Abstract
Metastasis is the leading cause of cancer-related death in almost all types of cancers, including colorectal cancer (CRC). Metastasis is a complex, multistep, dynamic biological event, and epithelial–mesenchymal transition (EMT) is a critical process during the cascade. Ajuba family proteins are LIM domain-containing proteins and are reported to be transcription repressors regulating different kinds of physiological processes. However, the expression and pathological roles of Ajuba family proteins in tumors, especial in tumor metastasis, remain poorly studied. Here, we found that JUB, but not the other Ajuba family proteins, was highly upregulated in clinical specimens and CRC cell lines. Ectopic expression of JUB induced EMT and enhanced motility and invasiveness in CRC, and vice versa. Mechanistic study revealed that JUB induces EMT via Snail and JUB is also required for Snail-induced EMT. The expression of JUB shows an inverse correlation with E-cadherin expression in clinical specimens. Taken together, these findings revealed that the LIM protein JUB serves as a tumor-promoting gene in CRC by promoting EMT, a critical process of metastasis. Thus, the LIM protein JUB may provide a novel target for therapy of metastatic CRC.
Keywords: Colorectal cancer, epithelial–mesenchymal transition, JUB, LIM protein, Snail
Metastasis accounts for approximately 90% of cancer-associated deaths.1 An estimated 1.2 million cases of colorectal cancer (CRC) were recorded worldwide in 2008. Approximately 35% of patients present with stage-IV metastatic disease when diagnosed with primary disease, and 20–50% of subjects with stage-II or- stage-III disease will progress to stage-IV disease.2,3 Mortality has declined by approximately 1.8% per year for stage-IV disease3 but the prognosis of metastatic CRC remains extremely poor (overall survival at 5 years <10%).4 Metastasis remains the most poorly understood component of the pathogenesis of cancer.
Epithelial–mesenchymal transition (EMT) is known to be fundamental for embryonic development, wound healing and fibrosis, but also for tumor invasion and metastasis.5,6 EMT is the initial process of tumor metastasis. Cells undergoing EMT usually acquire a mesenchymal phenotype, spindle-like morphology, high motility and invasiveness. Downregulation of E-cadherin is a hallmark of EMT. Control of transcription of the E-cadherin gene is the main mechanism accounting for downregulation of this protein.7 Several transcription factors have been reported to repress expression of E-cadherin: Snail, Slug, Twist and S1P,8–10 among which Snail was the first to be discovered. Snail has been reported to be a major transcription repressor of E-cadherin, frequently upregulated in breast cancer,8,11 esophageal squamous cell carcinoma12 and CRC.13
The LIM protein Ajuba family contains three members, JUB, WTIP and LIMD1, and is characterized by a unique N-terminal region, the preLIM region, and three tandem C-terminal LIM domains.14–16 This family of proteins function as scaffolds and have been reported to modulate many events in cells, such as cell proliferation and tissue size,17,18 DNA damage response,19 meiotic maturation of oocytes14 and embryonal cell proliferation and differentiation.20 However, the pathological roles of Ajuba family proteins in diseases remain unexplored. Langer et al.21 find that LIM protein Ajuba family proteins interact with the SNAG domain of the Snail family. They use Xenopus neural crest as a model of in vivo Snail-induced EMT and demonstrate that Ajuba LIM proteins contribute to neural crest development as Snail/Slug corepressors and are required for in vivo Snail/Slug function.21 Thus, we propose that Ajuba family proteins may also regulate EMT in cancer and promote metastasis.
In the present study, surprisingly, we find that the LIM protein JUB, but not the other two members of the Ajuba family, WTIP and LIMD1, was highly upregulated in CRC specimens and cell lines. Ectopic overexpression of JUB induces EMT and promotes migration and invasion in CRC cells. Silencing of JUB impairs EMT and inhibits migration and invasion in CRC cells. Further mechanistic study revealed that JUB promoting EMT depends on Snail, and JUB is also required for Snail to induce EMT, which is consistent with previous reports that JUB serves as a corepressor of Snail. The present study uncovered an important role of the LIM protein JUB in the pathological progress of tumorigenesis in CRC. Hereafter, JUB could be a novel therapy target for metastatic CRC.
Materials and Methods
Plasmids and antibodies
For overexpression of JUB, human JUB (538aa) was amplified by PCR from cDNA of SW620 cells and subcloned into a pSin-EF2-puro retroviral vector (Addgene). For depletion of JUB, two human shRNA sequences were cloned into the pSuper-retro-puro vector to generate pSuper-retro-JUB-RNAi(s). The target sequences were: RNAi#1, GGACCGGGATTATCACTTT, and RNAi#2, CCAAGTATACTGTGTCACC, as previously reported.22 Human Snail gene was amplified from cDNA and inserted into the EcoR I and Xho I sites of the pCDNA3.1 vector. For silencing of Snail, oligonucleotides were purchased from RiboBio (RiboBio, Guangzhou, Guangdong) and the target sequence is 5′-gctcggaccttctcccgaa-3′.
