Abstract
Gastric cancer (GC) develops through deregulation of gene expression and accumulation of epigenetic abnormalities, leading to tumor cell acquisition of malignant features. MicroRNAs (miRNAs) play a critical role in cancer development where they can act as oncogenes or oncosuppressors. To identify miRNAs that are associated with some clinicopathologic features of GC and/or participate in tumor progression, miRNA expression in 20 GC tissues and five corresponding non-neoplastic gastric mucosa was examined by miRNA microarray. Oligonucleotide array analysis was carried out for miRNA target prediction. The functions of candidate miRNAs and their target genes were also analyzed by quantitative RT-PCR, Western blotting, reporter gene assay, and cell invasion assay. Comparison of miRNA expression profiles revealed that downregulation of miR-148a was identified in most of the GC tissues. Downregulation of miR-148a was significantly correlated with an advanced clinical stage, lymph node metastasis, and poor clinical outcome. Custom oligonucleotide array analysis revealed that MMP7 expression was markedly downregulated in miR-148a-overexpressing GC cells; MMP7 was found to be a direct and functional target of miR-148a, participating in cell invasion. These results suggest that miR-148a contributes to the maintenance of homeostasis in normal stomach tissue and plays an important role in GC invasion by regulating MMP7 expression.
Keywords: Gastric cancer, microRNA, miR-148a, MMP7, cancer invasion
Gastric cancer (GC) remains a major public health issue as the fourth most common cancer and the second leading cause of cancer mortality worldwide.(1) Gastric cancer is assumed to originate from a sequential accumulation of molecular and genetic alterations to stomach epithelial cells.(2) A deeper understanding of the pathogenesis and biological features of GC is necessary to further inform and enhance early detection and treatment methods.
MicroRNAs (miRNAs) are non-protein-coding small RNAs in the size range of 19–25 nucleotides, which play important regulatory roles in posttranscriptional repression and fine-tune various essential biological processes.(3–5) Recent data clearly shows that miRNAs are abnormally expressed in various cancers, indicating novel roles as oncogenes or tumor suppressors in different tumor types.(6) Currently, miRNA target-prediction algorithms and publicly available databases are undergoing rapid and dramatic evolution. However, precision and sensitivity of state-of-the-art algorithms were estimated as ˜50% and 12%, respectively, when tested against proteomics-supported miRNA targets,(7) highlighting the necessity for mass experimental miRNA target validation.(8) As for GC, although there are several reports of aberrant expression of specific miRNAs, target predictions of identified miRNAs were carried out by several online databases in each study.(9,10) Each new finding of deregulated miRNA in GC and its target gene is a welcome addition to the body of knowledge; however, there is no denying the possibility that genuine targets can be overlooked using online algorithms.
In the present study, we examined miRNA expression in 20 GC cases and five corresponding non-neoplastic gastric mucosa using miRNA microarray. By comparing these expression profiles, we found downregulation of miR-148a in the majority of GC tissues. The correlation between the miR-148a expression level and clinicopathologic parameters of GC was also examined and we identified that lower expression levels of miR-148a correlate with worse clinicopathologic characters. To predict gene targets of miR-148a, we used an oligonucleotide array to generate a list of genes whose expression was deregulated by altered expression of miR-148a, and found that expression of MMP7 was suppressed by upregulation of miR-148a. It was confirmed that MMP7 was a direct and functional target of miR-148a using quantitative RT-PCR (qRT-PCR), Western blot analysis, and reporter gene assay. We also carried out target prediction of miR-148a using representative online databases. However, no lists of miR-148a target genes contained MMP7, thus validating the reliability of the conventional laboratory-based experimental strategy for miRNA target prediction, by which crucial target genes were detected-genes, which online target prediction could not detect.
Materials and Methods
MicroRNA microarray hybridization
Total RNA was isolated from frozen tissue with Isogene (Nippon Gene, Tokyo, Japan). Short-strand RNA was purified from total RNA with the RNeasy MinElute Cleanup Kit (Qiagen, Hilden, Germany). For the oligonucleotide array we used Genopal-MICH07 DNA chips (Mitsubishi Rayon, Tokyo, Japan) comprising 188 oligonucleotide DNA probes. Details are described in Data S1.
In situ hybridization for miR-148a
Biotin 3′-labeled locked nucleic acid-incorporated miRNA probe (miR-CURY LNA detection probe; Exiqon, Woburn, MA, USA) was used for visualization of miR-148a. Details are described in Data S1.
Cell lines
Nine cell lines derived from human GC were used. All cell lines were maintained as described previously.(11) Details are described in Data S1.
Oligonucleotide array construction, hybridization, detection, and data analysis
The oligonucleotide array, Genopal (Mitsubishi Rayon), was prepared as described previously.(12) Details of oligonucleotide array probes, total RNA isolation, RNA quantification and assessment of integrity, hybridization, detection, and data analysis were carried out as described previously.(13)
Quantitative RT-PCR and Western blot analysis
Quantification of MMP7 mRNA levels was carried out using real-time fluorescence detection as described previously.(14) For analysis of miR-148a and U6B, expression levels of miR-148a and RNU6B were measured and quantified as described previously.(15) Western blot analysis was carried out as described previously.(16) Details are described in Data S1.
CpG island determination, genomic DNA extraction, and bisulfite genomic DNA sequencing
A 20-kb genomic DNA fragment encompassing the miRNA coding sequences was analyzed with CpG plot software (http://bioweb2.pasteur.fr). We used the parameter values to report a CpG island as described previously.(17) Thermal cycling conditions were as described previously.(18) Methylation status of PCR products were confirmed as described previously.(19) Details are described in Data S1.
