Abstract
Understanding the developmental relationship between indolent and aggressive tumors is central to understanding disease progression and making treatment decisions. For example, most men diagnosed with prostate cancer have clinically indolent disease and die from other causes. Overtreatment of prostate cancer remains a concern. Here we use laser microdissection followed by exome sequencing of low- and high-grade prostate cancer foci from four subjects, and metastatic disease from two of those subjects, to evaluate the molecular relationship of coincident cancer foci. Seventy of 79 (87%) high-confidence somatic mutations in low-grade disease were private to low-grade foci. In contrast, high-grade foci and metastases harbored many of the same mutations. In cases in which there was a metastatic focus, 15 of 80 (19%) high-confidence somatic mutations in high-grade foci were private. Seven of the 80 (9%) were shared with low-grade foci and 65 (82%) were shared with metastatic foci. Notably, mutations in cancer-associated genes and the p53 signaling pathway were found exclusively in high-grade foci and metastases. The pattern of mutations is consistent with early divergence between low- and high-grade foci and late divergence between high-grade foci and metastases. These data provide insights into the development of high-grade and metastatic prostate cancer.
Keywords: Carcinoma/genetics, exomes, genetic heterogeneity, multiple primary neoplasms, prostatic neoplasms
Cancers are thought to progress from low grade to higher grade,(1) leading to efforts to detect and eradicate low-grade disease in order to prevent morbidity and mortality from high-grade disease. The grade of prostate cancer is characterized by the Gleason score on a scale of 2–10, with 10 representing the most poorly differentiated tumors. Although initially described as intermediate grade, Gleason 6 cancers are clinically indolent and have an overwhelmingly favorable prognosis.(2,3) For the purposes of this report, Gleason 6 foci are regarded as low grade.
Approximately 238 590 men were diagnosed with prostate cancer in the US in 2013, the majority of them with low-grade disease. It is estimated that 29 720 men will die from prostate cancer, which is among the lowest case/fatality ratio of any cancer.(4) Although prostate specific antigen (PSA) screening is able to identify early stage prostate cancer, most men diagnosed with prostate cancer die from competing causes. It is therefore unclear whether early detection associated with PSA screening decreases mortality.(5,6) Current clinical treatment guidelines support active surveillance for patients with low-grade prostate cancer for which the risk of tumor progression is low.(7) Recently some guidelines have even recommended against PSA screening.(8) Failure to follow these guidelines is likely due, in part, to concern that low-grade foci progress to higher grade and metastatic disease.
Several studies have contributed to a growing catalog of genetic variation in prostate cancer,(9)– (14) giving insight into single nucleotide variations, rearrangements and copy number variation prevalent in this disease. Aside from common rearrangements involving the ETS family of transcription factors, especially v-ets avian erythroblastosis virus E26 oncogene homolog (ERG), the mutational spectrum of prostate cancers is diverse.(10) Although increasing attention is being paid to the overall molecular heterogeneity of cancers within a single patient, there is limited data in this regard specifically for prostate cancer. This is especially relevant for prostate cancer, which is often multifocal.
More specifically, it is not clear if prostate cancer develops as a multifocal disease in which the foci are clonally related and share the vast majority of somatic mutations, or if it is a multicentric disease, with each focus having its own set of private somatic mutations and each representing an independent primary lesion. Recent studies have examined multifocal disease and found evidence of a monoclonal origin of coincident foci,(15) even when the foci have different Gleason patterns.(16,17) This conclusion is somewhat surprising, given the difference in clinical behavior of different Gleason patterns.
The present study targets multifocal disease, specifically with low-grade (Gleason 6) and high-grade (Gleason 8 or higher) foci, and compares the molecular relationship of these foci with each other and with synchronous metastatic disease. To expand the number of specimens available for the present study, prostatectomy samples that had been formalin fixed and paraffin embedded (FFPE) were used, with modifications made to standard protocols to increase the yield of material used for sequencing. Exome sequencing was used to achieve a comprehensive evaluation of all genes harboring somatic point mutations. The findings indicate that there is early divergence or complete independence of low-grade and high-grade disease, but late divergence of high-grade disease and synchronous metastases.
Materials and Methods
Tissue and laser capture microdissection (LMD)
Paraffin-embedded prostate and lymph node tissue that was 1–5 years old and matched FFPE and frozen normal prostate tissue was obtained from the Human Tissue Resource Center (HTRC) at the University of Chicago with the approval of the Internal Review Board. Tissue was reviewed by a genitourinary pathologist (J.B.T.) and regions of interest were marked for further evaluation. Four specimens had sufficient low- and high-grade disease for LMD. Two specimens (PrCa 6 and PrCa 18) also had sufficient lymph node metastatic disease. Tumor foci and histologically normal prostate were microdissected from 10-μm sections using a Leica Laser Microdissection (Wetzlar, Germany) system.