Antibodies, including anti-JUB (Cell Signaling Technology, Danvers, MA, USA), anti-WTIP (Sigma Aldrich, Santa Cruz, CA, USA), anti-LIMD1 (Sigma, Saint Louis, MO, USA), anti-β-actin (Sigma Aldrich), anti-E-cadherin (BD Biosciences, Bedford, MA, USA), anti-Vimentin (BD Biosciences) and anti-Snail (Cell Signaling Technology) were used for western blot analysis.
Cell culture and stable cell line establishment
Colorectal cancer cell lines, including SW480, SW620, KM12, HCT15, HCT116, Caco-2 and LoVo, were purchased from ATCC (Manassas, VA, USA) and maintained in RPMI-1640 medium (Invitrogen, Carlsbad, CA, USA) supplemented with 10% FBS (HyClone, Logan, Utah, USA), 1% penicillin/streptomycin and non-essential amino acids (HyClone) at 37° in 5% humidified CO2 atmosphere. SW620 and SW480 cells were selected for stable overexpression or knockdown of JUB.
For stable cell line establishment, the indicated plasmids (pSin-EF2-puro-retro-JUB and pSuper-puro-retro-JUB-RNAis) were packed into retrovirus in 293T cells. Then, the viruses were harvested and SW620 and SW480 cells were infected. After infection for 48 h, the cells were selected with medium containing puromycin (0.5 ug/mL) over 1 week.
RNA extraction, RT-PCR and real-time RT-PCR
Total RNA from cultured cells was extracted using Trizol reagent (Invitrogen) following the manufacturer's instructions. The cDNA was amplified and quantified using an ABI Prism 7500 Sequence Detection System (Applied Biosystems, Lab India, Haryana, India), with SYBR Green I dye (Molecular Probes, Invitrogen). β-actin was used as an internal control. Expression data were normalized to β-actin, and calculated as 2(-[(Ct of gene) − (Ct of beta-actin)]), where Ct represents the threshold cycle for each transcript. The primers for qRT-PCR were:
| JUB-Fwd | AGAGGCCAGGGAGGACTACT |
| JUB-Rev | GAGCAGCAAACAAAGCACTG |
| WTIP-Fwd | TGTGGGCTTGGCATCTACG |
| WTIP-Rev | TGCTGGAACCCGGAGTACAG |
| LIMD1-Fwd | TCACCCGAAGGCTGATTACT |
| LIMD1-Rev | AGGTGAAGCATGTGTCATGG |
| β-actin-Fwd | GCACAGAGCCTCGCCTT |
| β-actin-Rve | CCTTGCACATGCCGGAG |
Clinical tissue specimens and ethics statement
Tissue specimens were freshly collected from Zengcheng People's Hospital (BoJi-Affiliated Hospital of Sun Yat-Sen University), so that patients could be histopathologically and clinically diagnosed. All samples were obtained with prior written informed consents from the patients and approval from the Institutional Research Ethics Committees of Zengcheng People's Hospital (BoJi-Affiliated Hospital of Sun Yat-Sen University) ethics Committee.
Transwell assays
A total of 4 × 104 of each the indicated cells were suspended in 200 μL basic 1640 medium and seeded into the upper trans-well cell culture chambers (BD Biosciences) coated with Matrigel (for invasion assay) or without Matrigel (for migration assay). The lower chamber was loaded with 500 μL of 1640 medium with 10% FBS. After incubation for 48 h at 37°C in 5% CO2, cells were fixed with methanol and stained with 1% crystal violate. Cells presented on the lower surface of the membrane were quantified and photographed under a microscope. The average number of five randomly selected microscopic fields was presented.
3-D spheroid invasion assay
First, we coated the 24-well plates with 100% Matrigel (BD Biosciences) for the bottom layer. We incubated the coated plates at 37° for at least 30 min and then seeded 1 × 104 of the indicated cells mixed with 10% Matrigel on the top layer. The medium was refreshed every other day. Cells forming a 3-D spherical structure (spheres) were photographed at 2-day intervals for 2 weeks. Spheroids with outgrowth were considered to be invasive spheroids and the quantification of invasive spheroids was carried out as follows: high contrast images of spheroids were captured using AxioVision Rel.4.6 software (Carl Zeiss, Oberkochen, Germany) and 10 randomly selected fields of each well were analysed. The final data was presented as the percentage of invasive spheroids.