Plasmids
For constitutive expression of the MMP7 gene, commercially available MMP7 cDNA construct (Invitrogen, Carlsbad, CA, USA) was purchased. pCMV-MIR (Origene, Rockville, MD, USA) was used for constitutive expression of miR-148a. Details are described in Data S1.
Cell transfection
Transfection of cells was carried out with Lipofectamine RNAiMAX Reagent (Invitrogen) according to the manufacturer's instructions. Details are described in Data S1.
Reporter gene assay
Reporter gene assay was carried out as described previously.(20) Details are described in Data S1.
Tissue samples
A total of 81 primary tumor samples were collected from patients diagnosed with GC. Details are described in Data S1.
Cell growth and in vitro invasion assay
For the MTT assay, cells were seeded at a density of 2000 cells per well in 96-well plates. Cell growth was monitored after 1, 2, and 4 days by MTT assay.(21) Modified Boyden chamber assays were used to examine invasiveness as described previously.(20)
Statistical methods
Statistical differences between miRNA levels in non-neoplastic gastric mucosa and GC were evaluated using the Mann–Whitney U-test and Wilcoxon matched pairs test. The correlation between expression levels of miR-148a and clinicopathologic parameters was analyzed with Fisher's exact test. Univariate and multivariate Cox regression was used to evaluate the associations between clinical covariates and cancer-specific mortality. Hazard ratios and 95% confidence intervals were estimated from Cox proportional hazard models. P-values < 0.05 were considered to be statistically significant.
Results
MicroRNA microarray analysis
To identify GC-related miRNA, we carried out miRNA microarray profiling to compare between 20 cancer tissue and five corresponding non-neoplastic gastric mucosa. Through the comparison between these two profiles, six miRNAs showed significantly higher expression in corresponding non-neoplastic gastric mucosa than in cancer tissue (Table 1). Among six miRNAs, we focused on miR-148a because its expression was most downregulated in GC tissue. Further analysis of the same GC tissues by qRT-PCR also showed a significant downregulation of miR-148a (Fig. S1).
Table 1.
Summary of significantly downregulated miRNAs in gastric cancer tissue compared with non-neoplastic mucosa
| miRBase ID | Mature accession | Intensity |
Fold change | P-value | |
|---|---|---|---|---|---|
| Non-neoplastic mucosa | Tumor | ||||
| hsa-miR-148a | MIMAT0000243 | 892 | 279 | −0.31 | 0.0178 |
| hsa-miR-29a | MIMAT0000086 | 1626 | 932 | −0.57 | 0.0395 |
| hsa-miR-30a-3p | MIMAT0000088 | 681 | 412 | −0.60 | 0.0246 |
| hsa-miR-302b | MIMAT0000715 | 953 | 581 | −0.61 | 0.0207 |
| hsa-miR-30a-5p | MIMAT0000087 | 648 | 402 | −0.62 | 0.0443 |
| hsa-miR-125b-1 | MIMAT0000423 | 1257 | 804 | −0.64 | 0.0315 |
miRBase ID codes are available from http://www.mirbase.org.
Clinicopathologic characteristics of GC cases in which miR-148a is downregulated
To confirm the localization of miR-148a expression, in situ hybridization (ISH) was carried out using GC tissues. We could detect the miR-148a expression only in fundic glands and pyloric glands. However, neither cancer cells nor stromal tissues expressed miR-148a (Fig. 1a). Then, to further understand the relationship between miR-148a expression and clinicopathologic parameters, we examined the miR-148a expression levels in 61 formalin-fixed paraffin-embedded samples of primary GC and their corresponding non-neoplastic gastric mucosa by qRT-PCR. In this sample set, miR-148a expression levels were also significantly downregulated in GC tissue compared with corresponding non-neoplastic mucosa (Fig. S2). The expression level of miR-148a was evaluated using the tumor : normal ratio of miR-148a. The relationship between the miR-148a expression level and clinicopathologic parameters of GC is summarized in Table S1. We also evaluated the association between miR-148a expression levels and cancer-specific mortality. In all cases and in 41 advanced GC patients, the GC tissues with lower expression levels of miR-148a showed significantly worse prognosis than those with high expression (Fig. 1b,c). In order to further evaluate the association of miR-148a expression with cancer-specific mortality, we used both univariate and multivariate Cox proportional hazards analyses (Table 2). According to the univariate analysis, T grade, N grade, stage, and miR-148a expression were significantly associated with cancer-specific mortality. According to the multivariate model, expression of miR-148a was an independent prognostic classifier of cancer-specific mortality. These results indicate that loss of miR-148a may play important roles in GC progression.
Fig. 1.
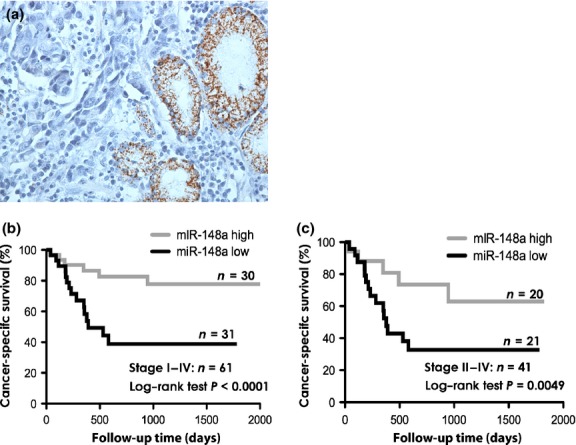
In situ hybridization for microRNA-148a (miR-148a) and relationship between miR-148a expression and gastric caner (GC) patient prognosis. (a) MiR-148a expression in GC tissue detected by in situ hybridization. (b) Log–rank test and Kaplan–Meier plots were constructed for the miR-148a high and miR-148a low groups. Cancer-specific survival of 61 patients with GC based on the expression levels of miR-148a (cut-off line = median miR-148a expression level). (c) Cancer-specific survival of 41 patients with advanced (stage II–IV) GC based on the expression levels of miR-148a (cut-off line = median miR-148a expression level in this group).