Exome library preparation
Genomic FFPE DNA was isolated using a QIAamp DNA FFPE Tissue Kit (Qiagen, Valencia, CA, USA) following the manufacturer's instructions, but increasing the incubation in Proteinase K to overnight and reducing the 90°C incubation to 30 min. DNA from frozen tissue was isolated using a QIAamp Mini Kit (Qiagen). DNA was fragmented using a Covaris S2 sonicator (Woburn, MA, USA), with settings of 10% duty cycle, intensity 4, 200 cycles per burst and 120 s. The fragmented DNA was blunt-end repaired using T4 Polymerase, Klenow and T4 PNK, and adenylated. Fragments were ligated to adaptors IS1_adapter_P5.F and IS3_adapter_P5 + P7.R(18) and an index tag added. The product was amplified and cleaned up using Agencourt AMPure XP beads (Beckman Coulter, Indianapolis, IN, USA).
Libraries were run on a BioAnalyzer (Agilent, Santa Clara, CA, USA) to confirm fragment size. The libraries from the metastases did not undergo further size selection. The remaining libraries underwent selection using an E-gel (Invitrogen, Grand Island, NY, USA) followed by PCR amplification.
Enrichment for exomes was performed using NimbleGen SeqCap EZExome v2.0 (Roche, Madison, WI, USA) following the manufacturer's instructions. The exome-enriched library DNA was amplified and run on a BioAnalyzer to confirm fragment size. The resulting libraries were run on a HiSeq 2000 (Illumina, San Diego, CA, USA) in a highly multiplexed 1 × 50 run and a subsequent 2 × 100 run.
Sequencing analysis and identification of somatic mutations
Overlapping or adapter-containing reads were clipped using SeqPrep-b83fd00. Reads were quality trimmed and aligned to hg18 using bwa 0.5.9(19) and merged with duplicates removed using Samtools 0.1.18.(20) Genome Analysis Toolkit(21) was used for local realignment and quality score recalibration. Somatic mutations were identified using VarScan v2.2.8.(22) High-confidence somatic mutations were defined as variants with coverage of at least 10×, no supporting reads in normal, at least four supporting reads in the tumor and with supporting reads on both strands. Germline single-nucleotide polymorphisms (SNPs) identified in dbSNP135, 1000 genomes and exomes of normal controls published by the Max Planck Institute (MPI)(23)– (25) were removed. SAMtools was used to determine the number of total and variant reads in matched libraries for positions mutated in low-grade foci. Sample ethnicity was determined by principle component analysis using common, germline, coding variants shared with samples from the 1000 genomes project.(24)
Custom capture and identification of previously identified mutations
DNA from seven additional foci from PrCa 6 were isolated using LMD (Leica Microsystems LMD6500). Custom capture probes targeting high-confidence mutations identified through exome sequencing were obtained from Roche. Sequencing libraries from the seven loci were prepared using the High-Throughput Genome Analysis Core at the University of Chicago, pooled, captured following the manufacturer's instructions, and sequenced on one lane of a HiSeq2000 (Illumina). Reads were analyzed with SeqPrep-b83fd00, aligned to hg18 using bwa, and merged with duplicates removed using Samtools 0.1.18. Samtools was used to determine the number of total and variant reads in each library for each variant position. Variants were considered present if >10% of reads supported the variant.
Confirmation of somatic mutations and immunohistochemisty (IHC)
Mutations were visualized using IGV 2.0.(26) Genomic DNA from each focus underwent amplification using GenomePlex WGA2 (Sigma, St. Louis, MO, USA) and primers targeting a subset of variants were designed using Primer3Plus and obtained from IDT (Coralville, IA, USA). The PCR products were sequenced in the University of Chicago DNA Sequencing and Genotyping Facility. ERG IHC was performed at the HTRC using anti-ERG antibody EPR3864 (Abcam, Cambridge, MA, USA).
Results
To evaluate the molecular relationship between low- and high-grade prostate cancer, we identified multifocal formalin-fixed, paraffin-embedded specimens with both low-grade and high-grade disease with sufficient mass for laser capture microdissection. The subjects from whom the specimens came were representative of patients with high-risk localized prostate cancer (Table 1).
Table 1.
Subject characteristics
| Case | Patient age (years) | Race | Stage | Gleason – gland† | Library | Gleason – focus‡ |
|---|---|---|---|---|---|---|
| PrCa 18 | 65 | Caucasian | pT3bN1Mx | 4 + 3 | Normal | NA |
| Low grade | 6 | |||||
| High grade | 8 | |||||
| Metastasis | NA | |||||
| PrCa 14 | 54 | African–American | pT3bN1Mx | 4 + 3 | Normal | NA |
| Low grade | 6 | |||||
| High grade | 8 | |||||
| PrCa 6 | 70 | Caucasian | pT3bN1Mx | 4 + 3 (5) | Normal | NA |
| Low grade | 6 | |||||
| High grade | 8 | |||||
| Metastasis | NA | |||||
| PrCa 25 | 59 | Asian | pT2cN0Mx | 3 + 4 | Normal | NA |
| Low grade | 6 | |||||
| High grade | 9 |
Gleason score assigned on pathological review of the prostatectomy specimen.
