Abstract
CD20 is expressed in most B-cell lymphomas and is a critical molecular target of rituximab. Some B-cell lymphomas show aberrant CD20 expression, and rituximab use in these patients is controversial. Here we show both the molecular mechanisms and the clinical significance of de novo diffuse large B-cell lymphomas (DLBCL) that show a CD20 immunohistochemistry (IHC)-positive and flow cytometry (FCM)- negative (IHC[+]/FCM[−]) phenotype. Both IHC and FCM using anti-CD20 antibodies L26 and B1, respectively, were analyzed in 37 of the 106 cases of de novo DLBCL; 8 (22%) of these cases were CD79a(+)/CD20(+) with IHC and CD19(+)/CD20(−) with FCM. CD20 (MS4A1) mRNA expression was significantly lower in IHC(+)/FCM(−) cells than in IHC(+)/FCM(+) cells (P = 0.0005). No genetic mutations were detected in MS4A1 promoter and coding regions. Rituximab-mediated cytotoxicity in the CDC assay using IHC(+)/FCM(−) primary cells was significantly lower than in IHC(+)/FCM(+) cells (P < 0.05); however, partial effectiveness was confirmed. FCM using rituximab detected CD20 more efficiently than B1. No significant difference was observed between IHC(+)/FCM(−) and IHC(+)/FCM(+) patients in overall survival (P = 0.664). Thus, lower expression of CD20 mRNA is critical for the CD20 IHC(+)/FCM(−) phenotype. Lower CD20 expression with FCM does not rule out rituximab use in these patients if expression is confirmed with IHC. FCM using rituximab may be more informative than B1 for predicting rituximab effectiveness in IHC(+)/FCM(−) cases.
Keywords: Diffuse large B-cell lymphoma, CD20, rituximab, immunohistochemistry, flow cytometry
CD20 is a cell surface antigen expressed specifically on most human B cells.1 Because CD20 is also expressed on more than 90% of B-cell lymphoma cells, CD20 has become a good molecular target for monoclonal antibody therapeutics.2,3 Rituximab is a mouse–human chimeric monoclonal antibody targeting CD20. Previous reports indicate that clinical outcomes in patients with B-cell lymphomas have been significantly improved with rituximab with conventional chemotherapies.4–6 However, the overall survival (OS) is still not satisfactory because more than 50% of B-cell lymphoma patients show relapse/recurrence of disease after several years.4 Thus, we believe that confirming the mechanisms of rituximab resistance7,8 is important for further improving the OS and progression free survival (PFS) of B-cell lymphoma patients.
Recently we reported that downregulation of CD20 protein expression after combination chemotherapy with rituximab is a critical reason for rituximab resistance.9–11 Other groups have indicated that abnormalities in CD20 expression because of shaving,12,13 genetic mutations or deletions,14–16 aberrant splicing,17 and internalization into the cytoplasm18,19 strongly correlate with lower sensitivity to rituximab treatment. Furthermore, lower expression of CD20 has been confirmed in even among patients with the same disease, such as diffuse large B-cell lymphoma (DLBCL).20–22 Previous reports regarding ADCC and CDC activity induced by rituximab indicate that lower protein expression is strongly correlated with the efficacy of anti-CD20 antibodies.23,24 Thus, knowing the level of CD20 protein expression may be very important in the clinical setting for predicting the outcome of anti-CD20 antibody therapy.
Although we and others recently recognized that some B-cell lymphoma patients show discrepancies in CD20 protein expression showing an immunohistochemistry (IHC)-positive and flow cytometry (FCM)- negative (IHC[+] and FCM[−]) phenotype,21,25 neither molecular mechanisms of this phenotype nor rituximab sensitivities have been elucidated. In this study, we analyzed the frequency of occurrence and clinical features of de novo DLBCL patients who showed the CD20 IHC(+)/FCM(−) phenotype and analyzed the molecular basis of the phenotype using primary clinical samples. In the present study we also examine the rituximab sensitivity of those cells compared with CD20 IHC(+)/FCM(+) B-cell lymphoma cells to determine whether rituximab can still be utilized in those patients in combination with conventional chemotherapies.
Materials and Methods
Patients and lymphoma tissue samples
Between January 2006 and May 2012 in Nagoya University Hospital, 106 patients were diagnosed with de novo DLBCL (Table1). All patients were treated with combination chemotherapy that included rituximab. The final follow up was on 22 November 2012. Lymphoma tissue was harvested and used for pathological analysis, and if a sufficient volume of tissue was obtained, FCM, chromosomal analysis, DNA, RNA and protein extraction, and cryopreservation were performed. Lymphoma tissues showing the CD20 IHC(+)/FCM(−) phenotype in the affiliated hospital were also sent to our laboratory as snap-frozen samples and utilized. These studies were conducted with institutional review board approval from the Nagoya University School of Medicine, and written informed consent was obtained from each patient analyzed in accordance with the Declaration of Helsinki.
Table 1.
Patients' characteristics of DLBCL with CD20 IHC(+)/FCM(−) phenotype
| Total | CD20 IHC(+)/FCM(+)‡ | CD20 IHC(+)/FCM(−)§ | P-value‡ vs§ | |
|---|---|---|---|---|
| Patients number (%) | 106 (100) | 29/37 (78) | 8/37 (22) | |
| Age | ||||
| Median [range] | 66 [26–88] | 65 [35–81] | 60 [52–77] | 0.394 |
| >60 y.o. | 80 (75) | 19 (66) | 3 (38) | 0.228 |
| Gender: male | 74 (70) | 20 (69) | 5 (63) | 1 |
| PS, >1 | 18 (17) | 7 (24) | 0 (0) | 0.308 |
| LDH, >UNL | 61 (58) | 17 (59) | 6 (75) | 0.683 |
| Extra nodal site(s), >1 | 23 (22) | 8 (28) | 2 (25) | 1 |
| Stage, III/IV | 57 (54) | 18 (62) | 6 (75) | 0.685 |
| IPI score at diagnosis | ||||
| 0, 1 | 33 (31) | 9 (31) | 2 (25) | 0.779 |
| 2 | 35 (33) | 8 (28) | 3 (38) | |
| 3 | 18 (17) | 4 (14) | 2 (25) | |
| 4, 5 | 20 (19) | 8 (28) | 1 (13) | |
| IHC classification† | ||||
| GCB | 34/72 (47) | 9/23 (39) | 3/5 (60) | 0.624 |
| Non-GCB | 38/72 (53) | 14/23 (61) | 2/5 (40) | |
| EBV status† | ||||
| EBER-ISH | 6/75 (8) | 0/22 (0) | 0/6 (0) | |
| Light chain restriction in FCM† | ||||
| Kappa | 13/28 (46) | 9/20 (45) | 4/8 (50) | 0.167 |
| Lambda | 6/28 (21) | 6/20 (30) | 0/8 (0) | |
| Negative | 9/28 (32) | 5/20 (25) | 4/8 (50) | |
The total patients' number examined in each analysis are indicated as denominators. ‡CD20 IHC(+)/FCM(+). §CD20 IHC(+)/FCM(−). EBV, Epstein–Barr virus; EBER-ISH, EBV-encoded RNA-in situ hybridization; GCB, germinal center B-cell type; IPI, international prognostic index; PS, performance status.
