Abstract
Targeting tumor angiogenesis is an established strategy for cancer therapy. Because angiogenesis is not limited to pathological conditions such as cancer, molecular markers that can distinguish between physiological and pathological angiogenesis are required to develop more effective and safer approaches for cancer treatment. To identify such molecules, we determined the gene expression profiles of murine tumor endothelial cells (mTEC) and murine normal endothelial cells using DNA microarray analysis followed by quantitative reverse transcription–polymerase chain reaction analysis. We identified 131 genes that were differentially upregulated in mTEC. Functional analysis using siRNA-mediated gene silencing revealed five novel tumor endothelial cell markers that were involved in the proliferation or migration of mTEC. The expression of DEF6 and TMEM176B was upregulated in tumor vessels of human renal cell carcinoma specimens, suggesting that they are potential targets for antiangiogenic intervention for renal cell carcinoma. Comparative gene expression analysis revealed molecular differences between tumor endothelial cells and normal endothelial cells and identified novel tumor endothelial cell markers that may be exploited to target tumor angiogenesis for cancer treatment.
Keywords: Antiangiogenic therapy, normal endothelial cell, tumor angiogenesis, tumor endothelial cell, tumor endothelial cell marker
Since the pioneering work of Judah Folkman, tumor blood vessels are recognized as an important target for cancer therapy.1–3 The discovery of bevacizumab, a humanized monoclonal antibody against vascular endothelial growth factor (VEGF), led to the use of antiangiogenic agents as a new approach for the treatment of cancer and hundreds of clinical trials involving antiangiogenic agents are currently underway.4–6 However, the benefits of antiangiogenic therapies are often marginal with harmful side-effects, largely because they inhibit normal as well as tumor-induced angiogenesis,7–10 and angiogenesis is required not only for tumor progression but also for normal physiological processes.11–14 Therefore, identification of novel therapeutic targets based on the difference between tumor and normal angiogenesis is crucial to prevent adverse effects associated with impaired physiological angiogenesis.
Tumor blood vessels differ from their normal counterparts in several ways, such as changes in morphology, altered blood flow and enhanced leakiness.15–17 These suggest that tumor endothelial cells (TEC), the main component of tumor vessels, are more relevant tools for developing antiangiogenic cancer therapy than normal endothelial cells (NEC). Some studies based on this concept focused on characterizing the gene expression profiles of TEC to identify molecules (TEC markers) associated with tumor angiogenesis.18–24 However, successful application of these TEC markers in the clinic has not been accomplished. This failure may be largely explained by impurities in the TEC during preparations, because isolated TEC were not cultured and their phenotypes were not verified.
Until recently, there were few reports describing the isolation and successful long-term culture of TEC. This is attributed to the technical difficulties caused by the small number of TEC that are enmeshed in a complex tissue that consists of vessel wall components, stromal cells and tumor cells. Moreover, isolated TEC may lose their specific phenotypes during in vitro culture. Therefore, most in vitro studies on tumor angiogenesis used NEC such as human umbilical vein endothelial cells, human dermal microvascular endothelial cells or bovine aortic endothelial cells.25
To address these issues, we developed a unique method to isolate highly purified murine tumor endothelial cells (mTEC) from human tumor xenografts or murine normal endothelial cells (mNEC) from dermal tissue of nude mice.26,27 Contrary to the stereotype that TEC may lose their specific phenotypes after dissociation from their tumor tissue, the isolated mTEC differed from mNEC in their phenotypic characteristics, including enhanced proliferation, motility, response to growth factors and resistance to chemotherapeutic drugs even after long-term culture.28–31 Thus, these mTEC maintain the specific characteristics of TEC in vivo and express molecular markers specific for tumor angiogenesis that can distinguish them from mNEC. This unique system for culturing endothelial cells (EC) encouraged us to seek novel molecules specifically associated with tumor angiogenesis.
Using the method described above,26,27 we purified and cultured three different types of mTEC and dermis-derived mNEC, compared their gene expression profiles using DNA microarray analysis and quantitative reverse transcription-polymerase chain reaction (qRT-PCR) assays, and identified 131 genes that were differentially upregulated in mTEC. We have already described the suitability of some of these genes including Bgn, Cxcr7 and Ptgir as TEC markers.32–34 Here, using RNAi techniques, we conducted functional screening of these 131 genes and identified five novel genes associated with the proliferation or migration of mTEC. To validate their applicability to cancer patients, we determined their expression levels in human TEC and tumor vessels isolated from human renal cell carcinoma (RCC) specimens.
Materials and Methods
Cell lines and culture conditions
The human oral squamous cell carcinoma cell line, HSC-3, was supplied by the Japanese Cancer Research Bank (Tokyo, Japan). The cells were cultured in Dulbecco's Modified Eagle Medium (DMEM; Sigma-Aldrich, St Louis, MO, USA) supplemented with 10% heat-inactivated fetal bovine serum (FBS). The human renal clear cell carcinoma cell line, OS-RC-2, was purchased from the RIKEN Cell Bank (Tsukuba, Japan) and cultured in RPMI1640 medium (Sigma-Aldrich) supplemented with 10% FBS. A375SM, a super-metastatic human malignant melanoma cell line, was provided by Dr Isaiah J. Fidler (MD Anderson Cancer Center, Houston, TX, USA).35 The cells were cultured in Minimum Essential Medium (GIBCO, Grand Island, NY, USA) supplemented with 10% FBS. These cells were cultured in a humidified atmosphere containing 5% CO2 at 37°C.
Antibodies
Antibodies purchased from commercial sources are as follows: mouse anti-human CD31 antibody (BD Pharmingen, San Diego, CA, USA); Alexa Fluor 647-mouse anti-human CD31 antibody (BioLegend, San Diego, CA, USA); anti-human CD105 antibody (BD Pharmingen); phycoerythrin-conjugated anti-human CD45 antibody (BD Pharmingen); fluorescein isothiocyanate-conjugated anti-human CD45 antibody (BioLegend); rabbit anti-human DEF6 (MBL; Nagoya, Japan); mouse anti-human TMEM176B (Abcam; Cambridge, MA, USA); and Alexa Fluor 594-conjugated anti mouse IgG, Alexa Fluor 488 goat anti-mouse IgG and Alexa Fluor 594 goat anti-rabbit IgG antibody (Invitrogen, Carlsbad, CA, USA).