Gleason score of the focus isolated by microdissection. PrCa 6 had a Gleason score of 4 + 3 with a minor component of 5. PrCa 14 had metastatic disease in a lymph node, which was not of sufficient mass to be isolated for sequencing. NA, not applicable.
Exome sequencing was performed on matched cancer foci and histologically normal prostate glands from four specimens (Fig. 1). To improve the yield of DNA for downstream sequencing, proteinase K digestion was increased to 18 h, 90° incubation was reduced to 30 min and size selection by gel extraction was replaced with size selection with an E-gel or eliminated altogether. On average, 88% of RefSeq coding bases were covered at 10× or greater. A mean of 30.1 somatic coding mutations was identified per cancer focus (Supporting Information Table S1).
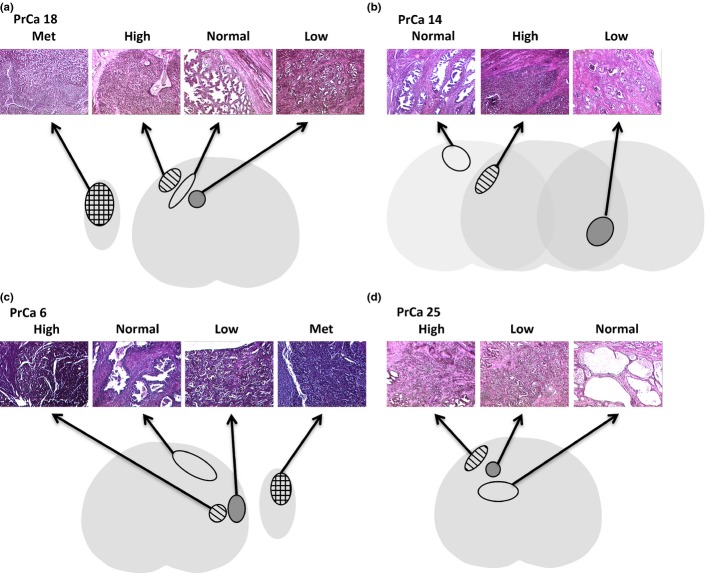
Fig. 1.
Histology of coincident prostate cancer foci. Representative H&E stains and illustrations representing the prostate cancer foci that were laser microdissected. Two cancer foci and uninvolved prostate glands were isolated from PrCa 18 (a), PrCa 14 (b), PrCa 6 (c) and PrCa 25 (d). In addition, a metastatic (Met) focus was isolated from PrCa 18 and PrCa 6. For PrCa 14 (b), the foci were from different levels of the prostate. For the other three specimens the foci were at the same level. Light gray, histologically normal prostate; dark gray, low-grade cancer focus; striped, high-grade cancer focus; checked, metastatic focus from a lymph node removed at the time of prostatectomy.
To evaluate potential bias introduced by formalin fixation, matched FFPE and frozen (Optimal Cutting Temperature compound [OCT]-embedded) samples of normal prostate were subjected to exome sequencing. The number of false positive variant reads was similar (Fig. S1A), as were the types of mutations (Fig. S1B), suggesting that there was little bias introduced from formalin fixation.
Three hundred and one somatic mutations were identified across 10 foci (Table S2). Of these, 71 were synonymous and unlikely to alter gene function (Fig. 2a). Of 174 unique genes harboring somatic mutations that were likely to be functional, 73 were not reported in five previous reports of mutations in localized and advanced disease.(9)– (12,14) The majority of variants identified were transitions leading to nonsynonymous mutations (Fig. 2b,c), as expected. A subset of 20 high-confidence somatic mutations was confirmed in tumor and uninvolved tissue with capillary sequencing (Fig. 2d). Those that were not confirmed by capillary sequencing had low variant allele frequency and were approximately the level of detection for capillary sequencing (˜20%). Nevertheless, the mutation (0.82/Mb) and validation rate (85%) using fixed prostate specimens in the present study are similar to previous reports (0.9/Mb and 91%, respectively) using frozen tissue.(9) The high fidelity of this sequencing supports the use of formalin-fixed tissue for next-generation sequencing studies.
Fig. 2.
