Abstract
The CKLF-like MARVEL transmembrane domain containing 3 (CMTM3) gene is a novel tumor suppressor with frequent epigenetic inactivation. In this study, we showed the role played by CMTM3 in gastric cancer cells as a tumor suppressor gene, and examined the correlation between CMTM3 expression and clinicopathological parameters using immunohistochemistry in gastric cancer patients with different pathological stages (n = 350). We found that CMTM3 expression was reduced or silenced by epigenetic regulation in gastric cell lines, and dramatically downregulated in primary gastric cancer tissues. Restoration of CMTM3 significantly affected migration and invasion of AGS and SGC-7901 cells (P < 0.001). In vivo experiments showed that peritoneal disseminated metastases were significantly suppressed by CMTM3 (P < 0.001). We further showed that the expression of MMP2 and the phosphorylation of Erk1/2 were decreased when CMTM3 was restored. In addition, by immunohistochemical staining, we found that the expression of CMTM3 was remarkably weaker in gastric cancer tissues than in normal mucosae (P = 0.008), and was significantly correlated with gender (P = 0.033), tumor depth (P = 0.049), stage (P = 0.021), and histological grade (P = 0.022). More importantly, CMTM3 expression was associated with prognosis in gastric cancer patients (P = 0.041), and was a significant independent prognostic indicator (hazard ratio = 0.704, 95% confidence interval, 0.498–0.994; P = 0.046). Our findings indicate that CMTM3 regulates migration and invasion of gastric cancer cells. Moreover, CMTM3 is a candidate marker for prognosis of gastric cancer in the clinic.
Keywords: CMTM3, gastric cancer, MARVEL, tumor suppressor gene, survival
Gastric cancer is the second leading cause of cancer-related death worldwide, accounting for 8% of all new cancer cases annually.1 Despite great efforts invested in promising tailored therapy, treatment was notoriously ineffective without prognostic markers for proper stratification of gastric cancer patients. Recently, accumulating evidence has shown that tumor suppressor genes (TSG) were critical to carcinogenesis of gastric cancer, and a large number of results showed that TSG functioned as potential prognostic factors.2 However, further study is required both in clinical and basic research for translating these TSG into clinical applications as markers for prognosis.
The CKLF-like MARVEL transmembrane domain (CMTM) family of proteins link classical chemokines and the transmembrane 4 superfamily with important functions in the immune system, male reproductive system, and tumorigenesis.3–10 In humans, the family includes nine members, CKLF and CMTM1–8. CMTM proteins contain a MARVEL domain that is involved in many essential cellular processes including tumor suppression.11,12 Our previous studies showed that CMTM3 CMTM5 are novel potential TSG with frequent epigenetic inactivation.6,9,13
Although widely expressed in normal tissues, CMTM3 is silenced or downregulated in many cancer cell lines and primary cancer tissues. Restoration of CMTM3 inhibits cell growth and leads to apoptosis with caspase-3 activation in CNE-2 cells.6 In detected cancer tissues including stomach, colon, and breast, the frequency of CMTM3 methylation is highest in gastric cancer.6 In addition, data from the Human Protein Atlas (http://www.proteinatlas.org/) shows that expression of CMTM3 is lower in stomach cancer than in normal tissues. These results indicate that CMTM3 may play important roles in gastric cancer, which requires further study.
In this paper, we showed that CMTM3 inhibited the migration and invasion of gastric cancer cells both in vitro and in vivo. Furthermore, we analyzed the association of CMTM3 expression with clinical features through immunohistochemical staining, and found that CMTM3 was associated with favorable prognosis in gastric cancer.
Materials and Methods
Cell lines, cell cultures, and treatment
Cell lines including KATOIII, SNU1, SNU16, NCI-N87, and AGS were obtained from ATCC (Manassas, VA, USA), and BGC823 and SGC-7901 were obtained from the Cell Research Institute (Shanghai, China). Cell lines MKN28 and MKN45 were from the Health Science Research Resources Bank (Tokyo, Japan). Cells were routinely grown in RPMI-1640 (NCI-N87, AGS, BGC823, SGC-7901, MKN28, and MKN45) or DMEM (KATOIII, SNU1, and SNU16) (Gibco BRL, Grand Island, NY, USA), supplemented with 10% (v/v) FBS (Gibco BRL) at 37°C in a humidified 5% CO2 atmosphere. Some cell lines were treated with 10 μM 5-aza-2′-deoxycytidine (Aza; Sigma, St. Louis, MO, USA) for 3 days, with or without further treatment with 100 nM trichostatin A (Sigma) for an additional 24 h.
Semiquantitative RT-PCR
Reverse transcription–PCR was carried out as previously described.6
DNA bisulfite treatment and methylation analysis
Bisulfite modification of DNA, methylation-specific PCR, and unmethylation-specific PCR were carried out as previously described.6
Adenovirus construction and cell infection
The construction, generation, and purification of ad5-null (vector containing adenovirus, defined as Mock) and ad5-CMTM3 was described previously.5
Protein extraction and Western blot analysis
Western blotting was carried out as described previously.5 For the detection of CMTM3, 120 μg total protein of each sample was loaded. Anti-MMP2, Erk1/2, and phosphorylated Erk1/2 antibody were purchased commercially (Cell Signaling Technology, Danvers, MA, USA).