Table 2.
Univariate and multivariate analysis of factors influencing survival in 61 patients with gastric cancer
| Parameter | Univariate analysis |
Multivariate analysis |
||||
|---|---|---|---|---|---|---|
| HR | 95% CI | P-value | HR | 95% CI | P-value | |
| Sex | ||||||
| Female | Ref. | |||||
| Male | 1.271 | 0.702–1.532 | 0.5896 | NA | NA | NA |
| Age, years | ||||||
| <60 | Ref. | |||||
| ≥60 | 1.122 | 0.442–1.483 | 0.8593 | NA | NA | NA |
| Mucin phenotype | ||||||
| I and N type | Ref. | |||||
| G and GI type | 0.903 | 0.214–3.802 | 0.1003 | NA | NA | NA |
| Histology† | ||||||
| Intestinal | Ref. | |||||
| Diffuse | 1.601 | 0.254–1.473 | 0.2805 | NA | NA | NA |
| T grade‡ | ||||||
| T1/2 | Ref. | Ref. | ||||
| T3/4 | 4.886 | 1.136–3.225 | 0.0030 | 13.22 | 3.371–50.747 | <0.0010 |
| N grade‡ | ||||||
| N0 | Ref. | Ref. | ||||
| N1/2/3 | 9.234 | 1.388–16.667 | <0.0010 | 5.063 | 0.931–94.529 | 0.0625 |
| Stage‡ | ||||||
| 0/I/II | Ref. | Ref. | ||||
| III/IV | 8.460 | 2.083–11.111 | <0.0010 | 34.579 | 6.293–222.270 | <0.0010 |
| miR-148a expression | ||||||
| High | Ref. | Ref. | ||||
| Low | 10.820 | 1.785–20.000 | 0.0041 | 3.076 | 1.216–8.776 | 0.0169 |
Histology was determined according to the Japanese Classification of Gastric Cancer (14th edition).
Tumor stage was classified according to the criteria of the International Union Against Cancer TNM classification of malignant tumors. CI, confidence interval; HR, hazard ratio; NA, not applicable; Ref., reference.
Analysis of MIR148A methylation in GC cell lines
To pursue the cause of miR-148a downregulation, we examined the methylation status in CpG islands of the MIR148a promoter region in GC cell lines. Expression levels of miR-148a measured by qRT-PCR are shown in Figure S3(a), and we carried out bisulfite sequencing of genomic DNAs from two GC cell lines with high miR-148a expression and two cell lines with low miR-148a expression. Results of bisulfite sequencing are shown in Figure S3(b). An inverse correlation was observed between frequency of methylation in MIR148a promoter-region CpG islands and expression levels of miR-148a. In addition, we ascertained that miR-148a expression was restored in five aza-dc-treated MKN-45 and MKN-74 cells (Fig. S3c). These results suggest that DNA methylation of CpG islands of MIR148A play an important role in transcriptional inactivation in GC cell lines.
Effect of deregulation of miR-148a expression on cell growth and invasive activity
To investigate the biological significance of miR-148a in GC, we carried out an MTT assay 4 days after altering miR-148a expression. In this experiment, we used MKN-1 cells transfected with miR-148a precursor and HSC-57 cells transfected with miR-148a inhibitor. MKN-1 and HSC-57 were selected as they possessed the lowest and highest endogenous miR-148a expression among nine GC cell lines, respectively. Successful overexpression and suppression of miR-148a in each treated cell line were confirmed by qRT-PCR (Fig. 2a). Cell growth of GC cells with deregulated miR-148a expression did not differ from that of cells transfected with control miRNA up to day 4 (data not shown). Next, a Transwell invasion assay was carried out. The invasiveness of miR-148a-overexpressing MKN-1 cells was reduced compared with the negative control miRNA-transfected MKN-1 cells (Fig. 2b). In contrast, the invasiveness of miR-148a-suppressed HSC-57 cells was greater than that of the negative control miRNA-transfected HSC-57 cells (Fig. 2c). These results are consistent with those of a previous report,(22) and indicate that miR-148a regulates some genes which in turn affect tumor invasion in GC cells.
Fig. 2.
