Abstract
Follicular lymphoma (FL) of the gastrointestinal tract, particularly duodenal follicular lymphoma (DFL), is a rare variant of FL with indolent clinical behavior, and this disease is included in the 2008 World Health Organization classification system. In contrast to nodal follicular lymphoma (NFL), DFL occurs most frequently in the second part of the duodenum, lacks follicular dendritic cell meshworks and has memory B-cell characteristics. However, its molecular pathogenesis is still unclear. In the present study, we examined 10 DFL, 18 NFL and 10 gastric MALT lymphoma samples using gene expression analysis. Quantitative RT-PCR experiments and immunohistochemical analysis for 72 formalin-fixed, paraffin-embedded tissues from an independent series, including 32 DFL, 19 gastric MALT lymphoma and 27 NFL samples, were performed for validation of microarray data. Gene expression profiles of the three lymphoma types were compared using 2918 differentially expressed genes (DEG) and results suggested that DFL shares characteristics of MALT lymphoma. Among these DEG, CCL20 and MAdCAM-1 were upregulated in DFL and MALT but downregulated in NFL. In contrast, protocadherin gamma subfamily genes were upregulated in DFL and NFL. Quantitative RT-PCR and immunohistochemical studies demonstrated concordant results. Double immunofluorescence studies revealed that CCL20 and CCR6 were co-expressed in both DFL and MALT. We hypothesize that increased expression of CCL20 and MAdCAM-1 and co-expression of CCL20 and CCR6 may play an important role in tumorigenesis.
Keywords: CCL20, duodenal follicular lymphoma, gene expression profile, MAdCAM-1, MALT lymphoma
Follicular lymphoma (FL) of the gastrointestinal tract, especially duodenal follicular lymphoma (DFL), is a rare variant of FL that is defined in the 2008 WHO classification system.1 At our institution, 136 of 957 (14.2%) FL diagnosed between 1989 and 2011 were gastrointestinal follicular lymphomas, suggesting that this entity may be more common in Japan than in other countries. The rarity of DFL, which accounts for only 1.0–3.6% of primary non-Hodgkin's lymphoma of the gastrointestinal tract, is a significant obstacle to the molecular investigation of this disease. We previously reported that DFL occurs most frequently in the second part of the duodenum, lacks activation-induced cytidine deaminase and follicular dendritic cell meshworks, has memory B-cell characteristics, and exhibits indolent clinical behavior as compared to nodal FL (NFL).2–5 Although the second part of the duodenum is the most frequent site for FL of the gastrointestinal tract, FL sometimes occurs in the gastrointestinal tract outside of the duodenum. Using double-balloon and/or capsule endoscopy, FL in the gastrointestinal tract primarily involve the duodenum and frequently spread throughout the small intestine. However, approximately 70% of FL are localized to the gastrointestinal tract or the adjacent lymph nodes.4
Various studies have investigated genetic variations contributing to the progression of FL. DFL harbors the t(14,18)(IGH-BCL2) translocation, which is also a common feature of NFL.3,6 While this translocation alone is considered necessary but insufficient to cause FL, it provides B cells with a survival advantage, allowing them to accumulate additional genomic alterations, host constitutional genetic variation, and survive in different microenvironments.7 The most common cytogenetic abnormalities in NFL are breakpoints in chromosome 1, deletions in the long arm of chromosome 6, trisomy 7, trisomy 12, presence of a derivative of chromosome 18, and duplication of chromosome X.8 Variations in the microenvironment, such as the follicular dendritic cell meshwork, macrophages and T-cell subsets, also play an important role in FL pathogenesis.9 Moreover, gene expression profiles (GEP) have been obtained for other low-grade B-cell lymphomas, such as FL, nodal marginal zone B-cell lymphoma and MALT lymphoma.10–12 However, the molecular pathogenesis of DFL is unknown, and no comprehensive studies investigating the contribution of gene expression to the pathogenesis of DFL have been published.
In the present study, we sought to clarify the molecular mechanisms of DFL pathogenesis using GEP.
Material and Methods
Patients and tissue samples
Fresh frozen tumor samples were obtained for 10 DFL, 18 NFL and 10 Helicobacter pylori-associated gastric MALT lymphomas. All DFL patients were clinical stage I or II1, and samples were positive for CD20, CD10 and BCL2 using immunohistochemistry. t(14;18/IGH-BCL2) translocations were detected by FISH analysis as previously described.3 Morphologic examination and CD20 immunostaining confirmed that the percentage of tumor cells exceeded 75% in all formalin-fixed, paraffin-embedded tissues (FFPET) tumor samples in this study. For comparison, 17 normal control samples were included; that is, 5 normal duodenal mucosa (NDU), 8 nodal reactive lymphoid hyperplasia (RLH) and 4 normal gastric mucosa (STCH) samples. Clinicopathological information is shown in Supplementary Tables S1–3. The clinical staging of NFL patients was determined according to Ann Arbor staging, and staging of DFL and MALT patients was determined according to the Lugano staging criteria.13
All patients were selected from routine and consultation files of the Department of Pathology, Okayama University Graduate School of Medicine, Dentistry and Pharmaceutical Sciences. Patient samples were diagnosed on the basis of morphology, immunophenotype and molecular findings according to the World Health Organization classification criteria. The study protocol was approved by the Institutional Review Board of Okayama University, Okayama, Japan, and written informed consent was obtained from each patient. All study procedures were conducted in accordance with the guidelines of the Declaration of Helsinki.