Wound healing assay
Wound healing assay was carried out as described previously.13
Cell migration and invasion assay
Cell migration and invasion assay was carried out as described previously.13
Peritoneal metastases animal model
Cells (2 × 106) were removed by trypsinization, washed twice with PBS, and then injected into the abdominal cavity of 6-week-old female nude mice provided by Peking Experimental Animal Center (Beijing, China), along with 200 mL PBS. There were eight mice in the Mock group and nine in the CMTM3 group. The animals were raised in specific pathogen-free experimental animal rooms with free access to water. After 4 weeks, mice were humanely killed, and any disseminated nodules ≥1 mm in diameter were counted.
Real-time PCR
Real-time PCR was carried out as previously described.4 Primers of MMP2 were (forward) 5′-ATAACCTGGATGCCGTCGT-3′ and (reverse) 5′-AGGCACCCTTGAAGAAGTAGC-3′. Univeral ProbeLibrary probe #70 was used for MMP2.
Gelatin zymography
Cells were incubated for 16 h in serum-free medium and 40 μL supernatants were used for gelatin zymography. Following electrophoresis, the gels were washed with renaturing buffer (2.5% Triton X-100) and then incubated for 24 h at 37°C in the developing buffer (10 mM CaCl2, 15 mM NaCl, 2 mM NaN3, and 50 mM Tris–HCl, pH 7.5). The gels were stained and destained at appropriate times. The Odyssey Infrared Imager (LI-COR Bioscience, Lincoln, NE, USA) was used to scan the gels.
Patients and tissue specimens
The study cohort consisted of 350 gastric cancer patients who underwent uneventful surgical resection, including 254 males and 96 females (mean age, 61 years; range, 22–87 years), between February 1998 and January 2007 in Peking University Cancer Hospital (Beijing, China). None of the patients received neoadjuvant chemotherapy or radiation therapy. The records of patients were reviewed with follow-up information. Gastric cancer was classified according to the American Joint Committee on Cancer's tumor staging system (7th edition) and the Japanese Classification of Gastric Carcinoma (3rd English edition).14,15 All patients were followed up until November 2007. Overall survival time was calculated from the date of initial surgery to date of death, counting death from any cause, or the last time of follow-up as the end-point. Fresh human tissues were fixed with 10% formalin in PBS for immunohistochemistry, or frozen in liquid nitrogen for RNA and protein extraction. This investigation was carried out after approval by the Ethics Committee of Peking University Cancer Hospital. Informed consent was obtained from each patient.
Immunohistochemistry analysis
Immunohistochemistry analysis was carried out as previously described.6
Statistical analysis
The statistical analysis was calculated from three independent experiments and presented as the mean ± SEM and P-value using Student's t-test. The χ2-test (Fisher's exact test) was carried out to analyze the correlations between CMTM3 expression and clinical pathological variables. Overall survival was plotted and calculated using the Kaplan–Meier method, and differences between groups were compared by the log–rank test. The Cox proportional hazard model (backward, stepwise) was used to estimate the influence of each variable on survival. P-values <0.05 (two-sided) were considered statistically significant. All analyses were performed by spss software version 13.0 (SPSS Inc., Chicago, IL, USA).
Results
Expression and methylation profile of CMTM3 in gastric cancer cell lines and primary tissues
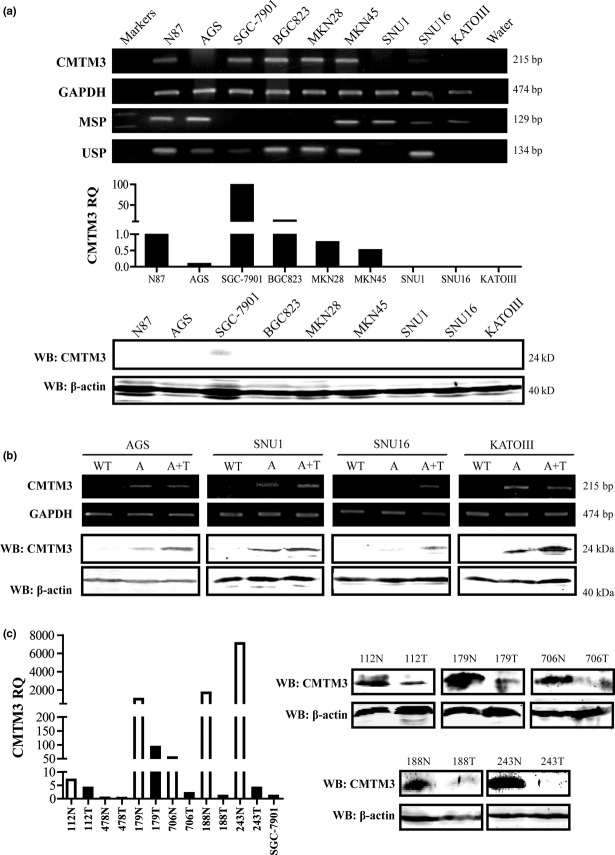
Our previous study showed that CMTM3 was silenced or downregulated in carcinoma cell lines with regulatory region methylation.6 In this study, we chose gastric cancer cell lines that have been widely used in gastric cancer research, especially in functional studies. We examined the expression of CMTM3 in nine cell lines by RT-PCR at the mRNA level. The result showed that CMTM3 was silenced in four of these cell lines (Fig.1a). Then we carried out real-time PCR and Western blot analyses to confirm the expression of CMTM3. As shown in Figure1(a), SGC-7901 had the highest mRNA level of CMTM3, and CMTM3 protein could be only detected in SGC-7901 by Western blotting (Fig.1a). To analyze the methylation status of CMTM3, methylation-specific PCR was carried out in nine cell lines. Full or partial methylation was found in the six of these (Fig.1a). Furthermore, we treated AGS, SNU1, SNU16 and KATOIII cells with demethylating agent Aza, or combined with histone deacetylase inhibitor trichostatin A (TSA). Indeed, CMTM3 expression was restored by Aza and TSA both on mRNA and protein levels (Fig.1b). However, in SNU16 cells, the expression of CMTM3 was not fully restored when treated with Aza. This result indicates that histone acetylation might play a more important role in controlling CMTM3 expression in SNU16 cells. These findings are consistent with our previous results, and indicate that epigenetic changes are correlated to CMTM3 silencing in gastric cancer cell lines.
