Abstract
Our previous study implied a correlation between inhibitors of differentiation-1 (Id-1) and cervical cancer development. However, how Id-1 contributes to cervical carcinogenesis is unknown. In the present study, we used an in vitro transformation model to investigate the role of Id-1 in the transformation of cervical cells. Human papillomavirus (HPV)-immortalized cervical epithelial cells (H8) were successfully transformed by exposure to the carcinogen N-nitrosopyrrolidine (NPYR). The expression of both Id-1 RNA and protein was significantly increased in transformed H8 cells, suggesting a possible role of Id-1 in cervical cell transformation. Ectopic expression of Id-1 in H8 cells potentiated NPYR-induced cell transformation. In contrast, silencing of Id-1 suppressed NPYR-induced H8 cell transformation. In addition, the expression of HPV E6 and E7 oncoproteins was upregulated while that of the tumor suppressors p53 and pRb was suppressed after H8 cell transformation. Our results suggest that Id-1 plays an oncogenic role in HPV-related cervical carcinogenesis, which sheds light on cervical cancer development mechanisms and implies that Id-1 is a potential target for cervical cancer prevention and therapy.
Keywords: Carcinogenesis, cervical cancer, human papillomavirus, inhibitors of differentiation-1, transformation
Cervical cancer is the third most common neoplastic disease afflicting women worldwide, with an estimation of 530 000 new cases and 275 000 deaths every year.1 Although the incidence and mortality in developed countries have declined due to cervical cytologic screening campaigns, the fact that more than 85% of the global cervical cancer patients reside in undeveloped countries should not be ignored.2 It is now clear that human papillomavirus (HPV) is the most important risk factor for cervical cancer development.3 More than 100 different HPV genotypes are detected in the female reproductive tract, among which at least 15 genotypes have high oncogenic potential.4 The most oncogenic HPV, type 16, is responsible for over 70% of cervical carcinomas and up to 90% of HPV-correlated extra-cervical carcinomas.5–7 By integration into host genomic DNA, the HPV encodes E6 and E7 proteins which promote cancer development by interacting with certain oncoproteins or tumor suppressors such as p53 and RB.8 However, the detailed mechanism for HPV-induced cervical carcinogenesis is not yet fully understood because the majority of HPV infections vanish before cancer development, partly due to immunological responses, while only 1% of HPV infections progress into carcinoma.9,10
Inhibitors of DNA binding proteins, also known as inhibitors of differentiation (Id) proteins, are helix-loop-helix (HLH) proteins and dominant inhibitors of the basic HLH transcription factors.11 The Id protein family consists of four components: Id-1, Id-2, Id-3 and Id-4, of which Id-1 is most intensively studied. Id-1 has multiple functions, such as regulating cell differentiation, inducing cell immortalization, inhibiting cell apoptosis and promoting tumor angiogenesis.11 Recent studies have demonstrated increased expression of Id-1 in several types of human tumors including gastric carcinoma, breast cancer, prostate carcinoma and ovarian cancer.12–15 Our previous studies have shown that Id-1 expression was increased in cervical carcinoma tissues, which was correlated to HPV infection, indicating a possible role of Id-1 in cervical cancer development.16,17 However, the detailed mechanism for HPV-induced cervical carcinogenesis and how Id-1 is involved in the whole carcinogenic process still remain unclear. It is critical to understand the molecular mechanisms involving Id-1 in the initiation and progression of cervical cancer, which will help to develop effective approaches for prevention and therapy of this malignancy.
The N-nitrosopyrrolidine (NPYR), a product of vaginal anaerobic metabolism and a possible co-factor for cervical carcinogenesis, is known to be a strong carcinogen.18–20 In this study, we established an in vitro cell transformation model with NPYR as a carcinogen to induce malignant transformation of HPV-16 immortalized human cervical epithelial H8 cell. With this model, we demonstrated a promoting role of Id-1 in the transformation of cervical epithelial cells and investigated possible mechanisms by which Id-1 contributes to HPV-related cervical cancer development.
Materials and Methods
Cell cultures
The HPV16 immortalized but not transformed human cervical epithelial cell line H8 originated from the Institute of Virology of Chinese Academy of Medical Sciences and was a generous gift from Professor Yong Zhao from the Department of Pathology of Chongqing Medical University. H8 cells cannot form tumors in nude mice or colonies in soft agar.21 The human cervical cancer-derived cell line used in the present study, SiHa, was from the American Type Culture Collection. Both H8 and SiHa cells were cultured in (DMEM) (Gibco/BRL, Grand Island, NY, USA) supplemented with 100 μg/mL of streptomycin, 100 U/mL of penicillin and 10% heat-inactivated FBS (Invitrogen, Carlsbad, CA, USA). The cultures were maintained in a humidified 5% CO2 incubator at 37°C. The medium was changed three times a week.