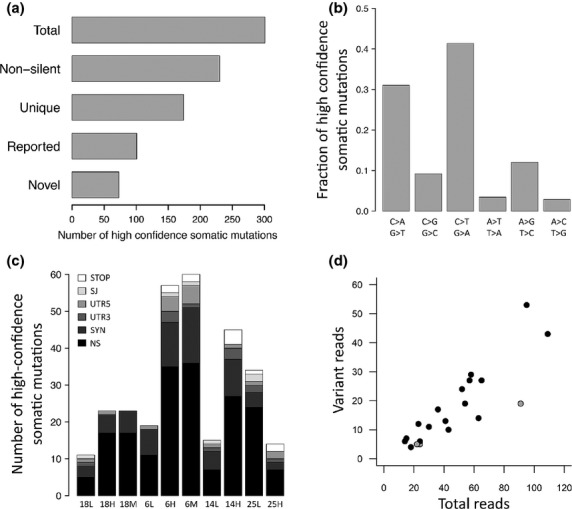
Mutation characteristics. (a) Number of high-confidence somatic mutations across all foci. Non-silent, non-silent mutations; Unique, number of unique genes harboring a non-silent mutation; Reported, gene reported to be mutated in references 9–12 and 14. (b) Spectrum of unique high confidence somatic mutations across all foci. (c) Number of high-confidence somatic mutation types in all prostate samples. L, low-grade focus; H, high-grade focus; M, metastatic focus; NS, nonsynonymous; SYN, synonymous; UTR3, 3′ untranslated region; UTR5, 5′ untranslated region; SJ, splice junction; STOP, premature stop codon. (d) Confirmation of high-confidence somatic mutations with capillary sequencing by number of total and variant reads (gray, not confirmed; black, confirmed). Confirmation rate, 0.85.
The vast majority of somatic mutations in low- and high-grade foci were private to the focus in which they were identified. Nine of 79 (11%) high-confidence somatic mutations in the low-grade foci were shared with a high-grade focus, and nine of 139 (7%) high-confidence somatic mutations in the high-grade foci were shared with a low-grade focus (Fig. 3). Capillary sequencing confirmed that these nine shared somatic mutations were not germline variants. In two subjects (14 and 18) all of the mutations were private. In the other two subjects (6 and 25) a small minority of mutations was shared. In contrast to the low-grade foci, 67 of 80 (84%) high-confidence somatic mutations identified in high-grade foci were shared with metastatic disease and metastatic foci had few mutations beyond those identified in high-grade disease (Fig. 3a,c).
Fig. 3.
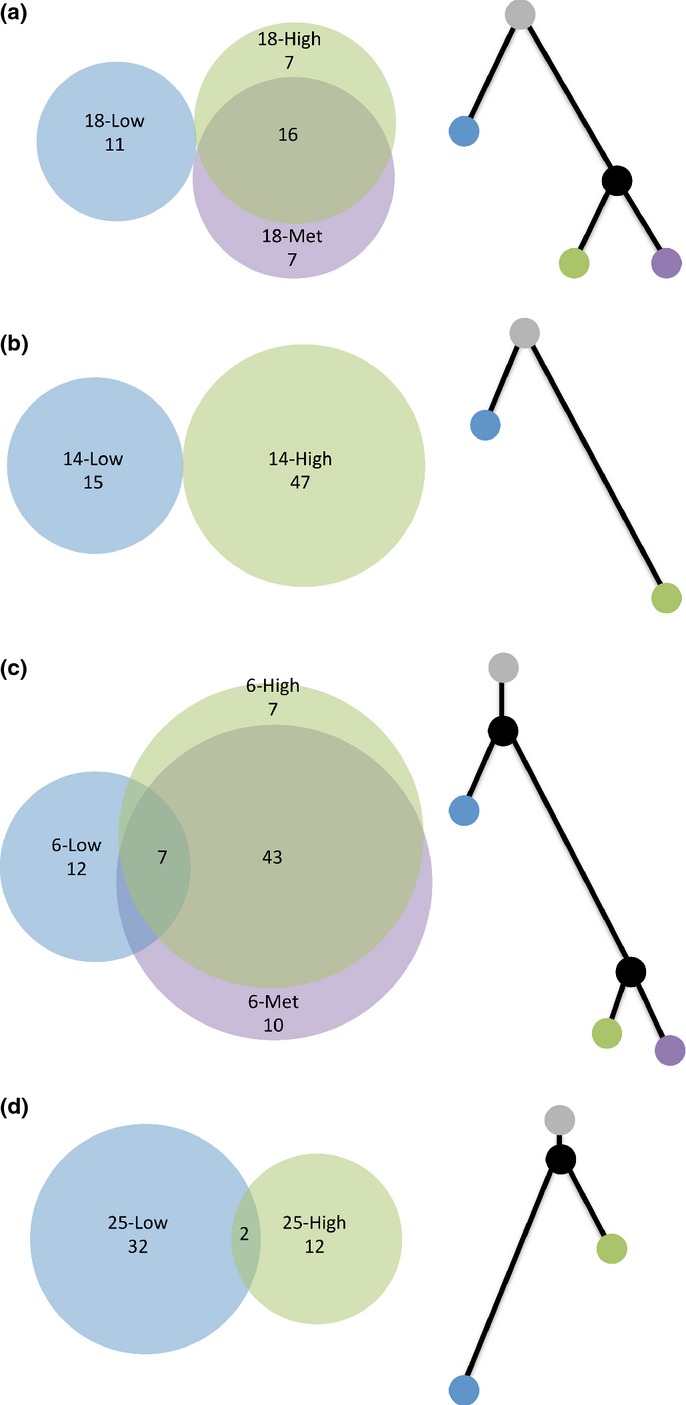
The molecular relationship of coincident foci. Venn diagrams (left) depicting the pattern of shared and private high-confidence somatic mutations and phylogenetic trees (right) depicting the relationship of coincident foci for PrCa 18 (a), PrCa 14 (b), PrCa 6 (c) and PrCa 25 (d). The number of high-confidence somatic mutations is labeled within each Venn diagram. In (c), there are no mutations that are shared between the low-grade focus and the metastatic focus and also not shared with the high-grade focus. Within each phylogenetic tree, branch length is proportional to the number of mutations. Gray, uninvolved prostate; blue, low-grade focus; green, high-grade focus; purple, metastatic focus; black, theoretical common progenitor.