Figure 1.
(a) Expression and methylation status of CMTM3 in cell lines was detected by RT-PCR, methylation-specific PCR (MSP), unmethylation-specific PCR (USP), and quantitative PCR. Western blotting (WB) confirmed the expression of CMTM3. RQ, relative quantity. (b) Expression of CMTM3 was detected by RT-PCR and WB after treatment with demethylation agent. A, cells treated with 5-aza-2′-deoxycytidine; A+T, cells treated with 5-aza-2′-deoxycytidine and trichostatin A; WT, untreated cells; (c) CMTM3 expression in gastric cancer paired tissues was detected by quantitative PCR and WB. Normal mucosae tissues (N) are shown as white columns; tumor tissues (T) are shown as black columns. Gastric cancer cell lines were also analyzed with the same standard, and the relative quantity of SGC-7901 was present.
In order to know the expression of CMTM3 in primary gastric cancer tissues and gastric mucosae tissues, we collected six paired tissues from patients and carried out real-time PCR and Western blot analysis of CMTM3. As shown in Figure1(c), CMTM3 expression was dramatically reduced in primary tumor tissues compared with that in normal mucosae tissues both at mRNA and protein levels, except for one pair (478N/T), in which CMTM3 protein could not be detected. More importantly, the expression of CMTM3 in SGC-7901, which had the highest CMTM3 expression in the detected cell lines, was much lower than that in most normal tissues (Fig.1c). These results indicated that CMTM3 expression was significantly reduced in gastric cancer, both in primary tissues and cell lines.
Restoration of CMTM3 suppresses migration and invasion of AGS and SGC-7901 in vitro and in vivo
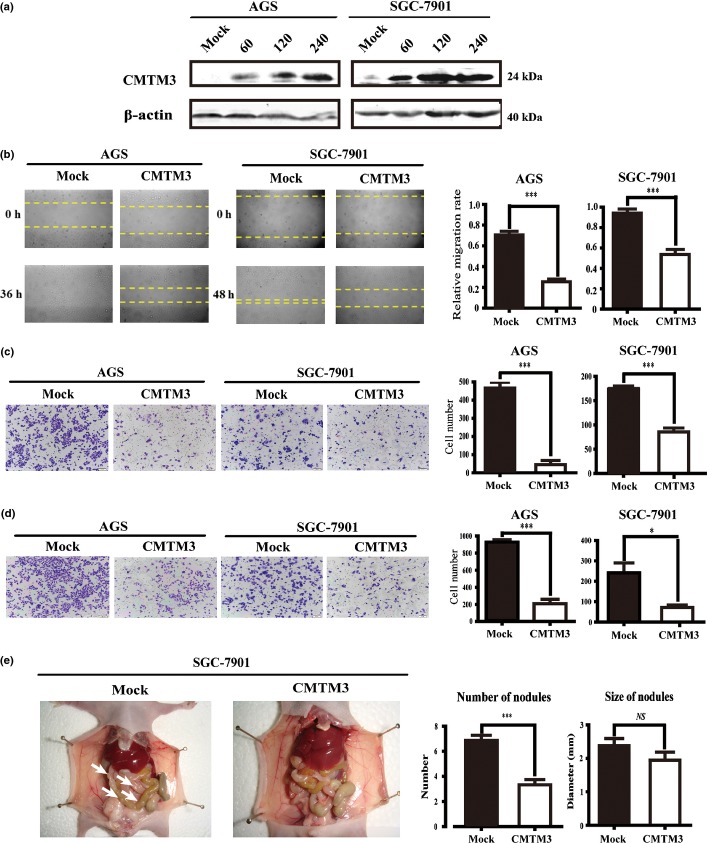
CMTM3 is downregulated in gastric cancer cell lines and primary tissues both at mRNA and protein levels. To know the function of CMTM3 in gastric cancer, we infected cells with adenovirus and confirmed the expression of CMTM3 in AGS and SGC-7901 cells by Western blotting (Fig.2a). We first tested the effect of CMTM3 on cell proliferation and apoptosis, and found that CMTM3 did not significantly affect cell growth or apoptosis in AGS and SGC-7901 cells (data not shown). To evaluate whether CMTM3 had any effect on cell mobility or invasion ability, we carried out the wound healing assay using AGS and SGC-7901 cells. After observing for 36–48 h after the scratch, we found that cells in the CMTM3 group were distinctively less migrated than those in the Mock group (Fig.2a). Strikingly, in SGC-7901 cells, the Mock group healed the wounded area in 36 h, whereas the CMTM3 group was unable to heal in the same time period. The relative migration rates were statistically significant (P < 0.001) (Fig.2b). These results showed that the migratory capacity of both cells lines was suppressed by CMTM3. To confirm this conclusion, we used a Transwell system with or without Matrigel to detect cell migration and invasion. As shown in Figure2(c,d), CMTM3 groups showed a markedly lower migratory and invasive capacity compared to the Mock groups, suggesting that CMTM3 might decrease the abilities of cell motility and invasion. Overall, the results suggest that CMTM3 inhibits migration and invasion of gastric cancer cells in vitro.
Figure 2.
