Abstract
Primary intraocular lymphoma (PIOL) is a rare lymphoma. Because of difficulties in obtaining tissue samples, little is known about the disease's genetic features. In order to clarify these features, we carried out single nucleotide polymorphism array karyotyping of IOL using genomic DNA extracted from vitreous fluid. We analyzed 33 samples of IOLs consisting of 16 PIOLs, 12 IOLs with a central nervous system (CNS) lesion at diagnosis (IOCNSL), and five secondary IOLs following systemic lymphoma. All were B-cell type. We identified recurrent copy number (CN) gain regions in PIOLs, most frequently on chromosome 1q followed by 18q and 19q. Chromosome 6q was the most frequent loss region. Although these CN gain regions of PIOL were in common with those of IOCNSL, loss of 6q22.33 containing PTPRK and 9p21.3 containing CDKN2A were more frequently deleted in IOCNSL. Large CN loss in 6q was detected in three of four PIOL patients who had early CNS development and short survival periods, whereas long-term survivors did not have such deletions. There was a correlation between gain of the IL-10 gene located on 1q and intravitreal interleukin-10 concentration, which was higher in IOL than in benign uveitis. The results suggest that IOCNSL is a highly malignant form of PIOL that infiltrates into the CNS at an early stage. They also indicate that genetic differences between PIOL and primary CNS lymphoma need to be clarified.
Keywords: Central nervous system, IL-10, intraocular lymphoma, single nucleotide polymorphism, vitreous body
Primary intraocular lymphoma (PIOL) is a lymphoma whose lesion is located in the eye ball, including the vitreous body, the retina, the choroid, the iris, and the ciliary muscle. It is a rare form of non-Hodgkin's lymphoma, accounting for less than 1% of all non-Hodgkin's lymphoma cases1 and of all intraocular neoplasms.2 The majority of PIOL are vitreoretinal lymphoma whose lesions are found exclusively in the retina and the vitreous body of the eye, resulting in the primary symptoms of blurred vision and decreased visual acuity.3 Ophthalmoscopic findings are a cloudy vitreous body and/or subretinal proliferative lesions due to tumor cell infiltration.3,4 Most PIOLs are B-cell lymphomas; T-cell lymphomas may occur, but are extremely rare.3
The diagnosis of IOL is quite difficult due to several reasons. First, it is hard to obtain sufficient biopsy material from the lesions. Retinal biopsy may cause visual field defects, and enucleation inflicts irreversible damage to visual acuity. Therefore, vitrectomy with vitreous sampling is used to obtain the specimen. Diagnosis is usually made by cytology for samples of vitreous fluid, although the detection rate is low (20–44.5%) because of degradation of the infiltrating cells or lack of cells in the samples.5,6 Polymerase chain reaction analysis using DNA extracted from vitreous fluid has been shown to have a much higher detection rate and has become the most persuasive procedure for the diagnosis of IOL.7 Using this method, we detected IgH rearrangements in 21 of 22 patients with IOL (95.5%).5 Diagnosis can be supported by the interleukin-10 (IL-10) : IL-6 cytokine ratio in the vitreous fluid.6,8,9 We also showed that 86% patients with an IL-10 : IL-6 ratio of >1.0 had IOL, whereas the majority of patients with benign uveitis had a ratio of <1.0.5 Using these methods, the accurate rate of IOL diagnosis has improved dramatically, and consequently, the number of PIOL patients has been increasing.
Many issues, however, relating to PIOL remain unresolved. According to previous reports, primary vitreoretinal lymphomas, which are the major types of PIOLs, are reported to be diffuse large B-cell lymphomas (DLBCL).10–12 In the clinical situation, however, the histological types of PIOLs are often undetermined because the material for diagnosis is usually vitreous fluid. Furthermore, the genetic features of PIOL have not yet been elucidated. In order to solve them, we carried out single nucleotide polymorphism (SNP) microarrays using genomic DNA extracted from the vitreous fluids.
Materials and Methods
Study design
The patients in this study were referred to the Tokyo Medical and Dental University (TMDU) or the Tokyo Medical University (TMU) hospital (both Tokyo, Japan) on suspicion of IOL between 2005 and 2011. After the diagnosis was made, they were treated at TMDU, TMU, or the primary hospitals from which they were referred.
The vitreous fluid of IOL patients was analyzed at TMDU, TMU, and The University of Tokyo (Tokyo, Japan). Patients who were HIV-positive were excluded. Written informed consent was obtained from patients except those who had already passed away. Obtaining the preserved samples with or without informed consent was approved by the ethics boards of these institutes.