To evaluate more broadly the clonal relationship of multifocal disease, custom capture baits targeting variants identified through exome sequencing of PrCa 6 were used to capture the DNA from seven additional cancer foci from that subject (Table S3). The average coverage depth of these variants was 208 and 78 previously identified variants were covered at >70× in each focus. Three high-grade foci harbored a median of 56 previously identified variants, three intermediate-grade foci harbored a median of 15 variants and one low-grade focus harbored three variants. As in the whole exome data a small minority of mutations was shared, with just two of 78 (2.6%) variants found to be ubiquitous among the seven cancer foci. Fourteen of 78 (17.9%) were not identified in any of the seven additional foci (Fig. S2). Eight of these 14 were among the 12 variants originally identified as private to the low-grade focus.
Rearrangements involving ERG are the most common genetic derangement in prostate cancer and are thought to occur early in tumorigenesis. These events are not typically captured by exome sequencing. Therefore we performed IHC for ERG to look for overexpression, which is typically restricted to cells harboring an ERG rearrangement. Of the two specimens exhibiting positive ERG staining, PrCa 6 showed consistently positive staining in all tumor foci (Fig. 4). In contrast, in PrCa 18 the low-grade focus was negative, the high-grade focus was equivocal with few cells positive for ERG and the metastatic focus was more widely focally positive.
Fig. 4.
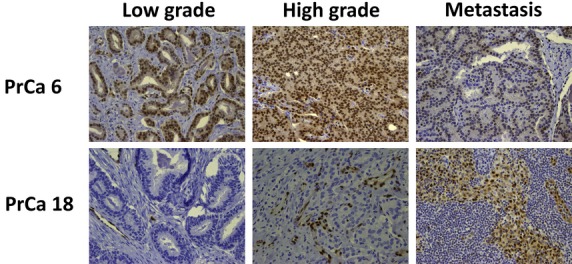
ERG expression. Immunohistochemical analysis for ERG in coincident foci from PrCa 6 and PrCa 18.
Together with the immunohistochemical analysis, the patterns of shared and private mutations indicate early divergence of low- and high-grade foci. In two cases no shared mutations were identified and independent origin of low- and high-grade disease cannot be ruled out. In contrast, there is late divergence of high-grade foci and metastatic disease. Consistent with this, there were no somatic mutations that were shared between low grade and metastatic disease and also not shared with high-grade disease.
Prostate tumor foci consist of an admixture of tumor cells and stromal cells. Despite microdissection of the tumor foci, the variant allele frequencies in the low-grade foci were modestly lower than the high-grade lesions (Fig. S3), suggesting higher contamination with non-tumor cells. The contaminating stromal cells may interfere with the ability to call somatic mutations, especially in the low-grade foci, leading to a falsely elevated number of apparently private mutations and obscuring the clonal relationship among concurrent foci. Therefore for each position identified as a private high-confidence somatic mutation in the low-grade foci, the number of sequencing reads supporting that mutation in the matched high grade and metastatic foci was determined, relaxing filters based on DNA strand and quality scores (Fig. S4). Of the 23 high-confidence somatic mutations in PrCa 18 and PrCa 6 identified as private mutations, none had two or more variant reads in high grade or metastatic foci. Thus, even with a less conservative definition of mutation (two variant reads), high-grade foci and metastases harbored few variants seen in low-grade foci and the majority of variants in the low-grade foci was private. This was corroborated with evidence from capillary sequencing. Three mutations private to low-grade foci and two shared with other foci were among the subset of mutations confirmed by capillary sequencing.