Primary B-cell lymphoma cells and cell lines
Primary B-cell lymphoma tissues were separated into single-cell suspensions in 10-cm culture dishes with RPMI1640 culture medium (Sigma–Aldrich, St. Louis, MO, USA). The B-cell lymphoma/leukemia cell lines SU-DHL4, SU-DHL-6, SU-DHL10, TMD8 and Daudi were used as positive controls for CD20 expression. RRBL19–11 and WILL226 are cell lines established from B-cell lymphoma patients showing CD20-negative phenotypic changes after repeated chemotherapy with rituximab.
Confirmation of CD20 protein expression with immunohistochemistry positive and flow cytometry analyses
For IHC analysis, CD20 protein expression was confirmed using mouse anti-CD20 antibody L26 (Dako, Carpinteria, CA, USA). A pan-B-cell marker CD79a expression for the detection of B-cell was confirmed by anti-CD79a antibody (Dako). FCM analysis was performed with a BD FACSAria III cell sorter (Becton Dickinson, Franklin Lakes, NJ, USA). For FCM, CD20 expression was confirmed with mouse anti-CD20 antibody B9E9 (a mouse monoclonal IgG2a antibody recognizing the B1 epitope [Beckman Coulter, Fullerton, CA, USA]) or B1 [Dako]). The percentages of negative and positive cells from FCM were determined after subtracting background from use of an isotypic control antibody (mouse IgG1 [Beckman Coulter]). B cell lymphoma cell population was basically confirmed by CD19 positivity in FCM analysis. FCM data of CD10, CD5, Igκ and Igλ were also referenced for lymphoma cell determination. If the percentage of CD20-positive cells in the tumor cell population was <12.5%, we considered those cells CD20 FCM negative. MFI of CD20 was measured with a BD FACSAria III cell sorter.
DNA, RNA and protein extraction from lymphoma tissues
Genomic DNA from tumor cells was extracted as described.10
Immunoblotting
Immunoblotting using whole-cell lysates of lymphoma cells was performed as described previously.9,10,27
In vitro CDC assay
For the CDC assay, 1.0 × 106 cells were resuspended in 500 μL normal human serum and the same amount of complete medium with 10 μg/mL rituximab at 37°C for 30 min. Normal human serum was obtained from healthy volunteer donors. Dead cells were evaluated with DAPI and Annexin V-FITC staining. Briefly, cells placed in 96-well plates were stained with 2 μg/mL DAPI and 2 μg/mL Annexin V-FITC for 15 min at room temperature in the dark and evaluated with FCM (FACSCalibur or FACSAriaII [BD]).
Detailed information of analytical procedures is also indicated in the Data S1 and S2.
Results
De novo diffuse large B-cell lymphoma patients with the CD20 IHC(+)/FCM(−) phenotype
CD20 protein expression was confirmed with IHC using L26 antibody for all de novo DLBCL patients diagnosed in Nagoya University Hospital (n = 106) (Table1). If sufficient lymphoma materials were harvested at diagnosis, FCM analysis was also performed (n = 37; 34.9%). Of those 37 cases, 8 (21.6%) were CD20-negative with FCM analysis, despite the CD20-positive phenotype with IHC. A CD20 IHC(−)/FCM(+) phenotype was not observed in this analysis. These results indicated that the CD20 IHC(+)/FCM(−) phenotype was not rare in de novo DLBCL patients.
Primary or cryopreserved lymphoma tissues showing the CD20 IHC(+)/FCM(−) phenotype obtained in Nagoya University Hospital (n = 8) and the affiliated hospitals (n = 4) were used for further analyses (Table2). Representatives of this phenotype are shown in Figure1(a) (IHC) and 1(b) (FCM). For IHC analysis, B cells were confirmed with anti-CD79a antibody, which recognizes a B-cell receptor component. CD20 protein expression was also confirmed in CD79a-positive B cells (Fig.1a). For FCM analysis, lymphoma cells were gated by side scatter and forward scatter or the CD45 expression level, and CD19-positive B-cell lymphoma populations were confirmed (Fig.1b). However, CD20 expression was not confirmed in these cell populations. B-cell light chain restriction was also confirmed with FCM, and 4 out of 12 cases (33.3%) expressed neither kappa-light chains nor lambda-light chains (Fig.1b and Table2). Interestingly, seven out of eight patients who expressed either the kappa or lambda chain expressed the kappa chain (87.5% of light chain-expressing patients). This percentage in CD20 IHC(+)/FCM(−) patients was higher tendency than that in CD20 IHC(+)/FCM(+) patients (nine out of 20 patients in Table1 [45.0%]).
Table 2.