Cell transformation
N-nitrosopyrrolidine (Sigma Chemical, St. Louis, MO, USA) was used to induce malignant transformation. To determine the dosage of NPYR to be used in the transformation assay, a preliminary cytotoxic test was performed as reported previously.22 Generally, cells were seeded at a density of 1 × 105 cells in 5 mL of culture medium per 60-mm dish (three dishes per group). After 3 or 4 days, when cells were in the log growth phase, the cells were fed 5 mL of fresh media and exposed to different concentrations of NPYR for 24 h. Cells were counted and cytotoxicity was determined by comparing viable cell numbers in exposed cultures to that of untreated controls. A dose of 80 μM NPYR that killed approximately 50% of cells was used for transformation assays following the procedure described in a previous report.23 Exponentially growing H8 cells were plated at a density of 1 × 105 cells per 60-mm dish and incubated for 24 h. The cells were exposed to NPYR for 48 h, followed by culture in fresh medium for another 48 h. This treatment cycle was repeated seven times followed by a subsequent incubation in fresh medium for 3–4 weeks before further experiments.
Soft agar assay
Anchorage-independent growth was determined by a soft agar assay as described previously.24 Briefly, a bottom layer of 1.4 mL (0.6%) was prepared with a 1:1 mixture of 1.2% low melting-point agarose (AMRESCO, OH, USA) (approximately 50°C) and warm 2 × DMEM, and was poured into each well of six-well plates. A top layer, which was a mixture of 500 μL of warm 2 × DMEM, 500 μL of the 0.6% base agar and cells at the density of 1 × 103 cells per well, was poured on top of the solidified bottom layer. The cultures were fed 1 mL of DMEM supplemented with 10% FBS, which was gently refreshed twice a week. The plates were kept in a cell culture incubator maintained at 37°C under 5% CO2 to allow colony growth. After 2 weeks of culture, the colony assay was terminated and cell colonies of more than 50 cells in a cluster were counted using a graduated eyepiece fitted in a transmission light microscope at a magnification of 40.25
Tumorigenic assays in nude mice
The tumorigenic ability of the transformed cells was examined in 6-week-old female nude mice obtained from the Animal Experimental Center of Sichuan University. The cells (1 × 106) were suspended in PBS (pH 7.4, 0.1 mL) and injected subcutaneously in the sub-axilla of the mice. Ten mice were used in each group. The mice were palpated every 3 days to detect tumor development. Tumor sizes were measured in two dimensions with a Vernier caliper and recorded in mm3 (length × width2).26 The mice were killed at week 8 and tumors were excised and weighed. All the experimental procedures were carried out in accordance with the Institutional Ethical Guidelines for Animal Experiments approved by the Animal Ethic Commission of the University.
Western blot
Cells were collected and whole cell lysates were prepared by lysing cells in RIPA lysis buffer containing a protease inhibitor cocktail (Sigma, St. Louis, MO, USA). Equal amounts of protein extracts (50 μg of protein) were separated in 15% polyacrylamide gels, electrotransferred to polyvinylidene fluoride membranes (Millipore Corporation, Bedford, MA, USA). The membranes were incubated overnight at 4°C in PBS containing 5% defatted milk powder, 0.1% Tween-20 and primary antibodies: Id-1 polyclonal rabbit antibody (1:500, Santa Cruz Biotechnology, Santa Cruz, CA, USA), HPV16 E6 polyclonal goat antibody (1:500, Santa Cruz Biotechnology), HPV16 E7 monoclonal mouse antibody (1:500, Santa Cruz Biotechnology), p53 monoclonal mouse antibody (1:500, Santa Cruz Biotechnology), Retinoblastoma (Rb) monoclonal mouse antibody (Bio-Rad Laboratories, Hercules, CA, USA) and β-actin monoclonal mouse antibody (1:500, Santa Cruz Biotechnology). The membranes were further incubated with secondary antibodies conjugated with HRP (Santa Cruz Biotechnology). A luminol reagent detection kit (Santa Cruz Biotechnology) was used for visual detection of proteins of interest.