Diagnosis and definition of IOL
Intraocular lymphoma was diagnosed using the following criteria: (i) typical eye involvement, a cloudy vitreous body and/or subretinal proliferative lesions; (ii) presence of lymphoma cells in the vitreous fluid; and (iii) clonality of the infiltrating lymphoma cells in the vitreous fluid using either PCR analysis of IgH or T-cell receptor gene rearrangements.5 Patients who had criterion (i) accompanied by either (ii) or (iii) were diagnosed with IOL.13
In this report, IOL confined to the eyes at diagnosis was defined as PIOL. Intraocular lymphoma accompanied by central nervous system (CNS) lesions at diagnosis was defined as IOL with CNS lesion (IOCNSL). As it could not be determined whether the eyes or the CNS was the primary site, we analyzed IOCNSL independently from PIOL. Patients with IOL who developed ocular lesions after detection of lymphoma of extraocular and extra-CNS sites were categorized as having secondary IOL (SIOL). Pathological examination and investigation of systemic involvement were carried out at the institutes from which patients were referred. Diagnosis of IOL was made at TMDU and TMU.
Measurement of intravitreal IL-10
To determine the concentrations of IL-6 or IL-10 in the vitreous fluids, 50 μL vitreous supernatant from each patient was applied for ELISA according to the manufacturer's instructions (Invitrogen, Camarillo, CA, USA). The lowest detection limits of the cytokines were 2.0 pg/mL for IL-6, and 1.0 pg/mL for IL-10.
Single nucleotide polymorphism microarray
Genomic DNA extracted from the vitreous fluid at the time of diagnosis was subjected to SNP array karyotyping using GeneChip 250K SNP arrays (Affymetrix, Santa Clara, CA, USA) as previously described.14 Details of the procedures are described in Data S1.
Statistical analysis
For statistical analyses, the Mann–Whitney U-test was carried out using GraphPad Prism 5 (GraphPad Software, La Jolla, CA, USA).
Results
Patients
Thirty-three IOLs were analyzed, including 16 PIOLs, 12 IOCNSLs, and five SIOLs. Their clinical information and laboratory findings of the vitreous fluids are summarized in Tables1 and S1, respectively.
Table 1.
Clinical information of IOL patients
| No | Age | Gender | PS | Extra-ocular lesions at diagnosis of IOL |
Initial treatment | Clinical course (time after diagnosis) | Survival period (after diagnosis) | |||
|---|---|---|---|---|---|---|---|---|---|---|
| Brain involvement | CSF infiltration | Systemic lesions | BM infiltration | |||||||
| PIOL-1 | 83 | M | 0 | (−) | (−) | (−) | (−) | VT, ivMTX | CNS development (4 Ms), Dead | 6 Ms |
| PIOL-2 | 71 | M | 0 | (−) | (−) | (−) | (−) | VT, ORT | CNS development (11 Ms), Alive | >17 Ms |
| PIOL-3 | 77 | F | 0 | (−) | (−) | (−) | (−) | VT, WBRT | LFU | Unknown |
| PIOL-4 | 57 | F | 0 | (−) | (−) | (−) | (−) | VT, sMTX, WBRT | CNS development (30 Ms), Dead | 48 Ms |
| PIOL-5 | 80 | M | 0 | (−) | (−) | (−) | ND | VT, enucleation | LFU | Unknown |
| PIOL-6 | 70 | F | 0 | (−) | (−) | (−) | ND | VT, ivMTX | Alive | >36 Ms |
| PIOL-7 | 65 | F | 1 | (−) | ND | (−) | ND | VT, itMTX | CNS development (ND), Dead | 26 Ms |
| PIOL-8 | 82 | F | 0 | (−) | ND | (−) | ND | VT, ivMTX | Alive | >52 Ms |
| PIOL-9 | 85 | F | 0 | (−) | ND | (−) | ND | VT, ivMTX | LFU | Unknown |
| PIOL-10 | 68 | F | 0 | (−) | ND | (−) | (−) | VT | CNS development (6 Ms), Dead | 8 Ms |
| PIOL-11 | 74 | F | 0 | (−) | (−) | (−) | ND | VT | CNS development (40 Ms), Alive | >53 Ms |
| PIOL-12 | 82 | M | 1 | (−) | ND | (−) | ND | VT, ivMTX | CNS development (4 Ms), Dead | 18 Ms |
| PIOL-13 | 51 | F | 0 | (−) | ND | (−) | ND | VT | Alive | >53 Ms |
| PIOL-14 | 55 | M | 0 | (−) | (−) | (−) | (−) | VT, ivMTX | CNS development (3 Ms), Dead | 12Ms |
