Abstract
Human REV7 (also known as MAD2L2 and MAD2B) is involved in DNA repair, cell cycle regulation, gene transcription, and carcinogenesis. In this study, we evaluated the expression of REV7 in epithelial ovarian cancer (EOC) and analyzed the association between its expression and chemosensitivity in ovarian clear cell carcinoma (CCC) cells. Expression of REV7 in human EOC tissues was assessed by immunohistochemical staining. Expression was detected in the majority of EOCs (92.0%) with especially high levels of expression frequently observed in CCCs (73.5%) compared with that of non-CCCs (53.4%). Enhanced immunoreactivity to REV7 was associated with poor prognosis represented by reduced progression-free survival in advanced stage (stage II–IV) EOC as assessed using Kaplan–Meier curves and log–rank tests. The effects of REV7 knockdown on cell proliferation and chemosensitivity in CCC cells were also analyzed in vitro and in vivo. Knockdown of REV7 in CCC cells decreased cell proliferation without affecting cell cycle distribution. Additionally, the number of apoptotic cells and DNA damaged cells were increased after cisplatin treatment. In a nude mouse tumor xenograft model, inoculated REV7-knockdown tumors showed significantly reduced tumor volumes after cisplatin treatment compared with those of the control group. These findings indicate that depletion of REV7 enhances sensitivity to cisplatin treatment in CCC, suggesting that REV7 is a candidate molecular target in CCC management.
Keywords: Apoptosis, chemosensitivity, cisplatin, DNA damage, ovarian clear cell carcinoma
Ovarian cancer is the fifth most common malignancy in women and the most common cause of gynecologic cancer-related death.1 The four main histological subtypes of epithelial ovarian cancer (EOC), serous, endometrioid, clear cell, and mucinous adenocarcinomas, are considered to be distinct diseases according to epidemiology, clinical characteristics, responses to treatment, prognosis, and molecular features.2–5 However, EOCs have been largely treated as a single disease. Compared with other EOC subtypes, clear cell carcinoma (CCC) is associated with poor outcome and greater resistance to platinum-based chemotherapy.6–9 Recent studies have shown that CCC carries either ARID1A or PIK3CA mutations as well as HNF-1β overexpression,10–12 but there are no biomarkers for chemotherapy response. Therefore, it is important to identify both the clinical indicators of CCC and the molecular pathways involved in its drug sensitivity.
A number of chemotherapeutic agents induce DNA damage in cells, whereby the DNA repair system is associated with their efficiency in cancer therapy. Deregulation of DNA repair proteins results in failure to repair DNA damage in cells and subsequent genetic instability and cell death. Thus, DNA repair inhibitors are expected to improve the outcome of cancer chemotherapy.13–15 REV7 (also known as MAD2L2 and MAD2B) is involved in translesion DNA synthesis (TLS), one of the damage tolerance processes, which completes DNA synthesis through DNA lesions to prevent DNA damage-induced cell death.16,17 REV7 makes a complex with REV3, forming DNA polymerase ζ, one of the specialized low fidelity polymerases, playing an important role in TLS.18–20 Involvement of REV7 in DNA damage tolerance in human cells has been clearly demonstrated. Inactivation of REV7 by RNAi technology increases sensitivity to DNA-damaging agents in nasopharyngeal carcinoma cells.21 Similarly, after siRNA-mediated REV7 depletion, human fibroblast cells show heightened sensitivity to UV-induced cytotoxicity and low sensitivity to UV-induced mutagenesis compared with control cells.22 It was also reported that REV7-depleted glioma cells show enhanced apoptotic response to ionizing radiation.23 Moreover, it was reported that mRNA levels of REV7 are significantly increased in human breast and colorectal cancers,24,25 and that REV7 interacts with cancer-related proteins PRCC (papillary renal cell carcinoma) and HCCA2 (hepatocellular carcinoma-associated gene 2).26,27 These findings suggest that REV7 expression is associated with cancer development and sensitivity to DNA-damaging agents.
In this study, we established the association between REV7 expression and the chemosensitivity of CCC using clinical materials and in in vitro and in vivo experiments. Our findings suggest that REV7 is a potential candidate for molecular target in CCC therapy.
Materials and Methods
Patients and tissue samples
One hundred and thirty-seven ovarian carcinoma tissue samples (47 serous adenocarcinomas, 19 mucinous adenocarcinomas, 22 endometrioid adenocarcinomas, and 49 CCCs) were obtained from patients who underwent surgical treatment at Nagoya University Hospital (Nagoya, Japan) between 1998 and 2003 following informed consent. The patients’ ages ranged from 23 to 82 years, with a median age of 54 years. The histological types were assigned according to the World Health Organization classification criteria. Clinical stage was assigned on the basis of the International Federation of Gynecology and Obstetrics staging system.
Immunohistochemical staining
Formalin-fixed and paraffin-embedded tissues were sliced at a thickness of 4 μm. For antigen retrieval, they were heated in Target Retrieval Solution pH 9.0 (Dako, Copenhagen, Denmark) for 40 min at 98°C. Endogenous peroxidase was inhibited using 3% H2O2 in methanol for 15 min. After blocking with 10% normal goat serum for 10 min at room temperature (RT), sections were incubated with primary antibodies for 90 min at RT and then incubated with the secondary antibody conjugated to HRP-labeled polymer (EnVision+ anti-rabbit; Dako) for 15 min at RT. Reaction products were visualized using diaminobenzidine (Dako), and nuclei were counterstained with hematoxylin. The staining intensity of REV7 was scored as 0 (negative), 1 (weak), 2 (medium), or 3 (strong) and then further classified into two categories: low, expression scores 0 and 1; or high, expression scores 2 and 3 (Fig.1a, see Data S1 for antibody information). The REV7 expression levels were evaluated by two independent blinded observers.
Figure 1.