Two genes exhibited recurrent nonsynonymous mutations or mutations in the untranslated region (Table S4). Low density lipoprotein (LDL) receptor-related protein 1B, which encodes an LDL-family receptor, is frequently deleted in multiple malignancies.(27) However, recent work suggests its high frequency of mutation may not imply a significant role in cancer.(28) ZNF717 is one of a number of zinc finger proteins and its role in malignancy is unclear.
Comparison of genes with nonsynonymous mutations in our data to the Catalogue Of Somatic Mutations In Cancer (COSMIC) cancer gene census revealed three genes in common (Table 2). These genes include TP53, the most frequently mutated gene in advanced cancers including prostate cancer,(12) as well as ATM and SS18L1. All of these three genes were mutated in high grade or metastatic disease, but not in low-grade disease. Gene ontology analysis(29) revealed enrichment for genes in the p53 signaling pathway among high-grade foci (adjusted P-value 0.037), including TP53 and ATM. Cell cycle genes were also enriched (adjusted P-value 0.054). No other pathways were statistically significantly enriched.
Table 2.
Overlap of mutated genes with the COSMIC cancer gene census
| Subject | Histology | Chromosome | Hg18 position | Reference | Variant | Symbol | GERP | Type | aa Change |
|---|---|---|---|---|---|---|---|---|---|
| 6 | High | 11 | 107675775 | G | A | ATM | 5.22 | STOP | W1710* |
| 6 | LN | 11 | 107675775 | G | A | ATM | 5.22 | STOP | W1710* |
| 6 | High | 20 | 60169915 | C | G | SS18L1 | 3.98 | NS | P87R |
| 6 | LN | 20 | 60169915 | C | G | SS18L1 | 3.98 | NS | P87R |
| 14 | High | 17 | 7517833 | C | A | TP53 | 4.78 | NS | C145F |
Three genes harboring a nonsynonymous mutation or a premature stop codon are also found in the COSMIC cancer gene census. All three were found in either high-grade foci or metastatic disease, but not in low-grade foci. GERP, genomic evolutionary rate profiling score; LN, lymph node metastasis; NS, nonsynonymous; STOP, introduction of premature stop codon; *, stop codon.
Discussion
Given the disparate outcomes between men with low-grade prostate cancer and those with high-grade disease, a fundamental question in prostate cancer management is the possible molecular relationship among grades and metastatic disease. Multiple lines of evidence suggest multifocal disease is often independent.(30,31) However, recent reports have demonstrated shared molecular derangements of coincident low- and high-grade foci.(16,17) The present study identifies mutations shared between low- and high-grade cancer foci, supporting the ability of a single progenitor to give rise to both low- and high-grade disease. However, the relationship between these foci was distant relative to the relationship between high-grade disease and synchronous metastases. In fact, in two of the four cases it cannot be ruled out that the foci arose from independent origins. The early divergence of low- and high-grade disease may help explain distinct differences in their clinical behavior. Indeed, we found no evidence of direct progression from low grade to metastatic disease. Instead, there was an overwhelming fraction of shared mutations between high grade and metastatic disease, which is consistent with late divergence of these high-grade foci (Fig. 5).
Fig. 5.
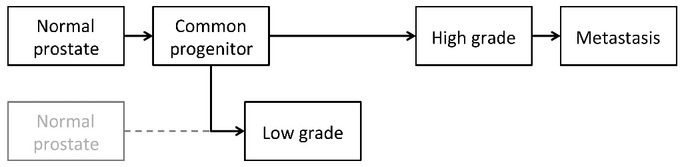
Model of prostate cancer evolution. Low-grade and high-grade cancer foci progress largely in parallel, diverging early from a common progenitor. Metastatic disease exhibits late divergence from high-grade disease. It cannot be ruled out that in some cases low- and high-grade diseases arise through independent origins.
For the cases in which there was evidence of a common progenitor for both low- and high-grade disease (PrCa 25 and PrCa 6), the nature of the common progenitor is not clear. Histological low-grade cancer, HGPIN or even histologically uninvolved prostate are all possible progenitors. If the progenitor is not histological low-grade cancer, high-grade and low-grade disease may have emerged simultaneously. Characterization of the common progenitor requires further study.
The present study demonstrates there is late divergence of high-grade cancer and synchronous metastatic disease, confirming high-grade disease as the precursor to metastasis and suggesting few if any additional mutations are required for malignant cells to gain the ability to metastasize. Given the difficulty in obtaining tissue from typical bone metastases in prostate cancer, more easily accessible prostate lesions might offer a substitute for identifying genetic derangements leading to primary resistance of metastatic disease. However, mechanisms of secondary resistance are likely induced or enriched by the selective pressure of therapy and thus are likely found at low levels, if at all, in the primary lesion.