| PIOL-15 | 79 | F | 1 | (−) | ND | (−) | ND | VT, ORT | LFU | Unknown |
| PIOL-16 | 57 | M | 0 | (−) | ND | (−) | ND | VT, ORT | Alive | >70 Ms |
| IOCNSL-1 | 70 | F | 1 | (+) | (−) | (−) | (−) | VT, enucleation, sMTX | Alive in CR | >27 Ms |
| IOCNSL-2 | 45 | F | 0 | (+) | ND | (−) | ND | VT, WBRT | LFU | Unknown |
| IOCNSL-3 | 52 | M | 0 | (+) | ND | (−) | (−) | VT, WBRT, sCT | LFU | Unknown |
| IOCNSL-4 | 47 | M | 0 | (+) | (−) | (−) | (−) | VT, WBRT, sCT | Alive | >36 Ms |
| IOCNSL-5 | 47 | M | 0 | (+) | (−) | (−) | (−) | VT, WBRT, sCT | Alive | >36 Ms |
| IOCNSL-6 | 66 | M | 3 | (+) | ND | (−) | ND | VT, ivMTX | LFU | Unknown |
| IOCNSL-7 | 52 | M | 4 | (+) | ND | (−) | ND | VT, ivMTX | PD, Dead | 29 Ms |
| IOCNSL-8 | 54 | M | 3 | (+) | ND | (−) | (−) | VT, ivMTX, sCT | PD, Dead | 8 Ms |
| IOCNSL-9 | 61 | M | 1 | (+) | ND | (−) | ND | VT, WBRT, sMTX | LFU | Unknown |
| IOCNSL-10 | 66 | M | 0 | (+) | (−) | (−) | (−) | VT, ivMTX, sCT | Alive | >69 Ms |
| IOCNSL-11 | 70 | M | 1 | (+) | (−) | (−) | (−) | VT, ivMTX | PD, Dead | 14 Ms |
| IOCNSL-12 | 88 | F | 2 | (+) | (−) | (−) | (−) | VT, ORT, sMT | LFU | Unknown |
| SIOL-1 | 75 | M | 0 | (−) | ND | (−) | ND | VT | LFU | Unknown |
| SIOL-2 | 73 | M | 0 | (−) | (−) | (−) | (−) | VT, sMTX | LFU | Unknown |
| SIOL-3 | 45 | M | 0 | (−) | (−) | (−) | (−) | VT | Alive | >70 Ms |
| SIOL-4 | 69 | M | 0 | (−) | (−) | (−) | (−) | VT, sMTX | CNS development (34 Ms), Alive | >99 Ms |
| SIOL-5 | 68 | F | ND | (−) | ND | (−) | ND | ND | LFU | Unknown |
+, Present; −, not present; BM, bone marrow; CNS, central nervous system; CR, complete response; CSF, cerebrospinal fluid; F, female; IOCNSL, IOL with central nervous system lesion at diagnosis; itMTX, intrathechal methotrexate injection; ivMTX, intravitreal methotrexate injection; LFU, lost to follow-up; M, male; Ms, months; ND, not described; ORT, ocular radiation therapy; PD, progressive disease; PIOS, primary IOS; PS, performance status, according to Eastern Cooperative Oncology Group criteria; sCT, systemic chemotherapy; SIOL, secondary IOL; sMTX, systemic methotrexate injection; VT, vitrectomy; WBRT, whole brain radiation therapy.
Characteristics of the copy number changes of IOLs
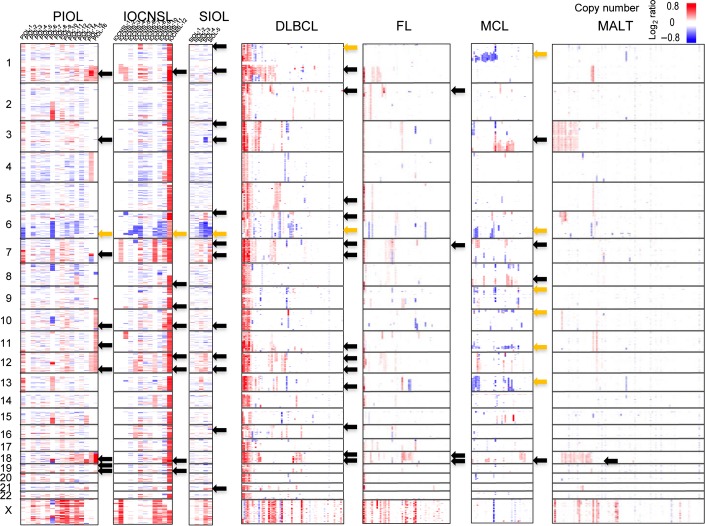
Copy number (CN) changes of all IOL samples are shown in Figures1 and 2. As in our previous report, we defined CN changes as CN gains, losses, as well as neutral loss of heterozygosity involving >3 Mb segments (Fig.2). As shown in Figures1 and 2(a), the most frequent CN gain region in PIOL was 1q, detected in 81% (13/16) of cases, followed by 18q and 19q in 69% (11/16), 12q in 63% (10/16), 7q and 11q in 56% (9/16), and 3q, 10q, and 19p in 50% (8/16). As shown in Figures1 and 2(b,c), the most frequent CN loss in PIOLs was on 6q (44%, 7/16), whereas uniparental disomy was detected on various regions in only patient.
Figure 1.
Distributions of copy number (CN) changes in intraocular lymphomas (IOLs) and different lymphoma type samples. The latter includes data of 238 primary B-cell lymphomas, including 64 samples of diffuse large B-cell lymphomas (DLBCLs), 52 follicular lymphomas (FL), 35 mantle cell lymphomas (MCLs), and 87 mucosa-associated lymphoid tissue (MALT) lymphomas reported in Kato et al.14 Genetic lesions are color-coded and plotted for each sample, as indicated. Samples were clustered in each lymphoma type. Regions with frequent CN change are indicated by arrows (black arrows, CN gains ≥50% in IOLs, ≥20% in the other lymphomas; orange arrows, CN losses). Note that genetic changes involving small regions are lost in this figure due to limited resolution. IOCNSL, IOL with a central nervous system lesion at diagnosis; PIOL, primary IOL; SIOL, secondary IOL.