Isolation of mTEC and mNEC
As described previously,27–29 mTEC were isolated from human tumor xenografts (oral carcinoma, renal carcinoma and melanoma) in nude mice and mNEC were isolated from the dermis as controls. The present study was approved by the Animal Care and Use Committee of Hokkaido University (approval ID, 08-0296) and all procedures for animal experiments were performed following the regulation on animal experimentation of Hokkaido University. All purified EC were cultured in EGM-2 MV and used between passages 15–25.
Microarray gene expression analysis
Total RNA was extracted from three types of mTEC (melanoma-derived EC, renal carcinoma-derived EC and oral carcinoma-derived EC) and mNEC using TRIzol (Invitrogen) according to the manufacturer's standard protocol. RNA was quantified using a RiboGreen RNA Quantitation Kit (Invitrogen) and RNA quality was confirmed using a 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). Gene expression profiles were obtained from 1.5 μg total RNA per sample using a GeneChip Mouse Genome 430 2.0 Array (Affymetrix, Santa Clara, CA, USA) according to the manufacturer's recommended protocol (GeneChip 3'-IVT Express Kit, P/N 702646 Rev. 7).
RT-PCR and qRT-PCR
Total RNA was extracted using the RNeasy Micro Kit (QIAGEN, Valencia, CA, USA). RT-PCR was performed with modifications. SYBR Green Real-time PCR Master Mix-Plus (Toyobo, Osaka, Japan) was used for qRT-PCR analysis. Cycling conditions followed the manufacturer's instructions according to Opticon Monitor version 3.0 (Bio-Rad, Hercules, CA, USA). Expression levels were normalized to those of genes encoding GAPDH or 18S rRNA. The primers for mouse Gapdh (mGAPDH), Cd31, Vegfr1, Vegfr2, Cd11b, Cd45, human HBEGF and human GAPDH (hGAPDH) were described previously.28 The primers for mouse Cd105 (Eng) are as follows: 5′-CTTCCAAGGACAGCCAAGAG-3′ and 5′-GGGTCATCCAGTGCTGCTAT-3′. The primers used for TEC markers are listed in Table1.
Table 1.
Primer information
| Species | Gene | Forward primer | Reverse primer |
|---|---|---|---|
| Mouse | Tmem176b | 5′- CTCCAAGTCTACTCCTCAAGCTCCA -3′ | 5′- CCAGAGTCCTACAGGAAAGCAGAGA -3′ |
| Pcdhb22 | 5′- ATCCGCAACCGAGGTGATG -3′ | 5′- AATGCGGATTTGCGAGGTG -3′ | |
| Nsg1 | 5′- GCCCTGATGGGTTTGTCTTGA -3′ | 5′- CACTGGAGTCTTGCTCCGTGTAGTA -3′ | |
| Enah | 5′- CACATTCAGAGTTGTGGGCAGA -3′ | 5′- TGCTGCCAAAGTTGAGACCATAC -3′ | |
| Def6 | 5′- CACCAACGTGAAACACTGGAATG -3′ | 5′- CGGGTCAGGCGCTTTAGAGA -3′ | |
| 18SrRNA | 5′- GGGAGCCTGAGAAACGGC -3′ | 5′- GGGTCGGGAGTGGGTAATTT -3′ | |
| Gapdh | 5′- TCTGACGTGCCGCCTGGAG -3′ | 5′- TCGCAGGAGACAACCTGGTC -3′ | |
| Human | TMEM176B | 5′- CCCTACCACTGGGTACAGATGGA -3′ | 5′- CTTCAAGACACAGACAGCCAGGA -3′ |
| PCDHB15 | 5′- GACCAGAGCCGAGTACAACATCAC -3′ | 5′- GTCCGACACCAGCACGGTTA -3′ | |
| NSG1 | 5′- CCGATGGGTTCGTCCTCAA -3′ | 5′- TCTTGCTCCGCGTAGTAGCTCTC -3′ | |
| ENAH | 5′- GTGGCTCAACTGGATTCAGCA -3′ | 5′- AGGAATGGCACAGTTTATCACGA -3′ | |
| DEF6 | 5′- CAGGGATACATGCCCTACCTCAAC -3′ | 5′- CAGCACAGCTCATCAAAGTGCTC -3′ | |
| GAPDH | 5′- ACAGTCAGCCGCATCTTCTT -3′ | 5′- GCCCAATACGACCAAATCC -3′ |
RNAi experiments
All siRNA (stealth siRNA) were purchased from Invitrogen and transfected at a final concentration of 3 nM using Lipofectamine RNAiMAX reagent (Invitrogen) according to the manufacturer's instructions. Total RNA for qRT-PCR analysis was extracted 48 h after transfection. For proliferation assays, 4 × 103 transfected cells were cultured in 96-well dishes and cell viability was measured 72 h after transfection using Alamar Blue reagent. For migration assays, transfected cells were maintained in six-well dishes for 48 h. After starvation with EBM-2 containing 0.2% bovine serum albumin for 2 h, cells were resuspended in EBM-2 containing 0.2% bovine serum albumin and transferred to the upper chambers of a BD BioCoat Angiogenesis System: Endothelial Cell Migration (BD Biosciences, Franklin Lakes, NJ, USA). Cell migration for 20 h toward the chemoattractant EGM-2MV was measured by labeling the migrated cells with Calcein AM Fluorescent Dye (BD Biosciences) according to the manufacturer's instructions.
Human tissue samples
Surgically resected tissues from three patients diagnosed with RCC (clear cell carcinomas; Table2) were analyzed. The specimens included tumor tissues and corresponding normal renal tissues 5–10 cm from the tumor. One portion of the sample was immediately snap-frozen in liquid nitrogen and stored at –80°C for immunohistological analysis and another was placed in Hank's Balanced Salt Solution (Life Technologies, Grand Island, NY, USA) on ice until EC isolation. Final diagnosis of RCC was confirmed by pathological examination of formalin-fixed surgical specimens. All protocols were approved by the Institutional (Hokkaido University) Ethics Committee (approval ID, 009-0148) and written informed consent was obtained from each patient before surgery.
Table 2.