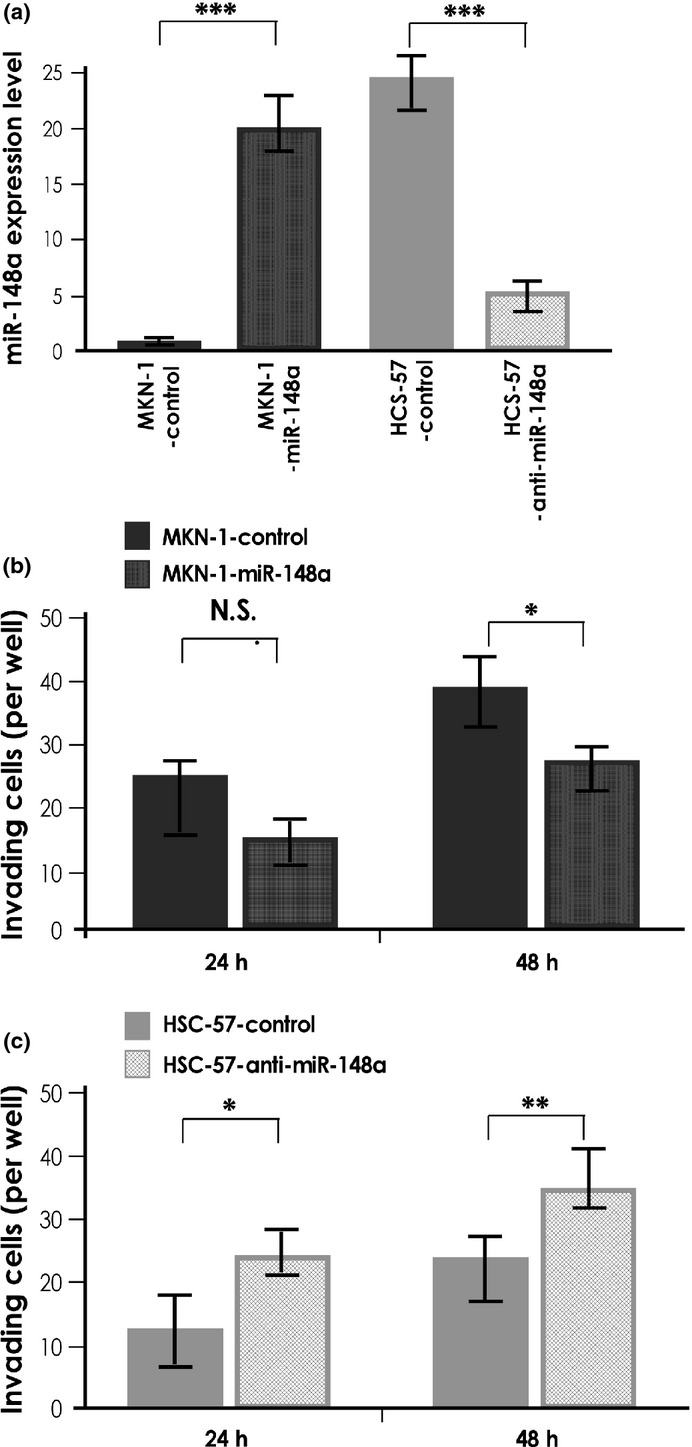
MicroRNA-148a (miR-148a) expression and functional analysis. (a) Quantitative RT-PCR analysis of miR-148a in MKN-1 gastric cancer cells transfected with pre-miR-148a and HSC-57 cells transfected with anti-miR-148a. (b) Effect of miR-148a downregulation on cell invasion of MKN-1 cells. MKN-1 transfected with negative control miRNA and pre-miR-148a were incubated in Boyden chambers. (c) Effect of miR-148a downregulation on cell invasion of HSC-57 cells. HSC-57 transfected with negative control miRNA and anti-miR-148a were incubated in Boyden chambers. Results are mean ± SD of triplicate measurements. *P < 0.05; **P < 0.01; ***P < 0.001. N.S., not significant.
MicroRNA-148a target prediction using custom oligonucleotide array
Although some reports have identified the target gene of miR-148a, these studies used online databases to derive predicted targets.(22–25) To identify novel GC invasion-related genes whose expression was directly regulated by miR-148a, we analyzed gene expression profiles from four GC cell lines showing deregulation of miR-148a alongside corresponding non-treated cells, using a custom oligonucleotide array. In each cell line, either overexpression or suppression of miR-148a was induced, according to their endogenous miR-148a expression level. The list of genes with expression altered more than twofold in miR-148a-deregulated cells is summarized in Table 3. Among these candidates, MMP7 was markedly downregulated by miR-148a overexpression both in MKN-45 and MKN-74 cell lines. Although target prediction for miR-148a was carried out using online databases including TargetScan, PicTar, and miRanda, these databases did not list MMP7 as a candidate miR148a target gene (data not shown). Therefore, further study of the correlation between miR-148a and MMP7 was undertaken.
Table 3.