Figure 2.

Copy number (CN) change is summarized in each type of intraocular lymphoma (IOL). Frequency of CN gains (a) and losses (b), as well as CN neutral loss of heterozygosity (c) involving >3 Mb segments were calculated and plotted for each IOL type. *Copy number changes detected in >20% of diffuse large B-cell lymphomas; **CN changes detected in >20% of follicular lymphomas; ***CN changes detected in >20% of mucosa-associated lymphoid tissue lymphomas; ****CN changes detected in >20% of mantle cell lymphomas. Black arrows indicate common frequent (>50%) gain regions in primary IOL (PIOL), and in IOL with a central nervous system lesion at diagnosis (IOCNSL). SIOL, secondary IOL.
Previously we investigated CN changes of genes in 238 primary B-cell lymphoma specimens of different histological types, including 64 samples of DLBCLs, 52 follicular lymphomas (FL), 35 mantle cell lymphomas (MCLs), and 87 mucosa-associated lymphoid tissue (MALT) lymphomas.14 The results of the present study were presented in comparison with them (Fig. 1). The most frequent CN gain region in DLBCL was 1q (>40%), and other frequent CN gains in DLBCL were found on 12q, 18q (>30%), and on 2p, 5q, 6p, 7p, 7q, 11q, 12p, 13q, 16p, and 18p (>20%), whereas CN losses were on 1p and on 6q (>20%). As shown in Figures1 and 2, 56% (5/9) of the CN gains found in >50% of PIOLs were also detected in >20% of DLBCLs, and were the most frequent gains. Conversely, the three most frequent gains found in DLBCL (>30%; 1q, 12q, and 18q) were detected in >63% of PIOLs (Figs.1,2a). In addition, the most frequent CN loss in PIOL was located on 6q, as in DLBCL (Fig.2b). In contrast, CN gains on 1q, which were the most frequent gains in PIOLs, were found infrequently (<10%) in FL, MALT lymphomas, and MCL.14 Among the nine most frequent gains in PIOLs (≥50%), only CN gains on18q were frequent in FL or MALT lymphomas (>20%), and only two gains, on 3q and on 18q, were frequent in MCL (>30%) (Fig.2a).
The most frequent CN gains in IOCNSLs were on 1q and 12p in 67% (8/12), followed by 8q, 9q, 10q, 12q, 18q, and 19q (6/12; 50%) (Fig.2a). In SIOLs, frequent gains (≥50%) were detected on 13 chromosomal lesions (Fig.2a). Among them, 50% (4/8) of gains in IOCNSLs and 55% (7/13) of gains in SIOLs were identical to those in DLBCL (>20%) (Fig.2a). Frequent CN losses were located on the 6q region in IOCNSLs and SIOLs, (58% and 80%, respectively) (Fig.2b).
Comparison between CN changes of IOLs
Next, we compared the genomic distribution of CN changes in each type of IOL. As shown in Figure2(a), among the nine most frequent (≥50%) gain regions in PIOLs, five (5/9; 56%) regions (1q, 10q, 12q, 18q, and 19q) were also frequent (≥50%) in IOCNSLs. The most frequent gain in PIOL (13/16; 81%) was located on 1q, which was also the most frequent gain (8/12; 67%) in IOCNSLs (Fig.2a). Concerning CN loss, 6q was the most frequent loss in PIOLs (7/16; 44%) and was also the most frequent (7/12; 58%) loss in IOCNSLs (Fig.2a). These data indicated that PIOL and IOCNSL had common frequent CN alterations.
Some genes that affect the prognosis of lymphoma have already been described. It has been reported that PCNSL with deletion of 6q22.33, which contains PTPRK, revealed an aggressive clinical course.15 As shown in Figure3(a,b), loss of 6q22.33 was detected in 50% (6/12) of IOCNSLs and in 25% (4/16) of PIOLs. In addition, loss of 9p21.3, which contains CDKN2A (p16), was detected in 6/12 (50%) IOCNSLs (Fig.3c), whereas it was deleted in 4/16 (25%) PIOLs (Fig.3d). Among them, homozygous deletion of CDKN2A had occurred in two IOCNSL patients (Fig.3e,f). Although statistical difference was not identified between them, loss of 6q22.33 and loss of 9p21.3 in IOCNSL seemed to be more common than that in PIOL.
Figure 3.