Clinical background of renal cell carcinoma specimens
| Case no. | Sex | Age (years) | TNM† | Subtype | Grade‡ | INF | Vascular invasion |
|---|---|---|---|---|---|---|---|
| 1 | F | 50 | T1b, N0, M0 | Clear Cell | G2 | INFa | V (−) |
| 2 | M | 60 | T1a, N0, M0 | Clear Cell | G2 | INFa | V (−) |
| 3 | F | 48 | T2b, N1, M0 | Clear Cell | G2 | INFa | V (+) |
According to the 7th edition of tumor-node-metastasis (TNM) staging guidelines.37
According to the Fuhrman system. F, female; INF, infiltration pattern; M, male; –, negative; +, positive.
Isolation of human renal TEC and NEC
Human TEC (hTEC) and human NEC (hNEC) were isolated from the excised human RCC and normal renal tissues, respectively, followed by flow cytometric analysis as described previously.33 All purified EC were plated and cultured in EGM-2MV (Lonza, Basel, Switzerland) and 15% FBS.
Immunohistochemistry
Frozen human tissue samples were cut into 8-μm thick sections. Immunofluorescence was performed as previously reported.33 Primary and secondary antibodies are described above. These samples were counterstained with diamidino-2-phenylindole (DAPI; Roche Diagnostics, Mannheim, Germany) and visualized using an Olympus FluoView FV1000 confocal microscope (Olympus, Tokyo, Japan).
Statistical analysis
All data are expressed as the mean ± standard deviation and subjected to two-sided Student's t-tests. Differences were considered significant for P < 0.05.
Results
Isolation and characterization of mTEC and mNEC
We first isolated and cultured three different types of mTEC (Melanoma-EC, Renal carcinoma-EC [Renal Ca-EC] and Oral carcinoma-EC [Oral Ca-EC]) from human tumor xenografts and mNEC (Skin-EC) from dermal tissues of nude mice as a normal control. These murine endothelial cells (mEC) were positive for the EC markers Cd31, Cd105, Vegfr1 and Vegfr2 and negative for the monocyte marker Cd11b and hematopoietic marker Cd45 using RT-PCR (Fig.1a). Human HBEGF, which is expressed in human tumor cells, was not detected in any of the mTEC (Fig.1a). These results excluded the possibility that these mEC were contaminated with non-EC such as monocytes, hematopoietic cells and human tumor cells. Furthermore, tube formation was observed when mEC were cultured on matrigel-coated plates (Fig.1b), indicating that these mEC maintained EC properties after isolation and culture. Thus, our isolation technique yielded highly pure and functional populations of mEC suitable for subsequent analyses.
Figure 1.
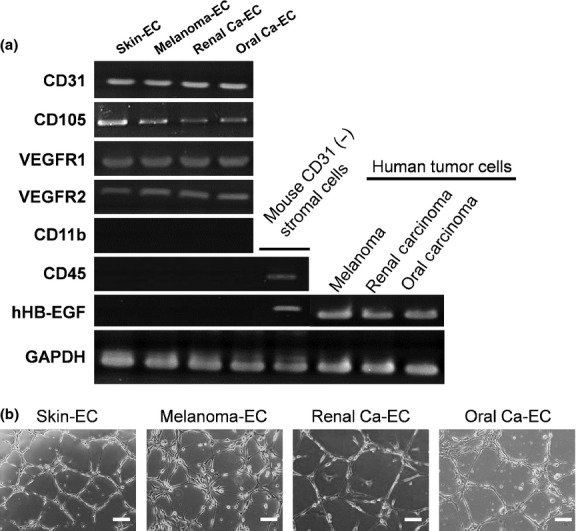
Characterization of murine tumor endothelial cells (mTEC) and murine normal endothelial cells (mNEC). (a) The expression of Cd31, Cd105, Vegfr1, Vegfr2, Cd11b, Cd45 and human HBEGF in mTEC (Melanoma-EC, Renal Ca-EC and Oral Ca-EC) and mNEC (Skin-EC) was analyzed using RT-PCR. CD31-negative non-EC fractions and human tumor cells (melanoma, renal carcinoma and oral carcinoma) were also analyzed. (b) Isolated and cultured mEC formed tubes on matrigel-coated plates. Bar, 100 μm. EC, endothelial cells; Renal Ca-EC, Renal carcinoma-EC; Oral Ca-EC, Oral carcinoma-EC; mEC, murine endothelial cells.
Expression profiling of isolated mTEC and mNEC
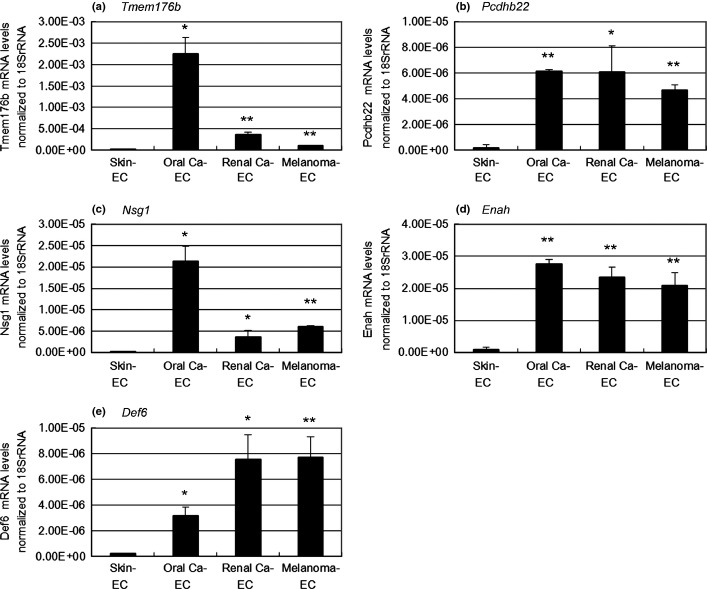
To identify novel markers of tumor endothelium by comparing the gene expression patterns between mTEC and mNEC, total RNA was extracted from eight independent mTEC populations derived from three types of human tumor xenografts (three melanomas, three renal carcinomas and two oral squamous cell carcinomas) and two populations of mNEC derived from two independent samples from the dermis of nude mice. RNA was used to probe an Affymetrix GeneChip Mouse Genome 430 2.0 array to determine transcriptional profiles. We focused on transcripts that were highly expressed in the three types of mTEC compared with mNEC. We detected 180 transcripts expressed in all mTEC with levels five times higher than that in mNEC. We excluded 19 genes with no human orthologs and 30 genes that were expressed less than five times higher by any mTEC compared with mNEC using qRT-PCR (Fig.2). The DNA microarray and RT-PCR analysis of representative genes including the five novel TEC markers are shown in Table3, Figure3 and Supporting Information Table S1. The levels of expression of these genes were higher in the three types of mTEC than that in mNEC. We selected 131 genes as potential TEC markers for functional screening using siRNA-mediated gene silencing (Fig.2).