RNA isolation and microarray procedures
Total RNA was extracted from fresh frozen samples using an miRNeasy Mini Kit (Qiagen, Valencia, CA, USA). The quality of the RNA was examined by 1% agarose electrophoresis and analysis of the 28s/18s ratio using an Agilent Bioanalyzer. Poor-quality RNA samples were eliminated from further analyses. The RNA was reverse transcribed to double-stranded cDNA using a poly dT-T7 promoter primer. Complementary RNA (cRNA) were then synthesized from the cDNA products using T7 RNA polymerase with Cy3 labeled-CTP. Labeled cRNA were synthesized and then hybridized to Whole Human Genome Agilent 8× 60K commercial oligo-DNA microarrays (Agilent Technologies, Santa Clara, CA, USA) according to the manufacturer's instructions. All microarray data can be viewed at the Gene Expression Omnibus under accession number GSE48047.
Microarray data analyses
After subtracting background noise, the intensity data were normalized using a 75th-percentile normalization in genespring software version 11.0.2 (Agilent Technologies, Santa Clara, CA, USA). Differentially expressed genes (DEG) for each lymphoma were then selected as compared with normal tissue or control disease data (duodenum, stomach, and RLH for DFL, MALT and NFL, respectively), using Student's t-tests to determine significance (corrected P-values <0.05) and fold changes to determine variations in expression (twofold up/down). After extracting DEG, the three lymphoma types were compared using hierarchical sample clustering of the DEG by the Euclidean distance and the average linkage method (MultiExperiment Viewer [MeV] version 4.8.1).14
For functional analysis of DEG, upregulated and downregulated DEG were separately subjected to gene set enrichment analysis (GSEA) with Gene Ontology (GO) terms using GeneSpring software version 11.0.2. The P-value for each GO term was calculated as an enrichment score described by a standard hypergenometric distribution. The Benjamini Yekutieli correction was used for multiple testing corrections. A second DEG set of known function was then selected if the gene was classified in the “biological process” category of GO terms.
For pathway analysis, we used a freely available database, GenMAPP 2.1 (http://www.genmapp.org/). To define gene sets for GSEA, twofold upregulated/downregulated genes for each lymphoma type were first selected as compared with normal tissue or control disease data as described above, but without the use of statistical tests. We then compared twofold upregulated/downregulated genes between DFL, MALT and NFL. Intersections and complements between these three sets in Venn diagrams were then defined as gene sets for GSEA, generating seven sets of upregulated/downregulated genes. Hypergeometric tests were conducted using a satellite tool, MAPPFinder2, to identify the pathways in which genes in a certain category for DFL, MALT and/or NFL were enriched. We calculated a nonparametric statistic based on 2000 permutations of the data in which the gene associations were randomized for each sample, generating a distribution of Z scores for each pathway, which were then used to assign P-values (the permute P). Alternatively, the P-value for each pathway was calculated, and multiple testing corrections were conducted using the Westfall–Young adjustment.
To determine whether samples harbored batch effects (systematic nonbiological differences) that could be derived from different dates of RNA isolation, we performed a principal component analysis using GeneSpring software version 11.0.2.
Quantitative real-time RT-PCR analysis
Quantitative RT-PCR experiments were performed on selected genes using TaqMan probes (Applied Biosystems, Foster City, CA, USA) as previously described.10 The relative degree of change for each gene was calculated using the RQ = 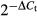 method (ΔCt = Ctgene – Ctbeta-actin), with beta-actin as the GEP endogenous control.
method (ΔCt = Ctgene – Ctbeta-actin), with beta-actin as the GEP endogenous control.
FISH
FISH for t(14;18)(q32;q21)/IGH-BCL2 translocations was performed using the LSI BCL2 FISH DNA fusion signal probe (Abbott Molecular, Wiesbaden, Germany) according to the manufacturer's instructions. We performed FISH directly on paraffin-embedded tissue sections and detected the hybridization signal as previously described.3,5
Immunohistochemical analysis
To validate the GEP of genes of interest selected from the microarray analyses (i.e. CCL20, MAdCAM-1, CCR6, and protocadherin gamma A3, A8 and B4), immunohistochemical studies were performed with 72 FFPET from an independent series, including 32 DFL, 19 gastric MALT lymphoma and 27 NFL samples. Heat-induced epitope retrieval or trypsin-induced retrieval, an avidin–biotin complex method, and an automated Bond-max autostainer (Leica Biosystems, Melbourne, Vic., Australia) were used for immunohistochemical staining, as described previously.5 For immunohistochemical evaluation, samples were scored as positive when 30% or more of lymphoma cells were positively stained. For MAdCAM-1, samples were positive when vascular endothelial cells were stained. The antibody panel used to assess these samples is shown in Supplementary Table S4.
Results
Gene expression profiles of the three tumor subtypes showed that duodenal follicular lymphoma and MALT lymphoma were closely related
We selected DEG from each lymphoma type (t-test corrected P-value <0.05 and twofold upregulated or downregulated; Suppl. Fig. S1). A total of 1078 downregulated genes and 1855 upregulated genes were selected. Thus, in total, 2918 downregulated and/or upregulated genes were selected as DEG (Suppl. Fig. S1c) and subjected to GEP analysis.