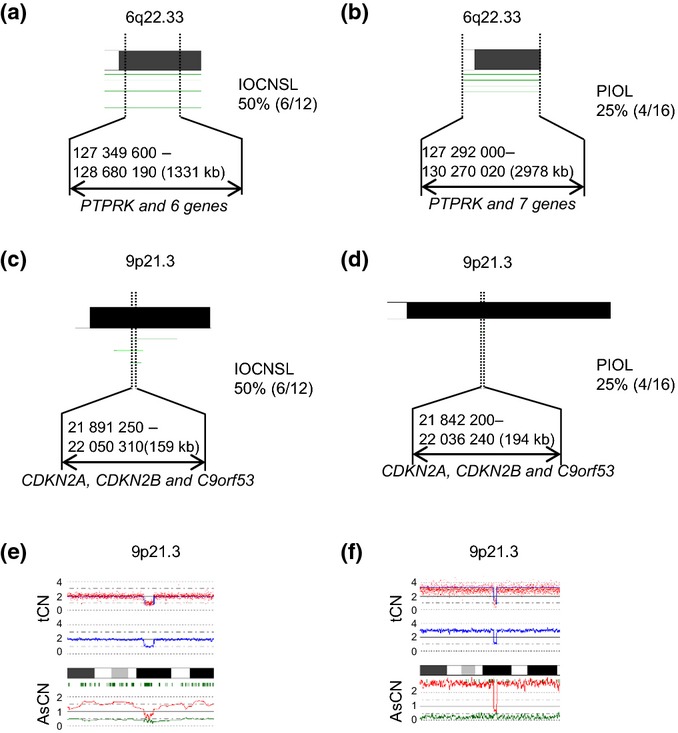
Outputs of deletions of 6q22.33 and 9p21.3 in each type of intraocular lymphoma (IOL), generated by CNAG software. The CNAG software is freely available (http://www.genome.umin.jp/). (a) Chromosome 6q22.33 shows recurrent copy number (CN) loss in IOL with a central nervous system lesion at diagnosis (IOCNSL). The frequency in IOCNSL was 50% (6/12). (b) Chromosome 6q22.33 shows recurrent CN loss in primary IOL (PIOL). The frequency in PIOL was 25% (4/16). (c) Chromosome 9p21.3 shows recurrent CN loss in IOCNSL. The frequency in IOCNSL was 50% (6/12). (d) Chromosome 9p21.3 shows recurrent CN loss in PIOL. The frequency in PIOL was 25% (4/16). In figure parts (a–d), each horizontal line represents a CN loss found in a single case. (e,f) CNAG outputs of homozygous deletions of 9p21.3 in IOCNSL-9 (e) and IOCNSL-12 (f). Presence of homozygous deletions is indicated by the biallelic reduction of allele-specific CN (AsCN). tCN, total CN.
Relation between genetic lesions and outcomes
The outcome of each patient is described in Table1. Survival of patients with PIOL and IOCNSL is shown in Figure S1. The median overall survival (OS) of patients with PIOL was 37 months, whereas the OS of patients with IOCNSL and SIOL could not be determined due to the limited number of patients. Differences in OS between PIOL and IOCNSL patients could also not be evaluated due to the small number of patients.
Among 16 patients with PIOL, outcome could be followed in 12. As shown in Figure S2, four patients (PIOL-1, 10, 12, and 14) who had early CNS development within 6 months of the time of diagnosis had large CN loss lesions in 6q. Among them, three patients (PIOL-1, 10, and 12) had deletion of 6q22.33. In contrast, four patients (PIOL-6, 8, 13, and 16) who were alive without progression for more than 36 months did not have large deletions in 6q. We could not find a relation between loss of 9p21.3, which was detected in four patients (PIOL-11, 13, 14, and 15), and outcome.
High-grade amplifications or homozygous deletions
We identified several high-grade amplifications (Fig.4). High-grade amplifications on eight lesions were found in PIOL-7 (Fig.4a–e), whereas they were found on three lesions in PIOL-15 (Fig.4f–h). High-grade amplification of 3p24.1-p23 was found in IOCNSL-11 (Fig.4i). They were not recurrent in our case series. The genes in the affected regions are shown in Figure4.
Figure 4.

Results of copy number analysis and outputs of high-grade amplifications in individual cases of intraocular lymphoma (IOL), generated by CNAG software. (a–e) Primary IOL (PIOL)-7. (f–h) PIOL-15. (i) IOL with a central nervous system lesion at diagnosis (IOCNSL)-11. Blue lines in the middle of each panel show the moving average of total copy numbers in five adjacent single nucleotide polymorphisms. The sample ID and possible gene targets are indicated in each panel.
Copy number gain of IL-10 gene and its concentration in vitreous fluid
The most frequent gain regions of PIOLs and IOCNSLs were located on 1q. It is noteworthy that the 1q32.1 region located on 1q contained the genes for IL-10, a cytokine whose concentration in the vitreous fluid is significantly high in IOL. The CN of the IL-10 gene located on 1q32.1 was increased in 69% (11/16) of PIOLs, in 58% (7/12) of IOCNSL, in 80% (4/5) of SIOLs, and in 67% (22/33) in total. In order to examine the relation between CN change and IL-10 concentration, we compared the intravitreal IL-10 concentration of IL-10 gain-positive patients to that of IL-10 gain-negative patients. As shown in Figure5(a), the IL-10 concentration of the gain-positive patients was significantly higher than that of the gain-negative patients in all IOLs. However, we could not find a significant difference between them in each IOL subtype (Fig.5b,c), probably due to the small number of samples.
Figure 5.