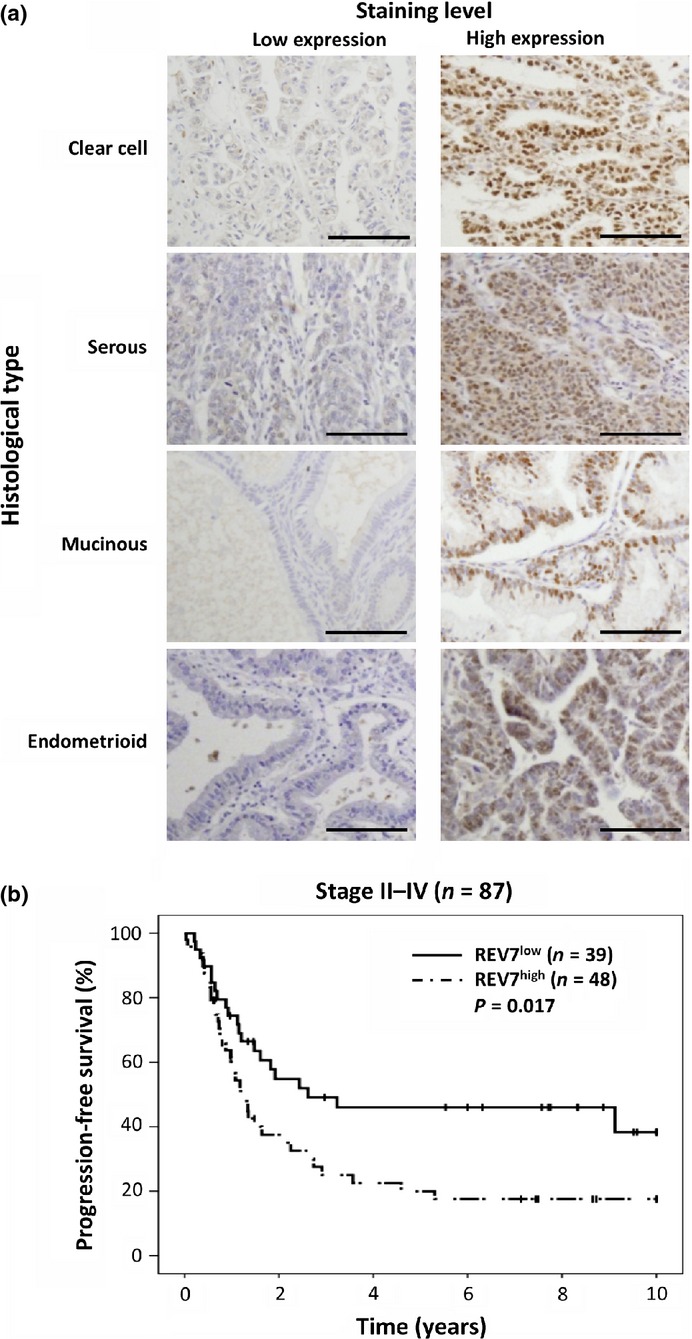
Immunohistochemical analyses of REV7 expression in epithelial ovarian cancer. (a) Representative images of immunoreactivity for REV7. Images of low REV7 staining levels, with a score of 1 (clear cell) or 0 (serous, mucinous, and endometrioid), are shown on the left; those with high REV7 staining levels, with a score of 3, are shown on the right. Scale bar, 100 μm. (b) Kaplan–Meier curves and log–rank tests for progression-free survival of patients with stage II–IV epithelial ovarian cancer.
Cell proliferation and viability assay
Cells were seeded in 96-well plates at a density of 2 × 103 cells in 100 μL medium. Twenty-four hours after seeding, the cell proliferation assay was carried out using WST-1 Reagent (Roche, Basel, Switzerland) according to the manufacturer's instructions. For the cell viability assay, 5 × 103 cells per well were seeded in 96-well plates and treated with the indicated concentrations of cisplatin (cis-diamminedichloroplatinum, CDDP) for 48 h. Cell viability was determined by the WST-1 assay. Absorbance was measured at 450 nm using a microplate reader (Tecan, Palm Springs, CA, USA).
Fluorescence immunocytochemistry
Cells were seeded in 96-well plates for cleaved caspase-3 staining and transferase-mediated biotin-16-dUTP nick-end labeling (TUNEL) assay or on coverslips for phospho-H2AX foci analysis. After CDDP treatment at the indicated concentrations for 24 h, the cells were fixed in 4% paraformaldehyde for 20 min at 4°C, washed in PBS, and permeabilized with 0.1% Triton X-100 in 0.1% sodium citrate for 2 min on ice. After blocking with 1% BSA, the cells were incubated with anti-cleaved caspase-3 or anti-phospho-H2AX antibody for 90 min at RT. The cells were then incubated with secondary antibody conjugated to Alexa Fluor 488 for 30 min at RT. Cellular DNA was counterstained with DAPI. The TUNEL assay was carried out following the manufacturer's instruction (In Situ Cell Death Detection Kit, Fluorescein; Roche). To assess the immunoreactivity of cleaved caspase-3 or TUNEL, the cells were counted using a Cellomics Array Scan VTI (Cellomics/Thermo-Fisher, Waltham, MA, USA). To assess the positivity for phospho-H2AX, the cells with more than 10 foci were counted using a fluorescence microscope (Olympus, Tokyo, Japan).
Mouse tumor xenografts
TOV-21G cells (1 × 107) with short-hairpin RNA (shRNA)-mediated REV7 depletion (shREV7) and control cells (shCont) in 200 μL PBS were injected s.c. into the right flank of eight 6-week-old female nude mice (Crlj:CD1-Foxn1nu; Charles River Laboratories, Yokohama, Japan). When tumor volumes reached approximately 100 mm3, the mice were given PBS or CDDP (2 mg/kg body weight) i.p. four times every 2 days (n = 4/group). We then measured the diameters of each tumor every 4 days after beginning treatment and calculated their estimated volumes using the following formula: volume = length ×width × width × 1/2. The mice were maintained in accordance with the institutional guidelines of Nagoya University Graduate School of Medicine, and experiments were carried out according to approved experimental protocols.
Statistical analysis
We used χ2-tests to analyze the associations between the REV7 status and clinicopathological parameters. The overall survival (OS) was defined as the time between the date of surgery and the last date of follow-up or date of death due to cancer. The progression-free survival (PFS) was defined as the time interval between the date of surgery and the date of progression/recurrence or date of last follow-up. Survival analysis was carried out according to the life tables and Kaplan–Meier methods using spss software (version 20; SPSS Inc., Armonk, NY, USA) and the log–rank test. A multivariate analysis was carried out using a Cox proportional hazards model. For data from in vivo and in vitro experiments, statistical comparisons among groups were carried out using non-paired Student's t-test or anova with Bonferroni corrections. Differences were considered significant at P < 0.05. Data are indicated as the mean ± SD.