A hierarchical sample clustering using 2918 DEG showed that the three lymphoma types (DFL, MALT and NFL) and the normal control tissues (NDU and STCH) were divided into two branches, while the control disease tissues (RLH) belonged to the former branch (Fig.1). Nine of 10 DFL and 8 of 10 MALT clustered in the same branch, indicating that DFL and MALT were more closely related than DFL and NFL. A heatmap of the 2918 DEG is shown in Supplementary Figure S2.
Figure 1.
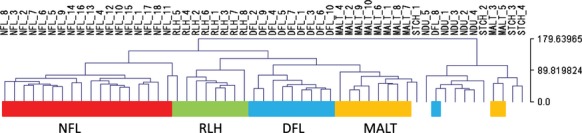
Hierarchical sample clustering was conducted using the 2918 differentially expressed genes (see Fig. S2). Although a few exceptions were observed, normal tissues (normal duodenal mucosa [NDU] and normal gastric mucosa [STCH]) were clustered together as one group, and the other group included lymphomas (duodenal follicular lymphoma [DFL], MALT and nodal follicular lymphoma [NFL]) and control disease samples (reactive lymphoid hyperplasia [RLH]). Clustering was conducted using MultiExperiment Viewer (MeV) version 4.8.1. The cluster for NFL is shown in red, RLH is in green, DFL is in blue, and MALT is in yellow. A clustering of the 2918 genes is shown in Supplementary Figure S3.
Gene ontology analysis
In order to summarize the biological features related to the transcriptomic alterations in lymphomas, 1708 downregulated and 1855 upregulated genes were separately subjected to GSEA with GO terms (Suppl. Table S5). GO terms representing immunity, such as “response to chemical stimulus” (corrected P-value, 1.015E-02), and, in another case, adhesion, such as “biological adhesion” (corrected P-value, 3.215E-02), were extracted for downregulated genes. For upregulated genes, GO terms related to immunity were listed in the top five as well. In subordinate positions, intriguingly, a term for “biological adhesion,” that is, “cell–cell adhesion” (corrected P-value, 3.837E-04), was listed (Suppl. Table S6). In the DFL versus NDU group, the top five upregulated genes were as follows: complement receptor 2, chemokine ligand 4 (CCL4), cannabinoid receptor 2 (macrophage), T-cell activation RhoGTPase activationg protein and interleukin 21 (IL-21). The top five downregulated genes were as follows: glutathione peroxidase 3, CDKN2B (p15), phospholipase A2 receptor 1, plasminogen activator and diacylglycerol lipase. In the NFL versus RLH group, the top five upregulated genes were protocadherin gamma subfamily B4, B3, B2, A5 and A3, and the top five downregulated genes were as follows: glucagon receptor, interferon-induced protein 44, discoidin domain receptor tyrosine kinase 1, claudin 3 and interferon alpha-inducible protein 6. In the MALT versus NDU group, the top five upregulated genes were as follows: Rho GTPase activating protein 17, Rap guanine nucleotide exchange factor 1, olfactory receptor family 2, transmembrane protein 40, and cholinergic receptor, nicotinic alpha 1. The top five downregulated genes were as follows: NADH dehydrogenase (ubiquinone) 1 alpha subcomplex 1, beta subcomplex 8, alpha subcomplex 12, alpha subcomplex 13 and succinate dehydrogenase complex. As described further below, protocadherin gamma subfamily genes (PCDHGB3, A5, B2, A3, B4, B1, A7, A9, A8, B6, B7, A12 and A2) are linked to “cell–cell adhesion.” In order to focus on genes of interest, we narrowed down the original 2918 DEG to a smaller set of DEG (i.e. the second DEG set). In this case, we selected 116 upregulated and 445 downregulated genes that were linked to GO terms in the “biological process” category (Fig.2a,b). We then examined whether these 116 upregulated and 445 downregulated genes retained a similar clustering pattern to that of the initial 2918 DEG; indeed, we obtained a dendrogram in which DFL and MALT were separated from NFL (Fig. S3).
Figure 2.