Intravitreal interleukin-10 (IL-10) levels in intraocular lymphomas (IOLs) with or without gain of the IL-10 gene. Data were analyzed for all patients (a), for primary IOL (PIOL) (b), and for IOL with a central nervous system lesion at diagnosis (IOCNSL) (c). The data represent the mean ± standard error of the mean.
Discussion
Primary intraocular lymphoma consists of various histological types, but the majority are DLBCL. Our results showed that PIOLs diagnosed by analyzing the vitreous fluid and DLBCL had common CN-altered regions in their chromosomes. Therefore, it is appropriate to plan treatment for PIOL as an aggressive lymphoma.
The most frequent CN gain found in PIOLs was located on 1q that contained the region for the IL-10 gene. Interestingly, there was correlation between gain of the IL-10 gene and intravitreal IL-10 concentration. The concentration of IL-10 in the vitreous fluid is significantly higher in IOL than that in benign uveitis, and is a useful tool to distinguish between the two conditions.5 In addition, IL-10 is an autocrine growth factor for B cells and promotes B-cell lymphoma development and proliferation.16,17 Our results indicated that IL-10 may be secreted by the tumor cells and promote disease progression.
In the present report, loss of 6q22.33 was detected in 25% of PIOLs and 50% of IOCNSLs. According to previously published reports, the frequencies of del(6)(q22) in PCNSLs was 45%,15 whereas that in systemic DLBCL was 25%.18 Loss of 6q22.33 might be common in IOCNSL as in PCNSL compared to PIOL or systemic DLBCL. 6q22.33 contains PTPRK. This gene encodes protein tyrosine phosphatase receptor κ (PTPRK), which belongs to the protein tyrosine phosphatase superfamily of enzymes and inhibits proliferation- or survival-promoting molecular signals mediated by tyrosine kinase. It can act as a tumor suppressor by inhibiting cell cycle progression. It was reported that deletion of 6q22 in PCNSL revealed an aggressive clinical course or poor prognosis.15,19 Interestingly, three of four patients who had early CNS development and short survival periods had large CN loss in 6q, whereas long-term survivors did not have such deletions. Statistical analysis could not be carried out because of the limited number of patients. Further investigation should be undertaken to clarify the relation between deletion of 6q and outcome, especially the role of 6q22.33.
In our study, 9p21.3, on which CDKN2A was located, was deleted in 25% of PIOLs and 50% of IOCNSLs. Although statistical difference was not determined between them, loss of 9p21.3 might be more common in IOCNSL than in PIOL. CDKN2A, located in the other frequent loss region in IOCNSL, 9p21.3, negatively regulates G1/S phase transition by inhibiting the kinase activity of CDK4/6. Thus, loss or inhibition of CDKN2A (p16) may result in increased tumor cell proliferation. In addition, it is well established that deletion of CDKN2A in DLBCL is associated with poor prognosis.18 These findings suggested that IOCNSL was a highly malignant form of PIOL that tends to spread into the CNS at an early stage. Further clinical survey on a large scale should be added to confirm the hypothesis.
Although we have clarified some issues about the genetic characteristics of IOL, the origin of PIOL cells remains unknown. Wallace et al. detected t(14;18) in 67% of PIOL.20 The high frequency of the translocation in PIOL suggests that the lymphoma cells originate from germinal center B-cells (GCBs) with high expression of BCL2.21 Consistent with this, we found that the BCL2 CN was increased in PIOL. The origin of PIOL cells, however, remains controversial. Lipford et al. reported that t(14;18) was determined in only 20% of PCNSL.22 Other investigators indicated a high somatic mutation load in the VH genes of PIOL cells.23 Furthermore, the immunophenotype of the PIOL tumor cells is MUM1/IRF4+;BCL-6+/−;CD10−. These results suggested that PIOL was an activated B-cell subtype of DLBCL.24 Further studies, including gene expression profiling, are required to confirm these results and to identify the cell of origin in PIOL lesions.
It has been reported that PIOL and PCNSL were closely related with each other. The majority of PIOL and PCNSL cases are pathologically diagnosed as DLBCL, and 60–82.5% of PIOL patients ultimately develop CNS lesions, whereas extra-CNS lesions are rare.6,11,25,26 In addition, the eyes are close to the CNS and are actually derived from the CNS during embryogenesis. For these reasons, PIOL and PCNSL are categorized as the same lymphoma in many reports,10,11 despite little genetic evidence to support this. As a result, potential differences between PIOL and PCNSL in their clinical features have not been evaluated. Our unpublished data, however, indicate that only one of 15 PCNSL patients diagnosed and treated during the last 10 years at our hospital developed eye lesions (Ayako Arai, unpublished data, 2013). In addition, the gain in chromosome 18, which was frequently detected in our PIOL series, was not described in the report by Sung and colleagues, the latest report for PCNSL analyzed by high-resolution array-based comparative genomic hybridization.27 These findings suggest that PIOL and PCNSL may have distinct clinical courses and genetic features. Contrary to the report by Sung et al., however, chromosome 18 was frequently gained in three early PCNSL reports.28–30 The CN alteration in PCNSL has been controversial. The difference in methods may be one of the reasons for the discrepancies, and further study on the same technology platform should be carried out in order to determine the relation between PIOL and PCNSL.