Figure 2.
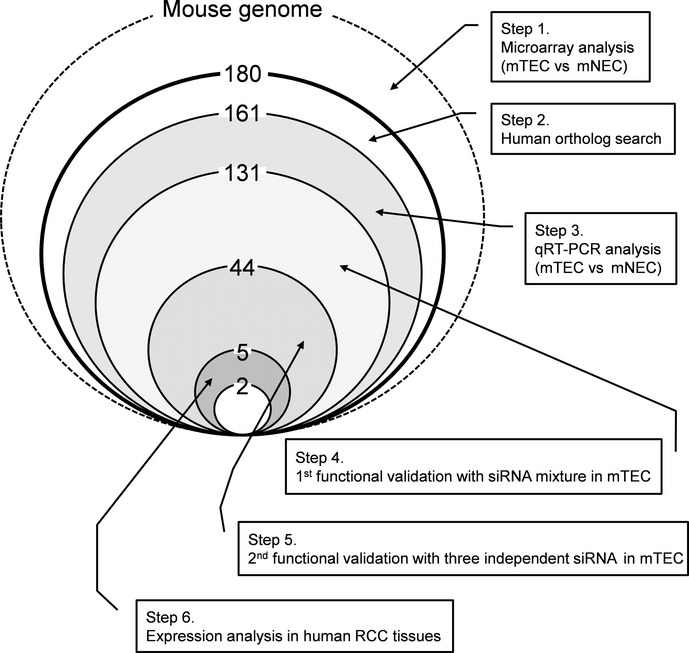
Schematic of tumor endothelial cell (TEC) marker selection. The strategy and results of TEC marker selection are summarized. We identified five novel TEC markers through analysis of gene expression profiles and functions in murine tumor endothelial cells (mTEC). mNEC, murine normal endothelial cell; RCC, renal cell carcinoma.
Table 3.
Representative data of DNA microarray analysis
| Probe (Mouse430_2) | Gene title | Symbol | Fold increase relative to Skin-EC |
||
|---|---|---|---|---|---|
| Oral Ca-EC | Renal Ca-EC | Melanoma-EC | |||
| 1418004_a_at | Transmembrane protein 176B | Tmem176b | 518.29 | 156.13 | 61.39 |
| 1418941_at | Protocadherin beta 22 | Pcdhb22 | 31.36 | 20.77 | 31.96 |
| 1423055_at | Neuron-specific gene family member 1 | Nsg1 | 147.51 | 28.57 | 53.99 |
| 1424800_at | Enabled homolog (Drosophila) | Enah | 23.66 | 14.78 | 21.43 |
| 1452796_at | Differentially expressed in FDCP 6 | Def6 | 5.02 | 8.36 | 14.09 |
EC, endothelial cells; Ca, carcinoma.
Figure 3.
Analysis of the transcription of tumor endothelial cell (TEC) marker genes. Using qRT-PCR, five novel TEC markers identified here were shown to be selectively upregulated in all types of murine TEC (mTEC) compared with murine normal endothelial cells. The relative expression levels of mRNA were normalized to that of 18S rRNA for each cell type (*P < 0.05; **P < 0.01). (a–e) mRNA expression of novel TEC markers (Tmem176b, Pcdhb22, Nsg1, Enah and Def6). EC, endothelial cells; Ca, carcinoma.
Functional validation of TEC markers using RNAi
Excessive angiogenesis occurs through a series of steps including enhanced EC proliferation and migration.36 Therefore, targeting proliferation and/or migration of EC is one of the most attractive and effective strategies for treating angiogenesis-dependent disorders. We reported that mTEC grow faster and migrate better than mNEC.28 These in vitro characteristics of mTEC represent enhanced tumor angiogenesis in vivo and the genes responsible for increased proliferation or migration of mTEC may serve as ideal targets for antiangiogenic therapy. To identify such molecules, we performed loss-of-function screening of the 131 potential TEC markers in Melanoma-EC, one of the mTEC that showed high activity in the proliferation and migration assays.28 We first cotransfected Melanoma-EC with three different sequences of siRNA per gene. Cell proliferation and/or migration were inhibited by >20% compared with mock-transfected cells using siRNA targeted to 44 genes (Fig.2). Subsequently, three different siRNA specific for each of the 44 genes were used to independently transfect Melanoma-EC. We performed migration and proliferation assay using siRNA and finally selected five genes (Def6, Nsg1, Enah, Tmem176b and Pcdhb22; Table3, Fig.3) whose respective siRNA (two or more per gene) inhibited cell proliferation or migration by >30% (Figs2, 4). Cell migration was inhibited by siRNA of Tmem176b, Pcdhb22, Nsg1 and Enah, but cell proliferation was not inhibited. In contrast, cell proliferation was inhibited by Def6 siRNA. These were considered potential regulators of proliferation or migration of mTEC. There was no gene whose two or more siRNA inhibited both cell proliferation and migration by >30% (data not shown). Knockdown of each gene was confirmed using qRT-PCR 48 h after transfection (Fig.4).
Figure 4.