Results
REV7 is frequently expressed in EOC, especially CCC
Ovarian cancer specimens from 137 patients were immunohistochemically stained for REV7. REV7 was mainly expressed in the nuclei of cancer cells, but not in the surrounding non-tumor cells (Fig.1a). The specific reaction of rabbit polyclonal anti-REV7 antibody was shown by comparison with negative control staining using the preimmune serum or normal rabbit IgG (Fig. S1). The immunohistochemical analyses revealed that 11 of 137 specimens (8.0%) were REV7-negative and 126 (92.0%) were REV7-positive. Fifty-four cases (39.4%) were classified into the low REV7 expression group (REV7low), and 83 cases (60.6%) were classified into the high REV7 expression group (REV7high) (Table1). The association between REV7 expression and clinicopathological parameters were statistically analyzed. All 49 CCC cases (100%) were REV7-positive, with 13 (26.5%) in the REV7low group and 36 (73.5%) in the REV7high group. In contrast, 77 of 88 non-CCC cases (87.5%) were REV7-positive, with 41 (46.6%) in the REV7low group and 47 (53.4%) in the REV7high group. The association between CCC histopathological type and high REV7 expression was statistically significant (P = 0.021), indicating that REV7 is highly expressed in CCC (Table1). However, REV7 expression was not associated with other clinicopathological parameters.
Table 1.
Association between REV7 expression and clinicopathological factors in patients with ovarian cancer (n = 137)
| Variables | Patients (n) | REV7 expression score |
REV7 expression level |
P-value | ||||
|---|---|---|---|---|---|---|---|---|
| 0 (n = 11) | 1 (n = 43) | 2 (n = 43) | 3 (n = 40) | Low (n = 54) | High (n = 83) | |||
| Age, years | ||||||||
| <60 | 96 | 7 | 35 | 24 | 30 | 42 (43.8) | 54 (56.2) | 0.112 |
| ≥60 | 41 | 4 | 8 | 19 | 10 | 12 (29.3) | 29 (70.7) | |
| FIGO stage | ||||||||
| I | 50 | 5 | 10 | 19 | 16 | 15 (27.8) | 35 (70.0) | 0.087 |
| II–IV | 87 | 6 | 33 | 24 | 24 | 39 (44.8) | 48 (55.2) | |
| Histological type | ||||||||
| Clear cell | 49 | 0 | 13 | 13 | 22 | 13 (26.5) | 36 (73.5) | 0.021 |
| Non-clear cell | 88 | 11 | 30 | 29 | 18 | 41 (46.6) | 47 (53.4) | |
| Serous | 47 | 4 | 15 | 17 | 11 | 19 (40.4) | 28 (59.6) | |
| Mucinous | 19 | 2 | 4 | 6 | 7 | 6 (31.6) | 13 (68.4) | |
| Endometrioid | 22 | 5 | 11 | 6 | 0 | 16 (72.7) | 6 (27.3) | |
| Residual tumor | ||||||||
| Absent | 96 | 8 | 30 | 29 | 29 | 38 (39.6) | 58 (60.4) | 0.951 |
| Present | 41 | 3 | 13 | 14 | 11 | 16 (39.0) | 25 (61.0) | |
| CA125, U/mL | ||||||||
| <50 | 27 | 1 | 6 | 6 | 14 | 7 (25.9) | 20 (74.1) | 0.109 |
| ≥50 | 110 | 10 | 37 | 37 | 26 | 47 (42.7) | 63 (57.3) | |
| Chemotherapy (platinum-based) | ||||||||
| Absent | 18 | 2 | 5 | 4 | 7 | 7 (38.9) | 11 (61.1) | 0.961 |
| Present | 119 | 9 | 38 | 39 | 33 | 47 (39.5) | 72 (60.5) | |
FIGO, International Federation of Gynecology and Obstetrics.
Next, we examined whether or not REV7 expression was associated with prognosis of EOC. When the association between REV7 expression and OS or PFS was assessed by Kaplan–Meier methods and the log–rank test, no significant association was found (OS, P = 0.711; PFS, P = 0.237). However, analysis of the association between REV7 expression and PFS in 87 cases with advanced stage EOC (stage II–IV) revealed that PFS in the REV7high group (n = 48) was significantly shorter than that in the REV7low group (n = 39) (P = 0.017, Fig.1b). Then, we analyzed the factors that contributed to the prognosis using a multivariate Cox proportional hazards model (Table S1). The results showed that REV7 expression was independently associated with PFS (P = 0.012). In addition, 1-year PFS was significantly associated with REV7 expression in cases with advanced stage EOC treated with platinum-based chemotherapy (P = 0.022, Table S2). These results indicate that REV7 expression is implicated in the prognosis of EOC at advanced stage.
Depletion of REV7 suppresses cell proliferation but does not affect the cell cycle in CCC cells
As it was revealed that REV7 is strongly expressed in CCC, we investigated the significance of REV7 expression in CCC biology. REV7-knockdown cells were generated using three CCC cell lines, ES-2, KOC-7C, and TOV-21G, by RNAi technology (Data S1). Expression levels of REV7 in the three CCC cell lines were similar and were upregulated compared with other cell lines derived from yolk sac tumor and serous adenocarcinoma (Fig. S2). Expression of REV7 was significantly suppressed in all CCC cell lines with shRNA targeting REV7 (Fig.2a). Then, we assessed the effects of REV7 knockdown on cell proliferation, in which shREV7 cells grew more slowly than shCont cells, indicating that REV7 affects cell proliferation (Fig.2b). Cell cycle distribution was also analyzed in shREV7 and shCont cells by flow cytometry, but it was not affected by REV7 depletion (Fig.2c, Data S1).
Figure 2.
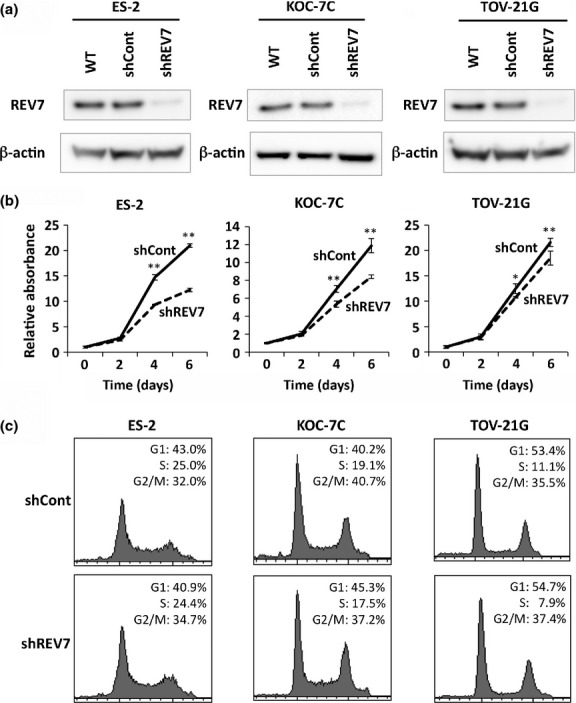
Knockdown of REV7 inhibits cell proliferation but does not affect the cell cycle of ovarian clear cell carcinoma cells. (a) Western blot images of REV7 expression in wild-type (WT), REV7-knockdown (shREV7), and control (shCont) clear cell carcinoma cells. The image of β-actin is indicated as an internal control. (b) Cell proliferation analysis of REV7-knockdown and control cells. The means ± SDs of relative cell counts are shown. *P < 0.05; **P < 0.01. (c) Graphical depiction of data obtained from flow cytometry analysis. Percentages of cell populations in each phase of the cell cycle are indicated.