Clinical characteristics and the molecular back grounds of the CD20 IHC(+)/FCM(−) patients
| CD20 expression |
||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| UPN | Age | Gender | Diag. | Stage | Patho. source | IHC | FCM | RT | CDS mutation | Promoter region mutation# | Karyotype | Light chain restriction | IPI (score) | Treatment | Response (alive or death) | Survival |
| 1 | 55 | F | DLBCL | IAE | LN | + | − | ↓ | − | − | Complex | Kappa | Low (0) | R-CHOP | CR (A) | 37M |
| 2 | 60 | M | DLBCL | IVB | LN | + | − | NT | NT | NT | Complex | − | High (4) | R-COP | CR (A) | 37M |
| 3 | 71 | M | DLBCL | IIA | LN | + | − | ↓ | − | − | Complex | Lambda | Low (1) | R-THP-COP | CR (A) | 32M |
| 4 | 66 | M | DLBCL | IIA | LN | + | − | ↓ | − | − | NE | Kappa | Low (1) | R-CHOP | CR (A) | 32M |
| 5 | 92 | F | DLBCL | IIA | LN | + | − | ↓ | − | − | Normal | Kappa | Low (1) | THP-COP | CR (D) | 11M |
| 6 | 51 | M | DLBCL | IA | LN | + | − | ↓ | − | − | NE | Kappa | Low (0) | R-CHOP | CR (A) | 25M |
| 7 | 66 | M | DLBCL | IIIA | LN | + | − | NT | NT | NT | Complex | Kappa | H-I (3) | THP-COP | PD (D) | 48M |
| 8 | 77 | F | DLBCL | IVB | GI, BM | + | − | ↓ | − | − | Normal | − | High (4) | R-CHOP | CR (D) | 13M |
| 9 | 52 | M | DLBCL | IVA | LN | + | − | ↓ | − | − | NE | − | L-I (2) | R-CHOP | CR (A) | 14M |
| 10 | 60 | F | DLBCL | IIIAE | LN | + | − | NT | NT | NT | NE | Kappa | L-I (2) | R-CHOP | CR (A) | 10M |
| 11 | 83 | M | DLBCL | IIIA | LN | + | − | ↓ | − | − | NE | − | H-I (3) | R-EPOCH | NA (A) | 9M |
| 12 | 67 | M | DLBCL | IIA | LN | + | − | ↓ | − | − | Normal | Kappa | L-I (2) | R-CHOP | CR (A) | 7M |
Black arrow, downregulated; BM, bone marrow; CDS, coding sequence of MS4A1 gene; Diag., diagnosis; GI, gastrointestinal; H-I, high-intermediate; L-I, low-intermediate; LN, lymphnode; NE, not evaluated; NT, not tested; Patho. Source, sources of tumor tissues for pathological analysis; R-CHOP, rituximab, cyclophosphamide, doxorubicin vincristine and prednisolone; RT, RT-PCR; THP, tetrahydropyranyl adriamycin; EPOCH, etoposide, vincristine, cyclophosphamide and prednisolone; #, 1000 bp upstream from the transcription start site (−1000 to +1) of MS4A1 gene.
Figure 1.
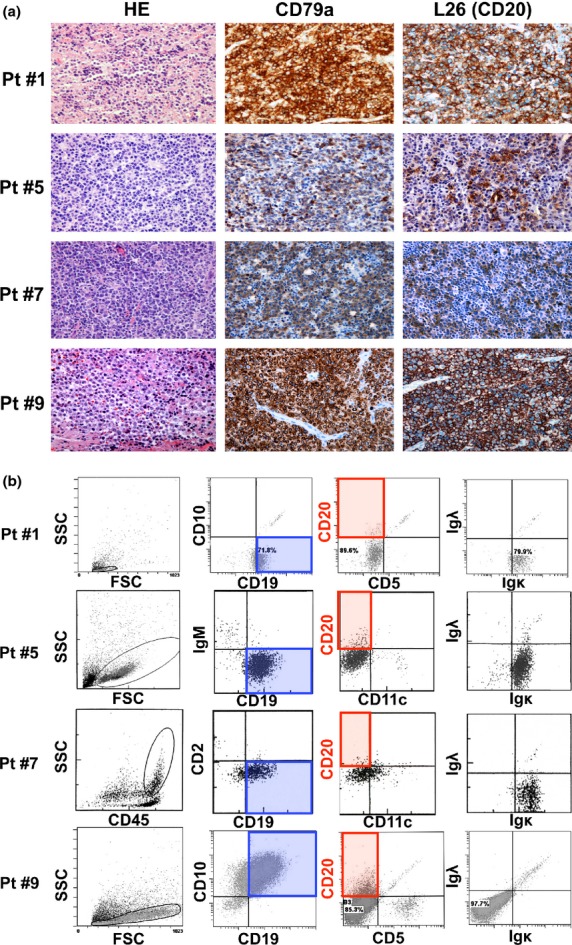
Immunohistochemistry (IHC) and flow cytometry (FCM) analysis of de novo diffuse large B-cell lymphoma (DLBCL) patients with the CD20 IHC(+)/FCM(−) phenotype. Representative data for four patients are indicated. (a) IHC analysis using anti-CD79a and L26 (anti-CD20) antibody. All those patients were diagnosed as CD79a(+) and CD20(+) de novo DLBCL. (b) FCM analysis of patients showing the CD20 IHC(+)/FCM(−) phenotype. B-cell lymphoma cells were confirmed by gating of SSC, FSC or CD45 expression levels, as well as the CD19-positive phenotype. CD20 expression in those cells was significantly low with FCM analysis. FSC, forward scatter; HE, hematoxylin–eosin staining; Ig, immunoglobulin; L26, anti-CD20 antibody for IHC; Pt #, patient number; SSC; side scatter. Original magnifications (a); ×200 (Olympus BX51TF microscope, Olympus, Tokyo, Japan, and Nikon DS-Fi1 camera, Nikon, Tokyo, Japan).
Lower expression of CD20 mRNA and protein in CD20 IHC(+)/FCM(−) B-cell lymphoma cells
Total RNA was prepared from CD20 IHC(+)/FCM(−) lymphoma cells for RT-PCR analysis. Semi-quantitative RT-PCR indicated that CD20 (MS4A1) mRNA expression was generally lower in CD20 IHC(+)/FCM(−) cells than that in CD20 positive control cells (Fig.2a). Quantitative RT-PCR was also performed (Fig.2b). Note that CD20 IHC(−)/FCM(−) cells were harvested from patients who showed a CD20-negative phenotypic change after repeated rituximab treatment and who showed clinical resistance to rituximab.9,10 CD20 mRNA expression was significantly lower in CD20 IHC(+)/FCM(−) cells than in IHC(+)/FCM(+) cells (P = 0.0005) and tended to be higher than in IHC(−)/FCM(−) cells (not significant).
Figure 2.