The most common gene fusion event in prostate cancer is the TMPRSS2-ERG fusion, leading to high levels of expression of ERG, which is otherwise expressed at very low levels in prostate cancer. We evaluated for the presence of this fusion through IHC for ERG and those findings supported the evidence from the exome sequencing data. A limitation of exome sequencing is the inability to identify additional chromosomal rearrangements. Although this is a limitation it was not required for the goal of the present study, which was to identify tumor lineages.
One goal of sequencing prostate tumors is to identify biomarkers that will supplement the prognosis given by clinical features alone. To that end we sought to identify mutated genes or pathways that were enriched in high grade and metastatic disease. TP53 is the most frequently mutated gene in advanced prostate cancer and we found the p53 signaling pathway to harbor mutations only in high grade and metastatic disease. Thus, members of the p53 signaling pathway are intriguing candidates for potential biomarkers of aggressive disease. Recently, expression of a panel of cell cycle progression-associated genes has been correlated with outcomes of those with low- or intermediate-grade prostate cancer.(32) We also found enrichment for cell cycle-related genes among those mutated in high-grade disease, although this was of borderline statistical significance in this small cohort.
Given the good outcomes of low-grade prostate cancer, we hypothesized that mutations not found in low-grade disease are more likely to be drivers of progression and metastasis and therefore more important potential therapeutic targets. Indeed, the three genes identified here that are also found in the COSMIC cancer gene census were identified exclusively in high grade and metastatic foci. Of note, all three mutations are in highly conserved regions as evidenced by the high genomic evolutionary rate profiling score and thus are likely to be biologically significant. Identification of driver genes through this strategy has the potential to guide the development of future therapies.
The present study contributes to the growing body of work demonstrating that next-generation sequencing from microdissected FFPE prostatectomy specimens is feasible. Based on these techniques, we found that coincident low- and high-grade prostate cancer can emerge through either independent progression or early divergence from a common progenitor. High-grade disease is characterized by mutations in known cancer-associated genes and the p53 signaling pathway. Metastatic disease is closely related to high-grade primary foci. These data can be leveraged to help identify potential biomarkers and drivers of progression of clinically significant disease.
Acknowledgments
The authors thank April Peterson for technical assistance in library preparation and A. Jason Grundstad, Dr Megan McNerney and Dr Thomas Stricker for advice on experimental design and data analysis. The present study is supported by the AACI Fellowship for Translational Cancer Research and DOD Prostate Cancer Research Program PRTA (D.J.V.W.) and the Chicago Cancer Genomes Project (K.P.W.).
Disclosure Statement
The authors have no conflicts of interest.
Supporting Information
Additional supporting information may be found in the online version of this article:
Fig. S1 Comparison of fixed and frozen.
Fig. S2 Variant allele frequency of variants in additional cancer foci.
Fig. S3 Variant allele frequencies.
Fig. S4 Frequencies of variants identified as private to the low-grade focus.
Table S1 Characteristics of sequencing libraries.
Table S2 High-confidence somatic mutations identified across all foci sequenced.
Table S3 Variants identified in additional cancer foci.
Table S4 Recurrently mutated genes.
References
- 1.Vogelstein B, Kinzler KW. The multistep nature of cancer. Trends Genet. 1993;9:138–41. doi: 10.1016/0168-9525(93)90209-z. [DOI] [PubMed] [Google Scholar]
- 2.Eggener SE, Scardino PT, Walsh PC, et al. Predicting 15-year prostate cancer specific mortality after radical prostatectomy. J Urol. 2011;185:869–75. doi: 10.1016/j.juro.2010.10.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ross HM, Kryvenko ON, Cowan JE, Simko JP, Wheeler TM, Epstein JI. Do adenocarcinomas of the prostate with Gleason score (GS) ≤6 have the potential to metastasize to lymph nodes? Am J Surg Pathol. 2012;36:1346–52. doi: 10.1097/PAS.0b013e3182556dcd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63:11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 5.Andriole GL, Crawford ED, Grubb RL, 3rd, et al. Mortality results from a randomized prostate-cancer screening trial. N Engl J Med. 2009;360:1310–9. doi: 10.1056/NEJMoa0810696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schröder FH, Hugosson J, Roobol MJ, et al. Prostate-cancer mortality at 11 years of follow-up. N Engl J Med. 2012;366:981–90. doi: 10.1056/NEJMoa1113135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mohler J, Bahnson RR, Boston B, et al. NCCN clinical practice guidelines in oncology: prostate cancer. J Natl Compr Canc Netw. 2010;8:162–200. doi: 10.6004/jnccn.2010.0012. [DOI] [PubMed] [Google Scholar]