Knockdown of REV7 enhances chemosensitivity and increases CDDP-induced apoptosis in CCC cells
Next, we investigated the effect of REV7 expression on chemosensitivity to DNA damaging agents in CCC cells. REV7-knockdown and control cells were treated with CDDP at various concentrations for 48 h, and cell viability was assessed. REV7 knockdown rendered cells more sensitive to CDDP, and the IC50 values were decreased by REV7 depletion compared with those in the shCont cells (1.61-fold, 1.98-fold and 1.93-fold decrease in IC50 in ES-2, KOC-7C, and TOV-21G cells, respectively) (Fig.3a). The chemosensitivity of REV7-knockdown cells was confirmed by colony formation assay, in which the number of colonies formed after CDDP treatment was decreased by REV7 depletion (Fig. S3, Data S1). In addition, chemosensitivity was also examined in shCont and shREV7 ES-2 cells with ectopic REV7 expression. Ectopic REV7 expression rescued enhanced chemosensitivity in shREV7 cells, however, its expression did not significantly affect chemosensitivity in shCont cells, although the IC50 of REV7-expressing shCont cells was elevated a little (Fig. S4), suggesting that endogenous REV7 expression is high enough for chemoresistance in ES-2 cells.
Figure 3.
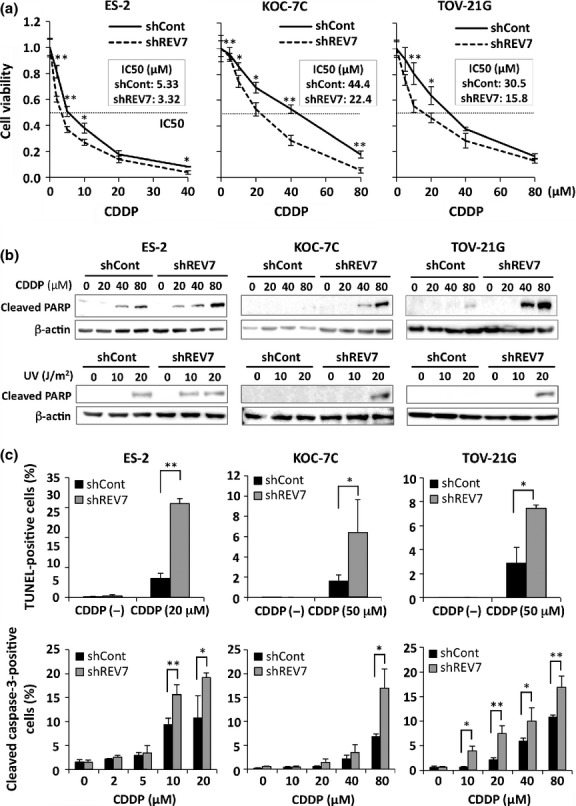
REV7 knockdown enhances sensitivity to cisplatin (CDDP) in ovarian clear cell carcinoma (CCC) cells. (a) Cell viability analysis of CDDP-treated CCC cells. The IC50 of each cell line is indicated. Experiments were carried out in triplicate and the means ± SDs of relative absorbance is shown. *P < 0.05; **P < 0.01. (b) Western blot analysis for cleaved poly(ADP-ribose) polymerase (PARP) in cells after DNA-damaging treatment. The REV7-knockdown (shREV7) and control (shCont) CCC cells were treated with CDDP for 48 h and used for Western blotting (upper panels). The shREV7 and shCont CCC cells were also treated with UV at the indicated doses and used for Western blotting 12 h after UV irradiation (lower panels). The blots of β-actin are indicated as internal controls. (c) Assessment of apoptosis in the shREV7 and shCont CCC cells after CDDP treatment. Immunoreactivity for TUNEL and cleaved caspase-3 in cells treated with CDDP was fluorescently analyzed. The means ± SDs of the percentage of TUNEL-positive (upper panels) or cleaved caspase-3-positive (lower panels) cells are shown. *P < 0.05; **P < 0.01.
We then assessed CDDP-induced apoptosis in REV7-knockdown and control cells. After the cells were treated with CDDP at concentrations of 0, 20, 40, and 80 μM for 48 h, the cleaved poly(ADP-ribose) polymerase (PARP) fragment could be detected in REV7-knockdown cells at lower concentrations of CDDP than in control cells (Fig.3b). Similar results were observed in cells treated with UV irradiation at doses of 0, 10, and 20 J/m2 (Fig.3b). To assess the enhancement of apoptosis in REV7-knockdown cells quantitatively, shREV7 and shCont cells were immunofluorescently stained for TUNEL and cleaved caspase-3 before and after CDDP treatment. The number of TUNEL-positive cells significantly increased in REV7-depleted cells after CDDP treatment (ES-2, 20 μM; KOC-7C and TOV-21G, 50 μM) when compared with that of control cells (Fig.3c). The percentages of cleaved caspase-3-positive cells after CDDP treatment dose-dependently increased in control cells and were significantly elevated by suppression of REV7 in all three cell lines (Fig.3c). These results indicate that CDDP-induced apoptosis is enhanced by REV7 depletion in CCC cells.
Depletion of REV7 causes accumulation of DNA double-strand breaks after CDDP treatment in CCC cells
As CDDP induces DNA damage of intrastrand and interstrand cross-links, which are repaired by nucleotide excision repair and homologous recombination repair machineries, it is possible that REV7 depletion causes dysfunction of DNA repair machinery and accumulation of DNA damage in cells, causing enhancement of apoptosis. DNA damage of double-strand breaks was assessed in shREV7 and shCont cells by immunofluorescence staining using anti-phospho-H2AX antibody. TOV-21G-derived shREV7 and shCont cells were treated with 50 μM CDDP for 24 h, and the cells were fluorescently immunostained for phospho-H2AX. Positive immunoreactivity, showing small foci formation in the nuclei, increased after treatment at a significantly high frequency in REV7-depleted cells compared with that of control cells (Fig.4). As a comparison, the cells treated with UV irradiation at 20 J/m2 were immunostained with anti-phospho-H2AX antibody 12 h after UV irradiation. Positive immunoreactivity was also detected in REV7-depleted cells at a significantly high frequency (Fig.4). These findings indicate that REV7 depletion results in accumulation of double-strand breaks in response to CDDP treatment.