Dual-luciferase reporter assay
A total of 4 × 104 cells were seeded in triplicate in 24-well plates. Twenty-four hours later, 500 ng of vector, pSin-EF2-puro-JUB or pSuper-puro-JUB-RNAi plasmids plus 150 ng E-cadherin-luciferase reporter construct and 5 ng pRL-TK Renilla plasmids were co-transfected into cells using Lipofectamine 2000 Reagent (Life Technologies, Carlsbad, CA, USA) according to the manufacturer's instructions. Forty-eight hours after transfection, reporter luciferase activity was measured using the Dual-Luciferase Reporter Assay (Promega), according to the manufacturer's instructions.
ChIP assay
Cells were grown to 70–90% confluence, fixed in 1% formaldehyde, and harvested by scratching. ChIP assays were performed using the EZ-CHIP kit (Upstate, Temecula, CA, USA) according to the manufacturer's instructions.21 Quantitative real-time PCR was performed on immunoprecipitated DNA. The primer sequences for detecting E-cadherin promoter are: forward, 5′-AGGTGAACCCTCAGCCAATC-3′; reverse, 5′-AGCGGGCTGGAGTCTGAA-3′.
Immunofluorescence staining
Cells were seeded on coverslips in 24-well plates (Costar; Corning Incorporated, Corning, NY, USA). After 24 h, the cells were washed with 1 × PBS and fixed with ice-cold methanol for 10 min at room temperature. Cell membrane was permeabilized with 0.2% Triton X-100 in PBS (PBS-T), blocked with 10% BSA in PBS-T for 30 min, then incubated with primary antibodies, E-cadherin, Vimentin (1:500, BD Biosciences), in 10% BSA for 1 h at room temperature. After three washes with PBS, the slides were incubated for 1 h in darkness with Rhodamine-conjugated secondary goat anti-mouse antibody (The Jackson Laboratory, Bar Harvot, Maine USA). After washing, the slides were counterstained with DAPI (Sigma Aldrich). Images were captured using the AxioVision Rel.4.6 computerized image analysis system.
Statistical analysis
Student's two-tailed t-test was used for the statistical analysis. Statistical analyses were performed using the SPSS 13.0 statistical software package. Data represent mean ± SD. Pearson correlation statistical analysis was used to analyze the correlation between JUB and E-cadherin expression. P-values less than 0.05 were considered statistically significant.
Results
JUB, but not the other members of the Ajuba family proteins, is upregulated in CRC
Until now, the expression of Ajuba family proteins had not been explored. First, we analyzed their expression, including JUB, WTIP and LIMD1, in a publicly-available dataset (http://www.ncbi.nlm.nih.gov/gds/). As shown in Figure 1a, we were surprised to find that JUB, detected by different probes, but not the other two members of the Ajuba family proteins, was highly upregulated in tumor tissues compared to the matched adjacent normal tissues (GSE32323, n = 34). Furthermore, we detected the expression of Ajuba family proteins, E-cadherin and Vimentin (Fig. S1), in normal colorectal epithelial cells and CRC cell lines. Compared to the normal colorectal epithelial cells, JUB was significantly overexpressed in both protein and mRNA levels; however, the expression of WTIP and LIMD1 showed no obvious changes (Fig.1b,c).
Figure 1.
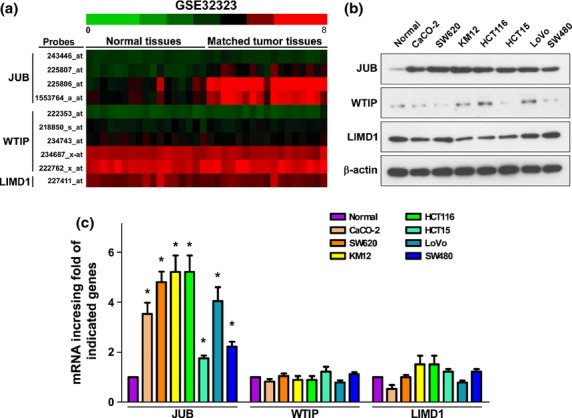
JUB is highly upregulated in colorectal cancer (CRC) specimens and cell lines. (a) Analysis of Ajuba family proteins transcriptional level in a published high-throughput microarray dataset (NCBI/GEO/GSE32323; n = 34). (b) Western blot analysis of the indicated proteins in CRC cell lines. β-actin was used as a loading control. (c) Real-time PCR analysis of Ajuba family proteins mRNA expression level. Expression data were normalized to β-actin and presented as mean ± standard deviation (SD) from three independent experiments. *P < 0.05 based on Student's t-test.
Ectopic overexpression of JUB induces epithelial–mesenchymal transition in colorectal cancer cells
Snail was proved to repress E-cadherin expression to induce EMT.23,24 Previous reports have shown that JUB functions as a corepressor of Snail,21 but whether it could also induce EMT has not been considered. Thus, we investigated the role JUB in EMT. As shown in Figure 2(a), western blot analsis revealed that ectopic expression of JUB in SW480 cells significantly reduced the expression of the epithelial marker, indicated by E-cadherin, and enhanced the expression of the mesenchymal marker, indicated by Vimentin. This was further confirmed by immunofluorescence staining (Fig.2b).