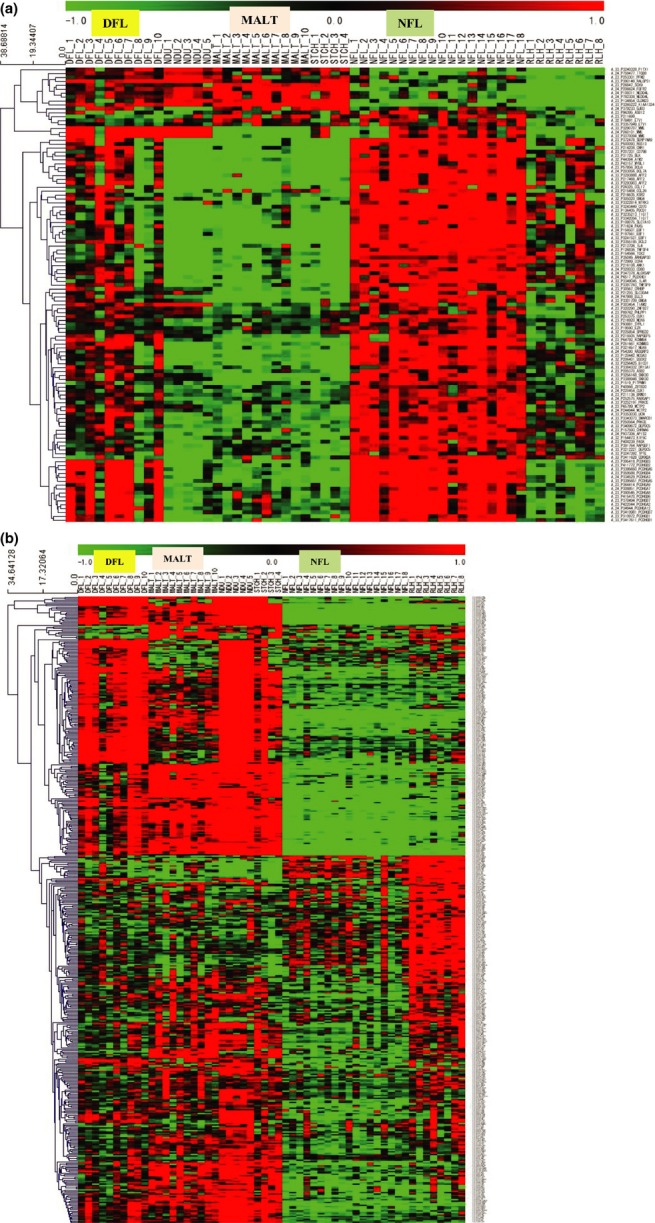
Hierarchical gene clustering using 116 upregulated genes (a) and 445 downregulated genes (b). Clustering was conducted using MultiExperiment Viewer (MeV) version 4.8.1.
Chemokines, interleukins and cell adhesion molecule signatures in the three lymphoma subtypes
Among the statistically significant DEG selected and described above, chemokine (C-C motif) ligand 20 (CCL20) and mucosal vascular addressin cell adhesion molecule 1 (MAdCAM-1) have been reported to be differentially expressed in several cancers,15–20 and, therefore, we focused on these genes in order to validate the expression profiles observed in the microarray analysis. Both CCL20 and MAdCAM-1 were upregulated in DFL and MALT, but downregulated in NFL in this study. We also focused on protocadherin gamma subfamily genes (PCDHGB3, A5, B2, A3, B4, B1, A7, A9, A8, B6, B7, A12 and A2), because these genes represented a prominent gene family in the present study and were upregulated in both DFL and NFL. We created a heatmap using CCL20, MAdCAM-1 and PCDHG family genes and lymphoma-related genes, as shown in Figure3. In this heatmap, we included not only DEG, but also the following non-DEG: IL2RG, ITGB2, MMP25, IL12RB1, TNFAIP8L2, TNFRSF8, ILDR2, CXCL10 and C3.
Figure 3.
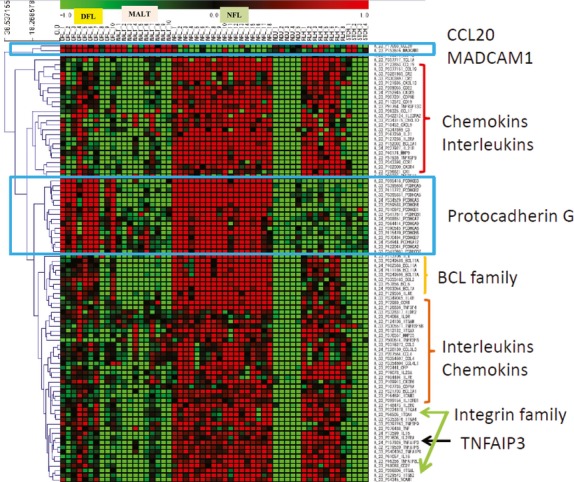
Heatmap of genes of interest, including 84 differentially expressed genes (DEG) and 9 non-DEG: IL2RG, ITGB2, MMP25, IL12RB1, TNFAIP8L2, TNFRSF8, ILDR2, CXCL10 and C3.
Validation of microarray data
The set of genes selected for validation by quantitative RT-PCR and immunohistochemistry included DEG, CCL20 (that was common in DFL and MALT, with opposite expression in NFL) and MAdCAM-1 (common in DFL and NFL, with opposite expression in MALT). Quantitative RT-PCR analysis showed significantly higher expression of CCL20 and MAdCAM-1 in DFL and MALT versus NFL. Coefficients of correlation with the microarray were 0.9245 for CCL20 and 0.7934 for MAdCAM-1 (Fig.4a,b). MAdCAM-1 was slightly upregulated in 5 of 18 NFL samples. These samples were all abdominal or mesenteric lymph nodes.
Figure 4.