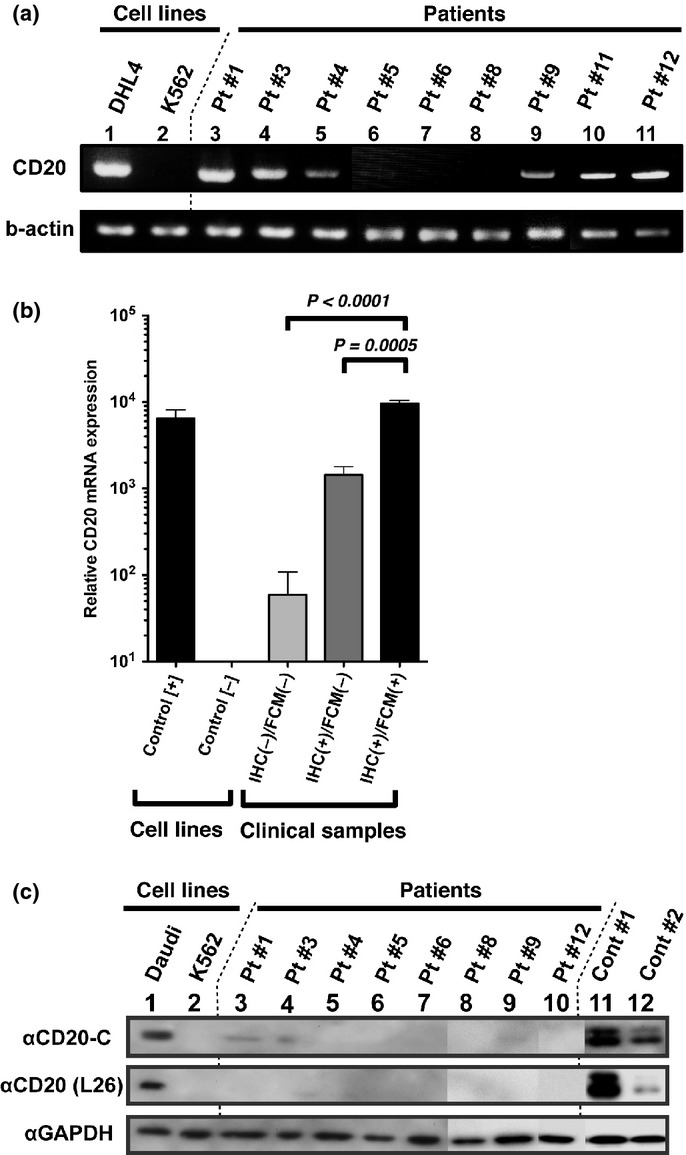
Confirmation of CD20 mRNA and protein expression with (a) semi-quantitative RT-PCR, (b) quantitative RT-PCR and (c) immunoblotting. Total mRNA and protein lysates were obtained from primary lymphoma samples for RT-PCR and immunoblotting. (a) The coding sequence of MS4A1 (CD20) mRNA was amplified using semi-quantitative RT-PCR. Beta actin mRNA was amplified as an internal control. (b) Quantitative RT-PCR for MS4A1 gene expression was performed. As an internal control, GAPDH expression was analyzed, and all data were normalized to its expression. (c) Immunoblotting was performed to confirm the CD20 protein expression. CD20-C recognizes the C-terminal region of the CD20 protein. The L26 antibody, which recognizes intracellular domains of the CD20 protein, was also used in this assay in addition to immunohistochemistry (IHC) analysis. Proteins from the Daudi and K562 cell lines were used as positive and negative controls, respectively. Cont #1 and #2 were derived from diffuse large B-cell lymphoma (DLBCL) clinical samples showing the CD20 IHC(+)/FCM(+) phenotype.
Immunoblotting analysis using two anti-CD20 antibodies that recognize different domains of the CD20 protein indicated that CD20 expression was generally lower in lymphoma samples showing the CD20 IHC(+)/FCM(−) phenotype than in positive control samples from patients showing the CD20 IHC(+)/FCM(+) phenotype (Fig.2c, lanes 3 to 10 vs lanes 11 and 12). Bands showing faint CD20 expression were confirmed with immunoblotting after a longer exposure (data not shown). These data suggest that lower MS4A1 gene expression may contribute to the lower CD20 protein expression in CD20 IHC(+)/FCM(−) cells, as seen with immunoblotting and FCM analyses. These results also indicate that CD20 protein accumulation in the cytoplasm is not a likely explanation for the CD20 FCM(−) phenotype.
Rituximab recognizes the CD20 cell surface antigen more readily than the B1 antibody with flow cytometry analysis
To confirm the rituximab effectiveness on CD20 IHC(+)/FCM(−) cells, we first performed FCM analysis using fluorescent (Alexa 488)-labeled rituximab in addition to a conventional anti-CD20 antibody B1 (Dako) (Fig3). We used primary B-cell lymphoma cells and cell lines showing the following phenotypes: CD20 IHC(+)/FCM(+) (primary; n = 10, cell lines; n = 3), IHC(+)/FCM(−) (primary; n = 5) and IHC(−)/FCM(−) after using rituximab (cell lines; n = 2). When using the B1 antibody (Fig.3a), the MFI of CD20 IHC(+)/FCM(−) cells was significantly lower than that of IHC(+)/FCM(+) cells (P = 0.03), consistent with the result of FCM analysis using the B9E9 antibody that recognized the B1 epitope of the CD20 protein (Fig.1b). Using the same cell samples, FCM analysis using Alexa 488-labeled rituximab was also performed (Fig.3b). The MFI of CD20 IHC(+)/FCM(−) cells showed a much lower tendency than that of IHC(+)/FCM(+) cells, but the difference was not significant (P = 0.21). Rituximab, as well as B1, did not detect CD20 expression in the IHC(−)/FCM(−) B-cell lines, RRBL1 and WILL2. These data suggest that CD20 protein is faintly expressed on the surface of CD20 IHC(+)/FCM(−) cells and that rituximab can detect CD20 on the cell surface more effectively than the B1 (B9E9) antibody, even when the expression is very faint.
Figure 3.
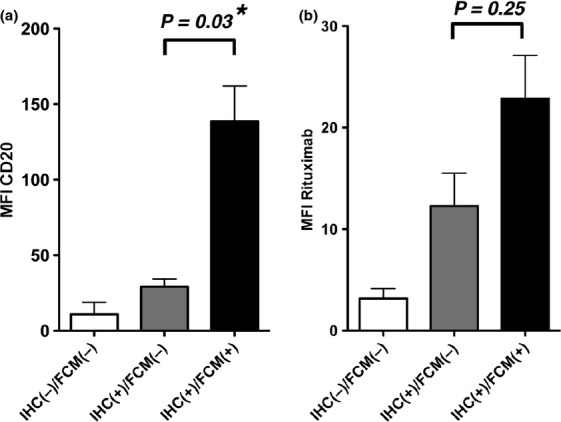
Flow cytometry (FCM) analyses using anti-CD20 B1 antibody and fluorescent-labeled rituximab. (a) FCM analysis using anti-CD20 B1 antibody was performed, and the MFIs of lymphoma cells were measured. RRBL1 and WILL2 cells were utilized as representative CD20 IHC(−)/FCM(−) samples. The P-value is shown, and the asterisk indicates a statistically significant difference. (b) The MFI value using Alexa 488-labeled rituximab was also analyzed in the same lymphoma samples as (a).
