the Japan Leukemia Research Fund awarded to Ms G.Y., and the National Natural Science Foundation of China (No. 81071951).
Disclosure Statement
The authors have no conflict of interest.
Supporting Information
Additional supporting information may be found in the online version of this article:
Table S1 Genetic imbalance result of diffuse large B-cell lymphoma (DLBCL).
References
- 1.Alizadeh AA, Eisen MB, Davis RE, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403:503–11. doi: 10.1038/35000501. [DOI] [PubMed] [Google Scholar]
- 2.Wright G, Tan B, Rosenwald A, Hurt EH, Wiestner A, Staudt LM. A gene expression-based method to diagnose clinically distinct subgroups of diffuse large B cell lymphoma. Proc Natl Acad Sci U S A. 2003;100:9991–6. doi: 10.1073/pnas.1732008100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hans CP, Weisenburger DD, Greiner TC, et al. Confirmation of the molecular classification of diffuse large B-cell lymphoma by immunohistochemistry using a tissue microarray. Blood. 2004;103:275–82. doi: 10.1182/blood-2003-05-1545. [DOI] [PubMed] [Google Scholar]
- 4.Haarer CF, Roberts RA, Frutiger YM, Grogan TM, Rimsza LM. Immunohistochemical classification of de novo, transformed, and relapsed diffuse large B-cell lymphoma into germinal center B-cell and nongerminal center B-cell subtypes correlates with gene expression profile and patient survival. Arch Pathol Lab Med. 2006;130:1819–24. doi: 10.5858/2006-130-1819-ICODNT. [DOI] [PubMed] [Google Scholar]
- 5.Tagawa H, Suguro M, Tsuzuki S, et al. Comparison of genome profiles for identification of distinct subgroups of diffuse large B-cell lymphoma. Blood. 2005;106:1770–7. doi: 10.1182/blood-2005-02-0542. [DOI] [PubMed] [Google Scholar]
- 6.Falini B, Fizzotti M, Pucciarini A, et al. A monoclonal antibody (MUM1p) detects expression of the MUM1/IRF4 protein in a subset of germinal center B cells, plasma cells, and activated T cells. Blood. 2000;95:2084–92. [PubMed] [Google Scholar]
- 7.Ota A, Tagawa H, Karnan S, et al. Identification and characterization of a novel gene, C13orf25, as a target for 13q31-q32 amplification in malignant lymphoma. Cancer Res. 2004;64:3087–95. doi: 10.1158/0008-5472.can-03-3773. [DOI] [PubMed] [Google Scholar]
- 8.Bea S, Zettl A, Wright G, et al. Diffuse large B-cell lymphoma subgroups have distinct genetic profiles that influence tumor biology and improve gene-expression-based survival prediction. Blood. 2005;106:3183–90. doi: 10.1182/blood-2005-04-1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Perez-Rosado A, Artiga M, Vargiu P, Sanchez-Aguilera A, Alvarez-Barrientos A, Piris M. BCL6 represses NF kappa B activity in diffuse large B-cell lymphomas. J Pathol. 2008;214:498–507. doi: 10.1002/path.2279. [DOI] [PubMed] [Google Scholar]
- 10.Lossos IS, Jones CD, Warnke R, et al. Expression of a single gene, BCL-6, strongly predicts survival in patients with diffuse large B-cell lymphoma. Blood. 2001;98:945–51. doi: 10.1182/blood.v98.4.945. [DOI] [PubMed] [Google Scholar]
- 11.Iqbal J, Greiner TC, Patel K, et al. Leukemia/Lymphoma Molecular Profiling Project. Distinctive patterns of BCL6 molecular alterations and their functional consequences in different subgroups of diffuse large B-cell lymphoma. Leukemia. 2007;21:2332–43. doi: 10.1038/sj.leu.2404856. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1 Genetic imbalance result of diffuse large B-cell lymphoma (DLBCL).