Quantitative real time-PCR
Total RNA was isolated from 1 × 107 cells with the RNeasy Mini kit (QIAGEN, Alameda, CA, USA) according to the manufacturer's instructions, and the quality of the RNA samples was confirmed by electrophoresis in 1% agarose gels. The RNA were reversely transcribed into cDNA using the iScript cDNA Synthesis Kit (Bio-Rad Laboratories) according to the manufacturer's instruction. Reactions were performed in a 10-μL SYBR GREEN PCR reaction volume in a 96-well optical reaction plate. The PCR mixture contained: 5 μL Eva Green Supermix (Bio-Rad Laboratories, Hercules, CA, USA), 1 μL forward and reverse primers, 2 μL ddH2O and 2 μL cDNA. B2M was detected as an internal control. The sequences of primers were as follows: for Id-1, forward: 5′-TCTACGACATGAACGGCTG-3′; reverse: 5′-GGTCCCTGATGTAGTCGAT-3′; for B2M, forward: 5′-TGCCGTGTGAACCATGTGA-3′; reverse: 5′-CCAAATGCGGCATCTTCAA-3′. PCR was performed on the CFX96 fluorescent quantitative PCR detection system (Bio-Rad Laboratories, Hercules, CA, USA) with the following parameters: initial denaturation at 98°C for 30 s, followed by 40 cycles of 95°C for 5 s and 65°C for 5 s, and then a final extension at 95°C for 10 s. All samples were performed in triplicate and the results were fitted to standard curves. The presence of a single specific PCR product was verified by melting curve analysis and confirmed by running an agarose gel. The amplification curves were analyzed and fold changes were determined using the comparative threshold cycle (Ct) method to quantify Id-1 mRNA expression, with the average 2−△△Ct values of normal H8 cells taken as one.27
Stable transducting H8 cells with viral vectors
The pWPI-hId1 (for Id-1 overexpression), the hId1-shRNA (for Id-1 silencing) and their respective control plasmids, pWPI and pGIPZ, were kindly provided by Dr Robert Benezra from the Department of Cancer Biology and Genetics, Memorial Sloan-Kettering Cancer Center, New York, USA. The validity of both hId1-shRNA and pWPI-hId1 was proved previously.28 CaPO4 precipitation was used for lentiviral particle production. Briefly, 2 × 106 HEK293T cells were co-transfected with 9 μg of target plasmids and helper vectors (9 μg of pDelta8.9 and 6 μg of pVSVg) using 60 μL of CaCl2 (2.5 M), 540 μL of H2O (double distilled) and 600 μL of 2 × BBS (50 mM BES, Calbiochem cat. 391334), 280 mM NaCl, 1.5 mM Na2HPO4). Sixteen hours after transfection, the medium was refreshed with 15 mL of DMEM containing 10% of FBS, and the cells were cultured at 37°C with 10% CO2. The first and second harvests of supernatant (viral preparation) were collected at 24 and 48 h, respectively, of incubation. Cell debris was removed by filtration with 0.45-μm filters. The viral preparation was added for 3 consecutive days to the H8 cell culture. As the transduced cells express GFP, the ratio of fluorescent versus nonfluorescent cells was calculated to determine transduction efficiency. As the pWPI-hId1 and controlled plasmid have no antibiotic selective marker, the stable transduced H8 cells were sorted and purified with a BD FACSAria Flow Cytometer (Becton-Dickinson Company, USA). To generate H8 cells with stable expression of shRNA, H8 cells transduced with either the pGIPZ vector (for generating a control) or the hId1-shRNA vector that were puromycin-resistant were selected by incubation with 5 μg/mL of puromycin for 15 days. The puromycin-resistant clones were pooled clones and the expression of Id-1 was determined by western blot to verify overexpression or silencing.
Statistics
All statistical analyses were carried out using the Student's t-test in the spss software package (version 13.0, SPSS Inc., Chicago, USA). Statistical significance was assumed at P < 0.05.
Results
Establishment of the N-nitrosopyrrolidine transformation model in H8 cells
The HPV-16 immortalized but not transformed H8 cells resemble normal human cervical epithelial cells but are not transformed by the HPV E6/E7 oncogene. We first confirmed that the H8 cells lack the capacities in anchorage-independent growth in soft agar and tumor-formation in nude mice, the malignant characteristics of cancer cells. The cervical cancer cell line SiHa was used as a positive control (Figs1 and 2). After treatment with NYPR for 4 weeks, the cells became capable of forming colonies in soft agar (Fig.1) and tumors in nude mice (Fig.2a). The tumor growth curve also showed a parallel trend between SiHa and transformed H8 groups (Fig.2b), suggesting that the cells were transformed by NPYR. Then the NYPR exposure protocol was used to assay the role of Id-1 in H8 transformation by NYPR.
Figure 1.