(a) Overexpression of CMTM3 in SGC-7901 and AGS gastric cancer cells was detected by Western blot analysis. Different MOIs (0, 60, 120, and 240) of adenovirus were used. (b) Effect of CMTM3 on cell migration was observed by wound healing assay. Photographs were taken at indicated time points after the scratch (magnification, ×100). (c, d) Effects of CMTM3 on cell migration (c) and invasion (d) were detected by Transwell assay (magnification, ×100). The statistical graph indicates the mean ± SD and P-value of the number of cells per five random high power fields (magnification, ×400) counted from three independent experiments. (e) Ability of cell migration and invasion in vivo was detected by a peritoneal spreading model in nude mice. Photographs were taken after the mice were killed. Arrows indicate tumor nodules. NS, not significant. *P < 0.05; ***P < 0.001.
To confirm the regulation effect of CMTM3 on cell migration and invasion in vivo, we used a peritoneal spreading model of gastric cancer using a total of 17 nude mice. SGC-7901 cells could successfully form tumor nodules on the peritoneum, whereas AGS cells failed to develop visible nodules on the peritoneum, as previously reported.16 As illustrated in Figure2(e), the number of peritoneal nodules was significantly lower in the CMTM3 group, compared with the Mock group (6.87 ± 0.398 vs. 3.33 ± 0.408, P < 0.001), whereas the difference in nodule size was not significant (2.380 ± 0.213 vs. 1.944 ± 0.239, P = 0.22). Combined with the in vitro findings, it is reasonable to conclude that CMTM3 is an inhibitor of migration and invasion in gastric cancer cells.
Inhibition of MMP2 expression and phosphorylation of Erk1/2 by CMTM3
To investigate the mechanism of CMTM3 as a tumor suppressor in gastric cancer cells, we collected AGS cells infected with Ad-Mock and Ad-CMTM3 and carried out the Tumor Metastasis RT Profiler PCR Array (SA Biosciences, Shanghai, China) which includes 84 genes known to be involved in cell migration and invasion. The PCR array results indicated that CMTM3 restoration may induce downregulation of several MMPs, especially MMP2 and MMP9. Then we measured the expression level of MMP2 and MMP9 in AGS cells and SGC-7901 cells by real-time PCR, and found that MMP2 expression was downregulated by CMTM3 significantly (Fig.3a), whereas the level of MMP9 was too low to be detected. Then gelatin zymography showed that CMTM3 inhibited the MMP2 level in cell supernatants and the gray density was significantly different (Fig.3b). To further confirm this result, we used Western blotting and found that the protein level of MMP2 from cell lysates was also reduced. These observations suggest that CMTM3 inhibits the expression and activity of MMP2. We also detected phosphorylated Erk1/2 (p-Erk1/2), a main signaling pathway regulating MMP2 expression. As shown in Figure3(c), p-Erk was remarkably reduced when CMTM3 was overexpressed, which indicated that CMTM3 might regulate MMP2 expression through the Erk1/2 signaling pathway.
Figure 3.
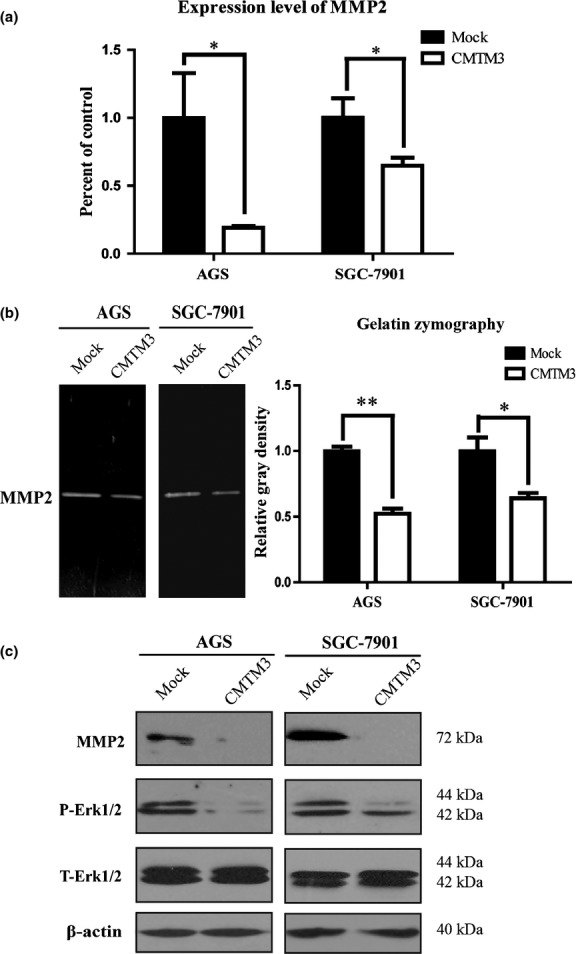
(a) Real-time PCR was carried out to detect MMP2 expression in AGS and SGC-7901 gastric cancer cells. Average percentage of control (Mock) with SEM is shown from three independent experiments. (b) MMP2 in AGS cell supernatants was observed by gelatin zymography. Average relative gray density with SEM is shown from three independent experiments. (c) MMP2, phosphorylated Erk1/2 (P-Erk1/2) and total Erk1/2 (T-Erk1/2) in two cell lines were observed by Western blotting. The experiment was repeated at least three times and one representative result is shown. *P < 0.05; **P < 0.01.