Figure 4.
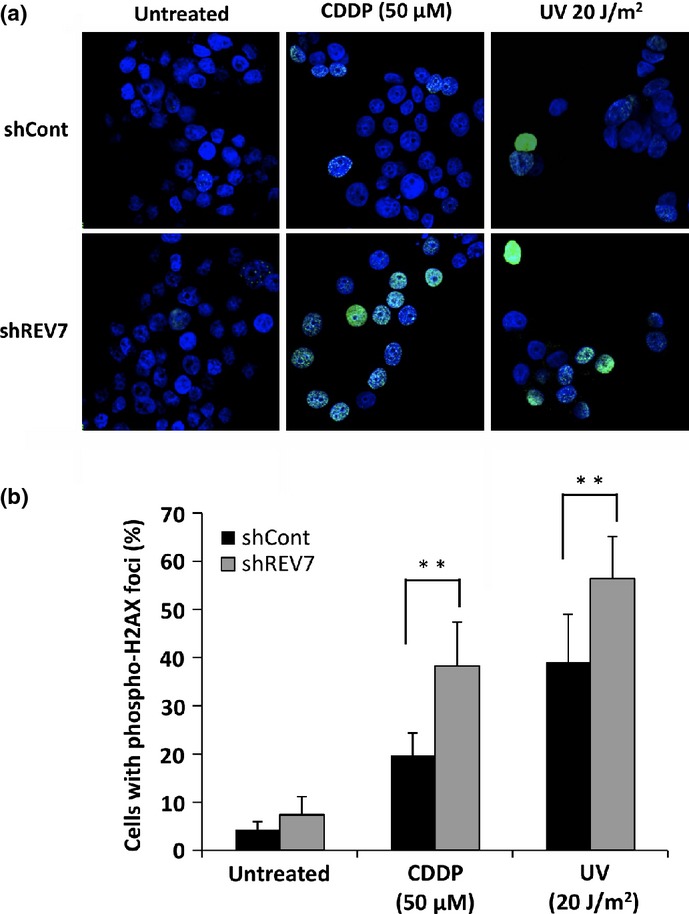
Depletion of REV7 enhances phosphorylation of H2AX after DNA-damaging treatment in ovarian clear cell carcinoma cells. (a) Fluorescence immunostaining for phospho-H2AX in control (shCont) or REV7-knockdown (shREV7) TOV-21G cells. Cells were treated with cisplatin (CDDP, 50 μM) or UV (20 J/m2), then fluorescently immunostained with anti-phospho-H2AX antibody (green) 24 h after beginning CDDP treatment (middle panels) or 12 h after UV irradiation (right panels). Cells without treatment were also stained (left panels). Nuclear counterstaining was carried out with DAPI (blue). (b) The percentages of cells with phospho-H2AX foci. Cells with more than 10 foci of phospho-H2AX were counted and their percentages were calculated. The means ± SDs of the percentages of phospho-H2AX-positive cells are shown. **P < 0.01.
Depletion of REV7 confers CDDP sensitization in vivo
To test whether REV7-depleted tumors are sensitive to systemic therapy of DNA damaging agents, a mouse-tumor model was established using shCont and shREV7 TOV-21G cells, and the effect of systemic CDDP treatment was assessed. We found that REV7-depleted tumors grew more slowly than control tumors, and CDDP treatment further drastically suppressed growth of REV7-depleted tumors (Fig.5a,b). Tumor volumes of control and REV7-depleted tumors at day 32 were reduced to 39.7 ± 8.9% and 15.3 ± 10.2% by CDDP treatment, respectively (P < 0.05) (Fig.5c). Immunohistochemical analyses of the tumor graft tissues at day 32 revealed that shREV7-TOV-21G-derived tumors showed low level expression of REV7 compared with shCont-TOV-21G-derived tumors (Fig.5d, Fig. S5) and the REV7-depleted tumors contained more cleaved caspase-3-positive cells and phospho-H2AX-positive cells than control tumors (P < 0.01) (Fig.5d–f). These results indicate that REV7 depletion confers enhanced sensitivity to CDDP treatment in CCC cells in vivo.
Figure 5.
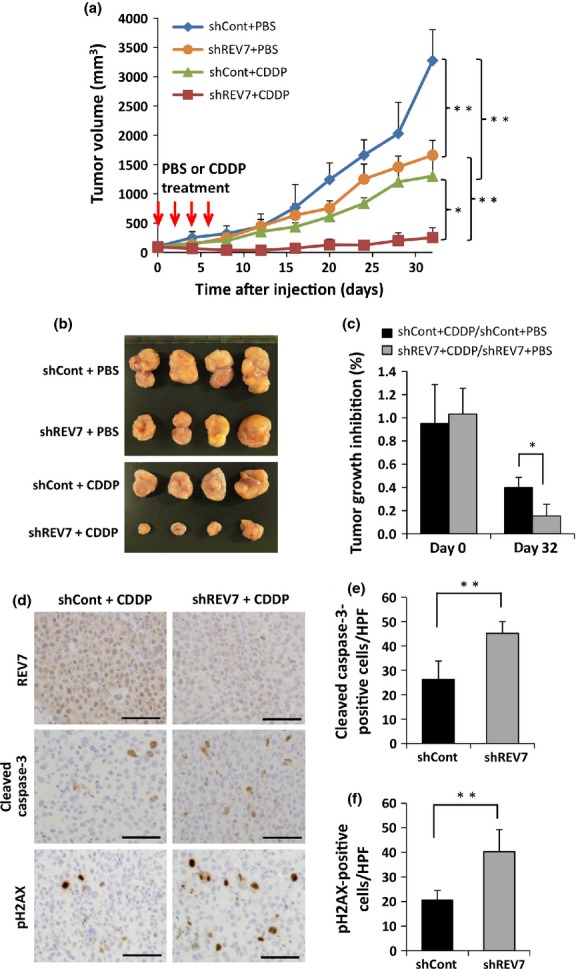
Suppression of REV7 enhances cisplatin (CDDP) sensitivity in vivo. (a) Growth of inoculated tumors in nude mice with and without systemic CDDP treatment. REV7-knockdown (shREV7) or control (shCont) ovarian clear cell carcinoma TOV-21G cells (1 × 107) were used in this analysis. The means ± SDs of tumor volume are shown. *P < 0.05; **P < 0.01. (b) Macroscopic images of transplanted tumors excised at day 32. (c) Reduction of tumor volume by CDDP treatment. Tumor volumes of the CDDP-treated group relative to the mean tumor volume of the PBS-treated group were calculated, and the means ± SDs of relative tumor volumes are shown. *P < 0.05. (d) Immunohistochemical analysis of excised tumors for REV7, cleaved caspase-3, and phospho-H2AX. Excised tumor tissues at day 32 were immunohistochemically stained with anti-REV7 (upper panels), anti-cleaved caspase-3 (middle panels), and anti-phospho-H2AX (lower panels) antibodies. Scale bar = 50 μm. (e, f) Quantitative assessment of cleaved caspase-3-positive cells (e) and phospho-H2AX-positive cells (f) in shREV7 and shCont tumors. Cells were counted under high power fields (HPF). The data obtained from five separate fields are shown as means ± SDs. **P < 0.01.