Figure 2.
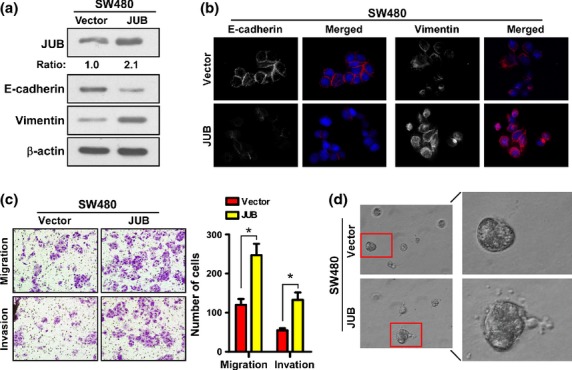
Ectopic expression of JUB induces epithelial–mesenchymal transition and enhances invasiveness in colorectal cancer cells. (a) Western blot analyses of E-cadherin and Vimnetin in the indicated cells. (b) Immunofluorescence staining of E-cadherin and Vimnetin in the indicated cells. DAPI was used to visualize nuclei. (c) Representative images (left panels) and quantification (right panel) of migrated and invaded SW480-JUB and vector cells. Data are the mean ± standard deviation (SD) from three independent experiments. (d) Representative images for 3-D culture of SW480-JUB and vector cells. All experiments were carried out at least three times. *P < 0.05 based on Student's t-test.
It is well known that EMT aids metastasis by augmenting the motility and invasiveness of tumor cells. Indeed, overexpression of JUB in SW480 cells led to more cells invading and migrating through the Matrigel-coated or non-coated membrane of the chamber compared to vector cells (Fig.2c). Strikingly, in the 3-D spheroid invasion assay, SW480-JUB cells displayed morphologies that were much more invasive, presenting more outward projections compared with vector cells (Fig.2d and Fig. S2a). These data demonstrate that JUB does promote EMT and invasiveness in CRC.
Silencing of endogenous JUB represses epithelial–mesenchymal transition in colorectal cancer cells
To further confirm the role of JUB in promoting EMT in the opposite way, SW620, a derivative of the SW480 cell line and a highly metastatic CRC cell line, which showed a high expression of JUB (Fig.1b), was selected for depletion of the endogenous expression of JUB. Indeed, silencing of JUB repressed EMT in SW620 cells. Knockdown of JUB reduced expression of E-cadherin and enhanced expression of Vimentin, indicated by both western blotting and immunofluorescence staining assays (Fig.3a,b). The motility and invasive ability were also impaired by silencing of JUB as fewer cells were presented in the under-surface of the transwell chamber membrane (Fig.3c). Consistent with the data detailed above, 3-D culture showed that silencing JUB reduced the invasiveness of SW620 cells, exhibiting fewer or no outward projections (Fig.3d and Fig. S2b). Taken together, the abovementioned data strongly suggest that JUB promotes EMT in CRC.
Figure 3.
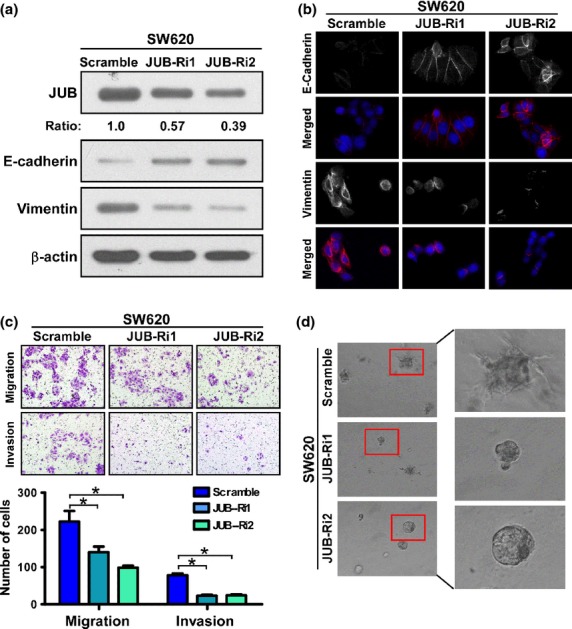
Silencing of endogenous JUB represses epithelial–mesenchymal transition and reduces invasion in colorectal cancer cells. (a) Western blot analyses of E-cadherin and Vimnetin in the indicated cells. (b) Immunofluorescence staining of E-cadherin and Vimnetin in the indicated cells. DAPI was used to visualize nuclei. (c) Representative images (Upper panels) and quantification (Lower panel) of migrated and invaded SW620-JUB-RNAi and vector cells. Data are the mean ± standard deviation (SD) from three independent experiments. (d) Representative images for 3-D culture of SW620-JUB-RNAi and vector cells. All experiments were carried out at least three times. *P < 0.05 based on Student's t-test.