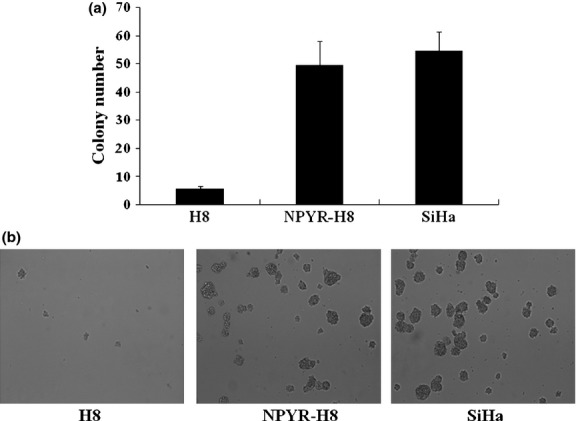
Establishment of N-nitrosopyrrolidine (NPYR)-induced transformation in H8 cells. Cells treated with NPYR were seeded in soft agar, and colony numbers were counted 2 weeks after seeding. Untreated H8 cells and SiHa cells were seeded as negative and positive controls, respectively. (a) Representative images taken under a light microscope (40×). (b) Colony numbers in each group counted in five fields (mean ± SD).
Figure 2.
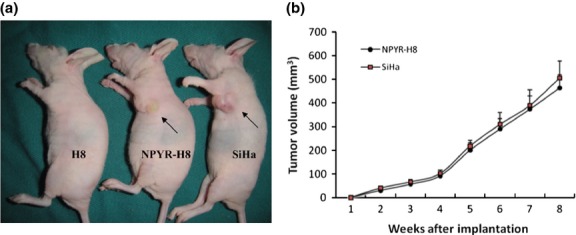
Tumorigenicity of N-nitrosopyrrolidine (NPYR)-transformed H8 cells in nude mice. Nude mice were injected (s.c.) with H8 cells, NPYR transformed H8 (NPYR-H8) cells or SiHa cells, and were photographed 8 weeks after injection. Tumor size of each group is shown (n = 10). (a) One representative mouse from each group is shown. Injection sites are indicated by arrowheads. (b) Tumor growth curves of the NPYR-H8 and SiHa groups.
Increased inhibitors of differentiation-1 expression during H8 transformation
We investigated Id-1 expression at both protein and RNA levels in parental and transformed H8 cells. SiHa cells were used as a positive control. A significant increase of Id-1 protein expression in transformed H8 cells compared to the parental H8 cells was detected by western blot (Fig.3a). By quantitative RT-PCR, the Id-1 RNA level in transformed H8 cells was found to be 3.8-fold higher than that in the untransformed H8 cells (Fig.3b, P < 0.05).
Figure 3.
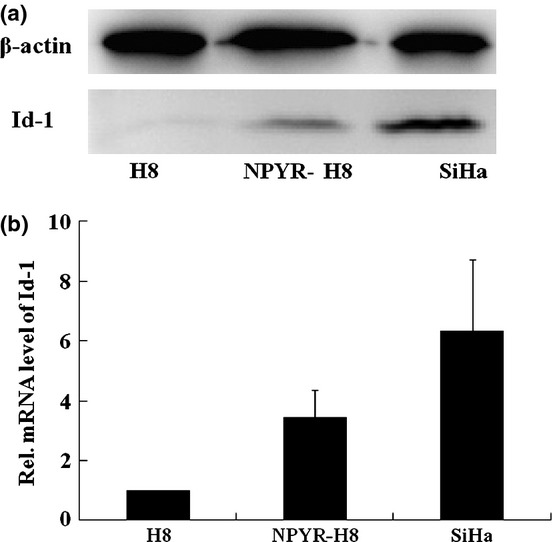
Increased inhibitors of differentiation-1 (Id-1) expression in N-nitrosopyrrolidine (NPYR)-transformed H8 cells. (a) Total proteins extracted from H8, NPYR-H8 and SiHa cells were subjected to 10% SDS-PAGE gels and assayed by western blot. (b) Id-1 RNA levels in H8, NPYR-H8 and SiHa cells were detected by qRT-PCR. Expression of Id-1 gene, in each sample was normalized against the B2M endogenous control. The normalized values were then calibrated against the H8 cell values that were arbitrarily set as 1.
Increased inhibitors of differentiation-1 overexpression increases colony formation in soft agar and tumor growth in nude mice
To determine whether Id-1 expression is required for transformation of H8 cells, we ectopically expressed Id-1 in H8 cells through lentivirus transduction. The stable Id-1-overexpressed H8 cells were sorted and purified using the BD FACSAria Flow Cytometer. Effective expression of Id-1 in pWPI-hId1-transduced cells was confirmed by western blot (Fig.4A). The cells were exposed to NPYR using our established protocol. As a result, H8 cells with a stable transduction of pWPI-hId1 showed an increased colony formation ability compared to the control plasmid pWPI (colony numbers: 75.4 ± 12.2 vs 48.4 ± 7.5, P < 0.05, Fig.4B,C). The results strongly support a promoting role of Id-1 in cervical epithelial cell transformation. We further investigated the biological consequence of Id-1 activation in tumorigenesis with the nude mouse xenograft model. pWPI-hId1 H8 cells developed larger tumors compared to the pWPI controls at the end of experiments (tumor size for pWPI-hId1 group: 1044 ± 142 mm3; control group: 427 ± 52 mm3, P < 0.05, Fig.5). Pathological analysis revealed that all the xenograft tumors were similar to human squamous cell carcinoma (SCC). However, tumors derived from Id1-overexpression cells (pWPI-hId-transfected) showed SCC with undifferentiated or low-grade differentiated types, while those from control cells (pWPI-transfected) revealed a better differentiation (Fig.6).