Discussion
The DNA repair system is required by cells to maintain genetic stability. Impairment of the DNA repair system causes increased susceptibility to DNA damaging agents, resulting in genetic instability and cell death.13,14 The REV7 protein is involved in TLS and homologous recombination repair,16–20,28 and Rev7-deficient mice show numerous apoptotic cells and accumulation of DNA damage in both germ cells and somatic cells,29 suggesting that REV7 dysfunction enhances cellular sensitivity to DNA damage.
In this study, we showed that most EOC types expressed REV7 protein and that high expression levels were frequently detected in CCC. Our results also showed that high REV7 expression was frequently observed in mucinous adenocarcinoma (Table1). Both CCC and mucinous adenocarcinoma tend to be resistant to antineoplastic agents.30 Moreover, we found that the PFS in the REV7high group was significantly shorter than that in the REV7low group in advanced EOC cases. In current treatment, platinum/paclitaxel-based chemotherapy is required for advanced cases, and the existence or development of chemoresistance is an important factor for poor prognosis. Our results indicate that high-level expression of REV7 contributes to poor prognosis of EOC at advanced stages, suggesting that REV7 expression may be associated with chemoresistance in EOC. However, no significant difference was observed between the PFS of REV7high and REV7low groups in only CCC cases at advanced stages (data not shown). A possible reason for this result is that a large portion of CCC cases were in early stage (stage I) and only 23 cases (46.9%) were in advanced stage (stage II–IV). To clarify the data, larger prospective studies for outcome analysis in CCC should be carried out in the future.
Development of chemoresistance is evoked by enhanced drug efflux, reduced drug uptake, aberrant apoptotic pathways, induction of drug-detoxifying mechanisms, and increased repair of DNA damage.15,31–33 Although it has been reported that p53 mutation, low expression of ARID1A, or upregulation of FGF1, annexin A3, bcl-2, or Ret finger protein can be responsible for platinum-based chemoresistance of EOC,34–39 the mechanisms of CDDP resistance in human ovarian cancer still remain unclear. Moreover, there are no promising biomarkers for predicting chemoresistance. It has been reported that REV7 affects the anticancer effect of DNA-damaging agents including CDDP, mitomycin C, and doxorubicin in nasopharyngeal carcinoma cells and of ionizing radiation in glioma cells.21,23 Therefore, we investigated the effect of REV7 expression on CDDP-induced anticancer effects in CCC cells. We showed that REV7 depletion suppressed proliferation of CCC cells without obvious alterations to the cell cycle, suggesting that REV7 depletion reduces cell proliferation by spontaneous stresses in the absence of DNA-damaging agents. In contrast to control cells, REV7-depleted cells subjected to CDDP treatment contained a large number of cells positive for cleaved caspase-3 and TUNEL and showed increased cleaved PARP by Western blotting. They also showed enhanced immunoreactivity for phospho-H2AX. These findings suggest that REV7 suppression renders CCC cells sensitive to endogenous and exogenous DNA damage and causes accumulation of DNA lesions and cell death after exposure to CDDP. This may be the cause of the reduced cell proliferation. At the mechanistic level, REV7 forms complexes with the TLS polymerases REV1 and REV3, and shRNA-mediated suppression of either REV1 or REV3 also leads to enhanced chemosensitivity to DNA-damaging agents.16,19,40,41 It is possible that the enhanced chemosensitivity in REV7-knockdown cells is caused by subsequent dysfunction of REV1 and REV3, which results in insufficient TLS processes. Thus, these data suggest that the status of the DNA damage tolerance system is an important factor for the efficiency of chemotherapy.
To show the effect of REV7 expression on systemic chemotherapy, we established a mouse-tumor model using shREV7 and shCont cells, and the anticancer effect of CDDP, delivered i.p., was analyzed. Our results indicated that REV7-depleted tumor growth was slow and was significantly suppressed by systemic CDDP treatment when compared with that of control tumors. In addition, immunohistochemical analysis revealed that REV7-depleted tumors showed enhanced immunoreactivity for cleaved caspase-3 and phospho-H2AX. These in vivo results were compatible with those obtained from in vitro experiments, suggesting that REV7 expression in CCC tumors may affect chemosensitivity to systemic CDDP treatment. Because REV7 expression in human cancer tissues can be detected easily by immunohistochemistry in comparison with REV1 and REV3, we propose that REV7 can be a good marker for chemosensitivity and a novel molecular target for CCC therapy. Further analyses of chemosensitivity in REV7-depleted tumors are necessary to confirm the association between REV7 expression and the outcome of chemotherapy in CCC.
Acknowledgments
We thank Ms. Kaori Ushida for technical assistance. This work was supported by Grants-in-Aid from the Global Center of Excellence (GCOE), and Scientific Research (A) (to MT, 23249020) and by Grants-in-Aid for Scientific Research (C) (to YM, 24590479), both commissioned by the Ministry of Education, Culture, Sports, Science and Technology of Japan.
Disclosure Statement
The authors have no conflict of interest.
Funding information
Global Center of Excellence. Ministry of Education, Culture, Sports, Science and Technology of Japan (23249020 and 24590479).
Supporting Information
Additional supporting information may be found in the online version of this article:
Supplementary materials and methods.