Genes downregulated by microRNA-148a (miR-148a) deregulation
| Symbol | Description | GenBank | Intensity |
Fold change | |
|---|---|---|---|---|---|
| Control | miR-148a | ||||
| MKN-1 | |||||
| KRTHB1 | Keratin 86 | NM_000584.2 | 168 | 72 | −2.33 |
| CDH17 | Cadherin 17, LI-cadherin | NM_001912 | 148 | 72 | −2.09 |
| APOE | Apolipoprotein E | NM_145804.1 | 118 | 57 | −2.03 |
| MKN45 | |||||
| MMP7 | Matrix metallopeptidase 7 | NM_002423.3 | 730 | 132 | −5.55 |
| MIA | Melanoma inhibitory activity | NM_006533.3 | 649 | 244 | −2.63 |
| MKN-74 | |||||
| MMP7 | Matrix metallopeptidase 7 | NM_002423.3 | 291 | 32 | −9.99 |
| NEIL | Nei endonuclease VIII-like 1 | NM_024608.3 | 2004 | 831 | −2.43 |
| MMP1 | Matrix metallopeptidase 1 | NM_002421.3 | 455 | 208 | −2.17 |
| HSC-57 | |||||
| MMP10 | Matrix metallopeptidase 10 | NM_002425.2 | 766 | 381 | −2.00 |
To confirm the regulation of MMP7 by miR-148a, MKN-45 and MKN-74 cells were transfected to induce miR-148a overexpression, then examined for MMP7 expression level by Western blotting. In both cell lines, MMP7 expression was significantly downregulated; levels were similar to that of MMP7-specific siRNA treated cells (Fig. 3a). The MMP7 mRNA levels determined by qRT-PCR were consistent with the MMP7 protein levels determined by Western blotting (Fig. 3b). It was also found that upregulation of MMP7 was inversely associated with miR-148a expression in GC cases analyzed in Figure 1(b) (Table S2). In addition, we examined the effect of miR-148a deregulation on the expression of epithelial–mesenchymal transition-related molecules. However, no significant alterations of these molecules were detected in miR-148a deregulated cells (Fig. S4a). As shown in Figure 2(b), overexpression of miR-148a repressed invasion of MKN-1 cells. However, MMP7 expression was not detected in MKN-1 cells (data not shown). It was reported that ROCK1 is a direct target of miR-148a and could be involved in GC invasion.(22) We checked expression of ROCK1 in MKN-1 and the alteration of its expression in miR-148a-deregulated cells. Actually, we detected that expression of ROCK1 was downregulated by miR-148a overexpression in MKN-1 cells (Fig. S4b). Furthermore, to prove that MMP7 is a direct target of miR-148a, we used a 3′-UTR sequence of MMP7 cloned into a reporter vector downstream of the luciferase complementary DNA (Fig. 3c). Transfection of this construct into three GC cell lines with high endogenous miR-148a expression levels led to suppression of MMP7 reporter activity, and mutation of the miR-148a binding site abolished the inhibitory effect of miR-148a on reporter activity (Fig. 3d). Conversely, transfection of this construct into three GC cell lines with low endogenous miR-148a expression levels did not lead to suppression of MMP7 reporter activity. However, cotransfection of this construct with miR-148a precursor into these cells showed that miR-148a suppressed the MMP7 reporter activity (Fig. S5). These results imply that miR-148a downregulates MMP7 expression by directly targeting its 3′-UTR.
Fig. 3.
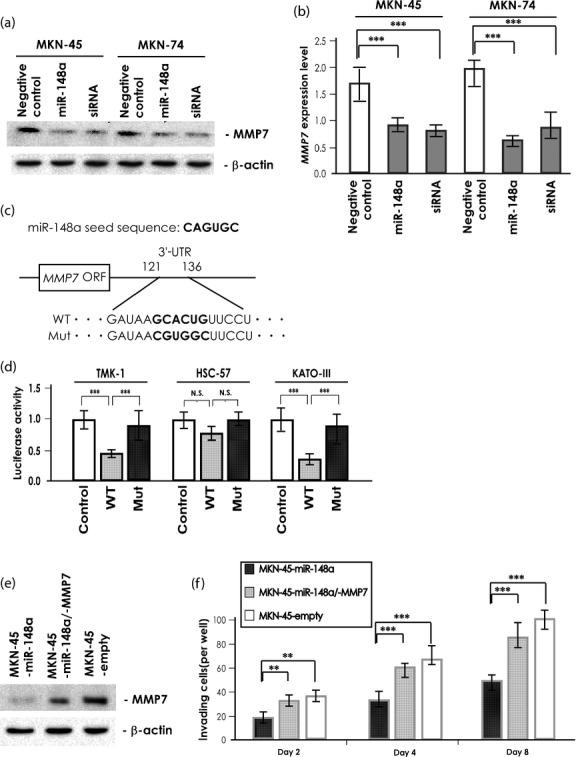
Effect of microRNA-148a (miR-148a) on MMP7 expression and gastric cancer (GC) cell invasiveness. (a) Western blot analysis of MMP7 in MKN-45 and MKN-74 cell lysates after treatment with negative control miRNA, pre-miR-148a, or MMP7-specific siRNA. A β-actin blot served as loading control. (b) Quantitative RT-PCR analysis of MMP7 in MKN-45 and MKN-74 cells after treatment with negative control miRNA, pre-miR-148a, or MMP7-specific siRNA. Results are mean ± SD of triplicate measurements. ***P < 0.001. (c) Schematic representation of miR-148a seed sequence and MMP7 3′-UTR, showing the putative miR-148a-binding site. The seed region is in bold text and the seed region is mutated in the reporter construct. (d) Luciferase activity of three GC cell lines cotransfected with reporter vector containing either control vector, wild-type (WT), or mutant miR-148a (Mut) 3-′UTR. Results are mean ± SD of triplicate measurements. ***P < 0.001. N.S., not significant. (e) Western blot analysis of MMP7 in MKN-45 cell lysates was carried out after stable transfection with the miR-148a construct, both miR-148a and MMP7 construct, or empty vector. β-Actin served as loading control. (f) Effect of miR-148a-stable transfection and restoration of MMP7 on cell invasion of MKN-45 cells. Boyden chambers were used for incubation of MKN-45 cells after stable transfection with miR-148a construct, both miR-148a and MMP7 construct, or empty vector. Results are mean ± SD of triplicate measurements. **P < 0.01; ***P < 0.001.