Snail is essential for JUB-induced epithelial–mesenchymal transition and JUB is required for Snail-induced epithelial–mesenchymal transition
Langer et al. (2008) demonstrate that JUB acts as a corepressor of Snail and targets the promoter region of E-cadherin to repress its expression.21 We tested whether this occurred in our current CRC model. First, we confirmed the interaction of JUB and Snail in CRC cells (Fig. S3a).Then, we measured the activity of an E-cadherin promoter containing reporter,21 which contains three Snail-binding E-boxes driving luciferase expression. Indeed, upregulation of JUB decreased while downregulation of JUB increased the reporter activity in both SW480 and SW620 cells (Fig. 4a). We noticed that overexpression of JUB increased while silencing JUB decresed the repression effect of Snail on E-cadherin promoter activity (Fig. S3b). In addition, JUB presented on the promoter of E-cadnerin as shown by JUB ChIP (Fig. S3c). Furthermore, we silenced Snail in SW480-JUB cells or reintroduced Snail-ORF into SW620-JUB-Ri cells. As shown in Figure4(b) and (c), silencing of Snail derepressed the expression of E-cadherin and repressed the motility and invasion abilities of SW480-JUB cells, which indicates that Snail is essential for JUB-induced EMT. However, overexpression of Snail in SW620-JUB-Ri cells only partially depressed E-cadherin expression and partially enhanced motility and invasion abilities (Fig.4b,c), which means that JUB is also required for Snail-induced EMT.
Figure 4.
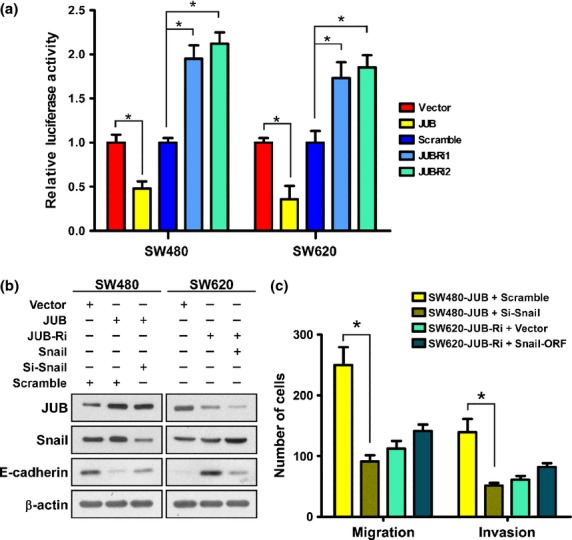
Snail is required for JUB-induced epithelial–mesenchymal transition. (a) Transient luciferase reporter assay using luciferase driven by the E-cadherin promoter in SW480 and SW620 cells. (b) Western blot of the indicated proteins in SW480 and SW620 cells transfected with indicated plasmids or siRNA. β-actin was used as a loading control. (c) Quantification of migrated and invaded cells of Transwell assay in the indicated cells. Data are the mean ± standard deviation (SD) from three independent experiments. *P < 0.05 based on Student's t-test.
Expression of E-cadherin inversely correlated with JUB expression in specimens
Finally, we examined whether JUB repressing E-cadherin expression to induce EMT identified in CRC cells is also evident in clinical CRC specimens. By analyzing 10 cases of freshly collected CRC tissue specimens, we found that specimens with higher expression of JUB showed lower expression of E-cadherin (Fig. 5a). Quantification and correlation analysis proved that expression of E-cadherin and JUB significantly correlated, inversely (r = −0.649, P = 0.042, Fig.5(b).
Figure 5.
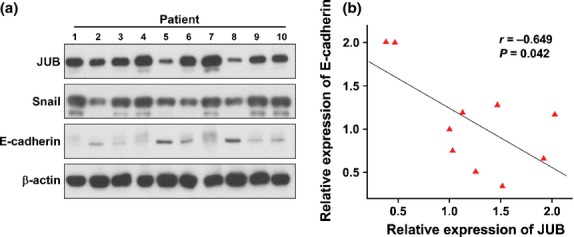
Reverse correlated expression between JUB and E-cadherin in clinical specimens. (a) Western blot analysis of JUB and E-cadherin in clinical specimens. (b) Quantification and Pearson correlation statistical analyzing of JUB and E-cadherin expression.