Expression of CMTM3 decreased in primary gastric cancer tissues and correlated with clinicopathological parameters
Next we aimed to examine whether CMTM3 had any clinical significance in gastric cancer. CMTM3 expression was detected in 350 gastric cancer samples and 222 normal gastric tissues by immunochemistry staining. The intensity of CMTM3 staining was remarkably weaker in primary gastric tissues than in normal mucosae (41.43% vs. 59.91%, P = 0.008). Its expression could also be observed significantly more frequently in patients with high grade well-differentiated tumor (Fig.4a).
Figure 4.
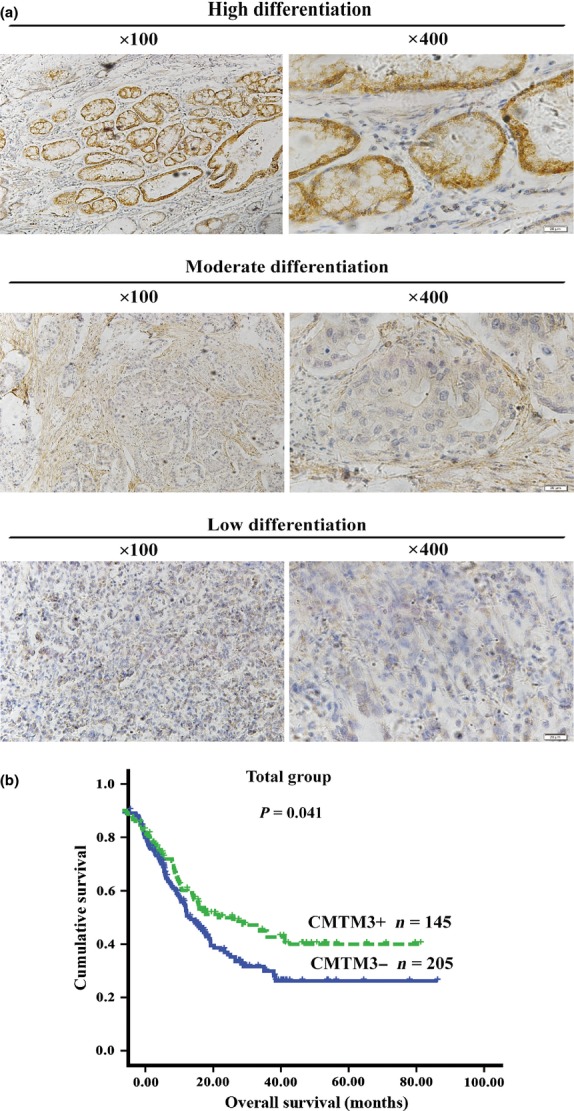
(a) Immunohistochemical staining showing the relationship between CMTM3 expression and gastric cancer by representative images (magnification, ×100 [left] and ×400 [right]). (b) Kaplan–Meier survival curve showing the association between CMTM3 expression and patients' survival. CMTM3+, CMTM3 positive; CMTM3−, CMTM3 negative; n, number of patients.
Table1 shows the correlation between CMTM3 expression and pathological parameters. Statistical analysis showed that CMTM3 expression was strongly associated with gender (P = 0.033), tumor depth (P = 0.049), stage (P = 0.021), and histological grade (P = 0.022). The association of between CMTM3 expression and tumor depth was analyzed statistically and showed that a direct relationship existed between CMTM3 expression and tumor invasiveness. Other features failed to reach statistically significant association with CMTM3 expression in our study (Table1).
Table 1.
Association between CMTM3 expression and clinicopathological characteristics in 350 gastric cancer patients
| CMTM3 expression |
|||||
|---|---|---|---|---|---|
| n | Negative (%) | Positive (%) | χ2 | P-value | |
| Gender | |||||
| Male | 254 | 140 (40) | 114 (32.57) | 4.551 | 0.033 |
| Female | 96 | 65 (18.57) | 31 (8.86) | ||
| Age, years (median, 61) | |||||
| ≤60 | 172 | 101 (28.86) | 71 (20.29) | 0.003 | 0.955 |
| >60 | 178 | 104 (29.71) | 74 (21.14) | ||
| Primary tumor location | |||||
| Cardia | 96 | 54 (15.47) | 42 (12.03) | 0.265 | 0.607 |
| Non-cardia | 253 | 150 (42.98) | 103 (29.51) | ||
| No record† | 1 | ||||
| Tumor size | |||||
| <5 cm | 183 | 104 (30.05) | 79 (22.83) | 0.542 | 0.461 |
| ≥5 cm | 163 | 99 (28.61) | 64 (18.92) | ||
| No record† | 4 | ||||
| Gastrectomy | |||||
| Total | 48 | 30 (8.57) | 18 (5.14) | 0.792 | 0.673 |
| Proximal | 103 | 57 (16.29) | 46 (13.14) | ||
| Distal | 199 | 118 (33.71) | 81 (23.14) | ||
| Borrmann type | |||||
| Borrmann I | 35 | 18 (5.64) | 17 (5.33) | 1.958 | 0.581 |
| Borrmann II | 97 | 59 (18.50) | 38 (11.91) | ||
| Borrmann III | 156 | 98 (30.72) | 58 (18.18) | ||
| Borrmann IV | 31 | 17 (5.33) | 14 (4.39) | ||
| Early | 25 | ||||
| No record† | 6 | ||||
| Tumor depth | |||||
| T1 | 25 | 11 (3.14) | 14 (4) | 7.852 | 0.049 |
| T2 | 37 | 16 (4.57) | 21 (6) | ||
| T3 | 50 | 28 (8) | 22 (6.29) | ||
| T4 | 238 | 150 (42.86) | 88 (25.14) | ||
| Lymph node metastasis | |||||
| N0 | 82 | 43 (12.29) | 39 (11.14) | 2.288 | 0.683 |
| N1 | 52 | 30 (8.57) | 22 (6.29) | ||
| N2 | 69 | 40 (11.43) | 29 (8.29) | ||
| N3a | 98 | 61 (17.43) | 37 (10.57) | ||
| N3b | 49 | 31 (8.86) | 18 (5.14) | ||
| Distant metastasis | |||||
| M0 | 313 | 181 (51.71) | 132 (37.71) | 0.675 | 0.411 |
| M1 | 37 | 24 (6.86) | 13 (3.71) | ||
| Stage | |||||
| I | 44 | 19 (5.43) | 25 (7.14) | 9.695 | 0.021 |
| II | 51 | 24 (6.86) | 27 (7.71) | ||
| III | 218 | 138 (39.43) | 80 (22.86) | ||
| IV | 37 | 24 (6.86) | 13 (3.71) | ||
| Hepatic metastasis | |||||
| Absent | 323 | 185 (52.86) | 138 (39.43) | 2.898 | 0.089 |
| Present | 27 | 20 (5.71) | 7 (2) | ||
| Peritoneal metastasis | |||||
| Absent | 327 | 191 (54.57) | 136 (38.86) | 0.054 | 0.817 |
| Present | 23 | 14 (4) | 9 (2.57) | ||
| Histological type | |||||
| Adenocarcinoma | 272 | 158 (45.14) | 114 (32.57) | 0.117 | 0.732 |
| Other types‡ | 78 | 47 (13.43) | 31 (8.86) | ||
| V/L invasion | |||||
| Absent | 162 | 97 (28.03) | 65 (18.79) | 0.050 | 0.823 |
| Present | 184 | 108 (31.21) | 76 (21.97) | ||
| No record† | 4 | ||||
| Histological grade | |||||
| Well | 12 | 6 (1.74) | 6 (1.74) | 9.673 | 0.022 |
| Moderately | 134 | 68 (19.71) | 66 (19.13) | ||
| Poorly | 174 | 109 (31.59) | 65 (18.84) | ||
| Undifferentiated | 25 | 20 (5.80) | 5 (1.45) | ||
| No record† | 5 | ||||
Data incomplete.