Several reports show that miR-148a regulates cancer cell invasion through suppressing its target genes.(22,23) To confirm the cellular level at which MMP7 contributes to GC cell invasion, the MKN-45 cell line was stably transfected with vector expressing miR-148a alone, and cotransfected with vectors expressing miR-148a and MMP7. MKN-45 cells were selected because they express high levels of MMP7 and low levels of miR-148a. In MKN-45-miR-148a, which stably expressed miR-148a, MMP7 expression was significantly suppressed, and in MKN-45-miR-148a/-MMP7, which stably expressed miR-148a and MMP7, the expression of MMP7 was 50% less than that of empty vector-transfected MKN-45 cells (Fig. 3e). Next, the invasive properties of these cells were examined by Transwell invasion assay. As shown in Figure 3(f), the invasiveness of MKN-45-miR-148a was approximately 50% less than that of MKN-45-empty vector at all endpoints. However, the invasiveness of MKN-45-miR-148a/-MMP7 was only approximately 10–20% less than that of the MKN-45-empty vector. These observations suggest that MMP7 plays a central role in GC invasion among miR-148a target genes.
Discussion
In this study, we showed frequent downregulation of miR-148a in GC tissue that was significantly correlated with lymph node metastasis, advanced clinical stage, and poor clinical outcome. We also established that MMP7 is a direct and functional target of miR-148a. This is the first report to identify miRNA that regulates MMP7 expression. Further studies revealed that overexpression of miR-148a suppressed GC cell invasion in vitro, and cotransfection of miR-148a and MMP7 restored invasiveness of GC cells. It has been reported that MMP7 plays a key role in invasive properties of various cancer cells, and MMP7 is well known as a representative cancer invasion-related gene whose expression is correlated with worse clinicopathologic characters.(26,27) Together with previous reports, our results suggest that miR-148a plays important roles in suppressing tumor progression in GC, and that the induction of MMP7 expression by downregulation of miR-148a contributes to GC progression.
MicroRNA microarray analysis and qRT-PCR revealed that miR-148a was significantly downregulated in GC tissue, and we also confirmed that hypermethylation of MIR148A CpG islands is correlated with suppression of miR-148a in GC cell lines. It has been reported that hypermethylation of MIR148A CpG islands is induced by DNA methyltransferase-1, however DNA methyltransferase-1, in turn, is conversely downregulated by miR-148a overexpression.(28) In healthy stomach tissue, next-generation sequencing reportedly confirms overexpression of miR-148a,(29) and downregulation of miR-148a has been reported in various cancers including GC.(30–32) In relation to miR-148a, almost all of its target genes were implicated in promoting cancer progression. These results suggest that miR-148a may play essential roles in maintaining homeostasis of normal stomach tissue, and when downregulated, sequential induction of some oncogenes may contribute to the development of gastric neoplasia. However, the points at which downregulation of miR-148a occurs in the process of carcinogenesis or tumor progression have not been fully elucidated. In GC, there is no report that has examined the timing of miR-148a downregulation. It is our current important task to determine when miR-148a is downregulated in gastric neoplasia.
A number of miRNA target-prediction databases have been developed and tested for accuracy and precision using both computational and laboratory techniques. However, when results from laboratory techniques were compared to those of computational approaches, analysis showed that most gene lists generated using computational target prediction contained high false-negative and false-positive results.(7,33) In addition, various miRNA target prediction programs, which use different algorithms of targeting, produce different lists of predicted targets. It is documented that these differences can arise from the source of 3′-UTR sequences. In the present study, using custom oligonucleotide array-based target prediction, we found that MMP7 expression is downregulated by miR-148a overexpression. Moreover, this was confirmed by qRT-PCR, Western blotting, and reporter gene assays. We determined the 3′-UTR of MMP7 by referring to the Gene database available from the National Center for Biotechnology Information (NCBI), and found that the 3′-UTR of MMP7 contains a putative binding site that is complementary to the miR-148a seed sequence. We compared the sequence of MMP7 coding region from the Gene database (NCBI: http://www.ncbi.nlm.nih.gov) with that from Ensembl (http://useast.ensembl.org) or UCSC (http://genome.ucsc.edu). Although slight differences were observed among these sequences, including the definition of the start point, most were almost completely matched. This result suggests that the extant online miRNA target prediction database did not detect MMP7 as a candidate target gene of miR-148a, due to a deficiency in their chosen algorithm. These considerations point to a need for further refinement of online databases, in order to effectively identify real target genes for any given miRNA. At this point in time, conventional laboratory techniques remain very valuable methods for effective identification of miRNA target genes.
In summary, we have shown that downregulation of miR-148a is an independent prognostic classifier in patients with GC. Furthermore, this study identified MMP7 as a direct target of miR-148a in GC cells and its restoration by downregulation of miR-148a in these cells increases tumor invasive activity. The present findings provide evidence for the contribution of miR-148a to the maintenance of homeostasis in normal stomach tissue and suggest an additional role for miR-148a in GC invasion through regulation of MMP7 expression. Elucidation of a fuller picture of the molecular mechanisms through which miR-148a is involved in gene deregulatory oncogenic processes could improve our understanding of carcinogenesis, particularly tumor progression in GC.
Acknowledgments
We thank Mr. Shinichi Norimura for excellent technical assistance. This work was carried out with the kind cooperation of the Research Center for Molecular Medicine, Faculty of Medicine, Hiroshima University (Hiroshima, Japan). We thank the Analysis Center of Life Science, Hiroshima University, for the use of their facilities. This work was supported by Grants-in-Aid for Research from the Ministry of Education, Culture, Science, Sports, and Technology of Japan, and, in part, by a Grant-in-Aid for the Third Comprehensive 10-Year Strategy for Cancer Control and for Cancer Research from the Ministry of Health, Labor, and Welfare of Japan, and from The National Institute of Biomedical Innovation (Program for Promotion of Fundamental Studies in Health Sciences). This work was also supported in part by a Research Fellowship of the Japan Society for the Promotion of Science and the National Cancer Center Research and Development Fund (23-A-9).