Figure 4.
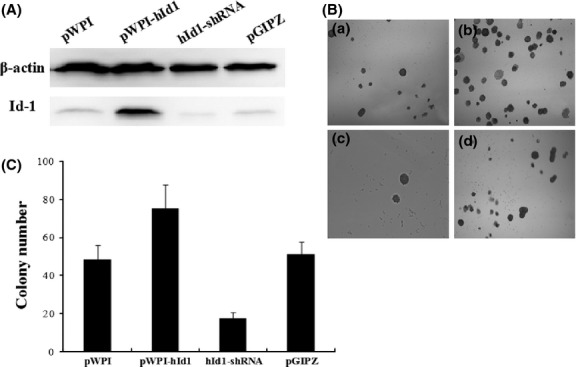
N-nitrosopyrrolidine (z)-induced transformation of H8 cells with increased inhibitors of differentiation-1 (Id-1) overexpression or silencing. (A) Total proteins extracted from the indicated H8 cells were subjected to 10% SDS-PAGE gels and assayed by western blot. (B) Cells treated with NPYR were seeded in soft agar, and colony numbers were counted at 2 weeks. Representative images were taken under a light microscope (40×). (a) NPYR transformed pWPI H8 cells; (b) NPYR transformed pWPI-hId1 H8 cells; (c) NPYR transformed hId1-shRNA H8 cells; (d) NPYR transformed pGIPZ H8 cells. (C) Quantification of colony numbers in each group counted in five fields (mean ± SD).
Figure 5.
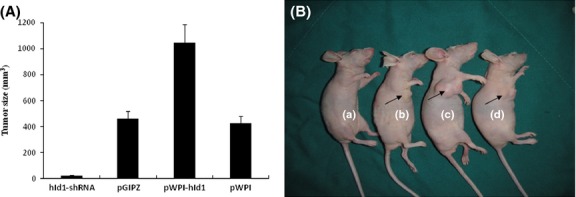
Xenografted tumor formations in nude mice. Nude mice were injected (s.c.) with (a) N-nitrosopyrrolidine (NPYR) transformed hId1-shRNA H8 cells, (b) NPYR transformed pGIPZ H8 cells (as control), (c) NPYR transformed pWPI-hId1 H8 cells, (d) NPYR transformed pWPI H8 cells (as control), and were photographed 8 weeks after injection. There were 10 mice in each group. Tumor size was presented as means ± SD. One representative mouse from each group is shown. Injection sites are indicated by arrowheads.
Figure 6.
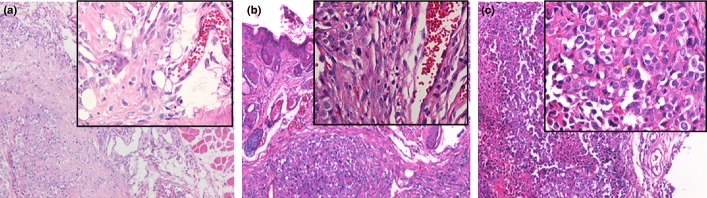
HE staining xenografted tumors in nude mice. (a) Tumors from N-nitrosopyrrolidine (NPYR) transformed pWPI H8 cells showed squamous cell carcionoma (SCC) with a better differentiation. (b) Tumors from NPYR transformed pWPI-hId H8 cells showed SCC with undifferentiated or low-grade differentiated type. (c) Tumors formed by SiHa cells showed SCC with undifferentiated type. Representative images (100× and 400× for the inserts) are shown.
Inhibition of Increased inhibitors of differentiation-1 expression via shRNA-mediated silencing inhibited colony formation in soft agar and tumor formation in nude mice
To further define the function of Id-1 in cervical epithelial transformation, we generated stable Id-1 gene silencing in H8 cells with shRNA (Id-1-shRNA H8 cells). Then we performed NPYR-induced transformation under the same conditions as in the previous experiments. The results showed that Id-1 gene silencing significantly decreased cell colonies in soft agar assay (hId1-shRNA, 17.4 ± 3.2; pGIPZ, 51.2 ± 6.8, P < 0.05) (Fig.4b,c). Id-1 gene silencing also decreased tumor sizes (at 8 weeks after injection, hId1-shRNA group: 23 ± 4 mm3; pGIPZ group, 460 ± 59 mm3, P < 0.05) (Fig.5). These results strongly suggest that Id-1 plays an important role in NPYR-induced transformation of HPV-16 immortalized cervical epithelial cells.