Signet-ring cell carcinoma, mucinous adenocarcinoma. V/L, vascular/lymphatic.
Expression of CMTM3 associated with favorable survival for gastric cancer patients as an independent prognostic marker
The analysis of survival and CMTM3 expression is depicted in Figures4(b) and S1. Kaplan–Meier survival curves showed that the high CMTM3 expression group had a significantly higher 5-year survival rate (48.77%) than the low CMTM3 expression group (34.642%; log–rank, 4.179; P = 0.041). A multivariate Cox proportional hazards model (Table2) using variables associated with survival in our study revealed that CMTM3 expression was a significant independent prognostic indicator (hazard ratio = 0.704; 95% confidence interval, 0.498–0.994; P = 0.046; Table2) associated with better survival among gastric cancer patients. Peritoneal metastasis, lymph node metastasis, distant metastasis, and adjuvant chemotherapy all had independent prognostic value in the multivariate analysis. Other clinical parameters had no prognostic value for survival in multivariate analyses. These results suggest that CMTM3 is an independent prognostic factor of gastric cancer.
Table 2.
Multivariate Cox analyses of factors associated with survival in gastric cancer patients by proportional hazards model
| HR | 95% CI | P-value | |
|---|---|---|---|
| CMTM3 expression | |||
| Positive versus negative | 0.704 | 0.498–0.994 | 0.046 |
| Peritoneal metastasis | |||
| Present versus absent | 1.807 | 1.076–3.037 | 0.025 |
| Lymph node metastasis | |||
| Present versus absent | 3.785 | 2.382–6.016 | <0.001 |
| Distant metastasis | |||
| Present versus absent | 3.104 | 2.028–4.753 | <0.001 |
| Adjuvant therapy | |||
| No versus yes | 2.122 | 1.515–2.973 | <0.001 |
CI, confidence interval; HR, hazard ratio.
Discussion
The tumor suppressor gene CMTM3 is located at 16q22.1, an important tumor suppressor locus with pathogenesis of multiple carcinomas.17 Our findings strongly indicate that CMTM3 plays important roles in migration and invasion of gastric cancer cells. Cell migration and invasion are two crucial steps in the progression of cancer, especially in cancer metastasis.18 Several TSGs involved in these two events have been discovered.19,20 Although expression of CMTM3 in gastric cancer patients did not correlate with distant metastasis, perhaps because of the limitation of the M1 stage sample size (37 patients), it was statistically related to the depth of primary tumor, which indicated that the invasion suppressive function of CMTM3 might happen during the early progression of the disease.
Compared with other cancers, patients with advanced stages of gastric cancer hardly have any symptoms, so identification of predictive markers is an important push to fight against this disease.21 Several important factors have been identified and linked with gastric cancer survival and these molecules participate in key processes involved in cancer development, including cell migration and invasion.22,23 In this paper, we found that CMTM3 expression was associated with favorable survival of gastric cancer patients, indicating that it might be a novel molecular prognostic marker. This finding not only expands the understanding of CMTM3 in cancer development, but also contributes to translating a TSG into clinical applications. In addition, expression of CMTM3 was found to be statistically associated with the depth of primary tumor. Previous reports also showed that the use of the TNM classification has predictive value for survival, even after changes to each edition.24
Another interesting finding in our study is that CMTM3 expression was associated with gender, and had a larger proportion of positive expression in males. In our previous study, CMTM3, which has the highest expression level in testes, could repress androgen receptor transactivation, perhaps through its typical leucine zipper and two LXXLL motifs.5 It is well known that the incidence of gastric cancer is higher in males than in females. In fact, there has been some evidence that androgen and its receptor play an important role in modifying the development of gastric cancer.25 Also, genes and proteins related to androgen synthesis and metabolism have been reported as correlating factors in gastric cancer.26 The exact function of CMTM3 and its relationship with androgen receptor in gastric cancer requires further study.