Disclosure Statement
The authors have no conflict of interest.
Supporting Information
Additional supporting information may be found in the online version of this article:
Fig. S1 Comparison between microarray and quantitative RT-PCR data for microRNA-148a (miR-148a) expression levels in the same gastric cancer (GC) sample set.
Fig. S2 Comparison between microRNA-148a (miR-148a) expression levels in non-neoplastic gastric mucosa and gastric cancer (GC).
Fig. S3 Quantitative RT-PCR and bisulfite genomic DNA sequencing analysis of MIR148A in gastric cancer (GC) cell lines.
Fig. S4 Effect of microRNA-148a (miR-148a) on epithelial–mesenchymal transition-related molecules and ROCK1 expression in gastric cancer cell lines.
Fig. S5 Effect of microRNA-148a (miR-148a) on MMP7 expression in gastric cancer cell lines.
Table S1 Correlation between microRNA-148a (miR-148a) expression level and clinicopathologic characters.
Table S2 Correlation between MMP7 and microRNA-148a (miR-148a) expression in gastric cancer tissue.
Data S1 Materials and methods.
References
- 1.Crew KD, Neugut AI. Epidemiology of gastric cancer. World J Gastroenterol. 2006;12:354–62. doi: 10.3748/wjg.v12.i3.354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yasui W, Sentani K, Sakamoto N, Anami K, Naito Y, Oue N. Molecular pathology of gastric cancer: research and practice. Pathol Res Pract. 2011;207:608–12. doi: 10.1016/j.prp.2011.09.006. [DOI] [PubMed] [Google Scholar]
- 3.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–66. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 4.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 5.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–5. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 6.Di Leva G, Croce CM. Roles of small RNAs in tumor formation. Trends Mol Med. 2010;16:257–67. doi: 10.1016/j.molmed.2010.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alexiou P, Maragkakis M, Papadopoulos GL, Reczko M, Hatzigeorgiou AG. Lost in translation: an assessment and perspective for computational microRNA target identification. Bioinformatics. 2009;25:3049–55. doi: 10.1093/bioinformatics/btp565. [DOI] [PubMed] [Google Scholar]
- 8.Thomson DW, Bracken CP, Goodall GJ. Experimental strategies for microRNA target identification. Nucleic Acids Res. 2011;39:6845–53. doi: 10.1093/nar/gkr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhao X, Dou W, He L, et al. MicroRNA-7 functions as an anti-metastatic microRNA in gastric cancer by targeting insulin-like growth factor-1 receptor. Oncogene. 2013;32:1363–72. doi: 10.1038/onc.2012.156. [DOI] [PubMed] [Google Scholar]
- 10.Tsukamoto Y, Nakada C, Noguchi T, et al. MicroRNA-375 is downregulated in gastric carcinomas and regulates cell survival by targeting PDK1 and 14-3-3zeta. Cancer Res. 2010;70:2339–49. doi: 10.1158/0008-5472.CAN-09-2777. [DOI] [PubMed] [Google Scholar]
- 11.Sakamoto N, Oue N, Noguchi T, et al. Serial analysis of gene expression of esophageal squamous cell carcinoma: ADAMTS16 is upregulated in esophageal squamous cell carcinoma. Cancer Sci. 2010;101:1038–44. doi: 10.1111/j.1349-7006.2009.01477.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nagao K, Togawa N, Fujii K, et al. Detecting tissue-specific alternative splicing and disease-associated aberrant splicing of the PTCH gene with exon junction microarrays. Hum Mol Genet. 2005;14:3379–88. doi: 10.1093/hmg/ddi369. [DOI] [PubMed] [Google Scholar]
- 13.Oue N, Sentani K, Sakamoto N, et al. Characteristic gene expression in stromal cells of gastric cancers among atomic-bomb survivors. Int J Cancer Suppl. 2009;124:1112–21. doi: 10.1002/ijc.24060. [DOI] [PubMed] [Google Scholar]
- 14.Kondo T, Oue N, Yoshida K, et al. Expression of POT1 is associated with tumor stage and telomere length in gastric carcinoma. Cancer Res. 2004;64:523–9. doi: 10.1158/0008-5472.can-03-1196. [DOI] [PubMed] [Google Scholar]
- 15.Shinmei S, Sakamoto N, Goto K, et al. MicroRNA-155 is a predictive marker for survival in patients with clear cell renal cell carcinoma. Int J Urol. 2013;20:468–77. doi: 10.1111/j.1442-2042.2012.03182.x. [DOI] [PubMed] [Google Scholar]
- 16.Yasui W, Sano T, Nishimura K, et al. Expression of P-cadherin in gastric carcinomas and its reduction in tumor progression. Int J Cancer Suppl. 1993;54:49–52. doi: 10.1002/ijc.2910540109. [DOI] [PubMed] [Google Scholar]
- 17.Herman JG, Graff JR, Myohanen S, Nelkin BD, Baylin SB. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci USA. 1996;93:9821–6. doi: 10.1073/pnas.93.18.9821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hanoun N, Delpu Y, Suriawinata AA, et al. The silencing of microRNA 148a production by DNA hypermethylation is an early event in pancreatic carcinogenesis. Clin Chem. 2010;56:1107–18. doi: 10.1373/clinchem.2010.144709. [DOI] [PubMed] [Google Scholar]