Increased expression of human papillomavirus E6/E7 proteins and decreased expression of p53 and pRb in transformed H8 cells
To explore the possible mechanism of Id-1 upregulation in NPYR-treated H8 cells, we examined the expression of the HPV E6 and E7 oncoproteins and their target tumor suppressors p53 and pRb in transformed H8 cells induced by NPYR. The results show that the expression of HPV E6 and E7 proteins was significantly increased, while that of p53 and pRb protein was decreased in the transformed cells (Fig.7).
Figure 7.
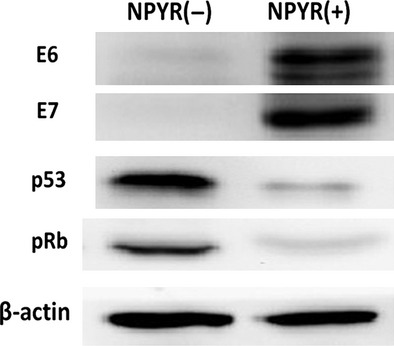
Evaluations of human papillomavirus E6, E7 and p53, and pRb during N-nitrosopyrrolidine-induced cell transformation. Total proteins from NPYR transformed or untreated H8 cells were subjected to 10% SDS-PAGE gels and followed by western blot. β-actin served as an input control.
Discussion
In our previous studies, we demonstrated a gradual increase of Id-1 expression that is associated with the progression of cervical carcinogenesis: cervical carcinoma (76%), high grade precancerous lesions (HSIL, 50%), low grade precancerous lesions (LSIL, 16%) and normal tissues (4%), which strongly suggest Id-1 to be a potential cervical cancer promoter.16 Id-1 knockdown in cervical cancer cells reduced anchorage-independent growth in vitro (Fig. S1). Other studies have also shown that in cervical carcinoma, expression of Id-1 protein is correlated to invasive and metastatic behaviors.29 This implies that Id-1 is a potential oncoprotein in HPV-related cervical cancer. However, the detailed mechanism by which Id-1 contributes to cervical cancer development is still elusive. We hypothesized that Id-1 could be involved in cancer development through promoting cervical epithelial transformation, the most important procedure in cancer development.
To test this hypothesis, we established an in vitro cervical epithelial transformation model. We transformed HPV-16 immortalized cervical H8 cells by NPYR, a metabolic product in the female genital tract with anaerobic vaginosis, which could be a potential carcinogen for cervical carcinogenesis.20 With both soft agar assay and tumorigenesis in nude mice, we successfully established an effective transformation of H8 cells. With this model, we focused on the role of Id-1 in the transformation process, which converts normal cervical epithelial cells to malignant cells.
First, we found that Id-1 expression was increased in transformed H8 cells and cancer cells compared to untransformed H8 cells. This result was consistent with what we have found in human cervical tissues, suggesting a relationship between Id-1 expression and cervical cancer development. Then, we overexpressed Id-1 through ectopic expression or suppressed Id-1 expression with shRNA-mediated knockdown in H8 cells. Id-1 overexpression increased while the Id-1 silencing decreased H8 cell transformation by NPYR. These findings are supportive to our hypothesis that Id-1 plays an important role in promoting epithelial cell transformations and contributes to cervical cancer development. However, Id-1 overexpression could not increase significant colony formation in soft agar assay alone (Fig. S2), which suggests that although Id-1 potentiated carcinogen-induced cervical epithelial cell transformation, itself it is insufficient to cause cell transformation. Thus, Id-1 is most likely an endogenous tumor promoter for cervical cancer development. These results may add new insight into the etiology of cervical cancer and suggest a potential target for therapy.
Human papillomavirus infection, particularly high risk HPV infection, has already been proved to be the most important etiology in cervical cancer development.8,9 Our previous study found that in carcinoma tissues, Id-1 expression was correlated to high risk HPV, especially HPV-16 infection.17 In the present study, we found the HPV E6 and E7 oncoproteins were upregulated in the Id-1 mediated cell transformation. This is consistent with a previous study showing that Id-1 expression is correlated to HPV E6/E7 expression in breast carcinoma tissues, and E6 and E7 induce breast cancer cell invasion and metastasis through induction of Id-1 overexpression.30 Because HPV E6 and E7 oncoproteins can promote cancer development through interacting with several tumor suppressors such as p53 and pRb, we hypothesize that Id-1 cooperates with HPV E6/E7 proteins, which involves inactivation of p53 and pRb in host cells, to promote cervical carcinogenesis. However, further studies are needed to verify this hypothesis.