CDC activity induced by rituximab is partially effective on CD20 IHC(+)/FCM(−) lymphoma cells
We next performed a rituximab-induced in vitro CDC assay using the same primary lymphoma cells and cell lines as in Figure3. Cells were cultured with or without rituximab for 30 min, and the dead cells were calculated by counting Annexin V- and PI- (or DAPI-) positive cells. Representative data are depicted in Figure4(a). Almost 100% of CD20-positive control SU-DHL4 cells were killed by rituximab-induced CDC activity, but CD20-negative K562 and WILL2 cells were not killed under the same conditions. For the CD20 IHC(+)/FCM(−) cells, partial cell death was observed (Fig.4a, #8). Because normal T cells and/or stromal cells were contaminating cell types in this assay when using primary lymphoma cells from lymphoma tissues, normalization to the B-cell population percentage estimated by determining the CD19-positive cell population was required (data not shown). This normalization for the percent of rituximab-induced cell death was performed for all data obtained from primary lymphoma samples. The relationship between CD20 MFI and the percent of cell death by rituximab-induced CDC activity is indicated in Figure4(b) (MFI; B1) and 4(c) (MFI; rituximab). From these data, a positive correlation was confirmed between the CD20 MFI level and the rituximab effectiveness, as reported previously.23,24 Importantly, rituximab was partially effective on CD20 IHC(+)/FCM(−) cells in vitro (cell death%; range 47–81%) compared to IHC(+)/FCM(+) cells (68–100%) (Fig.4b,c). Significantly lower efficacy of rituximab in the CDC assay in IHC(+)/FCM(−) cells compared with IHC(+)/FCM(+) cells was confirmed (P < 0.05) (Fig.4d). CDC activity was not observed in CD20 IHC(−)/FCM(−) RRBL1 and WILL2 cells (black diamonds in Fig.4b,c). These data suggest that rituximab-induced cytotoxicity can be observed with this CDC assay if the CD20 expression is confirmed with rituximab FCM analysis.
Figure 4.
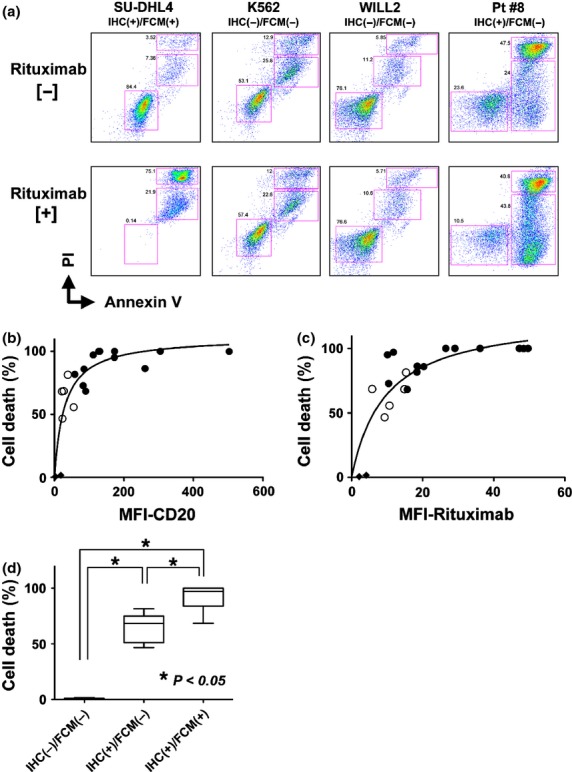
In vitro CDC activity induced by rituximab. (a) Annexin V-PI staining was performed with/without rituximab and human serum treatment in vitro. In this assay, living, pro-apoptotic, or dead cell populations were separated in a 2-dimensional graph, and the percentage of each group was calculated. The SU-DHL4 cell line was a positive control. The K562 and WILL2 cell lines were negative controls. Representative primary lymphoma cells showing the IHC(+)/FCM(−) phenotype were obtained from patient #8 and utilized in this assay. (b) The relationship between the percent of cell death with rituximab-induced CDC activity and the CD20-B1-MFI value (performed in Fig.3) was plotted in this graph. Primary lymphoma samples showing CD20 IHC(+)/FCM(+) (black circles) and IHC(+)/FCM(−) (white circles) were used. Each circle indicates one lymphoma sample from a corresponding patient. RRBL1 and WILL2 cells are indicated in black diamonds. (c) The same analysis using the rituximab-MFI values is shown. Nonlinear regression curve fitting is indicated as curved lines. (d) Cell death percentages were statistically compared using Turkey's multiple comparison test. Asterisks indicate significant differences.
No significant difference was observed in the overall survival rate between CD20 IHC(+)/FCM(+) and IHC(+)/FCM(−) patients
The OS rate was analyzed using Kaplan–Meier analysis (Fig.5). All DLBCL patients analyzed (n = 106) were treated with rituximab and CHOP-based combination chemotherapy at Nagoya University Hospital. The OS and PFS of all these patients at 3 years were 77% and 65.2%, respectively (Fig.5a,b). The OS and the PFS of each group classified by the IPI28 are indicated in Figure5(c,d). Patients with the IHC(+)/FCM(−) phenotype tended to show a lower survival rate than IHC(+)/FCM(+) patients, but no significant difference was found between these two groups (P = 0.664) (Fig.5e).
Figure 5.
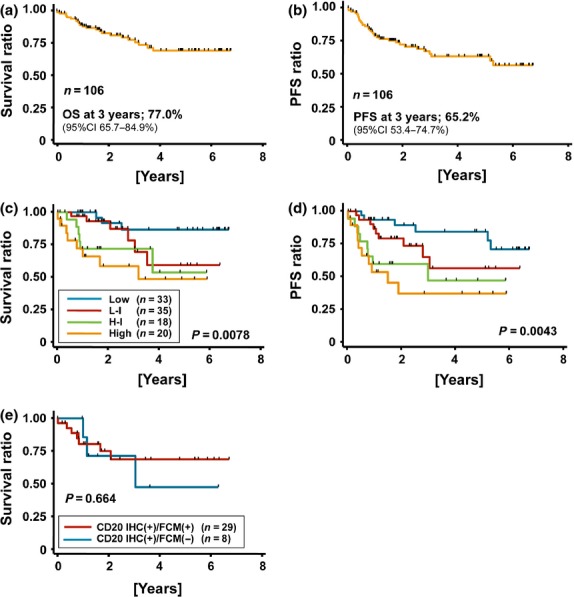
Prognosis of diffuse large B-cell lymphoma (DLBCL) patients with the CD20 IHC(+)/FCM(−) phenotype. (a) overall survival (OS) and (b) progression free survival (PFS) of DLBCL patients diagnosed in Nagoya University Hospital (n = 99). All patients were treated by combination chemotherapy with rituximab. These patients were classified by IPI, and the OS and PFS of each group are indicated in (c) and (d), respectively. (e) Comparison of OS of DLBCL patients who were diagnosed using both immunohistochemistry (IHC) and flow cytometry (FCM) (n = 36).