Discussion
In the current study, we investigated the role of Ajuba family proteins in CRC cells. First, we found, surprisingly, that only JUB but not WTIP or LIMD1 of the Ajuba family was highly upregulated in CRC in a publicly available dataset, and validated this in cell lines. Further investigation proved that JUB could modulate EMT program of CRC cells. SW480-JUB cells showed enhanced EMT, indicated by decreased expression of E-cadherin, increased expression of vimentin, and enhanced motility and invasiveness compared to vector cells. Silencing of JUB in SW620 cells dramatically impaired EMT, leading to increased expression of E-cadherin, decreased expression of vimentin, and impaired motility and invasiveness compared with control cells. Further investigation revealed that JUB requires Snail to induce EMT and is required for Snail-induced EMT. Correlation analysis between the expression of JUB and E-cadherin in clinical specimens validated the conclusion that JUB represses E-cadherin and promotes EMT. EMT is closely associated with stemness in several cancer models. Our preliminary experiment found that, in SW480 cells, overexpression JUB forms more and larger spheres as indicated by tumor sphere formation assay (Fig. S4). However, further confirmation and investigation is needed.
There are three members of Ajuba family proteins, JUB, LIMD1 and WTIP. Various reports have shown that Ajuba family proteins share a lot in common. They have been found to bind the same partners or the same complexes and to regulate the same cellular events, especially JUB and LIMD1.17,25–27 Ajuba family proteins share three tandem, homologous LIM domains at the COOH terminus, but contain divergent proline rich NH2 termini (preLIM region).14–16 Thus, it is possible and reasonable that Ajuba proteins possess specific roles for each other. For example, Witzel et al.28 report that JUB is the prominent Isl1-bingding partner while LIMD1 shows the weakest interaction with Isl1. Using LAST2 as a bait, only JUB of the family was identified as a binding partner.29 Here, we also found, surprisingly, that only JUB, but not LIMD1 or WTIP, was highly upregulated in the samples we obtained and in the publicly-available dataset, which indicats a specific role of JUB in CRC progression. However, it is still possible that WTIP and LIMD1 might also regulate EMT in CRC, but, absolutely, they do not play the prominent role.
According to various studies, JUB may regulate EMT via contrary mechanisms. Loss of cell–cell adhesion is a crucial aspect of EMT. JUB was originally found as a cytosolic protein. It was reported that JUB was recruited to cadherin-dependent cell junctions30 and was required for maintenance of E-cadherin adhesion.31 Thus, JUB seems to repress EMT rather than promote EMT. However, inhibition of PAK1, which directly phosphorylated Ajuba at Thr172, phenocopies depletion of Ajuba,30 which means that it is the phosphorylated JUB but not the un-phosphorylated JUB involving in E-cadherin adhesion maintenance. Recently it was proved that JUB contains a functional nuclear export signal and shuttles between cytoplasm and nucleus.20 Therefore, it is possible that phosphorylation of JUB modulated its subcellular localization, leading it to be recruited to cell junctions. JUB was proved to be a corepressor of Snail family in nucleus to induce EMT.21,26 JUB can recruit the effecter PRMT5 to Snail to repress Snail-target genes, such as E-cadherin.32 Our data show that, in CRC, JUB was dramatically upregulated and it repressed E-cadherin expression to induced EMT as a corepressor of Snail, which is consistent with previous studies. However, we cannot rule out the possibility that it is a consequence of the balance between the distribution of cytoplasmic and nuclear JUB, which requires further investigation.
In sum, in the present study we found an elevated expression of JUB in CRC. We proved that upregulated JUB induces EMT, leading to increased motility and invasiveness, by acting as a corepressor of Snail. Our current study indicates a novel and potential target for metastatic CRC.
Acknowledgments
We thank Rong Zhou, Xiaolan Xia,Yi Yang and Xiamin Ma for their contributions in the initial stages of these experiments. This work is supported by the Natural Science Foundation of China (81001111); the Natural Science Foundation of Guangdong (S2011040003696); Zengcheng People's Hospital Scholarship for Excellent Scientist (2013-YX-001).
Disclosure Statement
The authors have no conflict of interest.
Funding Information
Natural Science Foundation of China (81001111). Natural Science Foundation of Guangdong (S2011040003696). Zengcheng People’s Hospital Scholarship for Excellent Scientists (2013-YX-001).
Supporting Information
Additional supporting information may be found in the online version of this article:
Western blotting analysis of E-cadherin and Vimentin in colorectal cancer (CRC) cell lines.
Quantification of invasive spheroids. (a) Quantification of invasive spheroids in SW480-JUB and vector cells. (b) Quantification of invasive spheroids in SW620 JUB-RNA is and vector cells. Data presented as mean ± SD. *P < 0.05 based on Student's t-test.
JUB interacts with and functions as a corepressor of Snail. (a) Co-immunoprecipitation between JUB and Snail. IgG was used as a negative control. (b) Transient luciferase reporter assay using luciferase driven by the E-cadherin promoter in SW480. (c) ChIP analysis of JUB on the promoter of E-cadherin. Snail was used as a positive control. Data presented as mean ± SD. *P < 0.05 based on Student's t-test.
Tumor sphere formation assay analysis in SW480 cells.