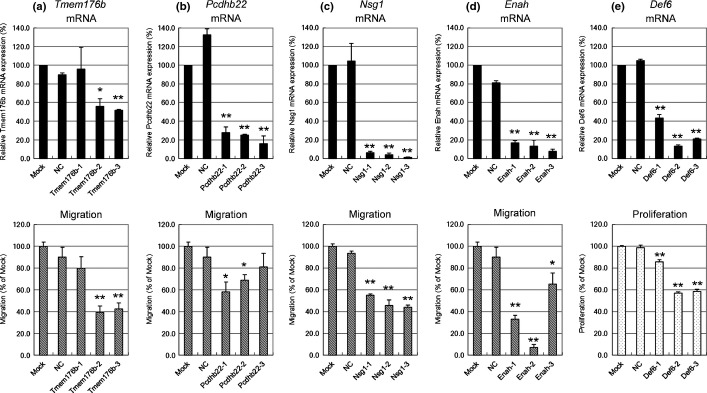
Functional analysis of tumor endothelial cell (TEC) markers using siRNA-mediated gene silencing. Melanoma-EC were transfected using three different siRNA for each gene. Silencing of each TEC marker 48 h after transfection was evaluated using qRT-PCR (upper panel). Cell proliferation was measured after 72 h using Alamar Blue (lower panel). Cell migration towards the chemoattractant EGM-2MV for 20 h was evaluated using the BD BioCoat Angiogenesis System: Endothelial Cell Migration (BD Biosciences) (lower panel). siRNA-mediated effects of each TEC marker are shown in (a–e) (**P < 0.01; *P < 0.05). (a) Tmem176b; (b) Pcdhb22; (c) Nsg1; (d) Enah; and (e) Def6. NC, Negative control siRNA.
Expression of TEC markers in human TEC in vitro and in vivo
The therapeutic potential of targeting candidate genes largely depends on whether their expression is upregulated in hTEC as well as in mTEC. Therefore, we analyzed the expression of the five putative TEC markers in hTEC isolated from RCC specimens and hNEC from normal renal tissues from the same patients, respectively. Because the EC population represents only a small percentage of the cells present in tumor tissue, sufficient quantities of specimens must be acquired for preparation of hTEC. This technical limitation forced us to choose RCC as the source of hTEC.
The hTEC and hNEC were obtained from three patients. The clinical backgrounds of patients with RCC who donated tissue specimens are shown in Table2. The binding of ulex europaeus agglutinin 1 (UEA-1 lectin), the expression of CD31 and CD105, and lack of expression of CD45 determined by flow cytometric analysis confirmed the high purity of the isolated human EC (hEC). Representative data are shown in Figure5(a). The expression levels of TMEM176B and DEF6 revealed by qRT-PCR analysis were significantly higher in hTEC than in hNEC for all paired samples (Fig.5b). In contrast, we were unable to detect upregulation of NSG1, ENAH or PCDHB15 (human ortholog of Pcdhb22) in hTEC (Fig. S1). These results indicate that two out of five TEC markers that we identified in mice were upregulated in mTEC (Fig. S2) and hTEC (Fig.5b). Furthermore, to determine the expression levels of TMEM176B and DEF6 in tumor blood vessels in RCC in vivo, we performed immunofluorescence double staining of the frozen sections of human renal tumors and normal kidney tissues (glomerulus) using anti-CD31 with either anti-TMEM176B or anti-DEF6 antibodies. TMEM176B and DEF6 were expressed in tumor blood vessels in renal cancer, but at much lower levels in normal blood vessels (Figs5c, S3). These results suggest that the transcription of these two genes was upregulated in hTEC in vivo and might be involved in tumor angiogenesis in cancer patients.
Figure 5.
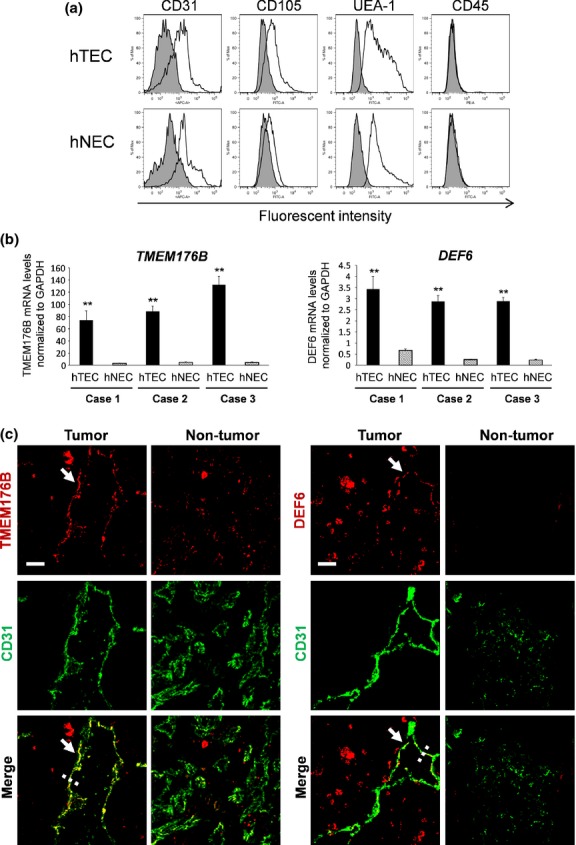
Analysis of TMEM176B and DEF6 expression in vitro and in vivo. (a) Verification of endothelial cells (EC) from a human sample. The binding of ulex europaeus agglutinin 1 (UEA-1 lectin), expression of CD31, CD105 and lack of expression of CD45 (white area) indicates high purity of the isolated human tumor endothelial cells (hTEC) and human normal endothelial cells (hNEC). The isotype control is shown in gray. (b) Upregulated expression of TMEM176B and DEF6 in hTEC. qRT-PCR analysis detected high levels of expression of both genes in hTEC compared with the corresponding hNEC in all three cases. Expression levels of the mRNA were normalized to that of GAPDH (**P < 0.01). (c) Both TMEM176B and DEF6 were strongly stained in tumor vessels using an anti-CD31 antibody in combination with an antibody against either TMEM176B or DEF6. In contrast, normal vessels (glomerular) of normal renal tissue were weakly stained. All samples were counterstained with DAPI. Profiles of immunofluorescence intensities along the dashed lines are shown in Figure S3(a,b). The signal intensities of TMEM176B or DEF6 in the CD31-positive area of whole sections were analyzed by ImageJ (NIH, Bethesda, MD, USA) quantitatively (Fig. S3c,d). Bar, 20 μm.
Discussion
In the present study, we isolated and cultured mTEC from three different types of human tumor xenografts and mNEC from the dermis of nude mice and compared their gene expression profiles. DNA microarray analysis and qRT-PCR analysis identified 131 genes that were upregulated in mTEC compared with mNEC. Functional analysis of these 131 genes using RNAi revealed that five were involved in the proliferation or migration of mTEC. Two, DEF6 and TMEM176B, were upregulated in hTEC and in vivo in tumor vessels of human RCC, suggesting that increased expression of these two proteins contributes to enhanced tumor angiogenesis in cancer patients. To the best of our knowledge, this is the first report that DEF6 and TMEM176B might be involved in tumor angiogenesis and might serve as targets for antiangiogenic therapy of cancer patients.