Mouse xenograft model of human diffuse large B-cell lymphoma with the CD20 IHC(+)/FCM(−) phenotype
A mouse xenograft model of human DLBCL was obtained by injecting primary DLBCL cells from the bone marrow of patient #8 (Table2) into the intra-peritoneal space of NOD/SCID mice (see Fig. S1).
Discussion
In this report, we showed that de novo DLBCL patients with the CD20 IHC(+)/FCM(−) phenotype are not rare, with a frequency of occurrence of 21.6% in patients analyzed with both IHC and FCM at diagnosis. Previous reports indicate the same phenomenon. Johnson et al. report that 16% of de novo DLBCL patients analyzed with both IHC and FCM (B9E9 antibody) showed reduced CD20 expression in FCM analyses despite a positive result with IHC analysis.25 Miyoshi et al. describe the relationship between the CD20 IHC-positive score and the FCM-positive (using B-Ly1 mouse monoclonal IgG1 antibody) rate in patients with de novo and relapsed DLBCL and follicular lymphoma.21 They also show that lower expression of CD20 with FCM is observed even with a higher IHC positive score. These reports and our data indicate that a discrepancy in protein expression analysis between IHC and FCM is a common phenomenon in DLBCL in the clinical setting.
Genetic mutations of MS4A1 have been speculated to be a molecular mechanism of the CD20 FCM-negative phenotype. Genetic mutations may lead to protein conformational changes in the CD20 protein. In particular, amino acid substitution in the large outer loop of CD20 may directly affect the effectiveness of antibody recognition,14,15,29,30 and mutations in the intracellular domain may lead to aberrant protein localization.14 We performed mutation analysis for the CD20 coding sequences (exons 3 to 8), and no mutations were found in patients with the CD20 IHC(+)/FCM(−) phenotype. Previous reports also indicate no significant missense or nonsense mutations in MS4A1.21,25 These data indicate that genetic mutations in MS4A1 are not the explanation for the CD20 IHC(+)/FCM(−) phenotype in de novo DLBCL. Nakamaki et al.16 report copy number loss of MS4A1 located at 11q12 in a specific patient who showed the CD20-negative phenotype after treatment with rituximab. Conventional chromosomal analysis showed that 11q12 genetic loss was not detected in patients with the IHC(+)/FCM(−) phenotype (data not shown). It remains possible that copy number loss of MS4A1 may, in part, be related to the lower CD20 mRNA expression in some de novo DLBCL patients with the CD20 IHC(+)/FCM(−) phenotype.
Quantitative RT-PCR analyses using primary lymphoma cells indicated that MS4A1 mRNA expression was significantly lower in CD20 IHC(+)/FCM(−) cells than in IHC(+)/FCM(+) cells (Fig.2b). Lower CD20 mRNA expression possibly meant that CD20 mRNA and protein expression was not confirmed in several samples from CD20 IHC(+)/FCM(−) cells in semi-quantitative RT-PCR (Fig.2a) and immunoblotting (Fig.2c). Because pan-B and C-terminal antibodies were used to detect CD20 protein in this assay, the possibilities of internalization of the protein into the cytoplasm and truncation of the protein can be mostly excluded as reasons for this phenotype. We did not examine why CD20 mRNA expression was repressed in those B cells, but possible explanations are as follows: (i) aberrant expression of transcription factors critical for MS4A1 expression such as IRF4, Pu.1, Pip11,31 and transforming growth factor-beta;32 (ii) abnormal epigenetic modulation by histone acetylation, methylation and DNA methylation at the MS4A1 promoter;11 and (iii) deregulation of normal cell differentiation into mature B cells. Using FCM analysis, 4 out of 12 patents with the CD20 IHC(+)/FCM(−) phenotype showed no light chain restrictions (Fig.1b and Table2). This finding suggests that some aberrant genetic and/or epigenetic mechanisms that downmodulate the light chain expression on lymphoma cells might correlate with this phenomenon. Further molecular analyses are required to demonstrate those possibilities.
An important question is whether significantly lower protein expression results in discrepancy in the data of IHC and FCM analyses. One likely explanation for this phenomenon is that the sensitivity for detecting CD20 protein is much higher with IHC using L26 than that with FCM using B9E9 and B1. If the expression is high enough, both analyses will indicate positive results, and if the CD20 mRNA level is almost 100 times lower than that in CD20 IHC(+)/FCM(+) cells, neither IHC nor FCM can detect CD20 protein expression, as seen in RRBL1 and WILL2 cells.9,26 If the CD20 mRNA expression level is almost 10 times lower than that in IHC(+)/FCM(+) cells, the anti-CD20 antibodies B9E9 and B1 in FCM may not sufficiently recognize the CD20 protein. Recent reports indicate that some newer generation antibodies such as ofatumumab,2,33 GA10134 and HuMab-7D824 show significantly higher cytotoxic activity than rituximab, even in the population of cells with lower CD20 protein expression. From these findings, using ofatumumab, GA101 and HuMab-7D8 may be a good strategy to overcome the partial rituximab resistance of CD20 IHC(+)/FCM(−) cells.
Interestingly, when using fluorescent-labeled rituximab in FCM, the difference in MFI between CD20 IHC(+)/FCM(−) and IHC(+)/FCM(+) cells was significantly smaller than that of FCM using the B1 antibody, indicating that the sensitivity of CD20 protein recognition by rituximab is much higher than that of the B1 and B9E9 antibodies. Because the partial efficacy of rituximab in inducing CDC activity was confirmed even in the CD20 IHC(+)/FCM(−) cells in the in vitro assay (Fig.3), utilization of rituximab for patients with the IHC(+)/FCM(−) phenotype may be still recommended in the clinical setting. Furthermore, using fluorescent-labeled rituximab in FCM at diagnosis may be much more informative than using B1/B9E9 to predict the rituximab effectiveness in vivo.