In conclusion, our study suggests that Id-1 promotes NPYR-induced cervical epithelial transformation, which sheds light on the mechanism of cervical cancer development and implies that Id-1 is a potential target for cervical cancer prevention and therapy.
Acknowledgments
We thank Dr Robert Benezra from the Department of Cancer Biology and Genetics, Memorial Sloan-Kettering Cancer Center, New York, USA for providing the pWPI-hId1, hId1-shRNA, pWPI, pGIPZ, pDelta8.9 and pVSVg plasmids, and Professor Yong Zhao from Department of Pathology of Chongqing Medical University for H8 cells. This work was supported by the grants from Nature Scientific Foundation of China (No. 81001158) and Chinese Governmental Scientific Fund for College (No. 2010SCU3003).
Disclosure Statement
The authors have no conflict of interest.
Funding information
Nature Scientific Foundation of China (81001158). Chinese Governmental Scientific Fund for College (2010SCU3003).
Supporting Information
Additional supporting information may be found in the online version of this article:
Soft agar assay of inhibitors of differentiation-1 knockdown in SiHa cells.
Soft agar assay of H8 cells with or without Id-1 overexpression.
References
- 1.Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2011;127:2893–917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- 2.International Agency for Research on Cancer (IARC) IARC Handbooks of Cancer Prevention: Cervix Cancer Screening. Lyon, France: IARC Press; 2005. Chapter 2. [Google Scholar]
- 3.You W, Dainty LA, Rose GS, et al. Gynecologic malignancies in women aged less than 25 years. Obstet Gynecol. 2005;105:1405–9. doi: 10.1097/01.AOG.0000163254.98217.91. [DOI] [PubMed] [Google Scholar]
- 4.Zheng ZM, Baker CC. Papillomavirus genome structure, expression, and post-transcriptional regulation. Front Biosci. 2006;11:2286–302. doi: 10.2741/1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.D'Souza G, Kreimer AR, Viscidi R, et al. Case-control study of human papillomavirus and oropharyngeal cancer. N Engl J Med. 2007;356:1944–56. doi: 10.1056/NEJMoa065497. [DOI] [PubMed] [Google Scholar]
- 6.Bulk S, Berkhof J, Bulkmans NW, et al. Preferential risk of HPV16 for squamous cell carcinoma and of HPV18 for adenocarcinoma of the cervix compared to women with normal cytology in the Netherlands. Br J Cancer. 2006;94:171–5. doi: 10.1038/sj.bjc.6602915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Castillo A, Koriyama C, Higashi M, et al. Human papillomavirus in upper digestive tract tumors from three countries. World J Gastroenterol. 2011;17:5295–304. doi: 10.3748/wjg.v17.i48.5295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Whiteside MA, Siegel EM, Unger ER. Human papillomavirus and molecular considerations for cancer risk. Cancer. 2008;113:2981–94. doi: 10.1002/cncr.23750. [DOI] [PubMed] [Google Scholar]
- 9.Porras C, Rodríguez AC, Hildesheim A, et al. Human papillomavirus types by age in cervical cancer precursors: predominance of human papillomavirus 16 in young women. Cancer Epidemiol Biomarkers Prev. 2009;18:863–5. doi: 10.1158/1055-9965.EPI-08-0951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Castle PE, Solomon D, Schiffman M, Wheeler CM. Human papillomavirus type 16 infections and 2-year absolute risk of cervical precancer in women with equivocal or mild cytologic abnormalities. J Natl Cancer Inst. 2005;97:1066–71. doi: 10.1093/jnci/dji186. [DOI] [PubMed] [Google Scholar]
- 11.Perk J, Iavarone A, Benezra R. Id family of helix-loop-helix proteins in cancer. Nat Rev Cancer. 2005;5:603–14. doi: 10.1038/nrc1673. [DOI] [PubMed] [Google Scholar]
- 12.Han S, Gou C, Hong L, et al. Expression and significances of Id1 helix-loop-helix protein overexpression in gastric cancer. Cancer Lett. 2004;216:63–71. doi: 10.1016/j.canlet.2004.07.035. [DOI] [PubMed] [Google Scholar]
- 13.Schoppmann SF, Schindl M, Bayer G, et al. Overexpression of Id-1 is associated with poor clinical outcome in node negative breast cancer. Int J Cancer. 2003;104:677–82. doi: 10.1002/ijc.11009. [DOI] [PubMed] [Google Scholar]