Specific immunohistochemical reaction of rabbit polyclonal anti-REV7 antibody.
Fig. S2 Expression of REV7 in ovarian cancer cell lines.
Fig. S3 Depletion of REV7 enhances chemosensitivity in colony formation assay.
Fig. S4 Expression of REV7 conferred resistance to cisplatin (CDDP).
Fig. S5 Immunohistochemical and immunofluorescence analyses of control (shCont) and REV7-knockdown (shREV7) tumors for REV7 expression.
Multivariate analysis of several clinicopathologic parameters in relation to survival of patients with advanced stage epithelial ovarian cancer.
Table S2 Association between REV7 expression and 1-year progression-free survival in patients with advanced stage epithelial ovarian cancer who received platinum-based chemotherapy.
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63:11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 2.Kobel M, Kalloger SE, Boyd N, et al. Ovarian carcinoma subtypes are different diseases: implications for biomarker studies. PLoS Med. 2008;5:e232. doi: 10.1371/journal.pmed.0050232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kurman RJ, Shih IM. Molecular pathogenesis and extraovarian origin of epithelial ovarian cancer–shifting the paradigm. Hum Pathol. 2011;42:918–31. doi: 10.1016/j.humpath.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mackay HJ, Brady MF, Oza AM, et al. Prognostic relevance of uncommon ovarian histology in women with stage III/IV epithelial ovarian cancer. Int J Gynecol Cancer. 2010;20:945–52. doi: 10.1111/IGC.0b013e3181dd0110. [DOI] [PubMed] [Google Scholar]
- 5.Prat J. New insights into ovarian cancer pathology. Ann Oncol. 2012;23(Suppl. 10):x111–7. doi: 10.1093/annonc/mds300. [DOI] [PubMed] [Google Scholar]
- 6.Chan JK, Teoh D, Hu JM, Shin JY, Osann K, Kapp DS. Do clear cell ovarian carcinomas have poorer prognosis compared to other epithelial cell types? A study of 1411 clear cell ovarian cancers. Gynecol Oncol. 2008;109:370–6. doi: 10.1016/j.ygyno.2008.02.006. [DOI] [PubMed] [Google Scholar]
- 7.Sugiyama T, Kamura T, Kigawa J, et al. Clinical characteristics of clear cell carcinoma of the ovary: a distinct histologic type with poor prognosis and resistance to platinum-based chemotherapy. Cancer. 2000;88:2584–9. [PubMed] [Google Scholar]
- 8.Takano M, Kikuchi Y, Yaegashi N, et al. Clear cell carcinoma of the ovary: a retrospective multicentre experience of 254 patients with complete surgical staging. Br J Cancer. 2006;94:1369–74. doi: 10.1038/sj.bjc.6603116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tan DS, Kaye S. Ovarian clear cell adenocarcinoma: a continuing enigma. J Clin Pathol. 2007;60:355–60. doi: 10.1136/jcp.2006.040030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Campbell IG, Russell SE, Choong DY, et al. Mutation of the PIK3CA gene in ovarian and breast cancer. Cancer Res. 2004;64:7678–81. doi: 10.1158/0008-5472.CAN-04-2933. [DOI] [PubMed] [Google Scholar]
- 11.Kobel M, Kalloger SE, Carrick J, et al. A limited panel of immunomarkers can reliably distinguish between clear cell and high-grade serous carcinoma of the ovary. Am J Surg Pathol. 2009;33:14–21. doi: 10.1097/PAS.0b013e3181788546. [DOI] [PubMed] [Google Scholar]
- 12.Wiegand KC, Shah SP, Al-Agha OM, et al. ARID1A mutations in endometriosis-associated ovarian carcinomas. N Engl J Med. 2010;363:1532–43. doi: 10.1056/NEJMoa1008433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bouwman P, Jonkers J. The effects of deregulated DNA damage signalling on cancer chemotherapy response and resistance. Nat Rev Cancer. 2012;12:587–98. doi: 10.1038/nrc3342. [DOI] [PubMed] [Google Scholar]
- 14.Curtin NJ. DNA repair dysregulation from cancer driver to therapeutic target. Nat Rev Cancer. 2012;12:801–17. doi: 10.1038/nrc3399. [DOI] [PubMed] [Google Scholar]
- 15.Johnson SW, Laub PB, Beesley JS, Ozols RF, Hamilton TC. Increased platinum-DNA damage tolerance is associated with cisplatin resistance and cross-resistance to various chemotherapeutic agents in unrelated human ovarian cancer cell lines. Cancer Res. 1997;57:850–6. [PubMed] [Google Scholar]
- 16.Murakumo Y, Roth T, Ishii H, et al. A human REV7 homolog that interacts with the polymerase zeta catalytic subunit hREV3 and the spindle assembly checkpoint protein hMAD2. J Biol Chem. 2000;275:4391–7. doi: 10.1074/jbc.275.6.4391. [DOI] [PubMed] [Google Scholar]
- 17.Murakumo Y. The property of DNA polymerase zeta: REV7 is a putative protein involved in translesion DNA synthesis and cell cycle control. Mutat Res. 2002;510:37–44. doi: 10.1016/s0027-5107(02)00250-6. [DOI] [PubMed] [Google Scholar]
- 18.Friedberg EC, Gerlach VL. Novel DNA polymerases offer clues to the molecular basis of mutagenesis. Cell. 1999;98:413–6. doi: 10.1016/s0092-8674(00)81970-4. [DOI] [PubMed] [Google Scholar]
- 19.Murakumo Y, Ogura Y, Ishii H, et al. Interactions in the error-prone postreplication repair proteins hREV1, hREV3, and hREV7. J Biol Chem. 2001;276:35644–51. doi: 10.1074/jbc.M102051200. [DOI] [PubMed] [Google Scholar]
- 20.Prakash S, Johnson RE, Prakash L. Eukaryotic translesion synthesis DNA polymerases: specificity of structure and function. Annu Rev Biochem. 2005;74:317–53. doi: 10.1146/annurev.biochem.74.082803.133250. [DOI] [PubMed] [Google Scholar]
- 21.Cheung HW, Chun AC, Wang Q, et al. Inactivation of human MAD2B in nasopharyngeal carcinoma cells leads to chemosensitization to DNA-damaging agents. Cancer Res. 2006;66:4357–67. doi: 10.1158/0008-5472.CAN-05-3602. [DOI] [PubMed] [Google Scholar]