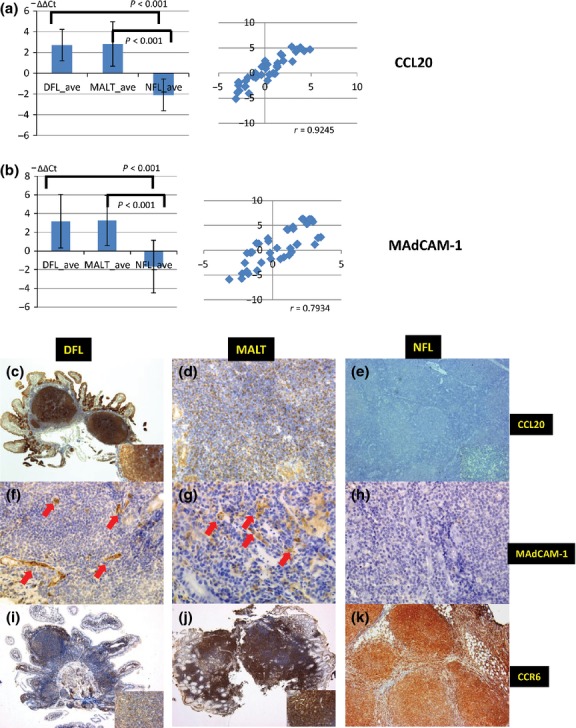
Validation of microarray data by quantitative RT-PCR and immunohistochemical studies. (a, b) Quantitative RT-PCR for CCL20 and MAdCAM-1, respectively. duodenal follicular lymphoma (DFL) and MALT showed higher expression levels of CCL20 and MAdCAM-1 than nodal follicular lymphoma (NFL) (P < 0.001). The coefficient of correlation with the microarray data was 0.925 for CCL20 and 0.7934 for MAdCAM-1. (c–k) Immunohistochemical results for CCL20, MAdCAM-1 and CCR6 in DFL, MALT and NFL. DFL and MALT were positive for CCL20 (c) and (d), while NFL was negative for CCL20 (e). Vascular endothelial cells were highly expressed in DFL and MALT (f) and (g), but not in NFL (h). DFL (i), MALT (j) and NFL (k) were positive for CCR6.
Immunohistochemical studies revealed that CCL20 protein expression was significantly higher in DFL and MALT samples than in NFL samples (P < 0.001). CCL20 was positive in 97% (31/32) of DFL samples and 89% (17/19) of MALT samples, but only 4% (1/27) of NFL samples (Fig.4c–e). MAdCAM-1 expression, present in vascular endothelial cells, was more frequently expressed in DFL (28/32, 88%) and MALT (18/19, 95%) samples than in NFL samples (5/27, 19%; Fig.4f–h). This difference was also statistically significant (P < 0.001). In contrast, the expression of PCDHGA8, A3 and B4 was significantly higher in DFL and NFL than in MALT (Fig.5a–i). A summary of the immunohistochemical results is shown in Table1.
Figure 5.
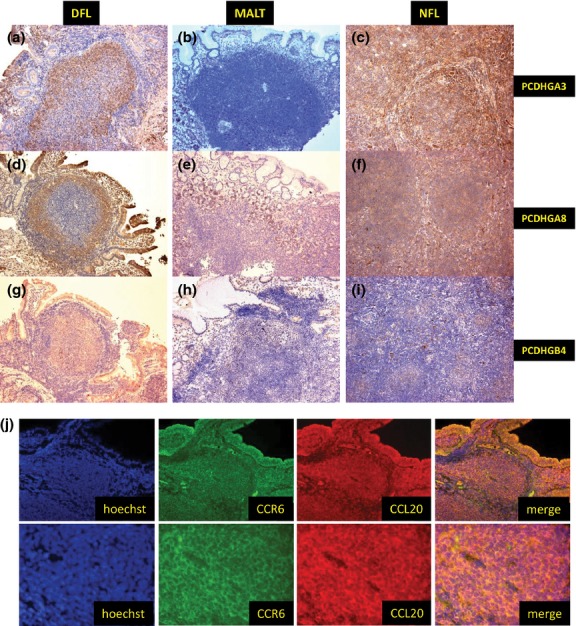
Validation of microarray data and double immunohistochemical staining for CCL20 and CCR6. (a–i) Immunohistochemical results for PCDHGA3, PCDHGA8 and PCDHGB4. Duodenal follicular lymphoma (DFL) and nodal follicular lymphoma (NFL) were positive for PCDHGA3 (a) and (c), PCDHGA8 (d) and (f), and PCDHGB4 (g) and (i), while MALT was negative for all three markers (b), (e) and (h). (j) Double immunohistochemical staining for CCL20 and CCR6 in DFL. Tumor cells co-expressed CCL20 and CCR6.
Table 1.
Summary of immunohitochemical results in DFL, MALT and NFL
| Antibodies | DFL | MALT | NFL | P (DFL, MALT vs NFL) | P (DFL, NFL vs MALT) |
|---|---|---|---|---|---|
| CCL20 | 31/32 (97.0) | 17/19 (89.0) | 1/27 (3.7) | <0.001 | NS |
| MAdCAM-1 | 28/32 (87.5) | 18/19 (94.7) | 5/27 (18.5) | <0.001 | NS |
| CCR6 | 28/32 (87.5) | 19/19 (100) | 14/27 (51.9) | 0.002 | NS |
| PCDHGA3 | 29/32 (90.6) | 1/19 (5.3) | 27/27 (100) | NS | <0.001 |
| PCDHGA8 | 28/32 (87.5) | 2/19 (10.5) | 27/27 (100) | NS | <0.001 |
| PCDHGB4 | 26/32 (81.2) | 2/19 (10.5) | 27/27 (100) | NS | <0.001 |
DFL, duodenal follicular lymphoma; NFL, nodal follicular lymphoma; NS, not significant.