- 8.Moyer VA. Screening for prostate cancer: U.S. Preventive Services Task Force recommendation statement. Ann Intern Med. 2012;157:120–34. doi: 10.7326/0003-4819-157-2-201207170-00459. [DOI] [PubMed] [Google Scholar]
- 9.Berger MF, Lawrence MS, Demichelis F, et al. The genomic complexity of primary human prostate cancer. Nature. 2011;470:214–20. doi: 10.1038/nature09744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barbieri CE, Baca SC, Lawrence MS, et al. Exome sequencing identifies recurrent SPOP, FOXA1 and MED12 mutations in prostate cancer. Nat Genet. 2012;44:685–9. doi: 10.1038/ng.2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grasso CS, Wu Y-M, Robinson DR, et al. The mutational landscape of lethal castration-resistant prostate cancer. Nature. 2012;487:239–43. doi: 10.1038/nature11125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kumar A, White TA, MacKenzie AP, et al. Exome sequencing identifies a spectrum of mutation frequencies in advanced and lethal prostate cancers. Proc Natl Acad Sci USA. 2011;108:17087–92. doi: 10.1073/pnas.1108745108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Taylor BS, Schultz N, Hieronymus H, et al. Integrative genomic profiling of human prostate cancer. Cancer Cell. 2010;18:11–22. doi: 10.1016/j.ccr.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Robbins CM, Tembe WA, Baker A, et al. Copy number and targeted mutational analysis reveals novel somatic events in metastatic prostate tumors. Genome Res. 2011;21:47–55. doi: 10.1101/gr.107961.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boyd LK, Mao X, Xue L, et al. High-resolution genome-wide copy-number analysis suggests a monoclonal origin of multifocal prostate cancer. Genes Chromosom Cancer. 2012;51:579–89. doi: 10.1002/gcc.21944. [DOI] [PubMed] [Google Scholar]
- 16.Sowalsky AG, Ye H, Bubley GJ, Balk SP. Clonal progression of prostate cancers from Gleason grade 3 to grade 4. Cancer Res. 2013;73:1050–5. doi: 10.1158/0008-5472.CAN-12-2799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kovtun IV, Cheville JC, Murphy SJ, et al. Lineage relationship of Gleason patterns in Gleason score 7 prostate cancer. Cancer Res. 2013;73:3275–84. doi: 10.1158/0008-5472.CAN-12-2803. [DOI] [PubMed] [Google Scholar]
- 18.Meyer M, Kircher M. Illumina sequencing library preparation for highly multiplexed target capture and sequencing. Cold Spring Harb Protoc. 2010;2010:pdb.prot5448. doi: 10.1101/pdb.prot5448. [DOI] [PubMed] [Google Scholar]
- 19.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–60. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li H, Handsaker B, Wysoker A, et al. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–9. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McKenna A, Hanna M, Banks E, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koboldt DC, Zhang Q, Larson DE, et al. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 2012;22:568–76. doi: 10.1101/gr.129684.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sherry ST, Ward MH, Kholodov M, et al. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–11. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Consortium T 1000 GP. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–73. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Burbano HA, Hodges E, Green RE, et al. Targeted investigation of the Neandertal genome by array-based sequence capture. Science. 2010;328:723–5. doi: 10.1126/science.1188046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Robinson JT, Thorvaldsdóttir H, Winckler W, et al. Integrative genomics viewer. Nat Biotechnol. 2011;29:24–6. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Prazeres H, Torres J, Rodrigues F, et al. Chromosomal, epigenetic and microRNA-mediated inactivation of LRP1B, a modulator of the extracellular environment of thyroid cancer cells. Oncogene. 2011;30:1302–17. doi: 10.1038/onc.2010.512. [DOI] [PubMed] [Google Scholar]
- 28.Lawrence MS, Stojanov P, Polak P, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013;499:214–8. doi: 10.1038/nature12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2008;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 30.Mehra R, Han B, Tomlins SA, et al. Heterogeneity of TMPRSS2 gene rearrangements in multifocal prostate adenocarcinoma: molecular evidence for an independent group of diseases. Cancer Res. 2007;67:7991–5. doi: 10.1158/0008-5472.CAN-07-2043. [DOI] [PubMed] [Google Scholar]
- 31.Lindberg J, Klevebring D, Liu W, et al. Exome sequencing of prostate cancer supports the hypothesis of independent tumour origins. Eur Urol. 2013;63:347–53. doi: 10.1016/j.eururo.2012.03.050. [DOI] [PubMed] [Google Scholar]
- 32.Cooperberg MR, Simko JP, Cowan JE, et al. Validation of a cell-cycle progression gene panel to improve risk stratification in a contemporary prostatectomy cohort. J Clin Oncol. 2013;31:1428–34. doi: 10.1200/JCO.2012.46.4396. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Comparison of fixed and frozen.
Fig. S2 Variant allele frequency of variants in additional cancer foci.
Fig. S3 Variant allele frequencies.
Fig. S4 Frequencies of variants identified as private to the low-grade focus.
Table S1 Characteristics of sequencing libraries.
Table S2 High-confidence somatic mutations identified across all foci sequenced.
Table S3 Variants identified in additional cancer foci.
Table S4 Recurrently mutated genes.