Our analysis showed no significant difference in OS between patients with CD20 IHC(+)/FCM(+) and IHC(+)/FCM(−) phenotypes (Fig.5), despite the significantly lower cytotoxic activity of rituximab on CD20 IHC(+)/FCM(−) cells compared to IHC(+)/FCM(+) cells (Fig.4d). Considering the in vitro CDC analysis, combination strategies with conventional chemo-regimens such as CHOP may improve the poor responsiveness to rituximab therapy, and, furthermore, ADCC and the direct signal transduction resulting in apoptosis can be induced in addition to CDC activity in vivo. Considering our clinical and in vitro data, rituximab utilization combined with chemotherapy is still recommended even for patients showing the CD20 IHC(+)/FCM(−) phenotype.
Acknowledgments
This work was supported in part by the National Cancer Center Research and Development Fund (23-A-17) and the Ministry of Education, Culture, Sports, Science and Technology (20591116, 24591388), Japan. We thank Naoe Goto and Takahiko Ito for providing the lymphoma clinical samples. We thank Takashi Sonoki for providing WILL2 cells. We thank Hiroshi Yamada and Mayumi Takatsu for preparation of the clinical samples and performing FCM. We also thank Chika Wakamatsu, Yukie Konishi, Manami Kira, Rie Kojima and Emi Kono for valuable laboratory assistance.
Disclosure Statement
H. Kiyoi had received research funding from Bristol-Myers Squibb, Novartis Pharma, Chugai Pharmaceutical Co., Ltd., and Kyowa Hakko Kirin Co., Ltd. T. Naoe had received research funding from Bristol-Myers Squibb, Novartis Pharma, Chugai Pharmaceutical Co., Ltd., Kyowa Hakko Kirin Co., Ltd., Dainippon Sumitomo Pharma, and Zenyaku Kogyo. T. Kinoshita had received research funding from Chugai Pharmaceutical Co., Ltd. and SymBio Pharmaceutical Co., Ltd.
Supporting Information
Mouse xenograft model of human DLBCL with the CD20 IHC(+)/FCM(−) phenotype.
Methods.
Results and legend.
References
- 1.Tedder TF, Engel P. CD20: a regulator of cell-cycle progression of B lymphocytes. Immunol Today. 1994;15:450–4. doi: 10.1016/0167-5699(94)90276-3. [DOI] [PubMed] [Google Scholar]
- 2.Maloney DG. Anti-CD20 antibody therapy for B-cell lymphomas. N Engl J Med. 2012;366:2008–16. doi: 10.1056/NEJMct1114348. [DOI] [PubMed] [Google Scholar]
- 3.Lim SH, Beers SA, French RR, Johnson PW, Glennie MJ, Cragg MS. Anti-CD20 monoclonal antibodies: historical and future perspectives. Haematologica. 2010;95:135–43. doi: 10.3324/haematol.2008.001628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Coiffier B, Thieblemont C, Van Den Neste E, et al. Long-term outcome of patients in the LNH-98.5 trial, the first randomized study comparing rituximab-CHOP to standard CHOP chemotherapy in DLBCL patients: a study by the Groupe d'Etudes des Lymphomes de l'Adulte. Blood. 2010;116:2040–5. doi: 10.1182/blood-2010-03-276246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Salles G, Seymour JF, Offner F, et al. Rituximab maintenance for 2 years in patients with high tumour burden follicular lymphoma responding to rituximab plus chemotherapy (PRIMA): a phase 3, randomised controlled trial. Lancet. 2011;377:42–51. doi: 10.1016/S0140-6736(10)62175-7. [DOI] [PubMed] [Google Scholar]
- 6.Hiddemann W, Kneba M, Dreyling M, et al. Frontline therapy with rituximab added to the combination of cyclophosphamide, doxorubicin, vincristine, and prednisone (CHOP) significantly improves the outcome for patients with advanced-stage follicular lymphoma compared with therapy with CHOP alone: results of a prospective randomized study of the German Low-Grade Lymphoma Study Group. Blood. 2005;106:3725–32. doi: 10.1182/blood-2005-01-0016. [DOI] [PubMed] [Google Scholar]
- 7.Smith MR. Rituximab (monoclonal anti-CD20 antibody): mechanisms of action and resistance. Oncogene. 2003;22:7359–68. doi: 10.1038/sj.onc.1206939. [DOI] [PubMed] [Google Scholar]
- 8.Rezvani AR, Maloney DG. Rituximab resistance. Best Pract Res Clin Haematol. 2011;24:203–16. doi: 10.1016/j.beha.2011.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tomita A, Hiraga J, Kiyoi H, et al. Epigenetic regulation of CD20 protein expression in a novel B-cell lymphoma cell line, RRBL1, established from a patient treated repeatedly with rituximab-containing chemotherapy. Int J Hematol. 2007;86:49–57. doi: 10.1532/IJH97.07028. [DOI] [PubMed] [Google Scholar]
- 10.Hiraga J, Tomita A, Sugimoto T, et al. Down-regulation of CD20 expression in B-cell lymphoma cells after treatment with rituximab-containing combination chemotherapies: its prevalence and clinical significance. Blood. 2009;113:4885–93. doi: 10.1182/blood-2008-08-175208. [DOI] [PubMed] [Google Scholar]
- 11.Sugimoto T, Tomita A, Hiraga J, et al. Escape mechanisms from antibody therapy to lymphoma cells: downregulation of CD20 mRNA by recruitment of the HDAC complex and not by DNA methylation. Biochem Biophys Res Commun. 2009;390:48–53. doi: 10.1016/j.bbrc.2009.09.059. [DOI] [PubMed] [Google Scholar]
- 12.Beum PV, Kennedy AD, Williams ME, Lindorfer MA, Taylor RP. The shaving reaction: rituximab/CD20 complexes are removed from mantle cell lymphoma and chronic lymphocytic leukemia cells by THP-1 monocytes. J Immunol. 2006;176:2600–9. doi: 10.4049/jimmunol.176.4.2600. [DOI] [PubMed] [Google Scholar]
- 13.Beum PV, Lindorfer MA, Taylor RP. Within peripheral blood mononuclear cells, antibody-dependent cellular cytotoxicity of rituximab-opsonized Daudi cells is promoted by NK cells and inhibited by monocytes due to shaving. J Immunol. 2008;181:2916–24. doi: 10.4049/jimmunol.181.4.2916. [DOI] [PubMed] [Google Scholar]