CCL20 and CCR6 co-expression in duodenal follicular lymphoma and MALT
CCR6 is a receptor for CCL20, and human CCR6 expression was originally demonstrated in dendritic cells, spleen, thymus, small intestine and peripheral blood leukocytes.21,22 We also examined CCR6 expression using immunohistochemistry. DFL (28/32, 88%) and MALT (19/19, 100%) showed strong expression of CCR6, while only approximately half of NFL samples (14/27, 52%) were positive for CCR6 (Fig.4i–k). Figure5(j) shows double immunohistochemical staining for CCL20 and CCR6 in DFL. Tumor cells in the follicle and duodenal villi co-expressed CCL20 and CCR6.
Discussion
We have previously reported that DFL shares some common characteristics with MALT lymphoma, such as immunoglobulin heavy chain usage, lack of follicular dendritic cell meshwork and memory B-cell characteristics.3,5 In the present study, clustering of the three tumor subtypes revealed that most of the DFL and MALT cases were clearly classified into the same GEP groups. Our previous study was only focused on the altered expression of a limited number of targets between NFL and DFL. However, from the present comprehensive study, hierarchical clustering data supported our previous data.
CCL20 (macrophage inflammatory protein-3α) is a recently described C-C chemokine identified by screening the GenBank database for expressed sequence tags for novel chemokine molecules.23 Its only known receptor, CCR6, was originally shown to be expressed in dendritic cells, the spleen, the thymus, the small intestine and the appendix.21 In the present study, CCL20 was strongly expressed in DFL and MALT lymphoma, and CCR6 was also frequently detected in DFL and MALT lymphoma. In a previous report, CCL20 and CCR6 autocrine and paracrine mechanisms were shown to play an important role in tumorigenesis in several types of cancer, such as pancreatic cancer and breast cancer.16,18 In the present study, CCL20 and CCR6 co-expression in tumor cells frequently occurred in DFL and MALT lymphoma. Therefore, CCL20 and CCR6 autocrine and paracrine mechanisms likely play an important role in the tumorigenesis of DFL and MALT lymphoma. Moreover, gastrointestinal FL occurs most frequently within the second part of the duodenum, as we reported previously.4,24 However, it is still unclear why the second part of the duodenum is the most involved site. Interestingly, CCL20 is selectively induced in the duodenum in a mouse model of sepsis.25 We speculate that chronic inflammation induced by CCL20 and CCR6 may be associated with the high frequency of gastrointestinal FL at the duodenum.
MAdCAM-1 is expressed on high endothelial venules in gut-associated lymphoid tissues and was shown to be overexpressed in DFL and MALT lymphoma in the present study. These results suggest that DFL may possess a common pathway of tumorigenesis and that MAdCAM-1 overexpression may be associated with the tendency of this tumor type to localize to the gastrointestinal tract.26,27
From analysis of other molecules, the PCDHG subfamily represents another important target. PCDH proteins are predominantly expressed in the nervous system and constitute the largest subgroup within the cadherin superfamily, and these proteins are required for normal development of the nervous system.28 In previous reports investigating non-small cell lung cancer, breast cancer, gastric cancer, Wilms' tumor and colorectal cancer, PCDH genes, and PCDHG subfamily genes in particular, were frequently silenced by epigenetic mechanisms.29–33 However, in the present study, PCDHG gene clusters were highly expressed in DFL and NFL tumor cells, and to the best of our knowledge, no reports have demonstrated that PCDHG family proteins are overexpressed in malignant lymphoma. Although further studies are required, our data suggest that PCDHG subfamilies may play an important role in lymphomagenesis.
In conclusion, our study demonstrated that DFL shared biological characteristics of both MALT lymphoma and NFL, but was more similar to MALT lymphoma than to NFL. Using an orthogonal technology we demonstrated increased expression of CCL20, and MAdCAM-1 distinguished DFL and MALT lymphoma from NFL and may play an important role in tumor localization within the duodenum. CCL20-CCR6 co-expression may also be involved in DFL tumorigenesis. Furthermore, our results suggest that an inflammatory background and antigen stimulation may underlie the pathogenesis of DFL.
Acknowledgments
This work was supported by a grant from the COE finance of Okayama University and the Japan Society for the Promotion Science (JSPS no. 19590348 and 24790350).
Disclosure Statement
The authors have no conflict of interest.
Funding information
COE finance of Okayama University. Japan Society for the Promotion Science (JSPS 19590348 and 24790350).
Supporting Information
Additional supporting information may be found in the online version of this article:
Venn-diagram of upregulated or downregulated genes/probes.
Fig. S2 Hierarchical sample and gene clustering for 2918 differentially expressed genes.
Fig. S3 Hierarchical sample clustering using 116 upregulated genes and 445 downregulated genes.