References
- 1.Chaffer CL, Weinberg RA. A perspective on cancer cell metastasis. Science. 2011;331:1559–64. doi: 10.1126/science.1203543. [DOI] [PubMed] [Google Scholar]
- 2.Jemal A, Siegel R, Ward E, et al. Cancer statistics. CA Cancer J Clin. 2008;58:71–96. doi: 10.3322/CA.2007.0010. [DOI] [PubMed] [Google Scholar]
- 3.Field K, Lipton L. Metastatic colorectal cancer-past, progress and future. World J Gastroenterol. 2007;13:3806–15. doi: 10.3748/wjg.v13.i28.3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jemal A, Murray T, Ward E, et al. Cancer statistics. CA Cancer J Clin. 2005;55:10–30. doi: 10.3322/canjclin.55.1.10. [DOI] [PubMed] [Google Scholar]
- 5.Thiery JP. Epithelial-mesenchymal transitions in development and pathologies. Curr Opin Cell Biol. 2003;15:740–6. doi: 10.1016/j.ceb.2003.10.006. [DOI] [PubMed] [Google Scholar]
- 6.Kalluri R, Neilson EG. Epithelial-mesenchymal transition and its implications for fibrosis. J Clin Invest. 2003;112:1776–84. doi: 10.1172/JCI20530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barbera MJ, Puig I, Dominguez D, et al. Garcia de Herreros, Regulation of Snail transcription during epithelial to mesenchymal transition of tumor cells. Oncogene. 2004;23:7345–54. doi: 10.1038/sj.onc.1207990. [DOI] [PubMed] [Google Scholar]
- 8.Martin TA, Goyal A, Watkins G, Jiang WG. Expression of the transcription factors snail, slug, and twist and their clinical significance in human breast cancer. Ann Surg Oncol. 2005;12:488–96. doi: 10.1245/ASO.2005.04.010. [DOI] [PubMed] [Google Scholar]
- 9.Yang J, Mani SA, Donaher JL, et al. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell. 2004;117:927–39. doi: 10.1016/j.cell.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 10.Rosivatz E, Becker I, Specht K, et al. Differential expression of the epithelial-mesenchymal transition regulators snail, SIP1, and twist in gastric cancer. Am J Pathol. 2002;161:1881–91. doi: 10.1016/S0002-9440(10)64464-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Parker BS, Argani P, Cook BP, et al. Alterations in vascular gene expression in invasive breast carcinoma. Cancer Res. 2004;64:7857–66. doi: 10.1158/0008-5472.CAN-04-1976. [DOI] [PubMed] [Google Scholar]
- 12.Usami Y, Satake S, Nakayama F, et al. Snail-associated epithelial-mesenchymal transition promotes oesophageal squamous cell carcinoma motility and progression. J Pathol. 2008;215:330–9. doi: 10.1002/path.2365. [DOI] [PubMed] [Google Scholar]
- 13.Roy HK, Smyrk YC, Koetsier J, Victor TA, Wali RK. The transcriptional repressor SNAIL is overexpressed in human colon cancer. Dig Dis Sci. 2005;50:42–6. doi: 10.1007/s10620-005-1275-z. [DOI] [PubMed] [Google Scholar]
- 14.Goyal RK, Lin P, Kanungo J, Payne AS, Muslin AJ, Longmore JD. Ajuba, a novel LIM protein, interacts with Grb2, augments mitogen-activated protein kinase activity in fibroblasts, and promotes meiotic maturation of Xenopus oocytes in a Grb2- and Ras-dependent manner. Mol Cell Biol. 1999;19:4379–89. doi: 10.1128/mcb.19.6.4379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Srichai MB, Konieczkowski M, Padiyar A, et al. A WT1 co-regulator controls podocyte phenotype by shuttling between adhesion structures and nucleus. J Biol Chem. 2004;279:14398–408. doi: 10.1074/jbc.M314155200. [DOI] [PubMed] [Google Scholar]
- 16.Sharp TV, Munoz F, Bourboulia D, et al. LIM domains-containing protein 1 (LIMD1), a tumor suppressor encoded at chromosome 3p21.3, binds pRB and represses E2F-driven transcription. Proc Natl Acad Sci USA. 2004;101:16531–6. doi: 10.1073/pnas.0407123101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reddy BV, Irvine KD. Regulation of Hippo signaling by EGFR-MAPK signaling through Ajuba family proteins. Dev Cell. 2013;24:459–71. doi: 10.1016/j.devcel.2013.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Das Thakur M, Feng Y, Jagannathan R, et al. Ajuba LIM proteins are negative regulators of the Hippo signaling pathway. Curr Biol. 2010;20:657–62. doi: 10.1016/j.cub.2010.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kalan S, Matveyenko A, Loayza D. LIM protein Ajuba participates in the repression of the ATR-Mediated DNA damage response. Front Genet. 