According to the present studies of CMTM3, it seems that this gene has tissue specificity, including expression profile, methylation status, and function. In gastric cancer, the downregulation of CMTM3 and the methylation of its regulatory region frequently happened. However, there was no methylation in any of the nine hepatocellular, 17 lung, and five prostate tumors we tested.6 In a study of non-small-cell lung cancer, methylation of CMTM3 was undetectable in 78 tumor tissues and matched normal tissues.27 In the respect of function, CMTM3 is a tumor growth inhibitor and an apoptosis inducer in CNE-2 cells.6 Nevertheless, in gastric cancer cells, it is a regulator of cell migration and invasion. These results indicate that the function of CMTM3 in tumorigenesis is one of tissue and cell specificity.
The mechanism of CMTM3 in migration and invasion of gastric cancer cells requires more study. Until now, a pattern of MARVEL proteins has been identified to contribute to tight junction regulation.28,29 These proteins are not only correlated with tight junction regulation, but also with cancer cell migration and invasion.30 Another CMTM family member, CMTM8, is also a regulator of the Erk1/2 signaling pathway.10 In this study, we found that CMTM3 may suppress cell migration and invasion through the inhibition of MMP2 expression and phosphorylation of Erk1/2, which provides us with a better understanding of the role of MARVEL proteins.
In sum, for the first time, our study shows the function and mechanism of CMTM3 and the relationship between its expression and clinical outcomes in gastric cancer. CMTM3 may be used as a marker for prognosis of gastric cancer, which would help to define initial medication to obtain a clinical benefit.
Acknowledgments
This study was supported by the National Key Technology R&D Program (2011ZX09307-001-05) and National Natural Science Foundation of China (30972780).
Disclosure Statement
The authors have no conflict of interest.
Funding information
National Key Technology R&D Program (2011ZX09307-001-05). National Natural Science Foundation of China (30972780).
Supporting Information
Additional supporting information may be found in the online version of this article:
Survival curves of different subgroups.
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Jankowski JA, Odze RD. Biomarkers in gastroenterology: between hope and hype comes histopathology. Am J Gastroenterol. 2009;104:1093–6. doi: 10.1038/ajg.2008.172. [DOI] [PubMed] [Google Scholar]
- 3.Han W, Ding P, Xu M, et al. Identification of eight genes encoding chemokine-like factor superfamily members 1-8 (CKLFSF1-8) by in silico cloning and experimental validation. Genomics. 2003;81:609–17. doi: 10.1016/s0888-7543(03)00095-8. [DOI] [PubMed] [Google Scholar]
- 4.Shao L, Li T, Mo X, et al. Expressional and functional studies of CKLF1 during dendritic cell maturation. Cell Immunol. 2010;263:188–95. doi: 10.1016/j.cellimm.2010.03.015. [DOI] [PubMed] [Google Scholar]
- 5.Wang Y, Li T, Qiu X, et al. CMTM3 can affect the transcription activity of androgen receptor and inhibit the expression level of PSA in LNCaP cells. Biochem Biophys Res Commun. 2008;371:54–8. doi: 10.1016/j.bbrc.2008.03.143. [DOI] [PubMed] [Google Scholar]
- 6.Wang Y, Li J, Cui Y, et al. CMTM3, located at the critical tumor suppressor locus 16q22.1, is silenced by CpG methylation in carcinomas and inhibits tumor cell growth through inducing apoptosis. Cancer Res. 2009;69:5194–201. doi: 10.1158/0008-5472.CAN-08-3694. [DOI] [PubMed] [Google Scholar]
- 7.Song HS, Shi S, Lu XZ, et al. Intracellular CMTM2 negatively regulates human immunodeficiency virus type-1 transcription through targeting the transcription factors AP-1 and CREB. Chin Med J (Engl) 2010;123:2440–5. [PubMed] [Google Scholar]
- 8.Plate M, Li T, Wang Y, et al. Identification and characterization of CMTM4, a novel gene with inhibitory effects on HeLa cell growth through Inducing G2/M phase accumulation. Mol Cells. 2010;29:355–61. doi: 10.1007/s10059-010-0038-7. [DOI] [PubMed] [Google Scholar]
- 9.Niu J, Li H, Zhang Y, et al. Aberrant expression of CKLF-like MARVEL transmembrane member 5 (CMTM5) by promoter methylation in myeloid leukemia. Leuk Res. 2011;35:771–6. doi: 10.1016/j.leukres.2010.11.023. [DOI] [PubMed] [Google Scholar]