Table S1 Clinical characteristics of DFL patients
Table S2 Summary of clinicopathology in MALT lymphoma patients
Table S3 Summary of clinicopathology in NFL patients
Table S4 Antibody panel for immunohistochemical study
Table S5 The top five gene ontology terms in downregulated or upregulated genes
Table S6 GO related to downregulated genes
References
- 1.Harris NL, Swerdlow SH, Jaffe ES. Follicular lymphoma. In: Harris NL, Swerdlow SH, Jaffe ES, Ott G, Nathwani BN, de Jong D, Yoshino T, Spagnolo D, et al., editors. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. 4th. Lyon: IARC; 2008. pp. 220–6. [Google Scholar]
- 2.Yoshino T, Miyake K, Ichimura K, et al. Increased incidence of follicular lymphoma in the duodenum. Am J Surg Pathol. 2000;24:688–93. doi: 10.1097/00000478-200005000-00007. [DOI] [PubMed] [Google Scholar]
- 3.Takata K, Sato Y, Nakamura N, et al. Duodenal and nodal follicular lymphomas are distinct: the former lacks activation-induced cytidine deaminase and follicular dendritic cells despite ongoing somatic hypermutations. Mod Pathol. 2009;22:940–9. doi: 10.1038/modpathol.2009.51. [DOI] [PubMed] [Google Scholar]
- 4.Takata K, Okada H, Ohmiya N, et al. Primary gastrointestinal follicular lymphoma involving the duodenal second portion is a distinct entity: a multicenter, retrospective analysis in Japan. Cancer Sci. 2011;102:1532–6. doi: 10.1111/j.1349-7006.2011.01980.x. [DOI] [PubMed] [Google Scholar]
- 5.Takata K, Sato Y, Nakamura N, et al. Follicular lymphoma lacks AID but express BACH2 and has memory B cell characteristics. Mod Pathol. 2013;26:22–31. doi: 10.1038/modpathol.2012.127. [DOI] [PubMed] [Google Scholar]
- 6.Shia J, Teruya-Feldstein J, Pan D, et al. Primary follicular lymphoma of the gastrointestinal tract: a clinical and pathologic study of 26 cases. Am J Surg Pathol. 2002;26:216–24. doi: 10.1097/00000478-200202000-00008. [DOI] [PubMed] [Google Scholar]
- 7.Relander T, Johnson NA, Farinha P, Connors JM, Sehn LH, Gascoyne RD. Prognostic factors in follicular lymphoma. J Clin Oncol. 2010;28:2902–13. doi: 10.1200/JCO.2009.26.1693. [DOI] [PubMed] [Google Scholar]
- 8.Johansson B, Mertens F, Mielman F. Cytogenetic evolution patterns in non-Hodgkin's lymphoma. Blood. 1995;86:3905–14. [PubMed] [Google Scholar]
- 9.Glas AM, Knoops L, Delahaye L, et al. Gene expression and immunohistochemical study of specific T-cell subsets and accessory cell types in the transformation and prognosis of follicular lymphoma. J Clin Oncol. 2007;25:390–8. doi: 10.1200/JCO.2006.06.1648. [DOI] [PubMed] [Google Scholar]
- 10.Arribas AJ, Martin YC, Abad CG, et al. Nodal marginal zone lymphoma: gene expression and miRNA profiling identify diagnostic markers and potential therapeutic targets. Blood. 2012;119:e9–21. doi: 10.1182/blood-2011-02-339556. [DOI] [PubMed] [Google Scholar]
- 11.Glas AM, Kersten MJ, Delahaye LJ, et al. Gene expression profiling in follicular lymphoma to assess clinical aggressiveness and to guide the choice of treatment. Blood. 2005;105:301–7. doi: 10.1182/blood-2004-06-2298. [DOI] [PubMed] [Google Scholar]
- 12.O'Rourke JL. Gene expression profiling in Helicobacter-induced MALT lymphoma with reference to antigen drive and protective immunization. J Gastroenterol Hepatol. 2008;23:S151–6. doi: 10.1111/j.1440-1746.2008.05553.x. [DOI] [PubMed] [Google Scholar]
- 13.Rohatiner A, d'Amore F, Coiffier B, et al. Report on a workshop convened to discuss the pathological and staging classifications of gastrointestinal tract lymphoma. Ann Oncol. 1994;5:397–400. doi: 10.1093/oxfordjournals.annonc.a058869. [DOI] [PubMed] [Google Scholar]
- 14.Saeed AI, Sharov V, White J, et al. TM4: a free open-source system for microarray data management and analysis. Biotechniques. 2003;34:374–8. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- 15.Durig J, Schmuker U, Duhrsen U. Differential expression of chemokine receptors in B cell malignancies. Leukemia. 2001;15:752–6. doi: 10.1038/sj.leu.2402107. [DOI] [PubMed] [Google Scholar]
- 16.Bell D, Chomarat P, Broyles D, et al. In breast carcinoma tissue, immature dendritic cells reside within the tumor, whereas mature dendritic cells are located in peritumoral areas. J Exp Med. 1999;190:1417–26. doi: 10.1084/jem.190.10.1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kimura T, Takeshima H, Nomiyama N, et al. Expression of lymphocyte-specific chemokines in human malignant glioma: essential role of LARK in cellular immunity of malignant glioma. Int J Oncol. 2002;21:707–15. doi: 10.3892/ijo.21.4.707. [DOI] [PubMed] [Google Scholar]