Taken together, our data suggest that IOCNSL is considered to be a highly malignant form of PIOL that infiltrates into the CNS at an early stage. Study should be continued to clarify the the origin of tumor cells of PIOL, genetic alteration predicting outcome, and the genetic similarity and difference between PIOL and other B-cell lymphomas.
Acknowledgments
We are grateful to Ms. Kaori Okada for excellent technical assistance. This work was supported by grants from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (23591375) and Japan Leukemia Research Fund.
Disclosure Statement
The authors have no conflict of interest.
Funding information
Ministry of Education, Culture, Sports, Science, and Technology of Japan (23591375). Japan Leukemia Research Fund.
Supporting Information
Additional supporting information may be found in the online version of this article:
Survival of patients with intraocular lymphoma (IOL). Differences in overall survival between patients with primary IOL (PIOL) and those with IOL with a central nervous system lesion at diagnosis (IOCNSL) could not be evaluated due to the small number of patients.
Copy number alterations on chromosome 6 in primary intraocular lymphoma.
Laboratory findings of the vitreous fluids of patients with intraocular lymphoma.
Procedures of single nucleotide polymorphism microarray.
References
- 1.Bardenstein DS. Intraocular lymphoma. Cancer Control. 1998;5:317–25. doi: 10.1177/107327489800500403. [DOI] [PubMed] [Google Scholar]
- 2.Choi JY, Kafkala C, Foster CS. Primary intraocular lymphoma: a review. Semin Ophthalmol. 2006;21:125–33. doi: 10.1080/08820530500350498. [DOI] [PubMed] [Google Scholar]
- 3.Coupland SE, Heimann H, Bechrakis NE. Primary intraocular lymphoma: a review of the clinical, histopathological and molecular biological features. Graefes Arch Clin Exp Ophthalmol. 2004;242:901–13. doi: 10.1007/s00417-004-0973-0. [DOI] [PubMed] [Google Scholar]
- 4.Cassoux N, Merle-Beral H, Leblond V, et al. Ocular and central nervous system lymphoma: clinical features and diagnosis. Ocul Immunol Inflamm. 2000;8:243–50. doi: 10.1076/ocii.8.4.243.6463. [DOI] [PubMed] [Google Scholar]
- 5.Sugita S, Takase H, Sugamoto Y, Arai A, Miura O, Mochizuki M. Diagnosis of intraocular lymphoma by polymerase chain reaction analysis and cytokine profiling of the vitreous fluid. Jpn J Ophthalmol. 2009;53:209–14. doi: 10.1007/s10384-009-0662-y. [DOI] [PubMed] [Google Scholar]
- 6.Kimura K, Usui Y, Goto H Japanese Intraocular Lymphoma Study Group. Clinical features and diagnostic significance of the intraocular fluid of 217 patients with intraocular lymphoma. Jpn J Ophthalmol. 2012;56:383–9. doi: 10.1007/s10384-012-0150-7. [DOI] [PubMed] [Google Scholar]
- 7.Shen DF, Zhuang Z, LeHoang P, et al. Utility of microdissection and polymerase chain reaction for the detection of immunoglobulin gene rearrangement and translocation in primary intraocular lymphoma. Ophthalmology. 1998;105:1664–9. doi: 10.1016/S0161-6420(98)99036-4. [DOI] [PubMed] [Google Scholar]
- 8.Chan CC, Whitcup SM, Solomon D, Nussenblatt RB. Interleukin-10 in the vitreous of patients with primary intraocular lymphoma. Am J Ophthalmol. 1995;120:671–3. doi: 10.1016/s0002-9394(14)72217-2. [DOI] [PubMed] [Google Scholar]
- 9.Whitcup SM, Stark-Vancs V, Wittes RE, et al. Association of interleukin 10 in the vitreous and cerebrospinal fluid and primary central nervous system lymphoma. Arch Ophthalmol. 1997;115:1157–60. doi: 10.1001/archopht.1997.01100160327010. [DOI] [PubMed] [Google Scholar]
- 10.Kluin PM, Deckert M, Ferry JA. Primary diffuse large B-cell lymphoma of the CNS. In: Jaffe E, Harris N, Stein H, editors. World Health Organization Classification of Tumors Pathology and Genetics of Tumours of Haematopoietic and Lymphoid Tissues. Lyon: IARC Press; 2008. pp. 240–1. [Google Scholar]
- 11.Coupland SE, Chan CC, Smith J. Pathophysiology of retinal lymphoma. Ocul Immunol Inflamm. 2009;17:227–37. doi: 10.1080/09273940903168696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chan CC, Sen HN. Current concepts in diagnosing and managing primary vitreoretinal (intraocular) lymphoma. Discov Med. 2013;15:93–100. [PMC free article] [PubMed] [Google Scholar]
- 13.Nakauchi Y, Takase H, Sugita S, et al. Concurrent administration of intravenous systemic and intravitreal methotrexate for intraocular lymphoma with central nervous system involvement. Int J Hematol. 2010;92:179–85. doi: 10.1007/s12185-010-0589-6. [DOI] [PubMed] [Google Scholar]