DEF6, also described as SLAT or IBP, is highly conserved in vertebrates and acts as a guanine nucleotide exchange factor for Rho-family GTPases, including RAC1, CDC42 and RHOA,38 which are involved in cytoskeletal organization, cell cycle progression and extracellular signal transduction, as well as in the proliferation, migration, invasion and metastasis of cancer cells.39–42 DEF6 is overexpressed in cancer cells (as suggested in Fig.5c) and may have an important function in tumor invasion and metastasis43,44; however, its role in tumor angiogenesis is unknown. In the present study, we show for the first time that DEF6 was upregulated in TEC compared with NEC and might mediate increased proliferation of TEC that enhance tumor angiogenesis. Its role in tumor endothelial function combined with its significance for tumor cell function makes it an appealing candidate as a target for cancer therapy.
TMEM176B, also known as LR8, belongs to the CD20/Fc-RI and membrane-spanning 4A (MS4A) family.45,46 It was discovered in human lung fibroblasts and is associated with human small cell lung carcinoma.47 Although several recent reports implicate human TMEM176B in cancer,48–50 no direct evidence is available regarding its function in cancer pathogenesis, including tumor angiogenesis. Here, we report for the first time overexpression of TMEM176B in TEC and further show using RNAi that TMEM176B mediates TEC migration. Moreover, immunohistochemical analysis revealed expression of TMEM176B in tumor vessels and in tumor cells (as suggested in Fig.5c) as reported previously.51 Although the contribution of TMEM176B in tumor cells to the malignant phenotype is unknown, it might serve as a target for cancer therapy. The physiological function of TMEM176B remains to be determined.
Unlike DEF6 and TMEM176B, we were unable to detect upregulation of NSG1, ENAH or PCDHB15 (human ortholog of Pcdhb22) in hTEC or tumor vessels of human RCC specimens. ENAH and PCDHB15 were expressed in vessels and in mesangial cells of normal tissues and NSG1 expression was not detected in tumor vessels (data not shown). Because the number of samples was limited and tumors other than RCC remain to be examined, we consider these genes to be worthy of future study.
The present study indicates the power of determining the differential expression of genes between TEC and NEC for identifying potential targets for antiangiogenic therapy. Although TEC markers such as ANTXR1 (TEM8), CD276 and JAG1 (Jagged1) were previously identified using this technique,18–24 the present study is the first, to our knowledge, to demonstrate upregulated expression of the genes encoding DEF6, TMEM176B, NSG1, ENAH and PCDHB15 in TEC. The major difference between the present and previous studies is our unique method for culturing mTEC, which overcomes the loss of the TEC phenotype after dissociation from their tumor tissue. These highly purified mTEC isolated from human tumor xenograft maintain the specific characteristics of TEC in vivo during long-term culture26–31 and therefore provide a more relevant system for tumor angiogenesis research and the identification of novel TEC markers.
In summary, here we report the identification of novel genes that are relevant to tumor angiogenesis by investigating the differences in gene expression patterns between mTEC and mNEC. Targeting these genes may lead to therapies that do not induce adverse effects associated with altering physiological angiogenesis. Further research will be required, particularly in vivo studies, to define the roles of the novel genes identified here in tumor angiogenesis, invasion and metastatic growth.
Acknowledgments
The authors thank Drs Osawa, Yamamoto, Kitayama, Kondoh, Matsuda and Kurosu for technical assistance. This work was supported in part by grant-in-aid for scientific research from the Ministry of Education, Science, and Culture of Japan (20390506 to K.H.) and by a research grant from Dainippon Sumitomo Pharma Co., Ltd.
Disclosure Statement
T.O., H.S., T. Kai, Y.M. and H.T. are employees of Dainippon Sumitomo Pharma Co., Ltd. The following genes are patent pending: Pcdhb22(PCT/JP2011/056631); Nsg1(PCT/JP2010/063124); Enah(PCT/JP2011/056639); Def6(PCT/JP2010/063669); and TMEM176b(JP2010/254205). This does not alter our adherence to all the Cancer Science policies on sharing data and materials.
Funding information
Ministry of Education, Science, and Culture of Japan (20390506). Dainippon Sumitomo Pharma Co., Ltd
Supporting Information
Additional supporting information may be found in the online version of this article:
Analysis of NSG1, ENAH and PCDHB15 expression in vitro.
Analysis of the transcription of Tmem176b and Def6 genes using qRT-PCR.
Immunofluorescence intensities of TMEM176B, DEF6 and CD31 in human RCC specimens.