- 14.Terui Y, Mishima Y, Sugimura N, et al. Identification of CD20 C-terminal deletion mutations associated with loss of CD20 expression in non-Hodgkin's lymphoma. Clin Cancer Res. 2009;15:2523–30. doi: 10.1158/1078-0432.CCR-08-1403. [DOI] [PubMed] [Google Scholar]
- 15.Mishima Y, Terui Y, Takeuchi K, et al. The identification of irreversible rituximab-resistant lymphoma caused by CD20 gene mutations. Blood Cancer J. 2011;1:e15. doi: 10.1038/bcj.2011.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nakamaki T, Fukuchi K, Nakashima H, et al. CD20 gene deletion causes a CD20-negative relapse in diffuse large B-cell lymphoma. Eur J Haematol. 2012;89:350–5. doi: 10.1111/j.1600-0609.2012.01838.x. [DOI] [PubMed] [Google Scholar]
- 17.Henry C, Deschamps M, Rohrlich PS, et al. Identification of an alternative CD20 transcript variant in B-cell malignancies coding for a novel protein associated to rituximab resistance. Blood. 2010;115:2420–9. doi: 10.1182/blood-2009-06-229112. [DOI] [PubMed] [Google Scholar]
- 18.Lim SH, Vaughan AT, Ashton-Key M, et al. Fc gamma receptor IIb on target B cells promotes rituximab internalization and reduces clinical efficacy. Blood. 2011;118:2530–40. doi: 10.1182/blood-2011-01-330357. [DOI] [PubMed] [Google Scholar]
- 19.Beum PV, Peek EM, Lindorfer MA, et al. Loss of CD20 and bound CD20 antibody from opsonized B cells occurs more rapidly because of trogocytosis mediated by Fc receptor-expressing effector cells than direct internalization by the B cells. J Immunol. 2011;187:3438–47. doi: 10.4049/jimmunol.1101189. [DOI] [PubMed] [Google Scholar]
- 20.Beers SA, French RR, Chan HT, et al. Antigenic modulation limits the efficacy of anti-CD20 antibodies: implications for antibody selection. Blood. 2010;115:5191–201. doi: 10.1182/blood-2010-01-263533. [DOI] [PubMed] [Google Scholar]
- 21.Miyoshi H, Arakawa F, Sato K, et al. Comparison of CD20 expression in B-cell lymphoma between newly diagnosed, untreated cases and those after rituximab treatment. Cancer Sci. 2012;103:1567–73. doi: 10.1111/j.1349-7006.2012.02307.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Horvat M, Kloboves Prevodnik V, Lavrencak J, Jezersek Novakovic B. Predictive significance of the cut-off value of CD20 expression in patients with B-cell lymphoma. Oncol Rep. 2010;24:1101–7. [PubMed] [Google Scholar]
- 23.van Meerten T, van Rijn RS, Hol S, Hagenbeek A, Ebeling SB. Complement-induced cell death by rituximab depends on CD20 expression level and acts complementary to antibody-dependent cellular cytotoxicity. Clin Cancer Res. 2006;12:4027–35. doi: 10.1158/1078-0432.CCR-06-0066. [DOI] [PubMed] [Google Scholar]
- 24.van Meerten T, Rozemuller H, Hol S, et al. HuMab-7D8, a monoclonal antibody directed against the membrane-proximal small loop epitope of CD20 can effectively eliminate CD20 low expressing tumor cells that resist rituximab-mediated lysis. Haematologica. 2010;95:2063–71. doi: 10.3324/haematol.2010.025783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johnson NA, Boyle M, Bashashati A, et al. Diffuse large B-cell lymphoma: reduced CD20 expression is associated with an inferior survival. Blood. 2009;113:3773–80. doi: 10.1182/blood-2008-09-177469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sonoki T, Li Y, Miyanishi S, et al. Establishment of a novel CD20 negative mature B-cell line, WILL2, from a CD20 positive diffuse large B-cell lymphoma patient treated with rituximab. Int J Hematol. 2009;89:400–2. doi: 10.1007/s12185-009-0295-4. [DOI] [PubMed] [Google Scholar]
- 27.Goto E, Tomita A, Hayakawa F, Atsumi A, Kiyoi H, Naoe T. Missense mutations in PML-RARA are critical for the lack of responsiveness to arsenic trioxide treatment. Blood. 2011;118:1600–9. doi: 10.1182/blood-2011-01-329433. [DOI] [PubMed] [Google Scholar]
- 28.Shipp MA. A predictive model for aggressive non-Hodgkin's lymphoma. The International Non-Hodgkin's Lymphoma Prognostic Factors Project. N Engl J Med. 1993;329:987–94. doi: 10.1056/NEJM199309303291402. [DOI] [PubMed] [Google Scholar]
- 29.Binder M, Otto F, Mertelsmann R, Veelken H, Trepel M. The epitope recognized by rituximab. Blood. 2006;108:1975–8. doi: 10.1182/blood-2006-04-014639. [DOI] [PubMed] [Google Scholar]
- 30.Niederfellner G, Lammens A, Mundigl O, et al. Epitope characterization and crystal structure of GA101 provide insights into the molecular basis for type I/II distinction of CD20 antibodies. Blood. 2011;118:358–67. doi: 10.1182/blood-2010-09-305847. [DOI] [PubMed] [Google Scholar]
- 31.Himmelmann A, Riva A, Wilson GL, Lucas BP, Thevenin C, Kehrl JH. PU.1/Pip and basic helix loop helix zipper transcription factors interact with binding sites in the CD20 promoter to help confer lineage- and stage-specific expression of CD20 in B lymphocytes. Blood. 1997;90:3984–95. [PubMed] [Google Scholar]
- 32.Kawabata KC, Ehata S, Komuro A, Takeuchi K, Miyazono K. TGF-beta-induced apoptosis of B-cell lymphoma Ramos cells through reduction of MS4A1/CD20. Oncogene. 2013;32:2096–106. doi: 10.1038/onc.2012.219. [DOI] [PubMed] [Google Scholar]
- 33.Teeling JL, Mackus WJ, Wiegman LJ, et al. The biological activity of human CD20 monoclonal antibodies is linked to unique epitopes on CD20. J Immunol. 2006;177:362–71. doi: 10.4049/jimmunol.177.1.362. [DOI] [PubMed] [Google Scholar]
- 34.Patz M, Isaeva P, Forcob N, et al. Comparison of the in vitro effects of the anti-CD20 antibodies rituximab and GA101 on chronic lymphocytic leukaemia cells. Br J Haematol. 2011;152:295–306. doi: 10.1111/j.1365-2141.2010.08428.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Mouse xenograft model of human DLBCL with the CD20 IHC(+)/FCM(−) phenotype.
Methods.
Results and legend.