- 10.Zhang W, Mendoza MC, Pei X, et al. Down-regulation of CMTM8 induces epithelial-to-mesenchymal transition-like changes via c-MET/extracellular signal-regulated kinase (ERK) signaling. J Biol Chem. 2012;287:11850–8. doi: 10.1074/jbc.M111.258236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lind GE, Ahlquist T, Kolberg M, et al. Hypermethylated MAL gene – a silent marker of early colon tumorigenesis. J Transl Med. 2008;6:13. doi: 10.1186/1479-5876-6-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Buffart TE, Overmeer RM, Steenbergen RD, et al. MAL promoter hypermethylation as a novel prognostic marker in gastric cancer. Br J Cancer. 2008;99:1802–7. doi: 10.1038/sj.bjc.6604777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shao L, Cui Y, Li H, et al. CMTM5 exhibits tumor suppressor activities and is frequently silenced by methylation in carcinoma cell lines. Clin Cancer Res. 2007;13:5756–62. doi: 10.1158/1078-0432.CCR-06-3082. [DOI] [PubMed] [Google Scholar]
- 14.Ajani JA, Barthel JS, Bekaii-Saab T, et al. Gastric cancer. J Natl Compr Canc Netw. 2010;8:378–409. doi: 10.6004/jnccn.2010.0030. [DOI] [PubMed] [Google Scholar]
- 15.Japanese Gastric Cancer Association. Japanese classification of gastric carcinoma. 3rd English edition. Gastric Cancer. 2011;14:101–12. doi: 10.1007/s10120-011-0041-5. [DOI] [PubMed] [Google Scholar]
- 16.Li Z, Zhan W, Wang Z, et al. Inhibition of PRL-3 gene expression in gastric cancer cell line SGC7901 via microRNA suppressed reduces peritoneal metastasis. Biochem Biophys Res Commun. 2006;348:229–37. doi: 10.1016/j.bbrc.2006.07.043. [DOI] [PubMed] [Google Scholar]
- 17.Carvajal-Carmona LG, Cazier JB, Jones AM, et al. Fine-mapping of colorectal cancer susceptibility loci at 8q23.3, 16q22.1 and 19q13.11: refinement of association signals and use of in silico analysis to suggest functional variation and unexpected candidate target genes. Hum Mol Genet. 2011;20:2879–88. doi: 10.1093/hmg/ddr190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Friedl P, Wolf K. Tumour-cell invasion and migration: diversity and escape mechanisms. Nat Rev Cancer. 2003;3:362–74. doi: 10.1038/nrc1075. [DOI] [PubMed] [Google Scholar]
- 19.Li X, Cheung KF, Ma X, et al. Epigenetic inactivation of paired box gene 5, a novel tumor suppressor gene, through direct upregulation of p53 is associated with prognosis in gastric cancer patients. Oncogene. 2012;31:3419–30. doi: 10.1038/onc.2011.511. [DOI] [PubMed] [Google Scholar]
- 20.Hou Q, Wu YH, Grabsch H, et al. Integrative genomics identifies RAB23 as an invasion mediator gene in diffuse-type gastric cancer. Cancer Res. 2008;68:4623–30. doi: 10.1158/0008-5472.CAN-07-5870. [DOI] [PubMed] [Google Scholar]
- 21.Cervantes A, Rosello S, Roda D, Rodriguez-Braun E. The treatment of advanced gastric cancer: current strategies and future perspectives. Ann Oncol. 2008;19(Suppl 5):v103–7. doi: 10.1093/annonc/mdn321. [DOI] [PubMed] [Google Scholar]
- 22.Hsu PI, Hsieh HL, Lee J, et al. Loss of RUNX3 expression correlates with differentiation, nodal metastasis, and poor prognosis of gastric cancer. Ann Surg Oncol. 2009;16:1686–94. doi: 10.1245/s10434-009-0428-2. [DOI] [PubMed] [Google Scholar]
- 23.Matsuoka J, Yashiro M, Sakurai K, et al. Role of the stemness factors sox2, oct3/4, and nanog in gastric carcinoma. J Surg Res. 2012;174:130–5. doi: 10.1016/j.jss.2010.11.903. [DOI] [PubMed] [Google Scholar]
- 24.Warneke VS, Behrens HM, Hartmann JT, et al. Cohort study based on the seventh edition of the TNM classification for gastric cancer: proposal of a new staging system. J Clin Oncol. 2011;29:2364–71. doi: 10.1200/JCO.2010.34.4358. [DOI] [PubMed] [Google Scholar]
- 25.Kominea A, Konstantinopoulos PA, Kapranos N, et al. Androgen receptor (AR) expression is an independent unfavorable prognostic factor in gastric cancer. J Cancer Res Clin Oncol. 2004;130:253–8. doi: 10.1007/s00432-003-0531-x. [DOI] [PubMed] [Google Scholar]
- 26.Freedman ND, Ahn J, Hou L, et al. Polymorphisms in estrogen- and androgen-metabolizing genes and the risk of gastric cancer. Carcinogenesis. 2009;30:71–7. doi: 10.1093/carcin/bgn258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang Y, Wang R, Song H, et al. Methylation of multiple genes as a candidate biomarker in non-small cell lung cancer. Cancer Lett. 2011;303:21–8. doi: 10.1016/j.canlet.2010.12.011. [DOI] [PubMed] [Google Scholar]
- 28.Raleigh DR, Marchiando AM, Zhang Y, et al. Tight junction-associated MARVEL proteins marveld3, tricellulin, and occludin have distinct but overlapping functions. Mol Biol Cell. 2010;21:1200–13. doi: 10.1091/mbc.E09-08-0734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mariano C, Sasaki H, Brites D, Brito MA. A look at tricellulin and its role in tight junction formation and maintenance. Eur J Cell Biol. 2011;90:787–96. doi: 10.1016/j.ejcb.2011.06.005. [DOI] [PubMed] [Google Scholar]
- 30.Kojima T, Takasawa A, Kyuno D, et al. Downregulation of tight junction-associated MARVEL protein marvelD3 during epithelial-mesenchymal transition in human pancreatic cancer cells. Exp Cell Res. 2011;317:2288–98. doi: 10.1016/j.yexcr.2011.06.020. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Survival curves of different subgroups.