Representative data of qRT-PCR analysis
References
- 1.Folkman J. Tumor angiogenesis: therapeutic implications. N Engl J Med. 1971;285:1182–6. doi: 10.1056/NEJM197111182852108. [DOI] [PubMed] [Google Scholar]
- 2.Folkman J. Angiogenesis in cancer, vascular, rheumatoid and other disease. Nat Med. 1995;1:27–31. doi: 10.1038/nm0195-27. [DOI] [PubMed] [Google Scholar]
- 3.Kerbel R, Folkman J. Clinical translation of angiogenesis inhibitors. Nat Rev Cancer. 2002;2:727–39. doi: 10.1038/nrc905. [DOI] [PubMed] [Google Scholar]
- 4.Hurwitz H. Integrating the anti-VEGF-A humanized monoclonal antibody bevacizumab with chemotherapy in advanced colorectal cancer. Clin Colorectal Cancer. 2004;4(Suppl. 2):S62–8. doi: 10.3816/ccc.2004.s.010. [DOI] [PubMed] [Google Scholar]
- 5.Sharma PS, Sharma R, Tyagi T. VEGF/VEGFR pathway inhibitors as anti-angiogenic agents: present and future. Curr Cancer Drug Targets. 2011;11:624–53. doi: 10.2174/156800911795655985. [DOI] [PubMed] [Google Scholar]
- 6.Young RJ, Reed MWR. Anti-angiogenic therapy: concept to clinic. Microcirculation. 2012;19:115–25. doi: 10.1111/j.1549-8719.2011.00147.x. [DOI] [PubMed] [Google Scholar]
- 7.Verheul HM, Pinedo HM. Possible molecular mechanisms involved in the toxicity of angiogenesis inhibition. Nat Rev Cancer. 2007;7:475–85. doi: 10.1038/nrc2152. [DOI] [PubMed] [Google Scholar]
- 8.Ratner M. Genentech discloses safety concerns over avastin. Nat Biotechnol. 2004;22:1004–198. doi: 10.1038/nbt1004-1198. [DOI] [PubMed] [Google Scholar]
- 9.Saif MW, Elfiky A, Salem RR. Gastrointestinal perforation due to bevacizumab in colorectal cancer. Ann Surg Oncol. 2007;14:1860–9. doi: 10.1245/s10434-006-9337-9. [DOI] [PubMed] [Google Scholar]
- 10.Kamba T, McDonald DM. Mechanisms of adverse effects of anti-VEGF therapy for cancer. Br J Cancer. 2007;96:1788–95. doi: 10.1038/sj.bjc.6603813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carmeliet P. Angiogenesis in life, disease and medicine. Nature. 2005;438:932–6. doi: 10.1038/nature04478. [DOI] [PubMed] [Google Scholar]
- 12.Risau W. Mechanisms of angiogenesis. Nature. 1997;386:671–4. doi: 10.1038/386671a0. [DOI] [PubMed] [Google Scholar]
- 13.Folkman J. Fundamental concepts of the angiogenesis process. Curr Mol Med. 2003;3:643–51. doi: 10.2174/1566524033479465. [DOI] [PubMed] [Google Scholar]
- 14.Chung AS, Lee J, Ferrara N. Targeting the tumor vascalature: insights from physiological angiogenesis. Nat Rev Cancer. 2010;10:505–14. doi: 10.1038/nrc2868. [DOI] [PubMed] [Google Scholar]
- 15.McDonald DM, Choyke PL. Imaging of angiogenesis: from microscope to clinic. Nat Med. 2003;9:713–25. doi: 10.1038/nm0603-713. [DOI] [PubMed] [Google Scholar]
- 16.Baluk P, Hashizume H, McDonald DM. Cellular abnormalities of blood vessels as targets in cancer. Curr Opin Genet Dev. 2005;15:102–11. doi: 10.1016/j.gde.2004.12.005. [DOI] [PubMed] [Google Scholar]
- 17.McDonald DM, Baluk P. Significance of blood vessel leakiness in cancer. Cancer Res. 2002;62:5381–5. [PubMed] [Google Scholar]
- 18.St Croix B, Rago C, Velculescu V, et al. Genes expressed in human tumor endothelium. Science. 2000;289:1197–202. doi: 10.1126/science.289.5482.1197. [DOI] [PubMed] [Google Scholar]
- 19.Carson-Walter EB, Watkins DN, Nanda A, Vogelstein B, Kinzler KW, St Croix B. Cell surface tumor endothelial markers are conserved in mice and humans. Cancer Res. 2001;61:6649–55. [PubMed] [Google Scholar]
- 20.Nanda A. St Croix B. Tumor endothelial markers: new targets for cancer therapy. Curr Opin Oncol. 2004;16:44–9. doi: 10.1097/00001622-200401000-00009. [DOI] [PubMed] [Google Scholar]
- 21.Seaman S, Stevens J, Yang MY, Logsdon D, Graff-Cherry C, St Croix B. Genes that distinguish physiological and pathological angiogenesis. Cancer Cell. 2007;11:539–54. doi: 10.1016/j.ccr.2007.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Buckanovich RJ, Sasaroli D, O'Brien-Jenkins A, et al. Tumor vascular proteins as biomarkers in ovarian cancer. J Clin Oncol. 2007;25:852–61. doi: 10.1200/JCO.2006.08.8583. [DOI] [PubMed] [Google Scholar]
- 23.Lu C, Bonome T, Li Y, et al. Gene alterations identified by expression profiling in tumor-associated endothelial cells from invasive ovarian carcinoma. Cancer Res. 2007;67:1757–68. doi: 10.1158/0008-5472.CAN-06-3700. [DOI] [PubMed] [Google Scholar]
- 24.van Beijnum JR, Dings RP, van der Linden E, et al. Gene expression of tumor angiogenesis dissected: specific targeting of colon cancer angiogenic vasculature. Blood. 2006;108:2339–48. doi: 10.1182/blood-2006-02-004291. [DOI] [PubMed] [Google Scholar]
- 25.Auerbach R, Akhtar N, Lewis RL, Shinners BL. Angiogenesis assays: problems and pitfalls. Cancer Metastasis Rev. 2000;19:167–72. doi: 10.1023/a:1026574416001. [DOI] [PubMed] [Google Scholar]
- 26.Hida K, Klagsbrun M. A new perspective on tumor endothelial cells: unexpected chromosome and centrosome abnormalities. Cancer Res. 2005;65:2507–10. doi: 10.1158/0008-5472.CAN-05-0002. [DOI] [PubMed] [Google Scholar]
- 27.Hida K, Hida Y, Amin DN, et al. Tumor-associated endothelial cells with cytogenetic abnormalities. Cancer Res. 2004;64:8249–55. doi: 10.1158/0008-5472.CAN-04-1567. [DOI] [PubMed] [Google Scholar]
- 28.Matsuda K, Ohga N, Hida Y, et al. Isolated tumor endothelial cells maintain specific character during long-term culture. Biochem Biophys Res Commun. 2010;394:947–54. doi: 10.1016/j.bbrc.2010.03.089. [DOI] [PubMed] [Google Scholar]