- 14.Kato M, Sanada M, Kato I, et al. Frequent inactivation of A20 in B-cell lymphomas. Nature. 2009;459:712–6. doi: 10.1038/nature07969. [DOI] [PubMed] [Google Scholar]
- 15.Cady FM, O'Neill BP, Law ME, et al. Del(6)(q22) and BCL6 rearrangements in primary CNS lymphoma are indicators of an aggressive clinical course. J Clin Oncol. 2008;26:4814–9. doi: 10.1200/JCO.2008.16.1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Beatty PR, Krams SM, Martinez OM. Involvement of IL-10 in the autonomous growth of EBV-transformed B cell lines. J Immunol. 1997;158:4045–51. [PubMed] [Google Scholar]
- 17.Moore KW, de Waal Malefyt R, Coffman RL, O'Garra A. Interleukin-10 and the interleukin-10 receptor. Annu Rev Immunol. 2001;19:683–765. doi: 10.1146/annurev.immunol.19.1.683. [DOI] [PubMed] [Google Scholar]
- 18.Jardin F, Jais JP, Molina TJ, et al. Diffuse large B-cell lymphomas with CDKN2A deletion have a distinct gene expression signature and a poor prognosis under R-CHOP treatment: a GELA study. Blood. 2010;116:1092–104. doi: 10.1182/blood-2009-10-247122. [DOI] [PubMed] [Google Scholar]
- 19.Nakamura M, Kishi M, Sakaki T, et al. Novel tumor suppressor loci on 6q22-23 in primary central nervous system lymphomas. Cancer Res. 2003;63:737–41. [PubMed] [Google Scholar]
- 20.Wallace DJ, Shen D, Reed GF, et al. Detection of the bcl-2 t(14;18) translocation and proto-oncogene expression in primary intraocular lymphoma. Invest Ophthalmol Vis Sci. 2006;47:2750–6. doi: 10.1167/iovs.05-1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alizadeh AA, Eisen MB, Davis RE, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403:503–11. doi: 10.1038/35000501. [DOI] [PubMed] [Google Scholar]
- 22.Lipford E, Wright JJ, Urba W, et al. Refinement of lymphoma cytogenetics by the chromosome 18q21 major breakpoint region. Blood. 1987;70:1816–23. [PubMed] [Google Scholar]
- 23.Coupland SE, Hummel M, Müller HH, Stein H. Molecular analysis of immunoglobulin genes in primary intraocular lymphoma. Invest Ophthalmol Vis Sci. 2005;46:3507–14. doi: 10.1167/iovs.05-0401. [DOI] [PubMed] [Google Scholar]
- 24.Coupland SE, Loddenkemper C, Smith JR, et al. Expression of immunoglobulin transcription factors in primary intraocular lymphoma and primary central nervous system lymphoma. Invest Ophthalmol Vis Sci. 2005;46:3957–64. doi: 10.1167/iovs.05-0318. [DOI] [PubMed] [Google Scholar]
- 25.Chan CC. Molecular pathology of primary intraocular lymphoma. Trans Am Ophthalmol Soc. 2003;101:275–92. [PMC free article] [PubMed] [Google Scholar]
- 26.Coupland SE, Damato B. Understanding intraocular lymphomas. Clin Experiment Ophthalmol. 2008;36:564–78. doi: 10.1111/j.1442-9071.2008.01843.x. [DOI] [PubMed] [Google Scholar]
- 27.Sung CO, Kim SC, Karnan S, et al. Genomic profiling combined with gene expression profiling in primary central nervous system lymphoma. Blood. 2011;117:1291–300. doi: 10.1182/blood-2010-07-297861. [DOI] [PubMed] [Google Scholar]
- 28.Boonstra R, Koning A, Mastik M, van den Berg A, Poppema S. Analysis of chromosomal copy number changes and oncoprotein expression in primary central nervous system lymphomas: frequent loss of chromosome arm 6q. Virchows Arch. 2003;443:164–9. doi: 10.1007/s00428-003-0836-9. [DOI] [PubMed] [Google Scholar]
- 29.Booman M, Szuhai K, Rosenwald A, et al. Genomic alterations and gene expression in primary diffuse large B-cell lymphomas of immune-privileged sites: the importance of apoptosis and immunomodulatory pathways. J Pathol. 2008;216:209–17. doi: 10.1002/path.2399. [DOI] [PubMed] [Google Scholar]
- 30.Harada K, Nishizaki T, Kubota H, Suzuki M, Sasaki K. Distinct primary central nervous system lymphoma defined by comparative genomic hybridization and laser scanning cytometry. Cancer Genet Cytogenet. 2001;125:147–50. doi: 10.1016/s0165-4608(00)00377-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Survival of patients with intraocular lymphoma (IOL). Differences in overall survival between patients with primary IOL (PIOL) and those with IOL with a central nervous system lesion at diagnosis (IOCNSL) could not be evaluated due to the small number of patients.
Copy number alterations on chromosome 6 in primary intraocular lymphoma.
Laboratory findings of the vitreous fluids of patients with intraocular lymphoma.
Procedures of single nucleotide polymorphism microarray.