- 14.Ouyang XS, Wang X, Lee DT, et al. Over expression of ID-1 in prostate cancer. J Urol. 2002;167:2598–602. [PubMed] [Google Scholar]
- 15.Schindl M, Schoppmann SF, Ströbel T, et al. Level of Id-1 protein expression correlates with poor differentiation, enhanced malignant potential, and more aggressive clinical behavior of epithelial ovarian tumors. Clin Cancer Res. 2003;9:779–85. [PubMed] [Google Scholar]
- 16.Li J, Jia H, Xie L, et al. Correlation of inhibitor of differentiation 1 expression to tumor progression, poor differentiation and aggressive behaviors in cervical carcinoma. Gynecol Oncol. 2009;114:89–93. doi: 10.1016/j.ygyno.2009.03.016. [DOI] [PubMed] [Google Scholar]
- 17.Li J, Xie L, Gan X, et al. Association of inhibitor of differentiation 1 expression with human papillomaviruses infections in cervical carcinoma. Int J Gynecol Cancer. 2011;21:1276–81. doi: 10.1097/IGC.0b013e31821f7452. [DOI] [PubMed] [Google Scholar]
- 18.Hebels DG, Jennen DG, Kleinjans JC, de Kok TM. Molecular signatures of N-nitroso compounds in Caco-2 cells: implications for colon carcinogenesis. Toxicol Sci. 2009;108:290–300. doi: 10.1093/toxsci/kfp035. [DOI] [PubMed] [Google Scholar]
- 19.Pavic N. Is there a local production of nitrosamines by the vaginal microflora in anaerobic vaginosis/trichomoniasis? Med Hypotheses. 1984;15:433–6. doi: 10.1016/0306-9877(84)90159-2. [DOI] [PubMed] [Google Scholar]
- 20.Barrington JW, Linton D, O'Leary A, et al. Anaerobic (bacterial) vaginosis and premalignant disease of the cervix. J Obstet Gynaecol. 1997;17:383–5. doi: 10.1080/01443619750112943. [DOI] [PubMed] [Google Scholar]
- 21.Kong L, Yu XP, Bai XH, et al. RbAp48 is a critical mediator controlling the transforming activity of human papillomavirus type 16 in cervical cancer. J Biol Chem. 2007;282:26381–91. doi: 10.1074/jbc.M702195200. [DOI] [PubMed] [Google Scholar]
- 22.Shimada T, Furukawa K, Kreiser DM, Cawein A, Williams GM. Induction of transformation by six classes of chemical carcinogens in adult rat liver epithelial cells. Cancer Res. 1983;43:5087–92. [PubMed] [Google Scholar]
- 23.Tsuchiya T, Tanaka-Kagawa T, Jinno H, et al. Inorganic arsenic compounds and methylated metabolites induce morphological transformation in two-stage BALB/c 3T3 cell assay and inhibit metabolic cooperation in V79 cell assay. Toxicol Sci. 2005;84:344–51. doi: 10.1093/toxsci/kfi082. [DOI] [PubMed] [Google Scholar]
- 24.Koleske AJ, Baltimore D, Lisanti MP. Reduction of caveolin and caveolae in oncogenically transformed cells. Proc Natl Acad Sci USA. 1995;92:1381–5. doi: 10.1073/pnas.92.5.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhao JJ, Pan K, Li JJ, et al. Identification of LZAP as a new candidate tumor suppressor in hepatocellular carcinoma. PLoS ONE. 2011;6:e26608. doi: 10.1371/journal.pone.0026608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yoshioka N, Wang L, Kishimoto K, Tsuji T, Hu GF. A therapeutic target for prostate cancer based on angiogenin-stimulated angiogenesis and cancer cell proliferation. Proc Natl Acad Sci U S A. 2006;103:14519–24. doi: 10.1073/pnas.0606708103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 28.Romero-Lanman EE, Pavlovic S, Amlani B, et al. Id1 maintains embryonic stem cell self-renewal by up-regulation of Nanog and repression of Brachyury expression. Stem Cells Dev. 2012;21:384–93. doi: 10.1089/scd.2011.0428. [DOI] [PubMed] [Google Scholar]
- 29.Darnel AD, Wang D, Ghabreau L, et al. Correlation between the presence of high-risk human papillomaviruses and Id gene expression in Syrian women with cervical cancer. Clin Microbiol Infect. 2010;16:262–6. doi: 10.1111/j.1469-0691.2009.02774.x. [DOI] [PubMed] [Google Scholar]
- 30.Yasmeen A, Bismar TA, Kandouz M, et al. E6/E7 of HPV type 16 promotes cell invasion and metastasis of human breast cancer cells. Cell Cycle. 2007;6:2038–42. doi: 10.4161/cc.6.16.4555. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Soft agar assay of inhibitors of differentiation-1 knockdown in SiHa cells.
Soft agar assay of H8 cells with or without Id-1 overexpression.