- 29.Ohga N, Hida K, Hida Y, et al. Inhibitory effects of epigallocatechin-3 gallate, a polyphenol in green tea, on tumor-associated endothelial cells and endothelial progenitor cells. Cancer Sci. 2009;100:1963–70. doi: 10.1111/j.1349-7006.2009.01255.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Akiyama K, Ohga N, Hida Y, et al. Tumor endothelial cells acquire drug resistance by MDR1 upregulation via VEGF signaling in tumor microenvironment. Am J Pathol. 2012;180:1283–93. doi: 10.1016/j.ajpath.2011.11.029. [DOI] [PubMed] [Google Scholar]
- 31.Ohga N, Ishikawa S, Maishi N, et al. Heterogeneity of tumor endothelial cells: comparison between tumor endothelial cells isolated from highly metastatic and low metastatic tumors. Am J Pathol. 2012;180:1294–307. doi: 10.1016/j.ajpath.2011.11.035. [DOI] [PubMed] [Google Scholar]
- 32.Yamamoto K, Ohga N, Hida Y, et al. Biglycan is a specific marker and an autocrine angiogenic factor of tumor endothelial cells. Br J Cancer. 2012;106:1214–23. doi: 10.1038/bjc.2012.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maishi N, Ohga N, Hida Y, et al. CXCR7: a novel tumor endothelial marker in renal cell carcinoma. Pathol Int. 2012;62:309–17. doi: 10.1111/j.1440-1827.2012.02792.x. [DOI] [PubMed] [Google Scholar]
- 34.Osawa T, Ohga N, Hida Y, et al. Prostacyclin receptor in tumor endothelial cells promotes angiogenesis in an autocrine manner. Cancer Sci. 2012;103:1038–44. doi: 10.1111/j.1349-7006.2012.02261.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kozlowski JM, Hart IR, Fidler IJ, Hanna N. A human melanoma line heterogeneous with respect to metastatic capacity in athymic nude mice. J Natl Cancer Inst. 1984;72:913–7. [PubMed] [Google Scholar]
- 36.Liekens S, De Clercq E, Neyts J. Angiogenesis: regulators and clinical applications. Biochem Pharmacol. 2001;61:253–70. doi: 10.1016/s0006-2952(00)00529-3. [DOI] [PubMed] [Google Scholar]
- 37.Sobin LH, Gospodarowicz MK, Wittekind C. The TNM Classification of Malignant Tumours. Hoboken, NJ: Union for International Cancer Control (UICC); 2009. 7th edn. [Google Scholar]
- 38.Mavrakis KJ, McKinlay KJ, Jones P, Sablitzky F. DEF6, a novel PH-DH-like domain protein, is an upstream activator of the Rho GTPases Rac1, Cdc42, and RhoA. Exp Cell Res. 2004;294:335–44. doi: 10.1016/j.yexcr.2003.12.004. [DOI] [PubMed] [Google Scholar]
- 39.Hall A. Rho GTPases and the actin cytoskeleton. Science. 1998;279:509–14. doi: 10.1126/science.279.5350.509. [DOI] [PubMed] [Google Scholar]
- 40.Ridley AJ. Rho family proteins: coordinating cell responses. Trends Cell Biol. 2001;11:471–7. doi: 10.1016/s0962-8924(01)02153-5. [DOI] [PubMed] [Google Scholar]
- 41.Vega FM, Ridley AJ. Rho GTPases in cancer cell biology. FEBS Lett. 2008;582:2093–101. doi: 10.1016/j.febslet.2008.04.039. [DOI] [PubMed] [Google Scholar]
- 42.Banyard J, Anand-Apte B, Symons M, Zetter BR. Motility and invasion are differentially modulated by Rho family GTPases. Oncogene. 2000;19:580–91. doi: 10.1038/sj.onc.1203338. [DOI] [PubMed] [Google Scholar]
- 43.Zhang Z, Wang Q, Li P, et al. Overexpression of the interferon regulatory factor 4-binding protein in human colorectal cancer and its clinical significance. Cancer Epidemiol. 2009;33:130–6. doi: 10.1016/j.canep.2009.05.004. [DOI] [PubMed] [Google Scholar]
- 44.Li P, Zhang Z, Wang Q, et al. The ectopic expression of IFN regulatory factor 4-binding protein is correlated with the malignant behavior of human breast cancer cells. Int Immunopharmacol. 2009;9:1002–9. doi: 10.1016/j.intimp.2009.04.008. [DOI] [PubMed] [Google Scholar]
- 45.Lurton J, Rose TM, Raghu G, Narayanan AS. Isolation of a gene product expressed by a subpopulation of human lung fibroblasts by differential display. Am J Respir Cell Mol Biol. 1999;20:327–31. doi: 10.1165/ajrcmb.20.2.3368. [DOI] [PubMed] [Google Scholar]
- 46.Zuccolo J, Bau J, Childs SJ, et al. Phylogenetic analysis of the MS4A and TMEM176 gene families. PLoS ONE. 2010;5:e9369. doi: 10.1371/journal.pone.0009369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gottschling S, Jauch A, Kuner R, et al. Establishment and comparative characterization of novel squamous cell non-small cell lung cancer cell lines and their corresponding tumor tissue. Lung Cancer. 2012;75:45–57. doi: 10.1016/j.lungcan.2011.05.020. [DOI] [PubMed] [Google Scholar]
- 48.Strelnikov V, Tanas A, Shkarupo V, Kuznetsova E, Gorban N, Zaletayev D. Non-microarray DNA differential methylation screening in breast cancer. Cancer Genet Cytogenet. 2010;203:93. [Google Scholar]
- 49.Hodo Y, Hashimoto S, Honda M, et al. Comprehensive gene expression analysis of 5′-end of mRNA identified novel intronic transcripts associated with hepatocellular carcinoma. Genomics. 2010;95:217–23. doi: 10.1016/j.ygeno.2010.01.004. [DOI] [PubMed] [Google Scholar]
- 50.Ryu SH, Kim KH, Kim HB, et al. Oncogenic Ras-mediated downregulation of Clast1/LR8 is involved in Ras-mediated neoplastic transformation and tumorigenesis in NIH3T3 cells. Cancer Sci. 2010;101:1990–6. doi: 10.1111/j.1349-7006.2010.01626.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cuajungco MP, Podevina W, Valluri VK, Bui Q, Nguyen VH, Taylor K. Abnormal accumulation of human transmembrane (TMEM)-176A and 176B proteins is associated with cancer pathology. Acta Histochem. 2012;114:705–12. doi: 10.1016/j.acthis.2011.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Analysis of NSG1, ENAH and PCDHB15 expression in vitro.
Analysis of the transcription of Tmem176b and Def6 genes using qRT-PCR.
Immunofluorescence intensities of TMEM176B, DEF6 and CD31 in human RCC specimens.
Representative data of qRT-PCR analysis