- 22.McNally K, Neal JA, McManus TP, McCormick JJ, Maher VM. hRev7, putative subunit of hPolzeta, plays a critical role in survival, induction of mutations, and progression through S-phase, of UV((254 nm))-irradiated human fibroblasts. DNA Repair (Amst) 2008;7:597–604. doi: 10.1016/j.dnarep.2007.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhao J, Liu S, Wang H, et al. Mitotic arrest deficient protein MAD2B is overexpressed in human glioma, with depletion enhancing sensitivity to ionizing radiation. J Clin Neurosci. 2011;18:827–33. doi: 10.1016/j.jocn.2010.11.009. [DOI] [PubMed] [Google Scholar]
- 24.Rimkus C, Friederichs J, Rosenberg R, Holzmann B, Siewert JR, Janssen KP. Expression of the mitotic checkpoint gene MAD2L2 has prognostic significance in colon cancer. Int J Cancer. 2007;120:207–11. doi: 10.1002/ijc.22155. [DOI] [PubMed] [Google Scholar]
- 25.Yuan B, Xu Y, Woo JH, et al. Increased expression of mitotic checkpoint genes in breast cancer cells with chromosomal instability. Clin Cancer Res. 2006;12:405–10. doi: 10.1158/1078-0432.CCR-05-0903. [DOI] [PubMed] [Google Scholar]
- 26.Li L, Shi Y, Wu H, et al. Hepatocellular carcinoma-associated gene 2 interacts with MAD2L2. Mol Cell Biochem. 2007;304:297–304. doi: 10.1007/s11010-007-9512-8. [DOI] [PubMed] [Google Scholar]
- 27.Weterman MA, van Groningen JJ, Tertoolen L, van Kessel AG. Impairment of MAD2B-PRCC interaction in mitotic checkpoint defective t(X;1)-positive renal cell carcinomas. Proc Natl Acad Sci U S A. 2001;98:13808–13. doi: 10.1073/pnas.241304198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Okada T, Sonoda E, Yoshimura M, et al. Multiple roles of vertebrate REV genes in DNA repair and recombination. Mol Cell Biol. 2005;25:6103–11. doi: 10.1128/MCB.25.14.6103-6111.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Watanabe N, Mii S, Asai N, et al. The REV7 subunit of DNA polymerase zeta is essential for primordial germ cell maintenance in the mouse. J Biol Chem. 2013;288:10459–71. doi: 10.1074/jbc.M112.421966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hess V, A'Hern R, Nasiri N, et al. Mucinous epithelial ovarian cancer: a separate entity requiring specific treatment. J Clin Oncol. 2004;22:1040–4. doi: 10.1200/JCO.2004.08.078. [DOI] [PubMed] [Google Scholar]
- 31.Fokkema E, Groen HJ, Helder MN, de Vries EG, Meijer C. JM216-, JM118-, and cisplatin-induced cytotoxicity in relation to platinum-DNA adduct formation, glutathione levels and p53 status in human tumour cell lines with different sensitivities to cisplatin. Biochem Pharmacol. 2002;63:1989–96. doi: 10.1016/s0006-2952(02)00983-8. [DOI] [PubMed] [Google Scholar]
- 32.Gottesman MM. Mechanisms of cancer drug resistance. Annu Rev Med. 2002;53:615–27. doi: 10.1146/annurev.med.53.082901.103929. [DOI] [PubMed] [Google Scholar]
- 33.Taniguchi K, Wada M, Kohno K, et al. A human canalicular multispecific organic anion transporter (cMOAT) gene is overexpressed in cisplatin-resistant human cancer cell lines with decreased drug accumulation. Cancer Res. 1996;56:4124–9. [PubMed] [Google Scholar]
- 34.Horio M, Kato T, Mii S, et al. Expression of RET finger protein predicts chemoresistance in epithelial ovarian cancer. Cancer Med. 2012;1:218–29. doi: 10.1002/cam4.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kassim SK, Ali HS, Sallam MM, et al. Increased bcl-2 expression is associated with primary resistance to chemotherapy in human epithelial ovarian cancer. Clin Biochem. 1999;32:333–8. doi: 10.1016/s0009-9120(99)00026-0. [DOI] [PubMed] [Google Scholar]
- 36.Katagiri A, Nakayama K, Rahman MT, et al. Loss of ARID1A expression is related to shorter progression-free survival and chemoresistance in ovarian clear cell carcinoma. Mod Pathol. 2012;25:282–8. doi: 10.1038/modpathol.2011.161. [DOI] [PubMed] [Google Scholar]
- 37.Reles A, Wen WH, Schmider A, et al. Correlation of p53 mutations with resistance to platinum-based chemotherapy and shortened survival in ovarian cancer. Clin Cancer Res. 2001;7:2984–97. [PubMed] [Google Scholar]
- 38.Smith G, Ng MT, Shepherd L, et al. Individuality in FGF1 expression significantly influences platinum resistance and progression-free survival in ovarian cancer. Br J Cancer. 2012;107:1327–36. doi: 10.1038/bjc.2012.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yan X, Yin J, Yao H, Mao N, Yang Y, Pan L. Increased expression of annexin A3 is a mechanism of platinum resistance in ovarian cancer. Cancer Res. 2010;70:1616–24. doi: 10.1158/0008-5472.CAN-09-3215. [DOI] [PubMed] [Google Scholar]
- 40.Doles J, Oliver TG, Cameron ER, et al. Suppression of Rev3, the catalytic subunit of Pol ζ, sensitizes drug-resistant lung tumors to chemotherapy. Proc Natl Acad Sci U S A. 2010;107:20786–91. doi: 10.1073/pnas.1011409107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xie K, Doles J, Hemann MT, Walker GC. Error-prone translesion synthesis mediates acquired chemoresistance. Proc Natl Acad Sci USA. 2010;107:20792–7. doi: 10.1073/pnas.1011412107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary materials and methods.
Specific immunohistochemical reaction of rabbit polyclonal anti-REV7 antibody.
Fig. S2 Expression of REV7 in ovarian cancer cell lines.
Fig. S3 Depletion of REV7 enhances chemosensitivity in colony formation assay.
Fig. S4 Expression of REV7 conferred resistance to cisplatin (CDDP).
Fig. S5 Immunohistochemical and immunofluorescence analyses of control (shCont) and REV7-knockdown (shREV7) tumors for REV7 expression.
Multivariate analysis of several clinicopathologic parameters in relation to survival of patients with advanced stage epithelial ovarian cancer.
Table S2 Association between REV7 expression and 1-year progression-free survival in patients with advanced stage epithelial ovarian cancer who received platinum-based chemotherapy.