2013;4:95. doi: 10.3389/fgene.2013.00095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kanungo J, Pratt SJ, Marie H, Longmore GD. Ajuba, a cytosolic LIM protein, shuttles into the nucleus and affects embryonal cell proliferation and fate decisions. Mol Biol Cell. 2000;11:3299–313. doi: 10.1091/mbc.11.10.3299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Langer EM, Feng Y, Zhaoyuan H, Rauscher FJ, 3rd, Kroll KL, Longmore GD. Ajuba LIM proteins are snail/slug corepressors required for neural crest development in Xenopus. Dev Cell. 2008;14:424–36. doi: 10.1016/j.devcel.2008.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haraguchi K, Ohsugi M, Abe Y, Semba K, Akiyama T, Yamamoto T. Ajuba negatively regulates the Wnt signaling pathway by promoting GSK-3beta-mediated phosphorylation of beta-catenin. Oncogene. 2008;27:274–84. doi: 10.1038/sj.onc.1210644. [DOI] [PubMed] [Google Scholar]
- 23.Cano A, Perez-Moreno MA, Rodrigo I, et al. The transcription factor snail controls epithelial-mesenchymal transitions by repressing E-cadherin expression. Nat Cell Biol. 2000;2:76–83. doi: 10.1038/35000025. [DOI] [PubMed] [Google Scholar]
- 24.Batlle E, Sancho E, Franci C, et al. The transcription factor snail is a repressor of E-cadherin gene expression in epithelial tumour cells. Nat Cell Biol. 2000;2:84–9. doi: 10.1038/35000034. [DOI] [PubMed] [Google Scholar]
- 25.Feng Y, Longmore GD. The LIM protein Ajuba influences interleukin-1-induced NF-kappaB activation by affecting the assembly and activity of the protein kinase Czeta/p62/TRAF6 signaling complex. Mol Cell Biol. 2005;25:4010–22. doi: 10.1128/MCB.25.10.4010-4022.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ayyanathan K, Peng H, Hou Z, et al. The Ajuba LIM domain protein is a corepressor for SNAG domain mediated repression and participates in nucleocytoplasmic shuttling. Cancer Res. 2007;67:9097–106. doi: 10.1158/0008-5472.CAN-07-2987. [DOI] [PubMed] [Google Scholar]
- 27.James V, Zhang Y, Foxler DE, et al. LIM-domain proteins, LIMD1, Ajuba, and WTIP are required for microRNA-mediated gene silencing. Proc Natl Acad Sci USA. 2010;107:12499–504. doi: 10.1073/pnas.0914987107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Witzel HR, Jungblut B, Choe CP, Crump JG, Braun T, Dobreva G. The LIM protein Ajuba restricts the second heart field progenitor pool by regulating Isl1 activity. Dev Cell. 2012;23:58–70. doi: 10.1016/j.devcel.2012.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Abe Y, Ohsugi M, Haraguchi K, Fujimoto J, Yamamoto T. LATS2-Ajuba complex regulates gamma-tubulin recruitment to centrosomes and spindle organization during mitosis. FEBS Lett. 2006;580:782–8. doi: 10.1016/j.febslet.2005.12.096. [DOI] [PubMed] [Google Scholar]
- 30.Marie H, Pratt SJ, Betson M, et al. The LIM protein Ajuba is recruited to cadherin-dependent cell junctions through an association with alpha-catenin. J Biol Chem. 2003;278:1220–8. doi: 10.1074/jbc.M205391200. [DOI] [PubMed] [Google Scholar]
- 31.Nola S, Daigaku R, Smolarczyk K, et al. Ajuba is required for Rac activation and maintenance of E-cadherin adhesion. J Cell Biol. 2011;195:855–71. doi: 10.1083/jcb.201107162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hou Z, Peng H, Ayyanathan K, et al. The LIM protein AJUBA recruits protein arginine methyltransferase 5 to mediate SNAIL-dependent transcriptional repression. Mol Cell Biol. 2008;28:3198–207. doi: 10.1128/MCB.01435-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Western blotting analysis of E-cadherin and Vimentin in colorectal cancer (CRC) cell lines.
Quantification of invasive spheroids. (a) Quantification of invasive spheroids in SW480-JUB and vector cells. (b) Quantification of invasive spheroids in SW620 JUB-RNA is and vector cells. Data presented as mean ± SD. *P < 0.05 based on Student's t-test.
JUB interacts with and functions as a corepressor of Snail. (a) Co-immunoprecipitation between JUB and Snail. IgG was used as a negative control. (b) Transient luciferase reporter assay using luciferase driven by the E-cadherin promoter in SW480. (c) ChIP analysis of JUB on the promoter of E-cadherin. Snail was used as a positive control. Data presented as mean ± SD. *P < 0.05 based on Student's t-test.
Tumor sphere formation assay analysis in SW480 cells.