- 18.Kleff J, Kusama T, Rossi DL, et al. Detection and localization of MIP-3α/LARC/Exodus, a macrophage proinflammatory chemokine, and its CCR6 receptor in human pancreatic cancer. Int J Cancer. 1999;81:650–7. doi: 10.1002/(sici)1097-0215(19990517)81:4<650::aid-ijc23>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- 19.Scarpino S, Stoppaciaro A, Ballerini F, et al. Papillary carcinoma of the thyroid: hepatocyte growth factor (HGF) stimulates tumor cells to release chemokines active in recruiting dendritic cells. Am J Pathol. 2000;156:831–7. doi: 10.1016/S0002-9440(10)64951-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shimizu Y, Murata H, Kashii Y, et al. CC-chemokine receptor 6 and its ligand macrophage inflammatory protein 3α might be involved in the amplification of local necroinflammatory response in the liver. Hepatology. 2001;34:311–9. doi: 10.1053/jhep.2001.26631. [DOI] [PubMed] [Google Scholar]
- 21.Graves DR, Wang W, Dairaghi DJ, et al. CCR6, a CC chemokine receptor that interacts with macrophage inflammatory protein 3α and is highly expressed in human dendritic cells. J Exp Med. 1997;186:837–44. doi: 10.1084/jem.186.6.837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baba M, Imai T, Nishimura M, et al. Identification of CCR6, the specific receptor for the CC chemokine LARC. J Biol Chem. 1997;272:14893–8. doi: 10.1074/jbc.272.23.14893. [DOI] [PubMed] [Google Scholar]
- 23.Iwasaki A, Kelsall BL. Localization of distinct Peyer's patch dendritic cell subsets and their recruitment by chemokines macrophage inflammatory protein (MIP)-3α, MIP-3β, and secondary lymphoid organ chemokine. J Exp Med. 2000;17:1381–93. doi: 10.1084/jem.191.8.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schmats AI, Streubel B, Kretschmer-Chott E, et al. Primary follicular lymphoma of the duodenum is a distinct mucosal/submucosal variant of follicular lymphoma: a retrospective study of 63 cases. J Clin Oncol. 2011;29:1445–51. doi: 10.1200/JCO.2010.32.9193. [DOI] [PubMed] [Google Scholar]
- 25.Esplugues E, Huber S, Gagliani N, et al. Control of TH17 cells occurs in the small intestine. Nature. 2011;475:514–8. doi: 10.1038/nature10228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu YX, Yoshino T, Ohara N, et al. Loss of expression of α4β7 integrin and L-selection is associated with high-grade progression of low-grade MALT lymphoma. Mod Pathol. 2001;14:798–805. doi: 10.1038/modpathol.3880393. [DOI] [PubMed] [Google Scholar]
- 27.Bende RJ, Smit LA, Bossenbroek JG, et al. Primary follicular lymphoma of the small intestine: alpha4beta7 expression and immunoglobulin configuration suggest an origin from local antigen-experienced B cells. Am J Pathol. 2003;162:105–13. doi: 10.1016/s0002-9440(10)63802-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen WV, Alvarez FJ, Lefebvre JL, et al. Funtional significance of isoform diversification in the protocadherin gamma gene cluster. Neuron. 2012;75:402–9. doi: 10.1016/j.neuron.2012.06.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Imoto I, Izumi H, Yokoi S, et al. Frequent silencing of the candidate tumor suppressor PCDH20 by epigenetic mechanism in non-small-cell lung cancers. Cancer Res. 2006;66:4617–26. doi: 10.1158/0008-5472.CAN-05-4437. [DOI] [PubMed] [Google Scholar]
- 30.Novak P, Jansen T, Oshiro MM, Watts GS, Kim CJ, Futscher BW. Agglomerative epigenetic aberrations are a common event in human breast cancer. Cancer Res. 2008;68:8616–25. doi: 10.1158/0008-5472.CAN-08-1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dallosso AR, Hancock AL, Szemes M, et al. Frequent long-range epigenetic silencing of protocadherin gene clusters on chromosome 5q31 in Wilms' tumor. PLoS Genet. 2009;5:e1000745. doi: 10.1371/journal.pgen.1000745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dallosso AR, Oster B, Greenhough A, et al. Long-range epigenetic silencing of chromosome 5q31 protocadherins is involved in early and late stages of colorectal tumorigenesis through modulation of oncogenic pathways. Oncogene. 2012;31:4409–19. doi: 10.1038/onc.2011.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang Y, Liu J, Li X, Li JC. PCDH17 gene promoter demethylation and cell cycle arrest by genistein in gastric cancer. Histol Histopathol. 2012;27:217–24. doi: 10.14670/HH-27.217. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Venn-diagram of upregulated or downregulated genes/probes.
Fig. S2 Hierarchical sample and gene clustering for 2918 differentially expressed genes.
Fig. S3 Hierarchical sample clustering using 116 upregulated genes and 445 downregulated genes.
Table S1 Clinical characteristics of DFL patients
Table S2 Summary of clinicopathology in MALT lymphoma patients
Table S3 Summary of clinicopathology in NFL patients
Table S4 Antibody panel for immunohistochemical study
Table S5 The top five gene ontology terms in downregulated or upregulated genes
Table S6 GO related to downregulated genes