- 19.Matsumura S, Oue N, Mitani Y, Kitadai Y, Yasui W. DNA demethylation of vascular endothelial growth factor-C is associated with gene expression and its possible involvement of lymphangiogenesis in gastric cancer. Int J Cancer Suppl. 2007;120:1689–95. doi: 10.1002/ijc.22433. [DOI] [PubMed] [Google Scholar]
- 20.Sakamoto N, Oue N, Sentani K, et al. Liver-intestine cadherin induction by epidermal growth factor receptor is associated with intestinal differentiation of gastric cancer. Cancer Sci. 2012;103:1744–50. doi: 10.1111/j.1349-7006.2012.02353.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alley MC, Scudiero DA, Monks A, et al. Feasibility of drug screening with panels of human tumor cell lines using a microculture tetrazolium assay. Cancer Res. 1988;48:589–601. [PubMed] [Google Scholar]
- 22.Zheng B, Liang L, Wang C, et al. MicroRNA-148a suppresses tumor cell invasion and metastasis by downregulating ROCK1 in gastric cancer. Clin Cancer Res. 2011;17:7574–83. doi: 10.1158/1078-0432.CCR-11-1714. [DOI] [PubMed] [Google Scholar]
- 23.Aprelikova O, Palla J, Hibler B, et al. Silencing of miR-148a in cancer-associated fibroblasts results in WNT10B-mediated stimulation of tumor cell motility. Oncogene. 2013;32:3246–53. doi: 10.1038/onc.2012.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cheng P, Chen C, He HB, et al. MiR-148a regulates osteoclastogenesis via targeting MAFB. J Bone Miner Res. 2013;28:1180–90. doi: 10.1002/jbmr.1845. [DOI] [PubMed] [Google Scholar]
- 25.Song H, Wang Q, Wen J, et al. ACVR1, a therapeutic target of fibrodysplasia ossificans progressiva, is negatively regulated by miR-148a. Int J Mol Sci. 2012;13:2063–77. doi: 10.3390/ijms13022063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fanelli MF, Chinen LT, Begnami MD, et al. The influence of transforming growth factor-alpha, cyclooxygenase-2, matrix metalloproteinase (MMP)-7, MMP-9 and CXCR4 proteins involved in epithelial-mesenchymal transition on overall survival of patients with gastric cancer. Histopathology. 2012;61:153–61. doi: 10.1111/j.1365-2559.2011.04139.x. [DOI] [PubMed] [Google Scholar]
- 27.Iguchi H, Kosaka N, Ochiya T. Versatile applications of microRNA in anti-cancer drug discovery: from therapeutics to biomarkers. Curr Drug Discov Technol. 2010;7:95–105. doi: 10.2174/157016310793180648. [DOI] [PubMed] [Google Scholar]
- 28.Zhu A, Xia J, Zuo J, et al. MicroRNA-148a is silenced by hypermethylation and interacts with DNA methyltransferase 1 in gastric cancer. Med Oncol. 2012;29:2701–9. doi: 10.1007/s12032-011-0134-3. [DOI] [PubMed] [Google Scholar]
- 29.Ribeiro-dos-Santos A, Khayat AS, Silva A, et al. Ultra-deep sequencing reveals the microRNA expression pattern of the human stomach. PLoS One. 2010;5:e13205. doi: 10.1371/journal.pone.0013205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ueda T, Volinia S, Okumura H, et al. Relation between microRNA expression and progression and prognosis of gastric cancer: a microRNA expression analysis. Lancet Oncol. 2010;11:136–46. doi: 10.1016/S1470-2045(09)70343-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen Y, Song Y, Wang Z, et al. Altered expression of MiR-148a and MiR-152 in gastrointestinal cancers and its clinical significance. J Gastrointest Surg. 2010;14:1170–9. doi: 10.1007/s11605-010-1202-2. [DOI] [PubMed] [Google Scholar]
- 32.Liffers ST, Munding JB, Vogt M, et al. MicroRNA-148a is down-regulated in human pancreatic ductal adenocarcinomas and regulates cell survival by targeting CDC25B. Lab Invest. 2011;91:1472–9. doi: 10.1038/labinvest.2011.99. [DOI] [PubMed] [Google Scholar]
- 33.Bentwich I. Prediction and validation of microRNAs and their targets. FEBS Lett. 2005;579:5904–10. doi: 10.1016/j.febslet.2005.09.040. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Comparison between microarray and quantitative RT-PCR data for microRNA-148a (miR-148a) expression levels in the same gastric cancer (GC) sample set.
Fig. S2 Comparison between microRNA-148a (miR-148a) expression levels in non-neoplastic gastric mucosa and gastric cancer (GC).
Fig. S3 Quantitative RT-PCR and bisulfite genomic DNA sequencing analysis of MIR148A in gastric cancer (GC) cell lines.
Fig. S4 Effect of microRNA-148a (miR-148a) on epithelial–mesenchymal transition-related molecules and ROCK1 expression in gastric cancer cell lines.
Fig. S5 Effect of microRNA-148a (miR-148a) on MMP7 expression in gastric cancer cell lines.
Table S1 Correlation between microRNA-148a (miR-148a) expression level and clinicopathologic characters.
Table S2 Correlation between MMP7 and microRNA-148a (miR-148a) expression in gastric cancer tissue.
Data S1 Materials and methods.


